Abstract
In the absence of telomerase, telomeres erode, provoking accumulation of DNA damage and death by senescence. Rare survivors arise, however, due to Rad52-based amplification of telomeric sequences by homologous recombination. The present study reveals that in budding yeast cells, postsenescence survival relying on amplification of the TG1-3 telomeric repeats can take place in the absence of Rad52 when overelongated telomeres are present during senescence (hence its designation ILT, for inherited-long-telomere, pathway). By growth competition, the Rad52-independent pathway was almost as efficient as the Rad51- and Rad52-dependent pathway that predominates in telomerase-negative cells. The ILT pathway could also be triggered by increased telomerase accessibility before telomerase removal, combined with loss of telomere protection, indicating that prior accumulation of recombination proteins was not required. The ILT pathway was dependent on Rad50 and Mre11 but not on the Rad51 recombinase and Rad59, thus making it distinct from both the type II (budding yeast ALT [alternative lengthening of telomeres]) and type I pathways amplifying the TG1-3 repeats and subtelomeric sequences, respectively. The ILT pathway also required the Rad1 endonuclease and Elg1, a replication factor C (RFC)-like complex subunit, but not Rad24 or Ctf18 (two subunits of two other RFC-like complexes), the Dnl4 ligase, Yku70, or Nej1. Possible mechanisms for this Rad52-independent pathway of telomeric repeat amplification are discussed. The effects of inherited long telomeres on Rad52-dependent recombination are also reported.
Genome stability in eukaryotic cells requires the integrity of the chromosome ends, the telomeres. Telomerase, a reverse transcriptase with a built-in RNA template specialized in the synthesis of telomeric DNA, is responsible for the maintenance of telomeres (2). In most mammalian somatic cells, telomerase is naturally inhibited and progressive shortening of telomeres provokes an irreversible cell cycle arrest called cellular senescence, which can also result from other cellular stresses (20, 49). Cell death eventually ensues unless a mechanism for telomere extension is reactivated. Reactivation of telomerase is a necessary step for immortalization in most tumors. In a subset of human cancers, mainly soft tissue sarcomas, osteosarcomas, and astrocytomas, tumor cells rely on an alternative pathway of telomere maintenance, the ALT (alternative lengthening of telomeres) pathway, for proliferation (10, 30). The ALT pathway is based on recombination between the repeated telomeric sequences (59). Indeed, telomeric DNA is composed of long stretches of TG-rich nucleotide repeats that are prone to recombination when left unprotected.
Yeast provides an attractive model for studying telomere-induced senescence. Like in mammals, following telomerase inactivation, maintenance of telomeres by telomerase is promptly replaced by an alternative pathway based on recombination (8, 48, 49, 56). Presumably, recombination occurs when the telomeres have lost their provision of telomere end protection proteins as telomeres shorten and become critically eroded, at 75 to 100 cell divisions after the onset of senescence in Saccharomyces cerevisiae (41). S. cerevisiae postsenescence survivors use two distinct pathways of recombination, both requiring Rad52, a DNA repair protein essential for basically all types of homologous recombination. A first class of survivors (type I), relying on Rad51, were found to amplify the subtelomeric Y′ elements and had very short terminal tracts of TG1-3 DNA, while Rad50-dependent type II survivors amplified the terminal TG1-3 sequences with no evidence for rearrangement of Y′ elements (38, 48, 68). Importantly, the nonessential subunit of DNA Polδ, Pol32, which is dispensable for replication and gene conversion, was essential for the generation of both these Rad51-dependent and Rad51-independent yeast survivors (51). Since Pol32 was also essential for break-induced replication (BIR), both BIR and the budding yeast telomeric recombination may function by establishment of a full replication fork by recombination in the absence of an origin of replication, as frequently suggested (51, 56). Type II recombination is interesting from a mechanistic point of view because it does not require the intervention of the Rad51 recombinase, the only budding yeast protein capable of strand invasion during mitotic homologous recombination (61, 67). Type II recombination generates highly heterogeneous terminal restriction fragments, which can attain 20 kb or more after telomere elongation, resembling those present in ALT cells (8, 30, 31). It has been proposed that type II recombination in budding yeast and the mammalian ALT pathway are probably mechanistically similar (50). A difference, however, may reside in the fact that type II recombination takes place before senescence (and represents a possible checkpoint response), while cells utilizing the ALT pathway had previously bypassed senescence by undergoing genomic rearrangements (50). Finally, S. cerevisiae telomerase-negative cells also utilize a recombination-independent survival pathway that involves repair of DNA double-strand breaks by palindromic DNA structures (55).
The present study aimed at further documenting the yeast telomerase-independent pathways of telomere maintenance. We set out to try to analyze a situation that had not been previously investigated in detail. The simple questions were as follows: once recombination is initiated at the telomeres, can we stop it, and if yes, how? We found that cells with unusually long telomeres can maintain viability without telomerase, as rare survivors, in the absence of Rad52, whereas cells lacking telomerase but beginning with normal-length telomeres require Rad52 for survival. This novel Rad52-independent pathway of postsenescence survival required Rad50, Mre11, the Rad1 endonuclease, and Elg1 but not the Rad51 recombinase or Rad59, a protein with homologies to Rad52. We propose to name this novel Rad52-independent pathway of telomeric recombination the ILT (inherited-long-telomere) pathway. Because the ILT survivors generated a type II-like pattern of recombination indistinguishable from that taking place in RAD52+ cells, we will frequently refer, for convenience, to type II telomeric recombination as type II-ALT (as argued above) to distinguish it from type II-ILT (or simply ILT). Genetic analysis suggested that the ILT pathway does not use nonhomologous end joining (NHEJ), single-strand annealing (SSA), or BIR. We propose mechanisms to explain the generation of the ILT postsenescence survivors, taking into account the requirement for the Rad1-Rad10 endonuclease and Mre11-Rad50-Xrs2 (MRX) complexes, together with the absence of strand invasion and of Rad52.
MATERIALS AND METHODS
Yeast strains and plasmids.
All strains used in this study were in the BF264-15D genetic background used in our laboratory (29). Strain origins, prior to backcrossing, were as follows. The rad51::KanMX4, rad52::KanMX4, rad59::KanMX4, rfa1::KanMX4/RFA1, dnl4::KanMX4, rad1::KanMX4, rad24::KanMX4, ctf18::KanMX4, nej1::KanMX4, and rif2::KanMX4 strains were purchased from Euroscarf (Frankfurt, Germany). The rad52-7::LEU2 strain was purchased from the Yeast Genetic Stock Center (Berkeley, CA) (strain record number XS560-1C-1D1). Noticeably, the rad52-7 null mutation corresponds to a disruption of the RAD52 open reading frame (ORF) resulting from insertion of LEU2 gene into the internal BglII site (located 402 bp downstream of the initiating ATG) of the RAD52 gene after introduction of digested pSM20 plasmid (63). On the other hand, the rad52::KanMX4 null mutation from Euroscarf represents a complete deletion of the whole RAD52 ORF (4, 72). The tlc1::LEU2 disruption construct was from the Gottschling laboratory (65). Construction of the yku80::TRP1 strain has been previously described (27). tlc1::TRP1 and tlc1::URA3 strains were obtained by disruption of the LEU2 marker by the TRP1 or URA3 marker in the backcrossed tlc1::LEU2 strain, and the yku80::LEU2 strain was obtained by disruption of TRP1 by LEU2 in the backcrossed yku80::TRP1 strain. The rad24::URA3 strain was from the Friedberg laboratory (64). The rad50::hisG-URA3-hisG strain was from the Haber laboratory (57). The mre11::LEU2 strain was from the Xiao laboratory (13). The stn1-13 mutation has been previously described (28, 29). The rfa1-t11 allele, from the Kolodner laboratory (70), was integrated at the RFA1 locus after cutting with NheI. The mre11-D16A (24) and mre11-H125L-D126V (9) alleles were expressed from a centromeric plasmid in the mre11Δ background. Yeast cells were grown in yeast extract-peptone-dextrose (YEPD) medium at 29°C unless otherwise indicated.
For construction of the elg1::URA3 deletion plasmid, URA3 was cloned between 305 bp of sequences located upstream of the ELG1 ATG and 308 bp downstream of its stop codon that had been initially cloned in pBluescript. The resulting PCR product of ELG1 ORF flanking sequences plus URA3 in the middle was directly transformed into yeast strains. Successful transformation resulted in deletion of the entire ELG1 ORF, as verified by Southern blotting. For construction of the rad1::URA3 disruption plasmid, URA3 was inserted at the endogenous BglII site of RAD1, located 2,346 bp downstream of the ATG, in a pBluescript plasmid that contained the last 1.9 kb of the RAD1 ORF (the total ORF is around 3.3 kb). The resulting PCR product of the last 1.9 kb of the RAD1 ORF sequences with inserted URA3 was directly transformed into yeast strains. Actual disruption of the RAD1 ORF was verified by Southern blotting. The CDC13-EST1 hybrid gene, cloned in a single-copy centromeric plasmid, allowed expression of an in-frame fusion protein under the control of the CDC13 natural promoter and terminated with the EST1 natural stop codon, as described previously (27). The codon for the first amino acid of Est1 in the hybrid construct directly followed the codon for the last amino acid of Cdc13 and was in frame with it.
Kinetics of senescence/survival and of growth rates.
The majority of the telomeric postsenescence survivors (∼ 90% in our strain background) are of type I and amplify the subtelomeric Y′ sequences, while the rest, of type II, amplify the TG1-3 repeats (48, 68) (Fig. 1C). Survivors can be isolated on agar-based culture plates as clones and identified by Southern blotting (see below). Indeed, in these “restreak assays,” performed on agar-based culture plates, restreaking of single colonies every 3 days (∼30 cell divisions per passage, at 29°C) allows the detection of both type I and type II survivors (48, 68). On the other hand, culturing the survivors among telomerase-negative mutants (here we used tlc1Δ mutants throughout, which were disrupted for TLC1, the RNA subunit of telomerase) in liquid medium rapidly leads to the generation (in 60 to 80 generations) of a population of survivors composed exclusively of type II survivors. This is due to the fact that the type II survivors grow much faster than the type I survivors and rapidly outgrow them in the liquid culture (68). In summary, type I survivor formation is favored by restreaking on agar but is undetectable in the liquid cultures in which type II survivors take over type I. Protocols for analyzing the occurrence of senescence and of postsenescence survival and growth rate have been published previously (25, 26).
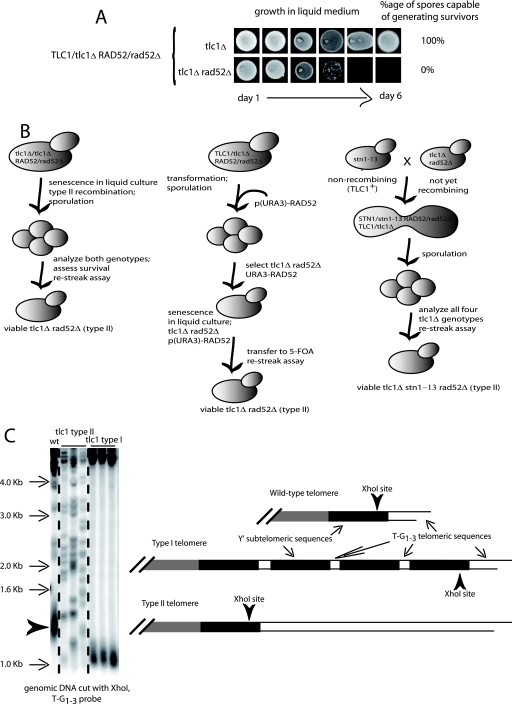
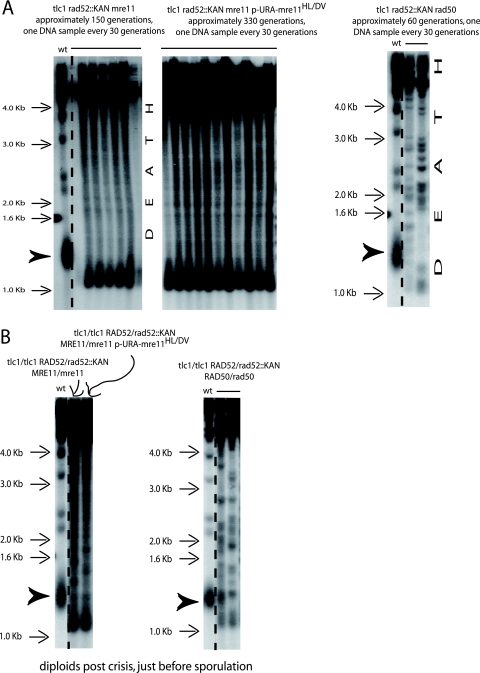
FIG. 1.
Telomeric recombination in budding yeast. (A) In the standard setup, a telomerase-negative (tlc1Δ) rad52Δ (::LEU2) double mutant issued from parents heterozygous for both TLC1 and RAD52 was unviable (lower row), unlike the tlc1Δ RAD52+ mutant (upper row). In this setup, the tlc1Δ mutant used to construct the original diploid was not yet recombining at the time of mating. Subsequently, following sporulation of the diploid and selection of the desired genotypes (day 1), cells were grown in liquid YEPD medium at 29°C and diluted down to 105 cells/ml every 24 h (one cell cycle is ∼90 min) for 6 continuous days. Aliquots of the cultures were dropped onto YEPD plates to assess viability every day (from one image to the next). (B) Schematic representation of three different methodologies used to overcome death by senescence in telomerase-negative cells (TLC1 encodes the RNA subunit of telomerase) in the absence of the RAD52 recombination protein. (C) Survival from telomerase inactivation generates two types of postsenescence telomeric recombination: type I, amplifying the Y′ subtelomeric sequences, and type II, amplifying the TG1-3 telomeric sequences, best identified after XhoI digestion as schematically explained. See also Materials and Methods.
Telomere organization and structure.
Genomic DNAs were prepared, separated, transferred, and hybridized with a 270-bp TG1-3 32P-labeled telomeric probe as described previously (29). Following digestion of genomic DNA with XhoI to cut within the Y′ regions of chromosomes (47), telomere tracts of wild-type cells appear as a broad band of ∼1.2 to 1.3 kb which represents the average length of most chromosomes, those containing Y′ subtelomeric regions. About one-third of S. cerevisiae chromosomes do not possess subtelomeric Y′, sequences. In these non-Y′ chromosomes, XhoI cutting typically generates fragments migrating at ∼2.1, 2.3, 3.3, and 3.9 kb in Southern blots (68). The disappearance of these non-Y′ fragments also attests to the fact that survivors have arisen by homologous recombination in senescing cells.
Type I and type II survivors were distinguished by Southern blot analysis following cutting of genomic DNAs with XhoI (26, 68). XhoI cleaves 0.9 kb from the 3′ end of the Y′ element, yielding ∼1.2 to 1.3 kb corresponding to these 0.9 kb plus ∼0.3 to 0.4 kb of terminal TG1-3 tracts. In type II survivors, XhoI cutting reveals the amplification of very long and heterogeneous TG1-3 sequences, located more distal than the single XhoI site that is present at the distal part of Y′ sequences. In contrast, in type I survivors, which amplify the Y′ subtelomeric sequences, the TG1-3 tracts have become very short, and since the amplified Y′ sequences are located more centromere proximal than the XhoI site, XhoI cutting reveals a fragment that is consistently ∼0.9 to 1.0 kb (Fig. 1C). The terminal parts of the Y′ elements contain short TG1-3 tracts, allowing type I survivors also to be conveniently detectable with a TG1-3 probe (68).
In some experiments, the genomic DNA was digested with a mixture of four restriction enzymes (AluI, HaeIII, HinfI, and MspI) using a 4-bp recognition sequence and the Southern blot was revealed with a TG1-3 probe. Since these enzymes cut within telomeric Y′ sequences but not within the TG1-3 sequences, they are currently used to confirm identification of type II telomeres (68). Results were analyzed using an FLA-5100 Fuji phosphorimager and the ImageGauge software.
The exact number of experiments for each mutant strain is provided in the corresponding figure legends. In general, for each mutant, at least three spores from three different crosses were selected and analyzed in terms of telomere organization, structure, and viability, under the conditions described above. Analysis of one spore's outcome represents one experiment.
RESULTS
Rad52-independent survival to telomeric senescence is possible when cells inherit telomeres already engaged in the process of recombination.
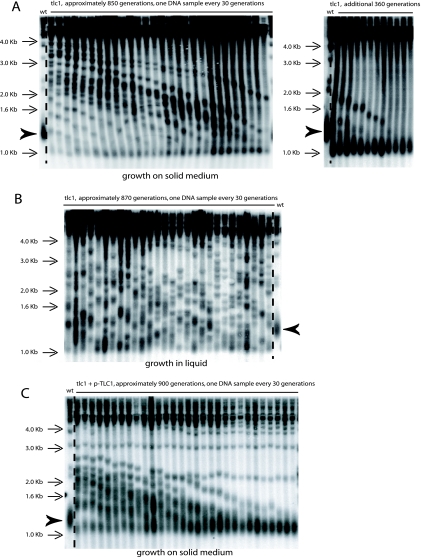
Telomerase-negative cells can escape death by senescence by elongating telomeres using an alternative pathway based on homologous recombination between repeated telomeric sequences. In the budding yeast Saccharomyces cerevisiae, Rad52 and either proteins of the Rad51 group or proteins of the MRX complex are essential for initiating this alternative pathway (8, 56). Accordingly, a telomerase-negative (tlc1Δ) rad52Δ double mutant was unviable (Fig. 1A), as previously established (48, 68). In a different setup, tlc1Δ/tlc1Δ RAD52+/rad52Δ (either rad52::LEU2 or rad52::KanMX4) diploids were initially propagated in liquid culture for ∼150 generations to induce telomeric recombination (Fig. 1B, left panel). Indeed, under such conditions, telomerase-negative cells, which die after 60 to 80 generations, have generated postsenescence survivors using the alternative pathway of telomere elongation by the end of this process of propagation in liquid culture (41). Importantly, all of these postsenescence survivors were of type II (amplification on the TG1-3 repeats), in contrast to type I survival, which amplifies the subtelomeric sequences (Fig. 1C). Indeed, in this protocol (Fig. 1B, left panel), we exploited the fact that the type II survivors grow much faster than the type I survivors and rapidly outgrow them in the liquid culture (68). Therefore, at this time of postsenescence growth, around 150 generations following telomerase inactivation, all of the tlc1Δ/tlc1Δ RAD52+/rad52Δ (rad52::LEU2 or rad52::KanMX4) diploids exhibited a type II pattern (data not shown) like the one illustrated in Fig. 1C (left panel). Following sporulation of these type II diploids, haploid mutants of the desired genotype, selected from tetrads with four viable spores, were propagated for extended periods of time by restreaking on agar-based plates prior to Southern analysis of telomeres. Importantly, restreaking cells at this point, rather than propagating them in liquid cultures, allowed recording of the occurrence of both types of recombination (see Materials and Methods). Strikingly, spores of the tlc1Δ rad52Δ (rad52::LEU2 or rad52::KanMX4) genotype issued from already recombining diploids generated survivors, thus contrasting with the situation described above in which all tlc1Δ rad52::LEU2 spores died. All of these tlc1Δ rad52Δ (rad52::LEU2 or rad52::KanMX4) survivors in this configuration, as well as in many different genetic contexts, had type II-like terminal restriction fragments (Fig. 2A and B and see below).
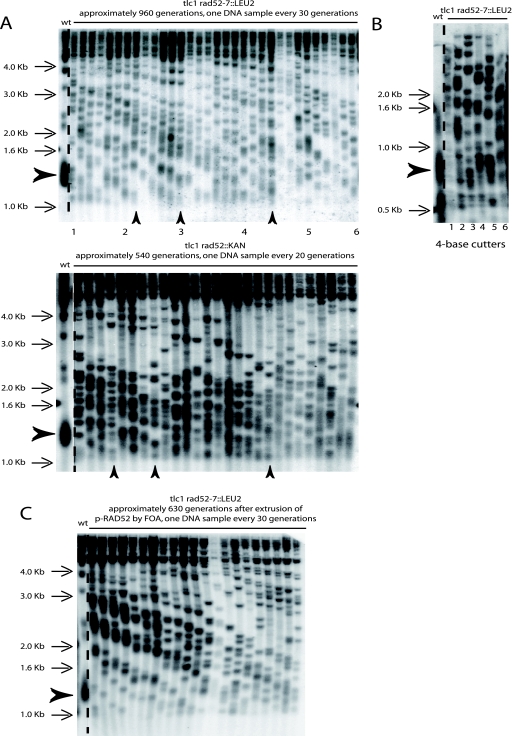
FIG. 2.
The ILT pathway of postsenescence survival amplifies telomeric TG1-3 repeats. (A) Representative Southern blots (XhoI digestion, TG1-3 probe) showing telomere organization in a tlc1Δ rad52Δ (rad52::LEU2 allele [top panel] or rad52::KanMX4 allele [bottom panel]) mutant obtained by sporulating a tlc1Δ/tlc1Δ rad52Δ/RAD52+ diploid already recombining in type II (following growth in liquid medium for ∼150 generations; see Fig. 1B, left panel). Cells were restreaked every 3 days on agar-based medium (YEPD, 29°C) for the indicated number of generations (around 30 generations per restreak under these conditions). The bulk size of wild-type telomeres, around 1.3 kb, is indicated by the arrowhead (lane 1). The three arrows at the bottom of each gel indicate three different times when critically short telomeres reelongated, indicated by the disappearance of the band with the lowest size at the next time point. Note that the slope of telomere shortening in the bottom panel is less than that in top panel because in the former, samples were taken every 20 generations, versus 30 generations in the latter. Ten experiments in total were performed with the rad52::LEU2 allele and eight with the rad52::KanMX4 allele. (B) Recutting samples 1 to 6 from the top panel in panel A with a mixture of restriction enzymes (AluI, HaeIII, HinfI, and MspI), using a 4-bp recognition sequence, which do not cut within the TG1-3 repeats (68; see also Materials and Methods) indicates that TG1-3 sequences are amplified in the Rad52-independent survivors. Rad52-dependent type I and II controls are shown in Fig. 4C. A TG1-3 probe was used. (C) Telomere organization in a tlc1Δ rad52::LEU2 strain (representative of a total of four experiments) which previously contained a RAD52-URA3 plasmid and was recombining in type II, after losing the RAD52 plasmid on 5-FOA (see Fig. 1B, middle panel). The same cells as those illustrated in Fig. 7B, first column, are shown. XhoI digestion and a TG1-3 probe were used.
The rad52::LEU2 mutation used in this study corresponds to a disruption of the RAD52 ORF, while in the rad52::KanMX4 mutant, the RAD52 ORF has been completely deleted (see Materials and Methods). At this point, it was important to make sure that when Rad52-independent recombination was assessed, the RAD52 gene was still in an inactivated or deleted form, like it was at the beginning of the experiment with the diploids. Southern analysis demonstrated the absence of the RAD52 ORF in all key tlc1Δ rad52::KanMX4 strains used in this study and its disruption in tlc1Δ rad52::LEU2 strains, after Rad52-independent survival had been recorded (see Fig. S1 in the supplemental material). Therefore, KanMX4 was still present in place of RAD52 at the RAD52 locus in these strains, and LEU2 was still inserted within the RAD52 ORF. Moreover, this analysis established that no other RAD52 ORF sequence was present elsewhere in the genomes of these strains. This ruled out the possibility that RAD52 might have been duplicated and inherited by the rad52::KanMX4 or rad52::LEU2 parent during the process of senescence. In additional controls, tlc1Δ rad52::LEU2 or rad52::KanMX4 strains were backcrossed several times to wild-type cells (nonrecombining TLC1+) (see Fig. S2 in the supplemental material). These successive crossing and sporulation steps had the effect of accelerating the process of resetting the telomere length at its wild-type level. Indeed, reintroducing wild-type-length telomeres by successive crosses had the effect of more rapidly “diluting” down, each time by half, the elongated telomeres. After checking that the TLC1+ rad52::LEU2 (or rad52::KanMX4) progeny had reacquired wild-type-length telomeres, these strains were next crossed to a nonrecombining tlc1Δ“young” strain. All tlc1Δ rad52::LEU2 and tlc1Δ rad52::KanMX4 selected spores failed to generate any postsenescence survivors and died, thus demonstrating that the recovered rad52::LEU2 and rad52::KanMX4 alleles had not reverted (see Fig. S2 in the supplemental material).
In summary, inheriting telomeres already engaged in the recombination process allows a significant percentage of cells to utilize a Rad52-independent survival pathway. We propose to call this new mechanism the ILT (for inherited-long-telomere) pathway of postsenescence survival. As explained in the introduction, we will frequently refer below to type II-ALT and type II-ILT to conveniently distinguish the two types of telomere amplification.
Growth characteristics of the ILT survivors.
It was interesting to see how the inherited type-II-like telomeres initially shortened in the haploid progeny, followed by sudden dramatic changes in the terminal restriction patterns, presumably at the time of each of the successive crises (Fig. 2A). To better characterize the ILT pathway, we performed quantitative time courses of postsenescence survival in liquid cultures by counting and diluting cells every day, together with replating assays to measure the ability of survivors to form colonies. The data (Fig. 3A and B) suggest that the ILT pathway was triggered when cell viability became very low, much lower than in the already-characterized type II-ALT and type I pathways. The telomere organization and structure of ILT cells (Fig. 2A) suggested that the sudden increase in cell viability after each of the troughs corresponding to crisis (Fig. 3A and B) was the direct result of excessive telomere shortening. In fact, although the telomere organization was recorded in survivors propagated on plates, while growth characteristics were from liquid cultures, it is likely that the phases in which ILT cells have short telomeres correspond to the slow-growth phases in liquid cultures.
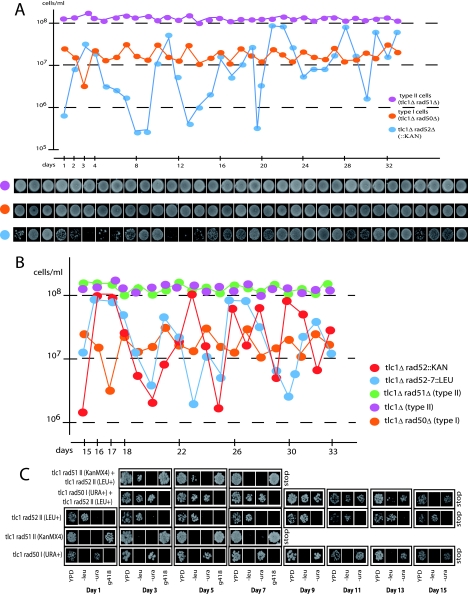
FIG. 3.
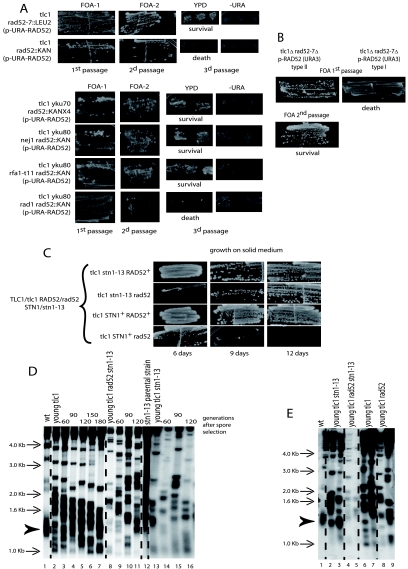
Growth characteristics of the ILT survivors compared with the Rad52-dependent type I and II-ALT survivors. (A) Cells of the indicated relevant genotype were grown in liquid cultures (YEPD, 29°C). The ILT survivors were obtained by sporulating an already-recombining tlc1Δ/tlc1Δ RAD52+/rad52::KanMX4 diploid that had grown for ∼150 generations in liquid culture to generate type II-ALT survivors (not shown). Type I survivors were stabilized as such in liquid culture because of the introduced RAD51 deletion. Cells of these three haploid mutants were counted and diluted every day to 105 cells/ml, and in parallel, 5 μl of each culture was dropped on agar medium, allowed to grow for 2 days, and photographed. (B) Another time course experiment, performed as for panel A, comparing in addition the rad52::KanMX4 and rad52::LEU2 strains. (C) Growth competition experiments between type I, type II-ALT, and type II-ILT survivors. Strains of the indicated relevant genotype (I and II refer to type I and type II survivors, respectively) were cultivated either separately (bottom three rows of cells) or as a mixture of two strains in the same flask (top two rows of cells) for 15 days, with dilution every day, in rich medium (YEPD) at 29°C (∼225 generations), and at intervals their growth was compared after spotting on selective media. To identify the strains having grown in the same culture, we took advantage of the nature of the auxotrophy conferred by the marker genes inserted at the disrupted loci, rad50::URA3, rad51::KanMX4, and rad52::LEU2. The ILT survivors (tlc1Δ rad52::LEU2) were rapidly outgrown by the Rad52-dependent type II survivors (tlc1Δ rad51::KanMX4) and competition stopped at day 7. On the other hand, the ILT and type I survivors were still competing after 11 days in culture, but the former then declined and disappeared. Southern analyses conducted in parallel established that these survivors were of the expected type of recombination (not shown). Only one experiment for each group of mutants (in panels A, B, and C) was performed.
To further characterize the ILT pathway, we performed growth competition experiments. The three different types of survivors, types I, II-ALT, and II-ILT, identified by the genetic markers of the mutations, were grown together as pairs in liquid culture, and every 2 days cells were diluted and plated out to identify which of the two strains had outgrown the other (Fig. 3C). In these experiments, the type II-ALT survivors completely outgrew the type II-ILT survivors after around 7 days (Fig. 3C). On the other hand, the ILT survivors grew just slightly worse than the type I survivors, as the latter cells completely took over in the mixed cultures only after 13 to 15 days of competition (Fig. 3C). Therefore, based on cell proliferation capacity, we suggest that the Rad52-independent (ILT) survival pathway is not a minor pathway of cell proliferation in the absence of telomerase.
Genetic requirements for the ILT pathway.
We next set out to further document the apparent similarity between the telomere amplification patterns in the ILT and type II-ALT survivors (Fig. 1C and 2A). Type I and type II-ALT postsenescence survivors, both resulting from Rad52-dependent telomeric homologous recombination, have been relatively well characterized in terms of their genetic requirements (14, 26, 38, 48, 68). For type I recombination on subtelomeric sequences, Rfa1 (RPA), Rad52, and Rad51 (Rad54, Rad55, Rad57) are essential and Rad50 and Rad59 are dispensable; for type II-ALT recombination on telomeric TG1-3 sequences, Rfa1 (RPA), Rad52, Rad50 (Mre11, Xrs2), and Rad59 are essential and Rad51 is dispensable. However, discrepancies in the literature exist in regard to Rad59, a recombination protein with homology to Rad52. Thus, in one study Rad59 was found to be essential for type II recombination (14), while in another study Rad59 facilitated type II recombination but its absence did not completely eliminate it (69). On the other hand, Rad50 was essential for long-term maintenance of type II recombination but not absolutely required for its initiation (14). In our strain background, in the presence of Rad52 and with the standard setup using non-previously recombining tlc1Δ strains, both Rad50 and Rad59 were essential for the initiation and maintenance of type II recombination, as established in previous studies (25, 26).
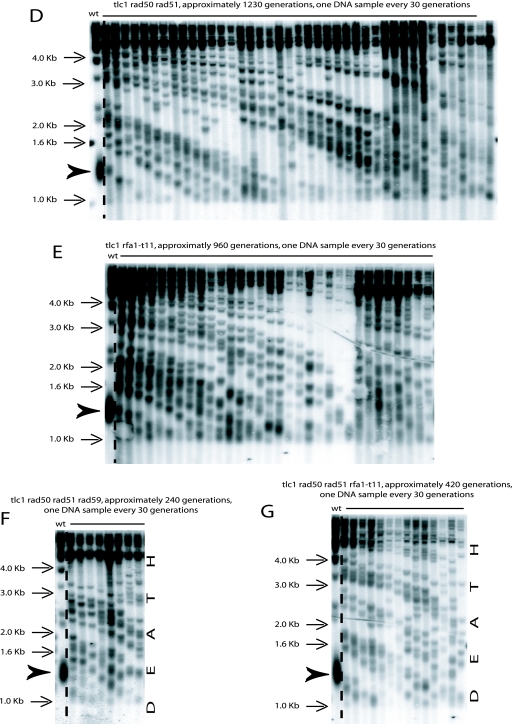
We found that in addition to Rad52, Rad51 and Rad59 were not required for the ILT pathway, using strains carrying either the rad52::LEU2 allele or the rad52::KanMX4 allele for rad51Δ and the rad52::LEU2 allele for rad59Δ (Fig. 4A and B; Table 1). In these strains, amplification of the telomeric sequences was also on the TG1-3 repeats (Fig. 4C), as observed above (Fig. 2B). On the other hand, Rad50 was essential for the ILT pathway (Fig. 5; Table 1). In the MRX complex, Mre11 is an ATP-stimulated nuclease that acts endonucleolytically and exonucleolytically on single-stranded DNA and hairpins and exonucleolytically on various types of double-stranded DNA ends. In initial experiments, we found that tlc1Δ rad52::LEU2 mre11-D16A mutants did not survive (data not shown). In parallel, we observed that the mre11-D16A mutation impaired type II recombination in the RAD52+ background (but not type I recombination, as expected) and that, as expected from earlier studies (24), the telomere size in telomerase-positive mre11-D16A cells was intermediate between that in mre11Δ cells and that in wild-type cells (data not shown). Like mre11-D16A, the mre11-H125L-D126V mutations confer a total lack of nuclease activity in vitro, but the Mre11-H125L-D126V mutant protein, unlike Mre11-D16A, retains the ability to interact with Rad50 (9, 24, 36, 58). As expected from the result with the mre11-D16A mutation, the tlc1Δ rad52::KanMX4 mre11Δ mutant (starting with overelongated telomeres) died (Fig. 5). Interestingly, survivors were recovered from the tlc1Δ rad52::KanMX4 mre11-H125L-D126V mutant strain (Fig. 5), indicating that Mre11 nuclease activity was not required for the ILT pathway. The absence of either MRX component, Rad50 or Mre11, led to early loss of viability in the ILT pathway, with, surprisingly, earlier death in tlc1Δ rad52::KanMX4 rad50Δ cells than in tlc1Δ rad52::KanMX4 mre11Δ cells (Fig. 5; Table 1).
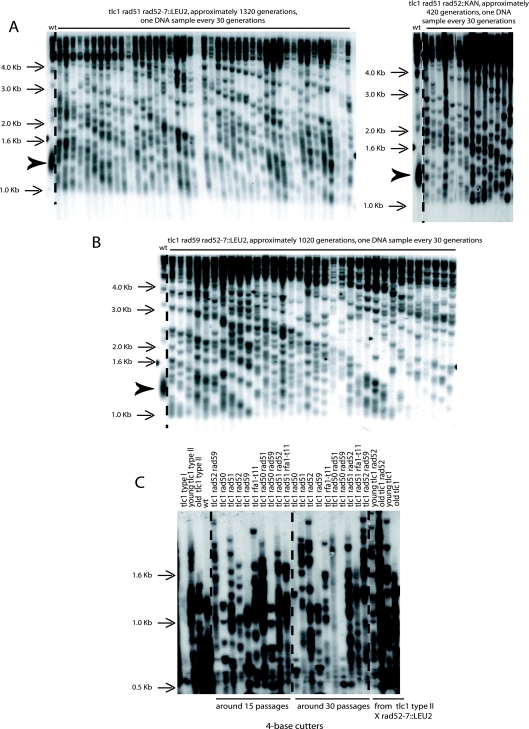
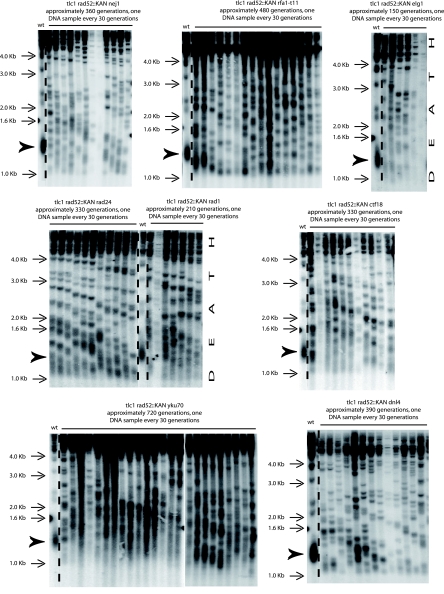
FIG. 4.
Telomere amplification can take place in the simultaneous absence of RAD51 and RAD52 (A) or of RAD59 and RAD52 (B) and is of type II in both cases, as attested here by the band pattern generated following XhoI digestion and use of a TG1-3 probe. Cells were restreaked on agar-based plates every 3 days for the indicated number of generations. Both strains originated from a diploid already recombining in type II. The bulk size of wild-type telomeres, around 1.3 kb, is indicated by the arrowhead (lanes 1). Totals of four, three, and four experiments were performed for the strains illustrated in panel A left, panel A right, and panel B, respectively. (C) Confirmation of the type II nature of telomeric recombination in various telomerase-negative (tlc1Δ) mutants. For this, genomic DNAs previously analyzed using XhoI cutting were recut with a mixture of restriction enzymes (AluI, HaeIII, HinfI, and MspI), using a 4-bp recognition sequence, which do not cut within the TG1-3 repeats. Unlike these type II survivors, in which cutting with these enzymes did not eliminate the presence of numerous telomeric bands of various high molecular weights, the type I survivors of the tlc1Δ rad50Δ (lanes 6 and 15), tlc1Δ rad59Δ (lanes 9 and 18), and tlc1Δ rad50Δ rad59Δ (lanes 12 and 21) strains did not exhibit fragments higher than the wild-type telomere bulk size, around 1.2 kb. All strains were isolated from already-recombining (in type II) tlc1Δ/tlc1Δ diploids. All rad52Δ mutants shown here were of the rad52::LEU2 background. DNA from a nonrecombining wild-type strain is shown in lane 4. A TG1-3 probe was used.
TABLE 1.
Genetic requirements of Rad52-dependent and Rad52-independent postsenescence survival pathways
| Relevant genotypea | Survivor typeb | Comments |
|---|---|---|
| tlc1 | I or II-ALT | Telomeres continuously shorten, as previously reported (68); after 700-800 cell divisions, telomeres convert from type II to type I most of the time (Fig. 9A) |
| tlc1 + TLC1 | Telomerase control | Normal telomere length regulation restored when TLC1 is reintroduced in type II survivors (Fig. 9C) |
| tlc1 rad50 | I | Rad59 essential for type II-ALT |
| tlc1 rad51 | II-ALT | Rad51 essential for type I |
| tlc1 rad52 (-7::LEU2 or ::KAN) | II-ILT | Evidence for existence of the ILT pathway (Fig. 2A) |
| tlc1 rad59 | I | Rad50 essential for type II-ALT |
| tlc1 rfa1-t11 | II-ALT | Rfa1 essential for type I |
| tlc1 rad50 rad51 | II-ALT | Rad50 dispensable for type II-ALT when Rad51 is absent |
| tlc1 rad50 rad52-7::LEU2 | — (6) | Rad50 essential for ILT |
| tlc1 rad50 rad52::KAN | — (3) | Rad50 essential for ILT |
| tlc1 rad50 rad59 | I | Rad50 and Rad59 essential for type II-ALT |
| tlc1 rad50 rfa1-t11 | — (6-15) | Rfa1 essential for type I |
| tlc1 rad51 rad52 (-7::LEU2 or ::KAN) | II-ILT | Rad51 dispensable for ILT |
| tlc1 rad51 rfal-tl1 | II-ALT | Rfa1 dispensable for type II-ALT |
| tlc1 rad59 rad52-7::LEU | II-ILT | Rad59 dispensable for ILT |
| tlc1 rfa1-t11 rad52::KANc | II-ILT | Rfa1 dispensable for ILT |
| tlc1 rad59 rfa1-tl1 | — (2) | Rfa1 essential for type I |
| tlc1 rad50 rad51 rad52-7::LEU2 | — (6) | Rad50 essential for ILT |
| tlc1 rad50 rad51 rad59 | — (15) | No possible survival in the absence of Rad50, Rad51, and Rad59 |
| tlc1 rad50 rad51 rfa1-t11 | — (15) | Rfa1 essential for type II-ALT in the absence of Rad50 |
| tlc1 rad50 rad59 rfa1-t11 | — (2) | Rfa1 essential for type I |
| tlc1 rad52::KAN | ||
| elg1d | — (7) | |
| rad1d | — (8) | |
| mre11d | — (6) | |
| rad50d | — (3) | |
| tlc1 rad52-7::LEU2 | ||
| rad50d | — (6) | |
| rad50 rad51d | — (6) | |
| tlc1 rad52::KAN | ||
| dnl4e | ||
| yku70e | ||
| nej1e | ||
| rad24e | ||
| ctf18e |
A “young” tlc1Δ RAD52+ mutant (prior to the onset of senescence) was mated to a “young” tlc1Δ mutant harboring one or several mutations in the indicated gene(s). The resulting diploid strain was then allowed to recombine during growth in liquid cultures for ∼150 generations, which yielded exclusively type II survivors. After sporulation, mutants of the desired genotype were selected and grown on agar-based plates. The cultures were then propagated by restreaking every 3 days (∼30 cell divisions per passage, at 29°C) for 90 to 130 days.
Survivor type (I or II) was determined after visualizing telomere structure (see Fig. 1C) by Southern blotting (see Materials and Methods). The samples were prepared after 90 days of growth on plates (following selection of the mutant as explained in footnote a), which corresponds to ∼900 cell divisions. In some cases, when phenotype confirmation was needed, cultures were further propagated for 14 additional passages. —, cell death at the passage indicated in parentheses.
Opposite results were obtained with the rad52-7::LEU2 background (see text).
Essential for ILT.
Dispensable for ILT.
FIG. 5.
Mre11 and Rad50 are essential to the ILT pathway, but Mre11 nuclease activity is dispensable for this process. (A) Left panel, deletion of MRE11 leads to the death of tlc1Δ rad52Δ cells which have undergone the senescence process with overelongated telomeres (see Fig. 1B, left panel). Middle panel, telomere organization in ILT survivors from tlc1Δ rad52Δ mre11Δ cells harboring the nuclease-deficient mre11-H125L-D126V allele on a centromeric plasmid (recombining in type II-ALT at the onset of senescence, at the beginning of the kinetics shown here). Right panel, the tlc1Δ rad52Δ rad50Δ cells that had undergone senescence in the presence of overelongated telomeres also died, but earlier than the mre11Δ mutant. The arrowheads indicate the size of the bulk of wild-type telomeres Three, four, and six experiments were performed in total for the strains illustrated in the left, middle, and right panels, respectively. (B) Since the pattern of telomere organization in the mre11-H125L-D126V ILT survivors was different from that in the other ILT survivors (see, for instance, Fig. 2A and 4A and B) and the pattern in mre11Δ cells was different from that in rad50Δ cells, as seen above, telomere organization in the parental diploid strains was analyzed. The mre11 pattern (darkened here, as well as in panel A, to visualize intermediate-size bands), characterized by a more abundant species of the smaller bands, was also present in the parental diploid strain (left panel), while the rad50 diploid exhibited a normal type II-like pattern (right panel). We assume that haploinsufficiency of MRE11 is probably the cause for this observed atypical type II pattern, with the smaller bands accounting for a tendency to type I recombination due to limiting amounts of Mre11 in the diploid (Mre11 is essential for type II-ALT recombination). A TG1-3 32P-labeled probe was used with XhoI cutting.
Since Rad59 is required for Rad52-dependent type II recombination, as previously shown (14; see above), but dispensable for the ILT pathway, as described above, we reasoned that mechanisms other than homologous recombination might underlie the ILT pathway. We selected several genes that play a major role in basic DNA repair mechanisms and asked whether they were required to sustain the ILT pathway. At this point, we noticed that the rfa1-t11 mutation prevented the generation of ILT survivors in the tlc1Δ strain carrying the rad52::LEU2 allele but not in that carrying the rad52::KanMX4 allele. A likely explanation was that in strains with the rad52::LEU2 disruption, the first 134 amino acids of Rad52 might still be produced (see Fig. S1 in the supplemental material) and interfere with other proteins, possibly trapping Rfa1 somewhere on the DNA breaks. We therefore decided, at this point, to exclusively use strains carrying the rad52::KanMX4 total deletion to continue our genetic analysis.
The Rad1-Rad10 endonuclease complex has been implicated in several pathways of DNA repair, and its main function is to remove 3′ flaps at single-stranded/duplex DNA junctions (67). Interestingly, deletion of RAD1 was found to inhibit the ILT pathway (Fig. 6; Table 1). Results were similar when a rad1::URA3 disruption instead of the rad1 total deletion was used. In contrast, RAD1 was dispensable for type II recombination in the presence of Rad52 (data not shown). An important pathway of double-strand break repair in which Rad1-Rad10 has been implicated is SSA. SSA, mainly restricted to breaks occurring between direct repeats, might potentially be used in the ILT pathway (see Discussion). RPA, a single-stranded DNA binding complex, is essential for SSA (67). Since all three subunits of RPA are essential, we used the best-documented recombination-deficient RPA allele, rfa1-t11 (70). rfa1-t11 mutants were proficient in the ILT pathway, as tlc1Δ rad52::KanMX4 rfa1-t11 originating from already-recombining cells with overelongated telomeres, as usual, generated survivors (Fig. 6). By comparison, tlc1Δ rfa1-t11 mutants have been shown to be defective in type II-ALT recombination (26). To confirm these important findings, we repeated the experiments with the rad1 and rfa1 mutants using the 5-fluoroorotic acid (5-FOA) protocol. In this protocol, recombining haploid cells containing a RAD52-URA3 plasmid are later challenged for plasmid loss using counterselection on 5-FOA medium (Fig. 1B, middle panel). In these experiments, tlc1Δ rad52::KanMX4 yku80Δ pRAD52-URA3 mutants (the yku80 mutation was used to improve the rate of the ILT pathway; see below) containing, in addition, either the rfa1-t11 or nej1Δ mutation and already recombining in type II, could survive the loss of the RAD52-URA3 plasmid on 5-FOA, while those containing the rad1Δ mutation could not lose the RAD52 plasmid. This confirmed that Rad1 is essential for the ILT pathway, while Nej1 (see below) and Rfa1 are not (Fig. 7A, bottom panel).
FIG. 6.
Elg1 and Rad1 are essential for the ILT pathway, while Dnl4, Rad24, Ctf18, Nej1, Yku70, and recombination-proficient RPA are dispensable. For all triple mutants of the indicated relevant genotype, the telomere structure is shown from the onset of senescence, triggered after sporulation of the diploids (which were already recombining in type II-ALT), to death (as indicated) or during the indicated number of generations. The arrowheads indicate the size of the bulk of wild-type telomeres. Three experiments were performed for the nej1Δ and dnl4Δ mutants, four for the rad24Δ and ctf18Δ mutants, five for the rfa1-t11 mutant, and six for the elg1Δ, rad1Δ, and yku70Δ mutants. A TG1-3 32P-labeled probe was used with XhoI cutting.
FIG. 7.
Loss of telomere capping promotes the ILT pathway. (A) Top panel, tlc1Δ rad52Δ (either ::LEU2 [first row] or ::KanMX4 [second row]) haploids (originating from a type II-ALT recombining diploid and harboring a centromeric plasmid carrying RAD52-URA3 marker [see Fig. 1B, middle panel]) were challenged for RAD52 plasmid loss on 5-FOA plates for URA3 counterselection. Survivors readily appeared on 5-FOA in the tlc1Δ rad52::LEU2 mutant (first passage, cells photographed 3 days after restreaking) and maintained viability upon subsequent passages (photographed 3 days after second passage). Actual loss of the RAD52-URA3 plasmid was verified on plates lacking uracil. Survivors also appeared in the tlc1Δ rad52::KanMX4 mutant but were not maintained and later died. These results were confirmed in two additional experiments for each strain. Bottom panel, deletion of YKU70 in the tlc1Δ rad52::KanMX4 mutant, illustrated above, led to extrusion of the RAD52-URA3 plasmid and survival (first row). Rows 2 to 4, tlc1Δ rad52::KanMX4 yku80Δ pRAD52-URA3 mutants (the yku80 mutation was used to improve the rate of the ILT pathway) containing, in addition, either the rfa1-t11 or nej1Δ mutation and already recombining in type II, could survive the loss of the RAD52-URA3 plasmid on 5-FOA, while those containing the rad1Δ mutation could not lose the RAD52 plasmid and died. For each of the strains illustrated in the top three rows, the results were confirmed in another experiment, and for the tlc1Δ yku80Δ rad1Δ rad52Δ mutant they were confirmed in three other experiments. (B) Same protocol as for panel A, using tlc1Δ rad52::LEU2 mutants previously recombining either in type II-ALT (left column) or in type I (right column), selected from an agar plate on the basis of their telomere organization by Southern blotting. In contrast to the type II survivors, type I survivors could not extrude the RAD52 plasmid and died (with no survivors appearing even during the first passage). Four experiments for each strain were performed. Telomere organization of the type II-ALT survivors is shown in Fig. 2C. (C) A mixed population of “young” tlc1Δ rad52::LEU2 haploid a cells (prior to senescence onset) and “old” stn1-13 haploid α cells (with telomeres equilibrated in the overelongated state [see panel D, lane 12]) were induced to sporulate without zygote isolation in order to gain time and avoid extensive shortening of the long stn1-13-induced telomeres before analysis (see Fig. 1B, right panel). Spores with the indicated relevant genotype were restreaked every 3 days on agar-based medium (YEPD, 29°C) in order to assess the appearance of postsenescence survivors. At the time the leftmost pictures were taken (6 days had then elapsed since the time of spore selection), the tlc1Δ stn1-13 RAD52+ (first row) and tlc1Δ STN1+ RAD52+ (third row) cells had already undergone a senescence crisis and recombined (not shown), while the tlc1Δ rad52Δ stn1-13 cells were still in senescence crisis (second row). The latter strain generated survivors at the next time point. Meanwhile, the tlc1Δ STN1+ rad52Δ cells (fourth row) died without generating survivors. Although telomeres were initially overelongated to a similar extent in all strains (see panel E), the presence of the stn1-13 mutation was required to allow postsenescence survival in the absence of Rad52 (second row). (D) Initially, telomeric patterns were similar for the tlc1Δ, tlc1Δ rad52Δ stn1-13, and tlc1Δ stn1-13 spores (lanes 2, 8, and 13 [“young”], respectively; the number of generations attained at the time of sample preparation is indicated above each lane). However, the tlc1Δ rad52Δ stn1-13 and tlc1Δ stn1-13 strains exhibited signs of telomeric recombination (lanes 9 to 11 and 14 to 16, respectively) while still having overelongated telomeres. In contrast, the overelongated telomeres of the tlc1Δ RAD52+ STN1+ strain progressively shortened without recombining (lanes 3 to 7). All three tlc1Δ strains contained shorter-than-wild-type telomeres, in addition to their stn1-13-elongated telomeres, inherited from their tlc1Δ rad52Δ parent. The bulk size of wild-type telomeres, around 1.3 kb, is indicated by the arrowhead (lane 1). (E) Telomeric patterns in the four tlc1Δ strains (patterns for two individual spores each are shown) illustrated in panels C and D. All four young tlc1Δ strains are basically undistinguishable from each other (with the exception of tlc1Δ rad52Δ stn1-13 strain, which generated much less material due to poor growth), with all containing both overelongated telomeres inherited from the “old” stn1-13 parent and shorter-than-wild-type telomeres due to the tlc1Δ mutation in the other parent. Data from panel C were confirmed in two other experiments; data from panels D and E were from the cells shown in panel C. A TG1-3 32P-labeled probe was used with XhoI cutting.
Elg1 plays a central role in genome stability by associating with the Rfc2 to -5 subunits of replication factor C (RFC) to form an RFC-like complex (RLC) and plays a role in telomere length regulation, albeit in an unknown manner (7, 66). ELG1-deleted cells did not support the ILT pathway, with death occurring during the seventh passage, after ∼200 generations (Fig. 6). In contrast, Elg1 was dispensable for telomeric recombination in the presence of Rad52 (data not shown). Rad24 and Ctf18 replace Elg1 in two additional RLCs (7). Deletion of either RAD24 or CTF18 did not inhibit the ILT pathway (Fig. 6; Table 1).
Deletion mutations of either the DNL4 ligase or YKU70 did not suppress the ILT pathway (Fig. 6; Table 1). Although Nej1, like Dnl4 and Yku70, is essential for NHEJ (16), we nevertheless analyzed the tlc1Δ rad52::KanMX4 nej1Δ mutant. Indeed, surprisingly, Nej1 represses the ligation function of Dnl4 at eroding telomeres and appears in fact to inhibit the lethal formation of circular chromosomes (46). Our data established that Nej1 was not required for the ILT pathway (Fig. 6 and 7A) and, in addition, did not appear to inhibit it (data not shown). Inactivation of the EXO1 exonuclease is required for a telomerase- and Rad52-independent pathway of immortalization in budding yeast, the so-called palindrome-dependent (PAL) mechanism (55). However, we note that here, the ILT pathway can take place in the presence of Exo1 in tlc1Δ rad52::KanMX4 mutants and is therefore distinct from the PAL mechanism (see Discussion).
Increasing telomerase accessibility together with weakening telomere capping can also trigger the ILT pathway.
Ku, a DNA repair complex functioning mainly in NHEJ in yeast, also has an important role in telomere end protection (21). Thus, yku70Δ tlc1Δ mutants exhibit accelerated senescence, while yku70Δ cdc13-1 mutants (Cdc13 binds telomeric DNA and together with its physical partner, Stn1, protects telomeres) undergo senescence even in the presence of functional telomerase. Already-recombining type II-ALT tlc1Δ rad52::LEU2 cells harboring a RAD52-URA3 plasmid (experimental setup shown in Fig. 1B, middle panel) could extrude the plasmid on 5-FOA medium, which was used for counterselection against URA3 (Fig. 2C and 7A, top panel, first row), while cells of the same genotype but bearing the rad52::KanMX4 allele extruded the RAD52 plasmid but died soon after (Fig. 7A, top panel, second row). Therefore, the rad52::LEU2 and rad52::KanMX4 constructs are not equivalent, as seen above. Importantly, RAD52 loss could be achieved in the tlc1Δ rad52::KanMX4 mutant (also starting with overelongated telomeres) when YKU70 had been deleted (Fig. 7A, bottom panel, first row). On the other hand, under the same conditions, type I tlc1Δ rad52::LEU2 p-RAD52 cells could not generate any survivors (Fig. 7B). Therefore, survivors already recombining on TG1-3 sequences can extrude Rad52 upon selective pressure, while those amplifying the subtelomeric Y′ sequences cannot. However, we note that type I survivors grow poorly, being continuously in a senescence crisis state due to their very short TG1-3 tracts. Therefore, this leaves open the possibility that the ILT pathway is also present in type I survivors but that nonspecific growth defects prevent its utilization.
It could be argued that postsenescence survival in the apparent absence of Rad52 could result from previous accumulation of Rad52 in the recombining parent of the tlc1Δ rad52Δ cells. To address this issue, we used the stn1-13 mutation, which provokes dramatic telomere elongation in a telomerase-dependent manner (27-29). tlc1Δ rad52::LEU2 stn1-13 triple mutant cells (originating from the cross between a “young,” nonrecombining tlc1Δ rad52::LEU2 mutant and an stn1-13 TLC1+ mutant with already-elongated telomeres but nonrecombining [Fig. 1B, right panel]) could generate postsenescence survivors in the restreak assay (Fig. 7C, second row). This did not occur in 100% of the spores, unlike in the presence of Rad52, but in only ∼60% of the spores (9 out of 15). Surprisingly, the tlc1Δ rad52::LEU2 STN1+ spores from the same mating did not generate survivors (Fig. 7C, fourth row). Yet, in these cells, born from one parent with stn1-13-elongated telomeres, half of the telomeres were still very long (Fig. 7E, lanes 8 and 9) because the stn1-13 mutant used to construct the diploid had telomeres stabilized in the maximally elongated state (Fig. 7D, lane 12). Therefore, we assume that altered telomere cap structure induced by the stn1-13 mutation occurring together with the presence of overelongated telomeres could trigger the ILT pathway. In support of this assumption, we noted that stn1-13 induced a very rapid disorganization of the telomeres, reminiscent of an accelerated senescence phenomenon. Indeed, the tlc1Δ stn1-13 (rad52::LEU2 or RAD52+) mutants recombined at as early as 60 generations (Fig. 7D, lanes 9 to 11 and 14 to 16), while in contrast, the tlc1Δ mutants from the same mating had not recombined yet after 180 generations (Fig. 7D, lanes 3 to 7) and eventually recombined only after their overelongated telomeres had become short again. This indicates a very strong telomere instability, which we propose can only be the result of the combination of the stn1-13 and tlc1Δ mutations and is directly at the origin of this early telomeric recombination, whether Rad52 is present or absent. Note that in this context, the tlc1Δ rad52::LEU2 mutant did not survive postsenescence (Fig. 7C) because the tlc1Δ rad52::LEU2 parent was a nonrecombining strain at the time of mating with the stn1-13 parent, as described above.
We attempted to mimic the effects of stn1-13 described above by using an alternative means to induce telomerase-mediated telomere elongation. Deletion of RIF2, which also provokes an increase in telomere length due to deregulation of Rap1 (73), did not allow postsenescence survival under the conditions used for stn1-13 (data not shown). Expression of a CDC13-EST1 fusion construct, which also provokes telomere overelongation as a result from direct tethering of telomerase to the telomeres (19, 27), did not induce the ILT pathway (data not shown). The rif2Δ mutation and the Cdc13-Est1 fusion not only provoke more moderate telomere elongation than the stn1-13 mutation but, unlike stn1-13, do not affect telomere end protection.
Characteristics of Rad52-dependent recombination starting with overelongated telomeres versus wild-type telomeres.
To further characterize the already well known Rad52-dependent type I and II (ALT-like) reactions, we analyzed haploid cells that initiated telomeric recombination with overelongated telomeres but this time in the presence of RAD52. Indeed, in all setups published to date, senescence was always induced in cells possessing wild-type-length telomeres prior to telomerase inactivation. Here, we compared the progeny originating from non-previously recombining diploids (one telomerase-positive parent) and therefore possessing wild-type-size telomeres with that originating from diploids previously induced to recombine in liquid culture (the two parents were telomerase negative) and therefore possessing overelongated telomeres of type II-ALT (Table 2).
TABLE 2.
Effects of telomere size (at the time of senescence) on telomeric recombination on solid medium in telomerase-negative (tlc1Δ) mutantsa
| Survivor fate and type of telomeric recombination initiated in the presence of wild-type-length telomeresb | Relevant genotype | Survivor fate and type of telomeric recombination initiated in the presence of overelongated telomeresc |
|---|---|---|
| I or II | tlc1Δ | II then Id |
| I | tlc1Δ rad50Δ | I |
| II | tlc1Δ rad51Δ | II |
| Death | tlc1Δ rad52Δ | II |
| I | tlc1Δ rad59Δ | I |
| I | tlc1Δ rfa1-t11 | II |
| Death | tlc1Δ rad50Δ rad51Δ | II |
| I | tlc1Δ rad50Δ rad59Δ | I |
| NDe | tlc1Δ rad50Δ rfa1-t11 | Death (6) |
| Atypical I | tlc1Δ rad51Δ rfa1-t11 | II |
| Death | tlc1Δ rad59Δ rfa1-t11 | Death (2) |
| Death | tlc1Δ rad50Δ rad51Δ rad59Δ | Death (8) |
| Death | tlc1Δ rad50Δ rad51Δ rfa1-t11 | Death (15) |
| Death | tlc1Δ rad50Δ rad59Δ rfa1-t11 | Death (2) |
A RAD52+ background was used throughout except for the tlc1Δ rad52Δ strain (row 4). Restreak assays were performed.
Numbers in parentheses indicate the number of passages before death occurred (∼30 generations per passage).
See Fig. 9.
ND, not determined.
A number of events were similar whether Rad52-dependent telomeric recombination had been initiated in the presence of wild-type telomeres or overelongated telomeres (Table 2). Thus, both Rad50 and Rad59 were essential for the initiation and maintenance of type II recombination in both setups (Fig. 8A, B, and C; Table 2). Similarly, Rad51 was essential for type I recombination in both setups (Table 2). Accordingly, as expected, the tlc1Δ rad50Δ rad51Δ rad59Δ cells died in both setups (Table 2). A difference arose in the case of Rad59. As reported above, both Rad50 and Rad59 are essential for the type II-ALT pathway. However, in cells initiating recombination with overelongated telomeres and when RAD51 had been deleted, Rad50 was no longer essential for type II-ALT and could then presumably be replaced by Rad59 (tlc1Δ rad50Δ rad51Δ strain) (Table 2; Fig. 8D).
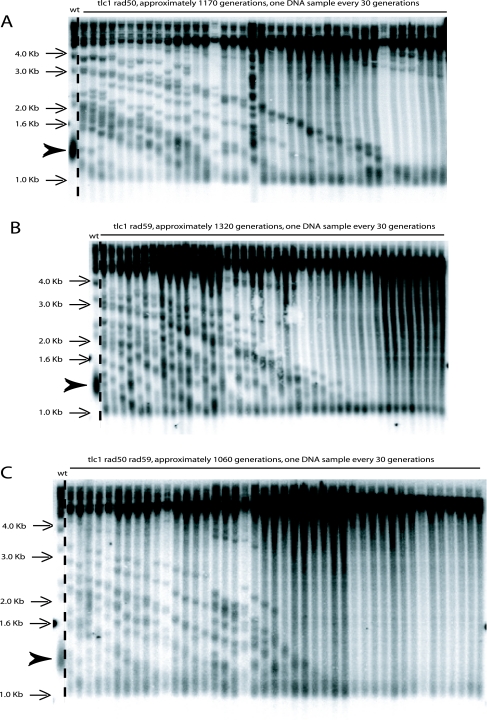
FIG. 8.
Kinetics of telomeric recombination in RAD52+ telomerase-negative (tlc1Δ) cells in the absence of Rad50 (A), of Rad59 (B), of both Rad50 and Rad59 (C), or of both Rad50 and Rad51 (D). Data obtained with the tlc1Δ rfa1-t11 strain (E) showed that the essentiality of RPA in telomeric recombination (26) could be relieved by overelongated telomeres. Rad50 and Rad59 are individually essential for type II-ALT recombination (A and B) (both strains end up with a type I pattern), but in the absence of Rad51, Rad50 becomes dispensable for type II-ALT recombination, presumably carried out by Rad59 (D). The tlc1Δ rad50Δ rad51Δ rad59Δ (F) and tlc1Δ rad50Δ rad51Δ rfa1-t11 (G) mutants died at the 8th and 15th passage, respectively. Note that all seven strains were RAD52+. In all seven representative examples shown here, tlc1Δ mutants were issued from a type II-ALT recombining diploid homozygous for tlc1Δ and also bearing one or several additional null, heterozygous mutations in the RAD50, RAD51, or RAD59 gene. After sporulation of the obtained diploid, mutants of the desired genotype were restreaked on agar-based plates every 3 days for the indicated number of generations (estimating that at the temperature of 29°C, ∼30 generations were produced every 3 days on plates). The intense band at 1.2 to 1.3 kb in each first lane (wt), indicated by an arrowhead, represents the average size of the bulk of wild-type telomeres. Digestion of the genomic DNA preparations with XhoI followed by Southern blotting with a TG1-3 probe allowed the two types of recombination to be distinguished. Type I recombination (on the Y′ subtelomeric regions) yielded an XhoI-restricted terminal fragment around 0.9 to 1.0 kb, while type II survivors amplifying the TG1-3 repeats exhibited many XhoI fragments of different sizes, as explained in Materials and Methods. For each of the mutants illustrated in panels A to E and G, four experiments were performed, while three experiments were done for the mutant shown in panel F.
The case of Rfa1 illustrated another difference between the two setups (wild-type length versus overelongated telomeres). Thus, impairment of the type II-ALT pathway by the recombination-defective rfa1-t11 mutation was relieved by the initial presence of overelongated telomeres (tlc1Δ rfa1-t11 strain) (Table 2; Fig. 8E). Type II-ALT in this strain relied on Rad50 (the tlc1Δ rad50Δ rfa1-t11 died [Table 2]). On the other hand, the initial presence of overelongated telomeres did not relieve the requirement of type I cells for Rfa1 under certain conditions, that is, in the absence of Rad50 or Rad59 (the tlc1Δ rad59Δ rfa1-t11 and tlc1Δ rad50Δ rad59Δ rfa1-t11 strains died in both setups [Table 2]).
We also observed that in some tlc1Δ RAD52+ mutants that were bound to die (because of a lack of particular recombination genes other than RAD52), death occurred only after a very long delay and at different times depending on the mutants (Table 2). Because the telomeres were already elongated at the time of genotype selection, a delay was expected, corresponding to “passive” telomere shortening with cell divisions, prior to senescence induction. However, surprisingly, that delay varied between 2 and 15 passages postsporulation (∼60 to 450 generations). Thus, the tlc1Δ rad50Δ rad51Δ rad59Δ and tlc1Δ rad50Δ rad51Δ rfa1-t11 mutants died after the 8th and 15th passages, respectively (Fig. 8F and G; Table 2). In contrast, two other tlc1Δ RAD52+ mutants, the tlc1Δ rad59Δ rfa1-t11 and tlc1Δ rad50Δ rad59Δ rfa1-t11 strains, died very early, at the second passage (Table 2). The lack of both Rad59 and functional RPA in these two strains was possibly responsible for this rapid death.
While performing control experiments to verify that reintroduction of TLC1 into a tlc1Δ strain with previously recombining telomeres restored normal telomerase-driven telomere length control (Fig. 9C), we observed an event that might be useful for further investigation of telomerase-independent survival. Telomeres of type II survivors continuously shortened when propagated on agar-based medium, as previously described (68), but were, in most cases, converted to an apparent type I pattern (Fig. 9A). Interestingly, this was also the case for tlc1Δ/tlc1Δ homozygous (but not for tlc1Δ/TLC1 heterozygous) diploids grown on agar-based medium (71). In contrast, when the tlc1Δ RAD52+ survivors were propagated in liquid culture, they conserved a type II pattern even for extended periods of time (Fig. 9B). Importantly, this phenotype of overall telomere shortening with time was observed whether recombination started with wild-type-length telomeres or with previously elongated telomeres. In contrast to this type I conversion taking place in old tlc1Δ RAD52+ survivors grown on semisolid medium, in the absence of Rad52 a type II pattern was conserved after extended periods of growth on agar-based plates (see, for instance, Fig. 2A). This was expected because the ILT pathway exhibited only type II-like terminal restriction fragments.
FIG. 9.
Evolution of postsenescence survival type with time in tlc1Δ RAD52+ cells. (A) In telomerase-negative mutants grown on solid (agar-based) medium, telomeres shortened progressively with passages (cells were restreaked, at 29°C, on agar-based plates every 3 days for the indicated number of generations). These survivors still exhibited a type II pattern until they attained their maximal shortening (left panel). At this point, telomeres in most of these old tlc1Δ mutants converted to a type I pattern (right panel), as seen in a total of 12 experiments. Indeed, the presence of an intense band at ∼0.9 to 1.0 kb following XhoI digestion, as well as the absence of any fragment above that size, suggested amplification of Y′ subtelomeric sequences (see Materials and Methods). In the remaining three cases, one or two additional bands remained visible over the ∼1.0-kb band (not shown). (B) When tlc1Δ mutants were propagated in liquid cultures, recombination on the telomeric TG1-3 sequences, indicated by the reappearance of numerous high-molecular-weight bands following XhoI digestion, persisted even after very long periods of time, as also seen in three additional experiments. The bulk size of wild-type telomeres, around 1.3 kb, is indicated by the arrowhead (lanes wt). A TG1-3 probe was used for panels A and B. (C) Pattern of Rad52-dependent telomeric recombination in a tlc1Δ mutant expressing TLC1 on a centromeric plasmid, introduced in the already-recombining tlc1Δ/tlc1Δ diploid. Progressively, the telomeric pattern varies from one typical of ongoing recombination (at the time of TLC1 introduction) to one indicating telomerase-based telomere length homeostasis, with, however, telomeres eventually stabilizing at a shorter-than-wild-type size, indicated by the arrowhead (wt, lane 1). Only one such experiment was performed. XhoI digestion and a TG1-3 probe were used.
DISCUSSION
The ILT pathway.
We report here on the identification of a novel mechanism of telomeric postsenescence survival, which we call the ILT pathway, that functions in the absence of the essential Rad52 homologous recombination protein. The differences between the ILT and the ALT-like pathways, both amplifying the TG1-3 repeats (type II), were an absence of a requirement for Rad52, Rad59, and functional RPA for the ILT pathway, in contrast to the type II-ALT pathway, which requires all three proteins. Gene conversion, which normally requires both Rad51 and Rad52, can be possible in the absence of Rad51 for ectopic events but, surprisingly, also for interchromosomal conversion (61, 62, 67). Allelic conversion is also possible in the absence of Rad52 but not in the absence of both Rad51 and Rad52 (62). It is assumed that each of these proteins can perform part of the other one's functions in its absence. Moreover, in a class of rad52 mutants deficient for double-strand break repair, but not in rad52Δ mutants, spontaneous homologous recombination was induced by DNA nicks and single-stranded gaps possibly generated at stalled replication forks (42). Finally, spontaneous and UV-induced Rad52-independent recombination, proceeding by yet-unidentified mechanisms requiring Rad50, functioned either through Rad51 and Dun1 or through Rad59 and Crt1 (15). Although the ILT mechanism has not been elucidated yet at the molecular level, there are several clues provided by our genetic analysis that strongly suggest that ILT survivors rely not on homologous recombination but rather on end-joining mechanisms (see our working model described below).
The ILT pathway appears to represent an important pathway because it is utilized by cells that experience loss of telomerase function, an important process linked to cancer biology. Cancerous cells may be able to use not only the only one alternative telomere maintenance mechanism to telomerase reactivation known to date, the ALT pathway, but also an equivalent of the budding yeast ILT pathway. This putative mammalian ILT pathway would be important, not only because it utilizes or not RAD51 and RAD52 but because it potentially functions on different telomeric DNA substrates than the ones identified to date. In this respect, we note that the budding yeast ILT survivors exhibited very deep troughs of loss of viability compared with those in type II-ALT and type I survivors (Fig. 3). This points out the possibility that the ILT mechanism might be intervening much less frequently than the ALT mechanism, due, for instance, to small amounts of a putative ILT protein substituting for Rad52. Alternatively, the particular telomeric substrate needed for the ILT pathway may take time to accumulate in the appropriate form.
The ILT pathway is distinct from the PAL pathway, a telomerase- and recombination-independent pathway recently identified also in budding yeast (55). The PAL survivors initially experienced chromosome loss starting at the telomeres and progressing toward the centromeres, followed by palindrome formation between inverted repeats, which then could serve as a template to resynthesize chromosomal DNA, possibly by a BIR-like mechanism (55). In contrast, the ILT survivors described here did not exhibit chromosome degradation, as far as we can tell from the Southern blots. Moreover, EXO1+ senescent cells never generated PAL survivors, presumably due to the Exo1 exonuclease being essential for generating single-stranded DNA required for cell cycle arrest (55), while here the ILT survivors occurred in the presence of Exo1. In addition, deletion of MRE11 increased the rate of PAL survivor formation (55). Here, in contrast, Mre11 was essential for the ILT pathway and mre11Δ mutants never formed ILT survivors.
The ILT survivors grew more slowly than those utilizing the Rad52 pathway. Possibly, DNA repair, which is less efficient in the absence of Rad52, leads to DNA damage being permanently left unrepaired. The ILT pathway is not a minor pathway in that it conferred only slightly less efficient cell proliferation than the Rad51- and Rad52-dependent major pathway (type I) of survival (Fig. 3). Note that although type II-ALT survivors overcome type I survivors in liquid culture, the latter is nevertheless a majority pathway, occurring in ∼65% of the recombination events (68) and in ∼90% in our strain background (25). This is in contrast to another, also type II, Rad50- and Rad51-independent, Rad52-dependent survival pathway that occurred at very low rates (25). On the other hand, rare survivors from a tlc1Δ rad52Δ strain that were not fully characterized (69) might potentially correspond to those described here. However, in that study, unlike in the present one, no survivors were generated from a tlc1Δ rad51Δ rad52Δ strain (69). It is not known yet whether the ILT pathway is a cryptic pathway. Indeed, although it was apparently inhibited in the presence of Rad52, one cannot eliminate the possibility that ILT survivors are generated and later disappear from the liquid cultures due to a growth disadvantage.
Wild-type cells can adapt to DNA damage by escaping from G2/M arrest. Potentially, the ILT survivors, harboring large amounts of DNA damage at several telomeres, might proliferate by adapting to DNA damage. However, since yku70Δ mutant cells are defective in DNA adaptation (40) but not in the ILT pathway, as shown here, this suggests that the ILT pathway is not likely to represent a classical case of adaptation to DNA damage.
Two recent papers have shown that telomeres in postsenescence survivors could be capped in an alternative manner that did not require the presence of the essential capping proteins, Cdc13 and Stn1 (37, 74). These (telomerase-positive) cdc13 null mutants either could live without Rad52 (but, importantly, had telomerase-maintained normal length telomeres) or underwent classical type II recombination when Rad52 was present (74). Therefore, the ILT pathway was not triggered in these cdc13Δ mutants, although it would be interesting to know whether it could be triggered in cdc13Δ tlc1Δ cells. We also note that the acquisition of genome stability in cdc13Δ or stn1Δ mutants appears to require partial abrogation of the cell cycle checkpoints (74), which is not the case for the ILT pathway.
One aspect of the present study is reminiscent of a mechanism by which telomerase-negative diploid yeast cells adopt the same telomeric type of recombination as the one present in the haploid ancestors, suggesting that cells use inherited telomere structures to elongate their telomeres (71). It is indeed noticeable that the ILT pathway, triggered by an inherited type II recombination, generates a type II-like pattern. However, this is a different situation, as, according to our working model (see below), the ILT mechanism probably does not represent homologous recombination, unlike the inherited type II-ALT.
Rad50 and Mre11 are essential for the ILT pathway.
Both Mre11 and Rad50 were essential for the ILT pathway, while the third member of the MRX complex, Xrs2, has not been tested. Although the MRX complex is generally thought to have modest roles in mitotic recombination (67), this is clearly not the case for telomeric recombination (14, 38, 56). While yeast strains with null mutations in any one of these three genes have very similar phenotypes (67), remarkably, here telomerase-negative rad52Δ strains died earlier in the absence of Rad50 than in the absence of Mre11. Although Mre11 is central in the formation of the MRX complex (Rad50 and Xrs2 fail to interact in its absence), it is probable that Rad50 is endowed with a more important function, linked to its capacity to bridge two chromosomes together (1, 17, 32, 67). One could therefore imagine that in the absence of Mre11, Rad50 is still organized around the DNA. Conversely, the absence of Rad50 would prevent any sort of cohesion between two DNA molecules, potentially explaining the more severe phenotype observed here in the rad50Δ background.
Both the mre11-D16A and mre11-H125L-D126V mutations are generally thought to confer a total lack of nuclease activity (9, 24, 36, 58). Lewis et al. (43) attributed the more severe defect of the mre11-D16A mutant compared with the mre11-H125N mutant to the presence of residual nuclease activity for the Mre11-H125N protein. However, Krogh et al. (36) argued from their measurements that nuclease deficiency was identical in both mutants. Moreover, although multimer formation of Mre11 was not affected by the mre11-D16A mutation (24), MRX complex formation was (36). On the other hand, the Mre11-H125L-D126V mutant protein, unlike Mre11-D16A and Mre11-H18L, retained the ability to interact with Rad50 (9, 24, 36, 58). Based on the previous findings that mre11-H125L-D126V mutations confer a deficiency in nuclease activity, one can conclude that lack of Mre11 nuclease activity does not prevent the ILT pathway.
Rad59 was dispensable for the ILT pathway yet indispensable for the Rad52-dependent ALT pathway. Rad59 has some homology with Rad52, but Rad59-mediated events still require Rad52 (3, 67). Our finding is not very surprising, as Rad59, which forms physical complexes with Rad51 and Rad52 and, separately, with Rad52 and RPA, is no longer associated with Rad51 or RPA in the absence of Rad52 (18). Therefore, the requirement of the type II-ALT pathway for Rad59 probably reflects the participation of Rad52 in these processes, while the ILT pathway functions in the absence of Rad52.
Rad1 and Elg1 are essential for the ILT pathway.
Our data on the absence of a role for Rad51, Rad52, and Rad59 in the ILT pathway cast serious doubts on the possibility that the ILT survivors used homologous recombination to amplify their telomeres. The finding that Rad1 was essential for the ILT pathway confirms this assumption, as the Rad1-Rad10 endonuclease has not been reported to function in homologous recombination.
The ILT pathway was not based on an NHEJ process, since telomerase-negative (tlc1Δ) rad52Δ mutants could perform telomeric recombination in the absence of either Dnl4, Yku70, or Nej1, which are all essential for NHEJ (16). Formally, BIR, which occurs when one end of a double-strand break undergoes strand invasion into a homologous chromosome and synthesizes new DNA up to the end of that chromosome, cannot be invoked as a potential mechanism underlying the ILT pathway, because it requires Rad52 (56). On the other hand, SSA, which can take place in the absence of Rad52 when flanking homologous sequences are long enough, as in the ribosomal DNA repeats (22, 60, 61), requires Rad1, which was found here to be indispensable for the ILT pathway. However, RPA, which is normally required for SSA, was found here to be dispensable for the ILT pathway. The eroded telomere that needs to undergo repair corresponds to a one-ended double-strand break for which the only possible repair, in the absence of strand invasion (due to deletion of Rad51 and Rad52), could be end joining with another broken telomere. This would represent an unusual example of SSA, which is normally restricted to double-strand breaks occurring between direct repeats. End joining with another telomere from another chromosome would result in chromosome breakage. On the other hand, end joining with extrachromosomal circular or linear DNA, present in postsenescence survivors (12, 37, 45), should be possible provided that these circles can be previously opened. During SSA, annealing of the complementary single strands is presumably accomplished by Rad52, and the 3′ heterologous tails left around each side of the annealed homologous regions are then removed by the Rad1-Rad10 endonuclease (61, 67). Therefore, in this scenario, during the ILT pathway, the MRX complex should be able to accomplish Rad52's task.
A very attractive possibility for the ILT pathway is that of the occurrence of microhomology-mediated end joining (MMEJ), which can repair double-strand breaks exhibiting only imperfect microhomology of 8 to 10 bp at the junctions (52). Again, this would involve end joining between the eroded telomere and extrachromosomal telomeric DNA. In budding yeast, MMEJ can take place in the absence of Rad52 and appears to be dispensable for Dnl4, possibly relying on Cdc9, another DNA ligase (52). Interestingly, MMEJ is strongly dependent on the Rad1-Rad10 endonuclease complex, as well as on Rad50 and Mre11 (39, 52). However, we note that MMEJ is dependent on Nej1 (39), while ILT is not. Interestingly, deletion of YKU70 stimulated MMEJ, possibly by accelerating nucleolytic degradation at the break (39), and improved triggering of the ILT pathway (Fig. 7A). One could be tempted to postulate that in this situation, loss of Yku function acts by weakening telomere end protection, much as we presume loss of Stn1 function does (Fig. 7 C to E). However, one should stress that it is not really known how the loss of Yku or Stn1 function affects telomere end protection (21, 29).
Rad1-Rad10 is also required for short-sequence homology, a repair process distinct from general recombination, which functions when homology sequences are less than 30 bp long (54). However, Rad52 is required for short-sequence homology (54).
Because the exact role of Elg1 in one of the three RLCs is not known yet at the molecular level, the reasons for its requirement for the ILT pathway may be numerous. Rad24 and Ctf18 in the other two RLCs were not essential for the ILT pathway. Elg1 plays a central role in genome stability by associating with the Rfc2 to -5 subunits of RFC (7). The RLCs may be required to carry out DNA polymerase exchanges at the replicating fork during DNA damage (7), and interestingly, Pol32, which is required for MMEJ (39), physically interacts with Elg1 but also with Rad24 and Ctf18 (5), and thus is presumably not pertinent to ILT. S phase in the elg1Δ mutant was very much delayed compared with that in the wild type (35) and might even not go to completion (6). The elg1Δ mutant may accumulate abnormal single-stranded gaps at its replication forks, possibly causing breaks that need to be repaired by recombination (6, 35). The pausing or even stalling of the replication forks in the telomeric regions of the chromosomes (23, 33, 53) may become dramatic in the type II survivors because of their overelongated TG1-3 repeats. A probable severe collapsing of the replicating forks at the telomeres of recombining elg1Δ cells, possibly repaired by the Rad52 machinery, might result in death in the absence of Rad52. Therefore, it is not clear yet whether or not Elg1 plays a pivotal role in the ILT pathway.
In conclusion, our working model begins with the observation that stn1-13 mutants recombined very rapidly despite the presence of overelongated telomeres (Fig. 7D). This somewhat rapid shortening of stn1-13 telomeres is reminiscent of a telomere rapid deletion (TRD) phenotype described for an stn1 mutant of Kluyveromyces lactis (34). Intriguingly, we observed that stn1-13 could trigger the ILT pathway. We propose that TRD, a mechanism of single-step nonreciprocal telomere deletion that can take place, albeit less efficiently, in the absence of Rad52 (44, 50), is triggered in the telomerase-negative cells by the inherited long telomeres or stn1-13. Elg1 would be required for the putative multiple rounds of DNA synthesis occurring within the putative D-loop that is formed by the overelongated telomere (50). Note that MRX is essential for TRD (11). When critically uncapped, a telomere would undergo MMEJ-like end joining with a previously TRD-released linear or circular telomeric DNA piece. Although the mre11-H125L-D126V mutant generated ILT survivors, it is formally possible that Mre11 nuclease activity still could be involved in the ILT process. Indeed, nuclease-deficient Mre11 cells appear to perform Ku-dependent NHEJ in substitution for MMEJ (39). In our hypothesis, MRX would mediate annealing of the processed broken ends, and unpaired 3′ flaps would be removed by the Rad1-Rad10 endonuclease. Finally, ligation of the broken ends, independent of Dnl4, may require Cdc9 or, alternatively, another enzyme with accessory ligase activities.
The experiments presented here have uncovered a novel pathway of telomere maintenance in the absence of telomerase and of the major homologous recombination proteins. A mammalian ILT pathway might exist and be relevant to cancer cells, as explained above. In parallel, the budding yeast ILT pathway needs complete elucidation at the molecular level and awaits further studies with the hypotheses proposed above as a starting point.
Supplementary Material
Acknowledgments
We thank Jim Haber, Richard Kolodner, Dan Gottschling, Kunihiro Ohta, Wei Xiao, and Errol Friedberg for gifts of strains and plasmids. We also thank Jim Haber for critical comments on the manuscript. We are very grateful to all three reviewers for excellent expertise and judicious comments and to one of them for coining the term “ILT pathway.”
This work was supported by grants from the Association pour la Recherche contre le Cancer.
Footnotes
Published ahead of print on 1 December 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Assenmacher, N., and K. P. Hopfner. 2004. MRE11/RAD50/NBS1: complex activities. Chromosoma 113157-166. [DOI] [PubMed] [Google Scholar]
- 2.Autexier, C., and N. F. Lue. 2006. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 75493-517. [DOI] [PubMed] [Google Scholar]
- 3.Aylon, Y., and M. Kupiec. 2004. DSB repair: the yeast paradigm. DNA Rep. 3797-815. [DOI] [PubMed] [Google Scholar]
- 4.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 213329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellaoui, M., M. Chang, J. Ou, H. Xu, C. Boone, and G. W. Brown. 2003. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J. 224304-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Aroya, S., A. Koren, B. Liefshitz, R. Steinlauf, and M. Kupiec. 2003. ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc. Natl. Acad. Sci. USA 1009906-9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Aroya, S., and M. Kupiec. 2005. The Elg1 replication factor C-like complex: a novel guardian of genome stability. DNA Rep. 4409-417. [DOI] [PubMed] [Google Scholar]
- 8.Bhattacharyya, M. K., and A. J. Lustig. 2006. Telomere dynamics in genome stability, Trends Biochem. Sci. 31114-122. [DOI] [PubMed] [Google Scholar]
- 9.Bressan, D. A., H. A. Olivares, B. E. Nelms, and J. H. J. Petrini. 1998. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryan, T. M., A. Englezou, L. Dalla-Pozza, M. A. Dunham, and R. R. Reddel. 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 31271-1274. [DOI] [PubMed] [Google Scholar]
- 11.Bucholc, M., Y. Park, and A. J. Lustig. 2001. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol. 216559-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cesare, A. J., C. Groff-Vindman, S. A. Compton, M. J. McEachern, and J. D. Griffith. 2008. Telomere loops and homologous recombination-dependent telomeric circles in a Kluyveromyces lactis telomere mutant strain. Mol. Cell. Biol. 2820-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chamankhah, M., T. Fontanie, and W. Xiao. 2000. The Saccharomyces cerevisiae mre11 (ts) allele confers a separation of DNA repair and telomere maintenance functions. Genetics 155569-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, Q., A. Iijpma, and C. W. Greider. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 211819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coïc, E., T. Feldman, A. S. Landman, and J. E. Haber. 2008. Mechanisms of Rad52-independent spontaneous and UV-induced mitotic recombination in Saccharomyces cerevisiae. Genetics 179199-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daley, J. M., P. L. Palmbos, D. Wu, and T. E. Wilson. 2005. Nonhomologous end joining in yeast. Annu. Rev. Genet. 39431-451. [DOI] [PubMed] [Google Scholar]
- 17.D'Amours, D., and S. P. Jackson. 2002. The MRE11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3317-327. [DOI] [PubMed] [Google Scholar]
- 18.Davis, A. P., and L. S. Symington. 2003. The Rad52-Rad59 complex interacts with Rad51 and replication protein A. DNA Rep. 21127-1134. [DOI] [PubMed] [Google Scholar]
- 19.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286117-120. [DOI] [PubMed] [Google Scholar]
- 20.Finkel, T., M. Serrano, and M. A. Blasco. 2007. The common biology of cancer and ageing. Nature 448767-774. [DOI] [PubMed] [Google Scholar]
- 21.Fisher, T. S., and V. A. Zakian. 2005. Ku: a multifunctional protein involved in telomere maintenance. DNA Rep. 41215-1226. [DOI] [PubMed] [Google Scholar]
- 22.Fishman-Lobell, J., N. Rudin, and J. E. Haber. 1992. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol. 121292-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fouché, N., S. Özgür, D. Roy, and J. D. Griffith. 2006. Replication fork regression in repetitive DNAs. Nucleic Acids Res. 346044-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furuse, M., Y. Nagase, H. Tsubouchi, K. Murakami-Murofushi, T. Shibata, and K. Ohta. 1998. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 176412-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grandin, N., and M. Charbonneau. 2003. Mitotic cyclins regulate telomeric recombination in telomerase-deficient yeast cells. Mol. Cell. Biol. 239162-9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grandin, N., and M. Charbonneau. 2007. Control of the yeast telomeric senescence survival pathways of recombination by the Mec1 and Mec3 DNA damage sensors and RPA. Nucleic Acids Res. 35822-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grandin, N., C. Damon, and M. Charbonneau. 2000. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol. Cell. Biol. 208397-8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grandin, N., C. Damon, and M. Charbonneau. 2001. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 201173-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grandin, N., S. I. Reed, and M. Charbonneau. 1997. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 11512-527. [DOI] [PubMed] [Google Scholar]
- 30.Henson, J. D., J. A. Hannay, S. W. McCarthy, J. A. Royds, T. R. Yeager, R. A. Robinson, S. B. Wharton, D. A. Jellinek, S. M. Arbuckle, J. Yoo, B. G. Robinson, D. L. Learoyd, P. D. Stalley, S. F. Bonar, D. Yu, R. E. Pollock, and R. R. Reddel. 2005. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin. Cancer Res. 11217-225. [PubMed] [Google Scholar]
- 31.Henson, J. D., A. A. Neumann, T. R. Yeager, and R. R. Reddel. 2002. Alternative lengthening of telomeres in mammalian cells. Oncogene 21598-610. [DOI] [PubMed] [Google Scholar]
- 32.Hopfner, K.-P., and J. A. Tainer. 2003. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr. Opin. Struct. Biol. 13:249-255. [DOI] [PubMed] [Google Scholar]
- 33.Ivessa, A. S., J. Q. Zhou, V. P. Schulz, E. K. Monson, and V. A. Zakian. 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 161383-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iyer, S., A. D. Chadha, and M. J. McEachern. 2005. A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol. Cell. Biol. 258064-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanellis, P., R. Agyei, and D. Durocher. 2003. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr. Biol. 131583-1595. [DOI] [PubMed] [Google Scholar]
- 36.Krogh, B. O., B. Llorente, A. Lam, and L. S. Symington. 2005. Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex stability in addition to nuclease activity. Genetics 1711561-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larrivée, M., and R. W. Wellinger. 2006. Telomerase- and capping-independent yeast survivors with alternate telomere states. Nat. Cell Biol. 8741-747. [DOI] [PubMed] [Google Scholar]
- 38.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, K., and S. E. Lee. 2007. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA breaks promotes microhomology-mediated end joining. Genetics 1762003-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94399-409. [DOI] [PubMed] [Google Scholar]
- 41.Lendvay, T. S., D. K. Morris, J. Sah, B. Balasubramanian, and V. Lundblad. 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 1441399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lettier, G., Q. Feng, A. A. de Mayolo, N. Erdeniz, R. J. Reid, M. Lisby, U. H. Mortensen, and R. Rothstein. 2006. The role of DNA double-stand breaks in spontaneous homologous recombination in S. cerevisiae. PloS Genet. 21773-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis, L. K., F. Storici, S. Van Komen, S. Calero, P. Sung, and M. A. Resnick. 2004. Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics 1661701-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li, B., and A. J. Lustig. 1996. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 101310-1326. [DOI] [PubMed] [Google Scholar]
- 45.Lin, C. Y., H. H. Chang, K. J. Wu, S. F. Tseng, C. C. Lin, C. P. Lin, and S. C. Teng. 2005. Extrachromosomal telomeric circles contribute to Rad52-, Rad50-, and polymerase delta-mediated telomere-telomere recombination in Saccharomyces cerevisiae. Eukaryot. Cell 4327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liti, G., and E. J. Louis. 2003. NEJ1 prevents NHEJ-dependent telomere fusions in yeast without telomerase. Mol. Cell 111373-1378. [DOI] [PubMed] [Google Scholar]
- 47.Louis, E. J., and R. H. Borts. 1995. A complete set of marked telomeres in Saccharomyces cerevisiae for physical mapping and cloning. Genetics 139125-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73347-360. [DOI] [PubMed] [Google Scholar]
- 49.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57633-643. [DOI] [PubMed] [Google Scholar]
- 50.Lustig, A. J. 2003. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat. Rev. Genet. 4916-923. [DOI] [PubMed] [Google Scholar]
- 51.Lydeard, J. R., S. Jain, M. Yamaguchi, and J. E. Haber. 2007. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448820-824. [DOI] [PubMed] [Google Scholar]
- 52.Ma, J. L., E. M. Kim, J. E. Haber, and S. E. Lee. 2003. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 238820-8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makovets, S., I. Herskowitz, and E. H. Blackburn. 2004. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol. Cell. Biol. 244019-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manthey, G. M., and A. M. Bailis. 2002. Multiple pathways promote short-sequence recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 225347-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maringele, L., and D. Lydall. 2004. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 182663-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McEachern, M. J., and J. E. Haber. 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 75111-135. [DOI] [PubMed] [Google Scholar]
- 57.Moore, J. K., and J. E. Haber. 1996. Cell cycle and genetic requirement of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 162164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moreau, S., J. R. Ferguson, and L. S. Symington. 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end-joining or telomere maintenance. Mol. Cell. Biol. 19556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neumann, A. A., and R. R. Reddel. 2002. Telomere maintenance and cancer—look, no telomerase. Nat. Rev. Cancer 2879-884. [DOI] [PubMed] [Google Scholar]
- 60.Ozenberger, B. A., and G. S. Roeder. 1991. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol. Cell. Biol. 111222-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pâques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pohl, T. J., and J. A. Nickoloff. 2008. Rad51-independent interchromosomal double-strand break repair by gene conversion requires Rad52 but not Rad55, Rad57, or Dmc1. Mol. Cell. Biol. 28897-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schild, D., I. L. Calderon, R. Contopoulo, and R. K. Mortimer. 1983. Cloning of yeast recombination repair genes and evidence that several are non-essential genes, p 417-427. In E. C. Friedberg and B. A. Bridges (ed.), Cellular responses to DNA damage. Alan R. Liss, New York, NY.
- 64.Siede, W., G. Nusspaumer, V. Portillo, R. Rodriguez, and E. C. Friedberg. 1996. Cloning and characterization of RAD17, a gene controlling cell cycle responses to DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 241669-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266404-409. [DOI] [PubMed] [Google Scholar]
- 66.Smolikov, S., Y. Mazor, and A. Krauskopf. 2004. ELG1, a regulator of genome stability, has a role in telomere length regulation and in silencing. Proc. Natl. Acad. Sci. USA 1011656-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Symington, L. S. 2002. Role of RAD52 epistastis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 198083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsukamoto, M., K. Yamashita, T. Miyazaki, M. Shinohara, and A. Shinohara. 2003. The N-terminal domain of Rad52 promotes RAD51-independent recombination in Saccharomyces cerevisiae. Genetics 1651703-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Umezu, K., N. Sugawara, C. Chen, J. E. Haber, and R. D. Kolodner. 1998. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics 148989-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wen, W. Y., H. J. Tsai, C. C. Lin, S. F. Tseng, C. W. Wong, and S. C. Teng. 2006. Telomere configuration influences the choice of telomere maintenance pathways. Biochem. Biophys. Res. Commun. 343459-466. [DOI] [PubMed] [Google Scholar]
- 72.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285901-906. [DOI] [PubMed] [Google Scholar]
- 73.Wotton, D., and D. Shore. 1997. A novel Rap1-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11748-760. [DOI] [PubMed] [Google Scholar]
- 74.Zubko, M. J., and D. Lydall. 2006. Linear chromosome maintenance in the absence of essential telomere-capping proteins. Nat. Cell Biol. 8734-740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.