Abstract
Bone morphogenetic protein 2 (BMP-2) is essential for postnatal bone formation and fracture repair. By screening chemical libraries for BMP-2 mimics using a cell-based assay, we identified inhibitors of microtubule assembly as stimulators of BMP-2 transcription. These microtubule inhibitors increased osteoblast differentiation in vitro, stimulated periosteal bone formation when injected locally over murine calvaria, and enhanced trabecular bone formation when administered systemically in vivo. To explore molecular mechanisms mediating these responses, we examined effects of microtubule inhibitors on the hedgehog (Hh) pathway, since this pathway is known to regulate BMP-2 transcription in osteoblasts and microtubules have been shown to be involved in Hh signaling in Drosophila. Here we show that in osteoblasts, inhibition of microtubule assembly increased cytoplasmic levels and transcriptional activity of Gli2, a transcriptional mediator of Hh signaling that we have previously shown to enhance BMP-2 expression in osteoblasts (M. Zhao et al., Mol. Cell. Biol. 26:6197-6208, 2006). Microtubule inhibition blocked β-TrCP-mediated proteasomal processing of Gli2 in osteoblasts. In summary, inhibition of microtubule assembly enhances BMP-2 gene transcription and subsequent bone formation, in part, through inhibiting proteasomal processing of Gli2 and increasing intracellular Gli2 concentrations.
Bone morphogenetic proteins (BMPs) were originally identified by their osteoinductive properties in ectopic sites (52). For more than 40 years, the osteoinductive properties of BMPs have been successfully applied to preclinical models to accelerate fracture healing and repair bone defects in various animal models. The prototype for this family is BMP-2, which induces osteoblast differentiation and bone formation (56). Subcutaneous or intramuscular implantation of BMP-2 induces massive amounts of new bone in rodents in a manner similar to that of endochondral bone formation (56, 59). Human recombinant BMP-2 has been used clinically for spinal fusion, fracture repair, and treatment of other bone defects (3, 15, 47). Expression of BMP-2 plays an important role in osteogenesis throughout embryonic skeletal development and adult bone remodeling. Conditional knockout (KO) studies have shown that tissue-specific inactivation of the BMP-2 gene in limbs results in spontaneous bone fractures, impaired fracture repair, and reduced bone mineral density in adult BMP-2 KO mice, indicating that BMP-2 is a critical factor for postnatal bone formation (51). BMP-2 was also recently recognized as an osteoporosis-associated gene by human polymorphism studies (49). Animal studies have found that both gene expression and anabolic activity of BMP-2 are significantly decreased in aging bones (11, 29, 31, 50, 58). These results suggest that BMP-2 gene expression is necessary for maintenance of postnatal bone formation, particularly during aging, and that progressive, age-related loss of BMP-2 function may be one of the molecular mechanisms involved in the development of osteoporosis.
Given its importance in physiological and pharmacologically induced bone formation, it is clearly important to understand the mechanisms involved in regulation of BMP-2 gene expression. We have previously identified multiple transcriptional mechanisms that are involved in regulating BMP-2 gene expression (8, 12, 61), as well as several classes of low-molecular-weight synthetic and naturally occurring compounds that stimulate BMP-2 gene expression, including statins and proteasome inhibitors. These compounds were identified using a cell-based screening assay based on a reporter linked to a murine BMP-2 promoter fragment. Systemic administration of statins and proteasome inhibitors led to an increase in bone formation in preclinical rodent models (12, 35). Thus, BMP-2 gene expression is a potential target for identifying compounds that stimulate bone formation. Using the same approach, we have now identified another specific class of compounds, inhibitors of microtubule assembly, as BMP-2-stimulating agents. Importantly, we found that these microtubule inhibitors have potent anabolic effects on bone formation in mice when administered locally or systemically.
Microtubules are major components of the cytoskeleton that play an essential role in a wide range of cellular processes, such as mitosis, cell motility, and intracellular trafficking (37). By providing scaffolding, sequestering, and delivery functions, microtubules are involved in various signaling pathways, such as sonic hedgehog (Shh)/Gli, Wnt/β-catenin, IκB/NF-κB, G-protein, and mitogen-activated protein kinase pathways, through diverse mechanisms (16, 23). These signaling pathways have all been implicated in skeletal development and homeostasis. In this study, we have focused on the hedgehog (Hh) pathway, since there is abundant evidence that microtubules are involved in the regulation of Hh signaling activity (38, 43, 48) and microtubule targeting drugs regulate osteoblast differentiation (17, 30). In the absence of Shh in flies, microtubules associate with a protein complex composed of cubitus interruptus (Ci) and other modulating proteins, leading to proteolytic processing of the Ci protein (6, 22, 53). The Gli family of transcription factors, which mediate Hh signaling, are vertebrate orthologs of Ci. We previously demonstrated that Gli2 is a powerful enhancer of BMP-2 expression in osteoblasts (61). Gli2 activity is known to be regulated by β-transducin repeat-containing protein (β-TrCP)-dependent proteasomal processing (40, 41). Since the Hh pathway is conserved from flies to vertebrates, microtubules may also be involved in the processing of the Hh mediator Gli2 in the proteasome. Microtubule filaments are highly dynamic cytoskeletal polymers comprised of α/β-tubulin heterodimers that undergo constant assembly at the positive end and disassembly from the negative end to maintain normal function (23, 36). Based on these observations, we hypothesized that inhibition of microtubule function by forced microtubule disassembly would inhibit Gli2 degradation and in turn increase Gli2 concentrations and lead to stimulation of BMP-2 gene expression and subsequent bone formation. In this study, we have provided in vivo evidence for anabolic effects of inhibition of microtubule assembly on bone. Utilizing the microtubule inhibitor TN16, we also have demonstrated potential mechanisms whereby microtubule inhibition enhances bone formation.
MATERIALS AND METHODS
DNA constructs and compounds.
β-TrCP cDNA was amplified by PCR using primers designed to amplify mouse β-TrCP (GenBank accession no. AF110396). A β-TrCP expression plasmid was made by inserting a Flag-tagged full-length β-TrCP cDNA into the pcDNA3 vector at the HindIII/BamHI sites. A His-tagged Gli2 expression plasmid (pcDNA3.1-His-Gli2) and a Gli-responsive reporter construct (8×3′Gli-BSδ5/LucII) that contains multiple copies of Gli response elements were kindly provided by Hiroshi Sasaki (44, 45). The hemagglutinin (HA)-ubiquitin expression plasmid was obtained from Xu Cao (University of Alabama at Birmingham). The mouse BMP-2 promoter luciferase reporter (−2712/+165-Luc) was constructed by linking the 5′ region of the mouse BMP-2 gene to firefly luciferase in the pGL3 vector. The Wnt signaling reporter TOP Flash and the β-catenin (pCI-Neo-β-catenin) and TCF4 (pCDNA-myc-hTCF4) expression plasmids were kindly provided by Bert Vogelstein (Johns Hopkins University, Baltimore, MD). The inhibitor of cAMP phosphodiesterases IBMX, inhibitors of microtubule assembly (TN16, colchicine, and nocodazole), and microtubule stabilizer toxal were all purchased from Calbiochem.
Cell culture, transfection, and reporter assay.
The osteoblast precursor 2T3 cells were derived from calvarial osteoblasts isolated from transgenic mice carrying the BMP-2-SV40 large-T-antigen transgene. These immortalized cells behave similarly to primary cultures of fetal rat calvarial osteoblasts with respect to their capacity to terminally differentiate into osteoblasts (13). 2T3 cells were cultured in α-minimal essential medium (α-MEM) supplemented with 10% fetal calf serum (FCS). Expression plasmids were transiently transfected into cells using the LipofectAmine Plus reagent, following the manufacturer's instructions (Invitrogen, Carlsbad, CA). For the luciferase reporter assays, 2T3 cells were plated in 24-well plates at a density of 4 × 104 cells per well in α-MEM with 10% FCS 18 to 24 h prior to transfection. Cells were incubated for 4 h at 37°C with 250 μl of opti-MEM transfection solution containing the LipofectAmine Plus reagent, 0.5 μg of reporter plasmids, 0.1 μg of the pSV-β-galactosidase β-galactosidase (β-Gal) expression vector (Promega, Madison, WI), and 0 to 0.5 μg of expression constructs. After 4 h of incubation, a fresh 250 μl of α-MEM medium containing 20% FCS was added. The cells were cultured for a further 24 to 48 h and then lysed with 100 μl of reporter lysis buffer (Promega). In compound treatment groups, cells carrying reporter plasmid were incubated with different compounds for 4 to 24 h. The luciferase activities of cell lysates were measured by using a luciferase reporter assay kit (Promega) and normalized with β-Gal activity.
KO mice.
Mice expressing one allele of Gli2 (Gli2−/+ mice) were a kind gift from Alexandra Joyner (New York University) (33). Mice were maintained as heterozygotes and crossed to generate mutant combinations. Fetal mice were genotyped by PCR analysis (34, 61). For the alkaline phosphatase (ALP) assay, Gli2 Western blotting, and BMP-2 mRNA PCR, primary calvarial cells were isolated from homozygous mutants (Gli2−/−) and their wild-type littermates (Gli2+/+) at embryonic day 20.5 (E20.5). The BMP-2 conditional KO mice (BMP2KO/BMP2flx;3.6Col1a1-Cre) were generated by crossing BMP-2 floxed mice with 3.6 Col1a1-Cre mice, by which the BMP-2 gene was inactivated by removal of exon 3 specifically in the osteoblastic lineage. These BMP-2 KO mice develop a remarkable osteopenic phenotype (57).
ALP activity assay.
2T3 cells or primary calvarial cells isolated from Gli2−/− and Gli2+/+ mice were seeded on 48-well plates in α-MEM medium with 10% FCS. The cells were treated with microtubule inhibitors or transfected with the Gli2 expression vector for 30 h in the same medium supplemented with 2.5% FCS. After lysing in 0.05% Triton X-100 buffer, cell lysates were analyzed for ALP activity in 96-well plates. Briefly, 10 μl of lysate was incubated with 90 μl of fresh AMP solution containing p-nitrophenyl phosphate substrate at 37°C for 30 to 60 min, following which 100 μl 0.5 N NaOH was added to stop the reaction. The plates were then read spectrophotometrically at 405 nm. ALP activity was determined using a p-nitrophenol standard curve and normalized to total cellular protein.
Cell proliferation assay.
2T3 cells were plated at a density of 8 × 103 cells/well in a 96-well plate in α-MEM medium with 10% FCS. After treatment with TN16 for 24 to ∼48 h, the culture medium was changed with 100 μl fresh medium and 20 μl of combined 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS)-phenazine methosulfate solution. The plates were incubated for 1 to 4 h at 37°C in a humidified, 5%-CO2 atmosphere. After incubation, the absorbance at 450 nm was read using an enzyme-linked immunosorbent assay plate reader.
Immunoblotting.
2T3 and C3H10T1/2 cells cultured in six-well plates were incubated with microtubule inhibitors or cotransfected with His-Gli2 and β-TrCP expression plasmids for 24 h, following which the cells were lysed with sodium dodecyl sulfate (SDS)-RIPA buffer containing the protease inhibitors phenylmethylsulfonyl fluoride (1 mM), aprotinin (10 μg/ml), and leupeptin (10 μg/ml). Sample were loaded on Mini-Protein II SDS-PAGE Ready gels (Bio-Rad, Hercules, CA), and proteins were electrophoretically separated under reducing conditions and then transferred onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad) in transblotting buffer (25 mM Tris, 192 mM glycine, and 20% [vol/vol] methanol, pH 8.3) at 4°C for 1 h. After blocking with 5% milk in Tris-buffered saline-Tween 20 (0.1% Tween 20) (TBS-T) was carried out for 1 h at room temperature, membranes were incubated with rabbit anti-mouse Gli2 for endogenous Gli2 (41) or rabbit Omni-probe anti-His antibody (Santa Cruz, CA) for His-Gli2 in 5% milk in TBS-T at 4°C overnight. After washes, membranes were incubated with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) antibody (Amersham Biosciences, Buckinghamshire, United Kingdom) at a 1:5,000 dilution at room temperature for 1 h and then washed six times with TBS-T buffer for 5 min each. Immunoblots were detected using the ECL (enhanced chemiluminescence) system (Amersham). The His-Gli2 protein signal was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein levels.
Immunoprecipitation.
The immunoprecipitation assay was performed using a Pierce kit (no. 45217). Briefly, the cells were lysed with the M-Per reagent. Two hundred microliters of rabbit Omni-probe anti-His antibody was added to 500 μl of cell lysate and incubated at room temperature for 1 h. Then, immune complex was added to the equilibrated immobilized protein G columm for 10 min, spun, washed with phosphate-buffered saline (PBS), and spun twice. One hundred ninety microliters of elution buffer was added, followed by a spin. Twenty microliters of eluted sample was mixed with 5 μl of sample buffer and boiled for 5 min. The immunoprecipitates were separated by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to a PVDF membrane, and immunoblotted with anti-α-tubulin antibody. The blotting was visualized using a horseradish peroxidase-coupled secondary antibody after enhancement by ECL.
Ubiquitination.
His-Gli2, HA-ubiquitin, and β-TrCP expression plasmids were cotransfected into C3H10T1/2 cells, and the cells were cultured for 24 h. Lysates were incubated with Omni-probe anti-His antibody and protein A beads at 4°C overnight. Immunoprecipitates were separated by SDS-PAGE, transferred onto a PVDF membrane, and immunoblotted with anti-HA antibody. Ubiquitination was confirmed as a smear of decreased protein bands.
Immnunofluorescence.
2T3 cells were seeded in an 8-well chamber slide at 1 × 104 cells per well and treated with microtubule inhibitors or IBMX or transfected with the β-TrCP vector for 24 h. Following fixation in cold methanol for 30 min at −20°C, cells were washed multiple times and then blocked with 5% bovine serum albumin-0.2% Triton X-100 for 1 h at room temperature. After probing with the appropriate primary antibodies (1:50 rabbit anti-mouse Gli2 [41] and/or mouse anti-α-tubulin antibodies) in 2% FCS solution overnight at 4°C, cells were washed with PBS and then secondary antibodies raised against rabbit or mouse IgG and conjugated with fluorescein isothiocyanate (FITC) or Cy3, respectively, were applied. After mounting, fluorescence was viewed using a Zeiss inverted LSM510 confocal microscope with fluorescence filters set accordingly: FITC green (488 nm) and Cy-3 red (543 nm). To detect the Smad signal by immunofluorescence in 2T3 cells, an anti-Smad1/5/8 antibody (Cell Signaling) was utilized.
Real-time PCR.
Total RNA was purified from 2T3 cells and reverse transcribed into cDNA. Quantitative PCR of mouse BMP-2 mRNA was performed using the cDNA template and mouse BMP-2 TaqMan primers/probe (Mm01340178_m1; Applied Biosystems) on the 7300 real-time PCR system (Applied Biosystems). Eukaryotic 18S rRNA detected using a VIC-MGB probe (4319413E; Applied Biosystems) served as an endogenous control.
Micro-computed tomography (microCT) analysis.
Three-month-old ICR Swiss mice were injected intraperitoneally with TN16 (in PBS) at 50 μl per injection twice daily for two consecutive days. The mice were sacrificed 1 month later, and tibiae were isolated and fixed in 70% ethanol. Each bone was scanned at an isotropic voxel size of 16 μM using a μCT 40 scanner (ScanCo Medical, Switzerland). Entire tibial metaphyses were obtained, and 200 slices right beneath the growth plate were analyzed using proprietary ScanCO μ40 software to measure the bone volume/total volume (BV/TV), trabecular number (Tb.N.), trabecular thickness (Tb.Th.,) and trabecular space (Tb.Sp.), and obtain high-resolution images of tibial trabecular bones.
BMD.
Femorae and tibiae isolated from 3-month-old TN16-treated ICR Swiss mice were scanned using a PIXImus bone densitometer (Lunar Corporation) with dual-energy X-ray absorptiometry technology. Bone mineral density (BMD) within a defined area in the metaphysis was analyzed using proprietary Lunar PIXI software.
Bone histology and histomorphometry.
Three-month-old ICR Swiss mice were subcutaneously injected over the calvariae or injected intraperitoneally with TN16 again at 50 μl per injection twice a day for 2 days. The mice were sacrificed 1 month after the injections. Calvariae, femorae, and tibiae were isolated, fixed, and embedded in paraffin. Four-micrometer-thick paraffin-embedded bone sections were stained with hematoxylin and eosin (H&E), and new bone thickness over the calvarial surface was measured. The trabecular bone volume in a defined area of the metaphysis beneath the growth plate was quantitated. To measure bone formation rates (BFR), 10 days before sacrifice, the mice were injected intraperitoneally with tetracycline and 4 days later with calcein. After sacrifice, bones were fixed in 70% ethanol and embedded in methylmethacrylate. Seven-micrometer-thick, double-labeled bone sections were viewed using fluorescence microscopy and data acquired using the “Osteomeasure” software program for bone histomorphometry (Osteometrics, Inc., Atlanta, GA) (12, 35). Using these data, the BFR was computed as follows: (i) labeled bone surface (BS) or mineralizing surface; (ii) mineral appositional rate (μm/day) = mean distance between two fluorescent labels divided by the number of days between labels; and (iii) BFR/BS (μm3/μm2/day) = mineralizing surface × mineral appositional rate/BS.
In situ hybridization.
A cRNA probe prepared from the full-length coding region of BMP-2 was labeled with digoxigenin using an RNA labeling kit (Roche Applied Science). The tibial sections were dewaxed, rehydrated, and fixed again with 4% paraformaldehyde. Sections were then treated with 2% glycine and proteinase K and acetylated using an acetic anhydride-Tris-EDTA acetate solution, followed by hybridization with the digoxigenin-labeled probe. Following two washes in 50% formamide, 5× SSC (1× SSC is 015 M NaCl plus 0.015 M sodium citrate), and 5% SDS for 30 min at 70°C and a third wash in 50% formamide and 2× SSC for 30 min at 65°C, sections were incubated with an antidigoxigenin-ALP antibody followed by Nitro Blue Tetrazolium-4-bromo-5-chloro-indolyl phosphate, which yields a purple-blue color. The sections were also counterstained with methyl green (nuclei).
RESULTS
Inhibition of microtubule assembly upregulates BMP-2 expression in osteoblasts.
Based on the known properties of BMP-2 to stimulate osteoblast differentiation and bone formation postnatally, we used a cell-based assay to search diverse chemical libraries for small molecules, which we successfully utilized to identify several classes of compounds BMP-2 mimics (12, 35). We have now identified another class of compounds, inhibitors of microtubule assembly, that unexpectedly stimulate BMP-2 transcription.
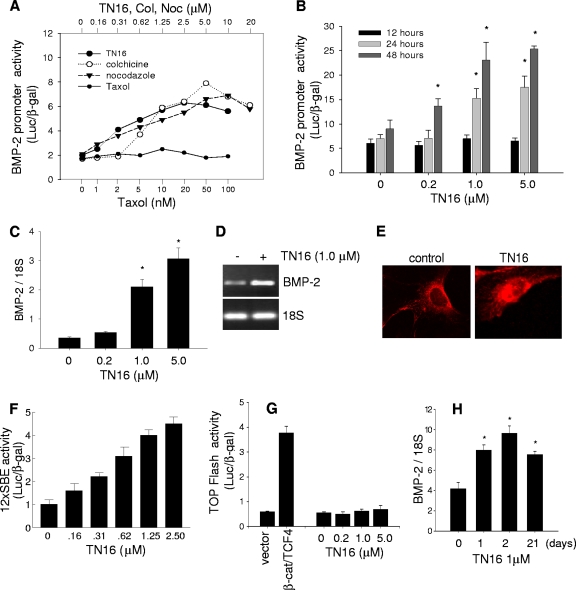
When immortalized mouse osteoblast precursor 2T3 cells (13) that had been stably transfected with a reporter construct of the mouse BMP-2 (−2712/+165) promoter (BMP-2-Luc) were cultured in the presence of the microtubule assembly inhibitors TN16 (3-anilinoethylidene-5-benzylpyrrolidine-2,4-dione), colchicine, and nocodazole at concentrations up to 20 μM for 24 h, there was a significant and dose-dependent increase in the transcriptional activity of the reporter. Maximal effects (two- to ∼fourfold increase) were observed at doses of the microtubule inhibitors between 1 and ∼10 μM (Fig. 1A). In contrast, treatment with the microtubule stabilizer taxol at doses up to 100 nM (Fig. 1A) had no effect on BMP-2 promoter activity. Further treatment with higher doses in the micromolar range similarly had no effect (data not shown). These results suggest that inhibition of microtubule assembly or forced microtubule disassembly has an enhancing effect on BMP-2 promoter activity while microtubule stabilization does not. In the following studies, we used TN16, as a representative of the class, to further address mechanisms mediating the stimulatory effect of microtubule inhibitors on BMP-2 expression. We also utilized TN16 in other in vivo and in vitro studies to characterize anabolic effects of microtubule inhibition on bone. First, we examined the time course of TN16 effects. The results showed that treatment of BMP-2-Luc cells with TN16 for 12 h had no effect while treatment for longer periods, up to 48 h, significantly enhanced BMP-2 promoter activity (Fig. 1B), suggesting that stimulation of BMP-2 transcription observed upon microtubule inhibition is an indirect effect. Stimulation of BMP-2 transcription was further confirmed by quantitative real-time PCR. Incubation with TN16 for 24 h markedly and dose-dependently increased BMP-2 mRNA levels in 2T3 cells compared with results for the vehicle control (Fig. 1C and D). We next sought to determine whether inhibition of microtubule assembly also increases BMP signaling. When 2T3 cells were treated with TN16, BMP-specific Smad1/5/8 translocation from the cytoplasm into the nuclei was enhanced (Fig. 1E), suggesting that TN16 treatment activates the BMP signaling pathway. In addition, using a BMP-specific reporter gene, 12SBE-Luc (60, 61), we found that TN16 dose-dependently increased BMP signaling reporter activity (Fig. 1F). To determine the specificity of such stimulation for the BMP pathway, we also examined the effects of microtubule inhibition on the Wnt/β-catenin pathway, which has been demonstrated to be important for postnatal bone formation, since microtubules regulate intracellular trafficking of β-catenin (2, 4, 21). Microtubule inhibition had no effect on canonical Wnt signaling activity in osteoblasts as determined using the β-catenin/TCF TOP-Flash reporter (Fig. 1G). Together these results suggest that inhibitors of microtubule assembly are powerful stimulators of BMP-2 gene expression in osteoblasts.
FIG. 1.
Effects of inhibition of microtubule assembly on BMP-2 transcription. (A) Effects of microtubule-targeting compounds on BMP-2 promoter activity. 2T3 cells transfected with the murine BMP-2 promoter reporter, −2712/+165-Luc (BMP-2-Luc), were incubated with TN16, colchicine (Col), or nocodazole (Noc) in a dose range from 0 to 20 μM (upper x axis) or with taxol at the doses of 0 to ∼100 nM (lower x axis) for 24 h. Luciferase activity of cell lysate was measured and normalized by activity of cotransfected β-Gal. (B and H) Time course of TN16 stimulation of BMP-2 promoter. The BMP-2-Luc cells were treated with TN16 at the indicated doses for 12, 24, or 48 h (B) or up to 3 weeks (H), and BMP-2 promoter reporter activity was measured as described above. (C and D) Effect of TN-16 on BMP-2 mRNA expression. 2T3 cells were treated with TN16 at the indicated doses for 24 h and total RNA purified from cell lysate and reverse transcribed to cDNA. Quantitative real-time PCR amplification was performed using Applied Biosystems’ primers (Mm01962381_s1 for mouse BMP-2 and 4319413E for 18S). (E) Immunofluorescence detection of Smads. 2T3 cells treated with TN16 at 1 μM for 24 h were fixed and incubated with an anti-Smad1/5/8 antibody (Cell Signaling) followed by a Cy3-conjugated second antibody. Fluorescence was visualized and captured with a confocal microscope system. (F) Effect of TN16 on BMP signaling. 2T3 cells were transfected with a 12×SBE-Luc reporter plasmid, containing multiple copies of Smad binding elements, and treated with TN16 at the indicated doses for 24 h. Luciferase activity was determined. (G) Lack of effect of TN16 on Wnt signaling. 2T3 cells were cotransfected with the TOP Flash construct (which has multiple binding sites for TCF) and with a β-catenin/TCF4 expression plasmid. The cells were then incubated without or with TN16 at the indicated doses for 24 h. The reporter activity was determined with β-Gal normalization. (H) Time course of TN16 stimulation of BMP-2 mRNA expression. 2T3 cells were seeded in the 48-well plate at a cell density of 3 × 103 cells per well and incubated with TN16 at 1 μM for 5 days. After a wash to remove TN16 with fresh medium, the cells were continuously cultured for up to 21 days. Total RNA was purified, and BMP-2 real-time PCR was performed with 18S as control. *, P < 0.05 versus results on day zero.
Inhibition of microtubule assembly stimulates bone formation in mice.
Previously we reported that systemic administration of statins and proteasome inhibitors to mice causes significant anabolic effects on bone, in part by stimulating BMP-2 gene expression in osteoblasts (12, 35). Since inhibitors of microtubule assembly also stimulate BMP-2 expression in osteoblasts, we hypothesized that these microtubule inhibitors would also exert an anabolic effect on bone.
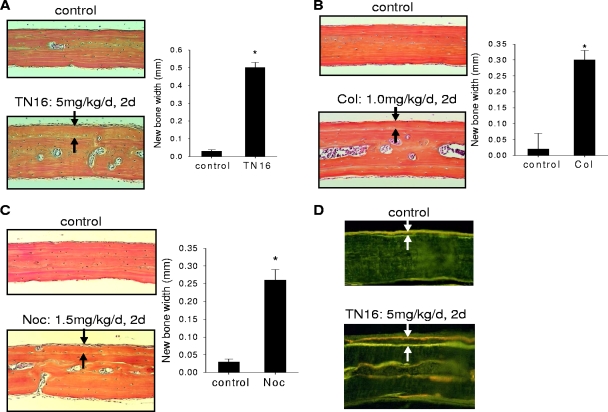
First, we examined the effects of local administration of microtubule inhibitors on calvarial new bone formation in vivo. Inhibitors of microtubule assembly were injected locally over calvariae of 1-month-old ICR Swiss mice. Histological sections of calvariae excised 4 weeks after injection showed that these drugs stimulated new periosteal bone formation on the calvarial bone surface compared with vehicle controls (Fig. 2A, B, and C, left). When the thickness of the newly formed woven bone between the new bone surface and the old bone was determined by histomorphometry, we found that administration of TN16 (5 mg/kg of body weight/day), colchicine (1 mg/kg/day), and nocodazole (1.5 mg/kg/day) for 2 days induced substantial new calvarial bone formation compared with results for vehicle controls (Fig. 2A, B, and C, right). To determine changes in BFR and mineral deposition rates, bones were labeled by sequential injections of tetracycline and calcein before sacrifice. BFR was significantly increased in calvarial bones from TN16-treated mice compared to results for those from vehicle-treated controls (Fig. 2D).
FIG. 2.
Effects of inhibition of microtubule assembly on periosteal bone formation. Microtubule inhibitors in a stock solution of dimethylsulfoxide were diluted with PBS and injected into subcutaneous tissue over calvariae of 1-month-old ICR Swiss mice (50 μl per injection twice a day for 2 days), which were then sacrificed 1 month later. The mice were also injected with tetracycline and calcein 10 and 6 days before sacrifice, respectively. After sacrifice, calvariae were processed and paraffin embedded for histology and H&E staining. New bone formation of the calvariae injected with TN16 (A), colchicines (Col) (B), or nocodazole (Noc) (C) is shown on the left, and the histomorphometry of thickness of new bone is shown on the right. Arrows indicate the newly formed woven bone between the new bone surface and the old bone. BFR in TN16-injected calvariae (D) were also measured by determining the distance between incorporated tetracycline and calcein double labels on bone surfaces in sections of plastic-embedded calvarial bones. Five to eight mice were used for each treatment. *, P < 0.01 versus results for control.
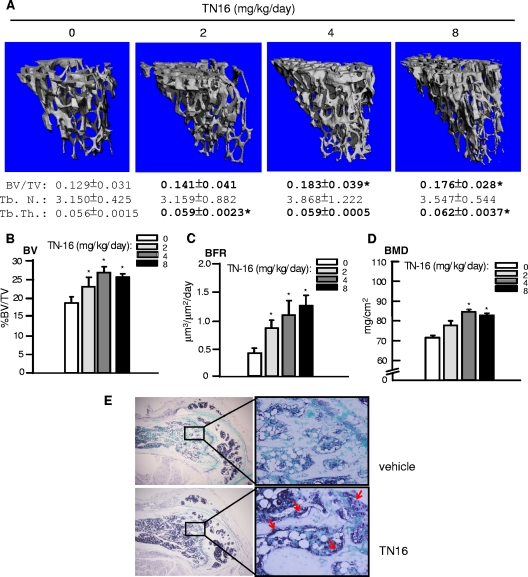
To investigate whether microtubule inhibitors also have anabolic effects on bone when administrated systemically, TN16 was injected intraperitoneally into 3-month-old ICR Swiss mice at different doses for two consecutive days, and parameters of bone formation, including BV/TV, BFR, and BMD, were analyzed 1 month after the last injection. MicroCT scanning and analysis showed that TN16 at 2 to ∼8 mg/kg/day increased the tibial trabecular bone volume significantly, with a maximal increase of 41.9% over that with the vehicle control (Fig. 3A). MicroCT analysis of the trabecular architecture revealed that the Tb.Th. of the tibial metaphysis was significantly elevated, although the Tb.N. and Tb.Sp. were not affected, suggesting an increase in trabecular bone formation. The marked increase of 30 to 50% in trabecular volume (Fig. 3B), determined histomorphometrically, in the groups treated with TN16 relative to results for the control was consistent with the microCT findings (Fig. 3A). This is also comparable to the increase in bone mass seen with proteasome inhibitor 1 (data not shown), a proteasome inhibitor used as a positive control and known to stimulate bone formation in rodents (15). Microtubule inhibitors also enhanced trabecular BFR in a dose-dependent manner as determined by double-labeling assays (Fig. 3C). In addition, we also measured the BMD and found that the tibial BMD was significantly increased upon forced microtubule disassembly (Fig. 3D). Since we have shown above that microtubule inhibitors are capable of stimulating BMP-2 expression in osteoblasts, we next attempted to examine BMP-2 levels in sections of femorae by in situ hybridization using a mouse BMP-2 probe. BMP-2 mRNA expression (purple-blue) in the osteoblasts along the trabecular bone surface and in chondrocytes in the growth plate (arrows) was increased in TN16-treated bones compared with results for controls (Fig. 3E).
FIG. 3.
Effects of inhibition of microtubule assembly on trabecular bone formation. Three-month-old ICR Swiss mice were injected intraperitoneally with TN16 at doses of 0, 2, 4, and 8 mg/kg/day (50 μl per injection twice a day for 2 days) and sacrificed 1 month later. The mice were also injected with tetracycline and calcein at 10 days and 6 days before sacrifice, respectively. At sacrifice, the tibiae were harvested for analyses. (A) MicroCT analysis. Tibial metaphysis was scanned by the ScanCO μCT 40 scanner with 417 slices. Two hundred slices just beneath the growth plate were analyzed using the ScanCO 40 software program to measure BV/TV, Tb.N., Tb.Th., and Tb.Sp. and to obtain high-resolution images of tibial trabecular bones. (B, C, and D) Tibial histomorphometry. Paraffin-embedded tibiae from TN16-injected mice given the above doses were processed and stained with H&E. BV/TV was measured on the trabecular area of these sections (B). BRF of tibia trabecular bones were measured by using double fluorescence of tetracycline and calcein (C). BMD of tibial metaphysis was measured using a PIXImus bone densitometer (D). (E) BMP-2 in situ hybridization. Hybridization was performed on sections of the femorae from the same mice using a mouse BMP-2 probe. Five to eight mice per group were used. *, P < 0.05 versus results for control.
Taken together, these data from in vivo studies show that microtubule inhibition compounds are effective stimulators of bone formation and this is a result of enhancement of BMP-2 expression in bone.
Inhibition of microtubule assembly enhances Gli2 protein concentrations in osteoblasts.
The above studies show that microtubule inhibitors are powerful stimulators of BMP-2 gene expression and bone formation. However, beyond our demonstration that enhanced BMP-2 transcription is likely involved, the mechanisms responsible for these effects are unclear.
Microtubules function as important cytoskeletal components for cell mitosis and are also involved in various cell signaling pathways (16, 37). We hypothesized that microtubule assembly may influence essential molecular components of the Hh pathway, since BMP-2 gene expression is dependent on this pathway, and we have shown that BMP-2 expression is regulated by the Hh mediator Gli2 (61). Microtubules are thought to provide a scaffold and associate with a protein complex composed of Ci proteins and other regulators, facilitating the proteolytic processing of the Ci protein (the fly homolog of vertebrate Gli proteins) (43, 53). Given that the Hh pathway is conserved from flies to vertebrates, Gli proteins might also associate with microtubules. We have shown that Gli2, a major transcriptional activator of Hh target genes, binds directly to the BMP-2 promoter and induces BMP-2 gene expression in osteoblasts and bones (61). Therefore, we hypothesized that the stimulation of BMP-2 expression by inhibition of microtubule assembly occurs because microtubule inhibition protects Gli2 from proteasomal processing and this in turn results in enhancement of Gli2 transcriptional activity.
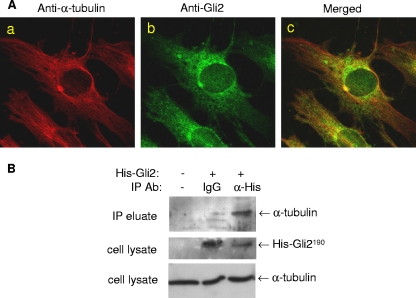
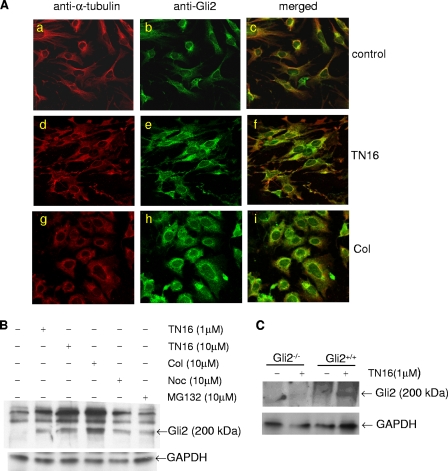
To test this hypothesis, we initially examined the interaction of microtubules with the Gli2 protein in osteoblasts by immunofluorescence using 2T3 cells double labeled with anti-α-tubulin and anti-Gli2 antibodies. Under the high magnification of an immunofluorescence microscope, we could clearly see the radial filaments of assembled microtubules which were stained with red Cy3 fluorescence in the cytoplasm (Fig. 4Aa). On the other hand, the Gli2 proteins were visualized by green FITC fluorescence, which was unevenly distributed over the cytoplasm and nuclei with a higher concentration surrounding the nuclei (Fig. 4Ab). Merged images revealed that the Gli2 immunofluorescent stain overlapped with the microtubule stain along the microtubule filaments in the same cytoplasmic compartment, particularly in the area surrounding the nuclei (Fig. 4Ac). Although not conclusive, this colocalization of microtubules and Gli2 is strongly suggestive of an association between these proteins. To provide further evidence of a physical interaction between Gli2 and microtubules, protein immunoprecipitation experiments were performed in which 2T3 cells were transfected with a His-Gli2 expression construct and an anti-His antibody was used to coprecipitate the cytoplasmic Gli2-microtubule complex. Western blots of the immunoprecipitate using an anti-α-tubulin antibody confirmed the presence of microtubules in the precipitated complex (Fig. 4B). These results indicate that microtubules recruit and associate with the Gli2 protein in 2T3 osteoblast precursor cells.
FIG. 4.
Interaction between microtubules and Gli2. (A) Immunofluorescence colocalization of microtubules and Gli2. 2T3 cells cultured in eight-well chamber slides were fixed and incubated with a 1:50 dilution of mouse anti-α-tubulin (Aa) or rabbit anti-mouse Gli2 (Ab) or both antibodies (Ac) and then with the appropriate secondary antibodies against rabbit and mouse IgG, conjugated with FITC and Cy3, respectively. Fluorescence on the mounted slides was captured with a confocal microscope system. (B) Immunoprecipitation of microtubules and Gli2.Cell lysates from 2T3 cells transfected with a His-Gli2 vector were incubated with rabbit Omni-probe anti-His antibody (lane 3), IgG (lane 2), or no Ab (lane 1) and then loaded onto an immobilized protein G column. The eluted immunoprecipitated complex was separated by SDS-PAGE and blotted with anti-α-tubulin antibody (upper panel). The expression of His-Gli2 (middle) or α-tubulin (bottom) as a control in cell lysates was detected by blotting with anti-His or anti-α-tubulin antibody (middle).
Next, we examined the effects of the microtubule inhibitors on the microtubule network and on Gli2 expression in the cytoplasm. 2T3 cells were treated with TN16 (1.0 μM) or colchicine (10 μM), and double immunofluorescence labeling was performed. Treatment with these inhibitors of microtubule assembly altered the immunofluorescence pattern of microtubules in the cytoplasm. While a normal microtubule architecture, demonstrated by clear filaments in the cytoplasm, was maintained in control vehicle-treated 2T3 cells (Fig. 5Aa), the microtubule network in the cells treated with either TN16 or colchicine was markedly disrupted, as evidenced by a replacement of the normal radial pattern of the filaments with a more diffuse pattern (Fig. 5Ad and g). In addition, there was a distinct change in the shape of 2T3 cells treated with microtubule inhibitors from a dendritic to an irregular (TN16) or rounded (colchicine) shape. This provides further evidence that microtubules are one of the cytoskeletal components responsible for maintaining cell shape. More importantly, we found that inhibition of microtubule assembly increased the immunofluorescence intensity of Gli2-FITC expression in the cells (Fig. 5Ab, e, h, c, f, and i), supporting the notion that microtubule inhibition-induced accumulation of the Gli2 protein most likely occurs as a result of inhibition of Gli2 proteolytic degradation. Although endogenous Gli2 was barely detectable in untreated 2T3 cells by Western blotting, treatment with microtubule inhibition drugs, including TN16 (1 μM), colchicine (10 μM), or nocodazole (10 μM), substantially increased the Gli2 protein level to a similar degree to that seen in proteasome inhibitor PS1-treated cells (Fig. 5B). To further confirm the specificity of the anti-Gli2 antibody, we have compared the effects of TN16 on the expression of endogenous Gli2 in wild-type and Gli2-deficient cells. When primary calvarial osteoblasts isolated from Gli2 knockout (Gli2−/−) and wild-type (Gli2+/+) mice (61) and treated with TN16 at μM for 24 h were compared, we found as expected that TN16 clearly induced a protein band (∼200 kDa) (Fig. 5C, lane 4) that was recognized by the anti-Gli2 antibody in the wild-type calvarial cells. But contrast, TN16 had no such effect in the Gli2-deficient cells (Fig. 5C, lane 2).
FIG. 5.
Effects of inhibition of microtubule assembly on Gli2 expression. (A) Immunofluorescence colocalization of microtubules and Gli2. 2T3 cells seeded in eight-well chamber slides were treated with vehicle (Aa, b, and c), TN16 at 1.0 μM (Ad, e, and f), or colchicine (Col) at 10 μM (Ag, h, and i) for 24 h. The fixed cells were stained with a 1:50 dilution of mouse anti-α-tubulin (Aa, d, and g) or rabbit anti-mouse Gli2 (Ab, e, and h) or both antibodies (Ac, f, and i), followed by the appropriate secondary antibodies conjugated with Cy3 (Aa, d, g, c, f, and i) or FITC (Ab, e, h, c, f, and i). After mounting, fluorescence was captured by a confocal microscope system. (B) Western blot of Gli2. 2T3 cells were treated with microtubule inhibitors (TN16, 1.0 and 10 μM; colchicine [Col], 10 μM; nocodazole [Noc], 10 μM; MG132, 10 μM) for 24 h. Proteins in cell lysates were resolved by SDS-PAGE, transferred onto a PVDF membrane, and blotted with a rabbit anti-mouse Gli2 antibody. GAPDH served as a loading control. (C) Western blot of Gli2 of Gli2-deficient cells. The primary calvarial osteoblasts were isolated from Gli2 knockout (Gli2−/−) or wild-type (Gli2+/+) newborn mice and treated with TN16 at 1 μM for 24 h. The endogenous Gli2 protein levels were examined by Western blotting as described above.
Together, these observations suggest that inhibition of microtubule assembly increases Gli2 protein levels in osteoblasts and this occurs because microtubule inhibition protects Gli2 from degradation.
Inhibition of microtubule assembly inhibits β-TrCP-mediated proteasomal degradation of Gli2.
Microtubules recruit a protein complex which facilities ubiquitination and proteasomal processing of Ci/Gli proteins (6, 22, 38, 48, 53). Recently β-TrCP was identified as the specific E3 ubiquitin ligase that mediates Gli2 proteolysis in nonosteoblastic cells, such as HEK 293 cells and mouse embryonic fibroblasts (40, 41, 55). However, the role of β-TrCP in Gli2 processing in osteoblasts has not been characterized. To determine the effects of microtubule inhibition on β-TrCP-mediated Gli2 degradation, we next addressed whether β-TrCP is critical for Gli2 degradation in osteoblasts.
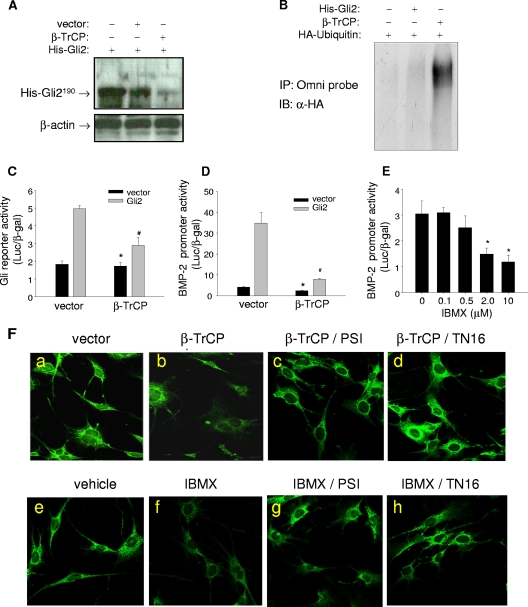
First, we examined the effect of overexpression of β-TrCP on Gli2 degradation in osteoblasts. Previously, using osteoblast precursor C3H10T1/2 cells, we showed that protein kinase A (PKA)-mediated phosphorylation is involved in the proteolytic processing of the His-Gli2 protein (61). Here we showed that cotransfection of β-TrCP and full-length His-Gli2 (His-Gli2190) in C3H10T1/2 cells resulted in a marked reduction in protein concentrations of His-Gli2190 compared with controls, suggesting that β-TrCP mediates proteolytic processing of the Gli2 protein (Fig. 6A). Next, His-Gli2, HA-ubiquitin, and β-TrCP expression plasmids were cotransfected into C3H10T1/2 cells, cell lysates immunoprecipitated with anti-His-Gli2 antibody, and the resulting precipitate immunoblotted with an anti-HA antibody. Overexpression of β-TrCP caused a smear of the HA-ubiquitin stain (Fig. 6B, lane 3) compared with results for the controls (lanes 1 and 2), indicating that β-TrCP induces ubiquitination of the Gli2 protein. We then examined the effects of β-TrCP on Gli2 transcriptional activity. Again, overexpression of β-TrCP in osteoblastic cells significantly decreased both basal and Gli2-enhanced luciferase activity of a Gli response reporter (Fig. 6C). Similar inhibitory effects of β-TrCP were also observed in the BMP-2 promoter reporter assay (Fig. 6D). Since β-TrCP-mediated Gli2 degradation relies on prior Gli2 phosphorylation, we examined the effects of activation of PKA on Gli2 expression in osteoblasts, since PKA has been shown to phosphorylate Gli2. 2T3 cells that carry the BMP-2 promoter reporter gene were incubated with the cAMP/PKA activator IBMX at 0.1 to 10 μM for 24 to 48 h, followed by a luciferase assay. We found that treatment with IBMX for 24 h slightly increased the luciferase activity (data not shown). However, if the cells were treated for 48 h, IBMX dose-dependently and significantly inhibited the luciferase reporter activity (Fig. 6E). Together, these results suggest that the E3 ubiquitin ligase β-TrCP and PKA pathway are responsible for proteasomal processing of Gli2, which downregulates BMP-2 expression in osteoblasts.
FIG. 6.
Effects of inhibition of microtubule assembly on β-TrCP-mediated Gli2 degradation. (A) Western blot of Gli2. C3H10T1/2 cells were cotransfected with His-Gli2 and β-TrCP expression plasmids, and after 24 h, cells were lysed and proteins were separated by SDS-PAGE and probed with a rabbit Omni-probe anti-His antibody. β-Actin is the loading control. (B) Ubiquitination of Gli2. C3H10T1/2 cells were cotransfected with His-Gli2, HA-ubiquitin, and β-TrCP plasmids. Cell lysates were incubated with an Omni-probe anti-His antibody and protein A beads. The immunoprecipitate (IP) was separated by SDS-PAGE and blotted (immunoblot [IB]) with anti-HA (α-HA) antibody. (C and D) Effects of β-TrCP on luciferase reporter activity. 2T3 cells were cotransfected with Gli2, β-TrCP plasmids, and a Gli response reporter construct, 8×3′Gli-BSδ5/LucII (C) or the BMP-2 promoter reporter, −2712/+165-Luc (D). Luciferase activity was measured with β-Gal normalization. (E) Effects of IBMX on BMP-2 promoter activity. The BMP-2-Luc 2T3 cells were incubated with IBMX at doses from 0.1 to 10 μM for 48 h. The reporter luciferase activity was determined. (F) Immunofluorescence of Gli2. 2T3 cells seeded in eight-well chamber slides with different treatments were processed for immunofluorescence assay using anti-mouse Gli2 antibody and FITC-labeled secondary antibody. Controls (Ea and e), β-TrCP transfection (Eb), or IBMX (2 μM) incubation (Ef), β-TrCP with 100 nM PSI (Ec), β-TrCP with 1 μM TN16 (Ed), IBMX with PSI (Eg), or IBMX with TN16 (Eh) is shown. *, P < 0.05 versus results with vector (black); #, P < 0.05 versus results with vector (gray).
Next, we determined the effects of microtubule inhibition on β-TrCP-mediated Gli2 degradation. We found that β-TrCP overexpression substantially decreased the fluorescence intensity of Gli2-FITC in the cytoplasm compared with the vector control (Fig. 6Fa and b). Treatment with IBMX at 2 μM reduced Gli2-FITC fluorescence in the cells (Fig. 6Fe and f). Importantly, PKA/β-TrCP-induced Gli2 degradation in osteoblasts is also proteasome dependent, since addition of PS1 to the culture prevented Gli2 degradation (Fig. 6Fc and g). To evaluate the effects of inhibition of microtubule assembly, β-TrCP-transfected or IBMX-treated cells were simultaneously coincubated with TN16 at 1 μM for 24 h. As expected, TN16 treatment markedly attenuated both the IBMX- and β-TrCP-mediated reduction in Gli2 fluorescence in the cytoplasm (Fig. 6Fd and h). Together, these results suggest that microtubules are involved in β-TrCP-mediated Gli2 degradation and that inhibition of microtubule assembly protects Gli2 from degradation through this pathway.
Inhibition of microtubule assembly enhances Gli2 transcriptional activity.
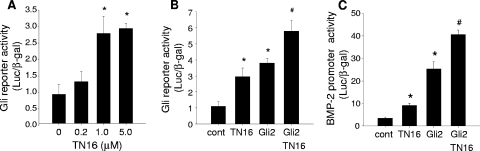
Gli2 transactivates target genes through consensus Gli response elements (44, 45). Since inhibition of microtubule assembly upregulates Gli2 protein expression in osteoblasts, we next determined the effects of a microtubule inhibitor on the transcriptional activity of Gli2. When 2T3 cells transfected with a Gli-responsive reporter construct, 8×3′Gli-BSδ5/LucII, which contains multiple copies of Gli response elements, were incubated with TN16 at different doses, not only was there a dose-dependent enhancement in Gli reporter activity (Fig. 7A) but TN16 and Gli2 had an additive effect in transactivating the Gli reporter gene (Fig. 7B). Furthermore, we found that microtubule inhibition also potentiated the effect of Gli2 to stimulate BMP-2 promoter activity (Fig. 7C). These results strongly support our hypothesis that disruption of microtubules inhibits Gli2 degradation, which in turn increases Gli2 transcriptional activity.
FIG. 7.
Effects of inhibition of microtubule assembly on Gli2 transcriptional activity. (A) Dose response of TN16. 2T3 cells transfected with a Gli reporter gene, 8×3′Gli-BSδ5/LucII, were treated with TN16 at the indicated doses for 24 h. Reporter luciferase activity was quantitated and normalized to activity of cotransfected β-Gal. (B and C) Potentiation of TN16 effects by Gli2. 2T3 cells were cotransfected with a Gli2 expression vector and the 8×3′Gli-BSδ5/LucII reporter (B) or −2712/+165-Luc reporter (C) and then treated with TN16 (1.0 μM) for 24 h. Luciferase activity of the Gli reporter (B) or BMP-2 promoter reporter (C) were measured with β-Gal normalization. *, P < 0.01 versus control; #, P < 0.05 versus Gli2.
Inhibition of microtubule assembly promotes osteoblast differentiation.
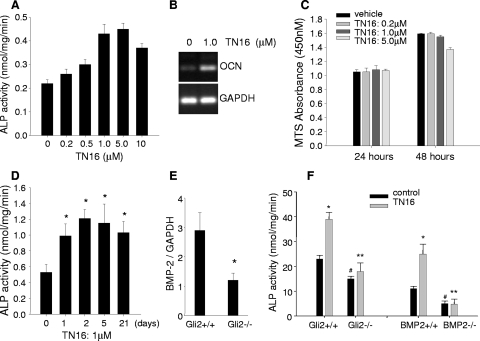
Since inhibition of microtubule assembly enhances BMP-2 expression in osteoblasts, we examined the direct effects of microtubule inhibition on osteoblast differentiation in vitro. In 2T3 cells incubated with TN16 at doses from 0 to 10 μM for 48 h, ALP activity increased dose dependently (Fig. 8A). We also examined the effect of TN16 on expression of the osteocalcin gene, which is known as an osteoblast-specific marker gene. After 48 h of treatment with TN16 at 1.0 μM, the osteocalcin mRNA expression level was significantly elevated over that with vehicle control (Fig. 8B). In addition, we found that microtubule inhibitors did not have a significant effect on proliferation of 2T3 cells as assessed by the MTS assay (Fig. 8C). These data suggest that microtubules are involved in the regulation of osteoblast function and that the anabolic effects of microtubule inhibition on bone are mediated primarily by promoting osteoblast differentiation but not by inducing osteoblast proliferation. To explain the long-term effect of TN16 on bone in vivo after injections for only 2 days, we conducted an experiment with a long-term cell culture. 2T3 cells were seeded in the plate at a lower density and incubated with TN16 at 1 μM for 5 days. After a wash to remove TN16, the cells were continuously cultured. We found that up to week three, the levels of both BMP-2 mRNA expression (Fig. 1H) and ALP activity (Fig. 8D) in these osteoblasts were still significantly higher than those with vehicle control. These results may provide useful information for the mechanisms of the long-term effect of microtubule inhibitors on bone.
FIG. 8.
Effects of inhibition of microtubule assembly on osteoblast differentiation. (A) Effects of TN16 on ALP activity. 2T3 cells were incubated with TN16 at the indicated doses for 48 h, and ALP activity of cell lysates was measured using a Sigma ALP kit and normalized to total cellular proteins. #, P < 0.05; *, P < 0.01 versus results for control. (B) Effects of TN16 on osteocalcin expression. 2T3 cells were treated with TN16 at 1.0 μM for 24 h. Reverse transcription-PCR of mouse osteocalcin mRNA was performed with GAPDH as an internal control. (C) Effects of TN16 on cell proliferation. 2T3 cells were treated with TN16 at the indicated doses for 24 to 48 h. Cell proliferation was measured spectrophotometricaally at 450 nm using a Promega MTS kit. (D) Time course of TN16 stimulation of ALP activity. 2T3 cells were seeded in the 48-well plate at a cell density of 3 × 103 cells per well and incubated with TN16 at 1 μM for 5 days. After a wash to remove TN16 with fresh medium, the cells were continuously cultured for up to 21 days. The ALP activity of the cell lysates were then measured as described above. *, P < 0.05 versus results for day zero. (E) BMP-2 mRNA expression in Gli2-deficient osteoblasts. Primary osteoblast cells were isolated from calvariae of Gli2+/+ or Gli2−/− embryos (E20.5). Total RNA was extracted from the cell lysate, and the BMP-2 mRNA level was quantitated by real-time reverse transcription-PCR with mouse BMP-2 primer. Data presented are normalized to GAPDH. *, P < 0.05 versus results for Gli2+/+ cells. (F) Effects of TN16 on ALP activity in osteoblasts null for Gli2 (left panel) or BMP-2 (right panel). Calvarial osteoblastic cells were isolated from Gli2+/+ and Gli2−/− littermates at E20.5, and bone marrow mesenchymal stem cells (BMSCs) were flushed from the conditional BMP-2 knockout (BMP2−/−) or wild-type (BMP2+/+) mice (2 months old). These primary osteoblastic cells were cultured in α-MEM in the presence of TN16 at 1.0 μM for 24 h, and ALP activity of cell lysates was determined. *, P < 0.01 versus results with vehicle; #, P < 0.05 versus results with Gli2+/+ or BMP-2+/+ cells; **, P < 0.01 versus results for Gli2+/+ or BMP-2+/+ cells with TN16 treatment.
Previously we showed that a Gli2 null mutation in mice reduces BMP-2 expression in bone and this in turn inhibits osteoblast differentiation, suggesting that Gli2 plays an essential role in maintaining osteoblast function (61). Therefore, here we determined whether the effects of microtubule inhibition in promoting osteoblast differentiation are Gli2 dependent. Primary osteoblastic cells from calvariae of E20.5 wild-type (Gli2+/+) and Gli2 null mutant (Gli2−/−) embryos were isolated, and the effects of TN16 on ALP activity were examined. First, we confirmed by using real-time PCR that BMP-2 gene mRNA expression in the Gli2-deficient cells was significantly decreased compared with that in wild-type calvarial cells (Fig. 8E). TN16-induced microtubule inhibition increased ALP activity in wild-type cells but not in Gli2-deficient cells (Fig. 8F, left panel). These results provide in vivo evidence that inhibition of microtubule assembly stimulates osteoblast differentiation and this is mediated through Gli2. In this study, we also utilized BMP-2-deficient cells to determine if the effects of microtubule inhibitors on osteoblasts are dependent on BMP-2. Recently we have established an osteoblast-specific BMP-2 knockout mouse model by crossing BMP-2 floxed mice with Col1a1-Cre mice (57). The bone marrow mesenchymal stem cells (BMSCs) were isolated from the conditional BMP-2 knockout (BMP2−/−) and control (BMP2+/+) mice and cultured under osteogenic conditions. We found that the basal level of ALP activity was reduced in the BMP2−/− BMSCs compared with that in the control cells. We also found that while treatment with TN16 significantly increased ALP activity in the control BMSCs, TN16 lost this effect in the BMP-2 KO cells (Fig. 8F, right panel). This provides evidence that BMP-2 is a major player in effects of microtubule inhibitors on bone.
DISCUSSION
BMP-2 has been shown to be an important growth factor for postnatal bone formation and is an important molecular target for drug discovery. Our group has previously characterized the 5′ flanking promoter region of the mouse BMP-2 gene (9, 10, 14, 19, 20) and established a cell-based BMP-2 promoter reporter assay that enables high-output screening. Using this assay system, we screened low-molecular-weight-compound libraries and independently identified statins and proteasome inhibitors as stimulators of BMP-2 expression and bone formation (12, 35). In the present studies, we describe another class of BMP-2 stimulators, inhibitors of microtubule assembly, which were identified using the same approach.
The dynamic assembly and disassembly of microtubules have made microtubules a molecular target in cancer drug discovery (23). Microtubule-targeting drugs, including microtubule assembly inhibitors and microtubule stabilizers, suppress microtubule dynamics rather than increasing or decreasing microtubule-polymer mass (23). Through screening assays, we found that inhibitors of microtubule assembly, such as TN16, colchicine, and nocodazole, enhance BMP-2 transcription, while taxol, a microtubule-stabilizing drug, has no effect (Fig. 1), suggesting that inhibition of microtubule assembly mediates upregulation of BMP-2 gene expression. In these studies, we have particularly characterized the effects of TN16 on bone formation and its mechanisms both in vivo and in vitro, since TN16 is known as a typical microtubule inhibitor which binds to colchicine-sensitive sites of tubulins, inhibits microtubule assembly, and prevents the stabilization of microtubules (1).
Since microtubule inhibitors increase BMP-2 expression in osteoblasts, we determined the effects of these drugs and specifically of TN16 on bone formation. We found that TN16, administered either locally over the calvaria or systemically in mice, has a robust anabolic effect on bone. An increase in calvarial periosteal bone formation and an increase in the trabecular bone volume of long bones were evident after a month even though TN16 was administered only for the first 2 days (Fig. 2 and 3). This suggests that transient treatment with microtubule inhibitors enhanced BMP-2 expression at an early stage and that this transient stimulation is sufficient to initiate a process resulting in osteoblastic differentiation and bone formation. This notion was further supported by the in vitro observation in long-term cell culture (Fig. 1H and 8D).
In cancer therapy, microtubule-targeting drugs have been reported to have tolerable toxicities (18, 28, 32). In our experiments with mice, we did not observe obvious signs of toxicity (weight loss or changes in behavior and eating) using TN16, colchicine, and nocodazole at the doses indicated. In addition, we did not see any evidence of toxicity either at necropsy or in bone histology sections. Nevertheless, we cannot completely exclude potential drug toxicity in other organs and tissues. This obviously needs to be further characterized, and mechanisms for specific targeting to bone may need to be developed.
To determine molecular mechanisms by which microtubule inhibition activates BMP-2 gene transcription, we focused on the Hh signaling mediator Gli2, since microtubules are known to play a role in this pathway (6, 22, 38, 43, 48) and we have previously identified Gli2 as a powerful transcriptional enhancer of the BMP-2 gene (61). Gli2 mutations result in skeletal developmental abnormalities in humans and mice (33, 34, 42). We have previously reported that Gli2 directly binds to the BMP-2 promoter and transactivates the BMP-2 gene in osteoblasts (61). As with other mediators of the Hh pathway, such as Ci and Gli3, Gli2 activity is modulated by Hh signaling activity. In the absence of Hh signaling, Gli2 undergoes ubiquitin-dependent proteasomal degradation (40, 41). In contrast, with Hh activation, Gli2 degradation is inhibited, resulting in Gli2 accumulation in the cytoplasm, nuclear translocation, and activation of target genes (27). Microtubules are thought to associate with the Ci complex and facilitate Ci proteolytic processing. Therefore, we hypothesized that microtubule inhibition by forced disassembly may protect Gli2 from degradation and thereby enhance BMP-2 transcription. In immunoprecipitation-and-immunofluorescence experiments (Fig. 4), we found that microtubules and Gli2 associate each other and colocalize within the same cytoplasmic compartment. However, these data do not indicate unequivocally whether such an association is direct. A possible model in which microtubules would function as a scaffold in the Hh pathway is recruitment of a protein complex composed of the Costal 2 (Cos2), Fused, Suppressor of Fused, and Ci/Gli proteins and facilitation of the proteolytic processing of Ci/Gli proteins (6, 22, 38, 43, 48, 53). Microtubule-organized complexes are known to associate with the 20S proteasome for protein degradation (54), and there is evidence that this association is mediated by Cos2, which serves as a bridge between microtubules and other components in the complex, including the Ci protein (24, 25, 37). Further investigation is required to identify the role of Cos2 in microtubule function and regulation of Gli2 by microtubule disassembly in osteoblasts.
Studies with flies have suggested that without Shh, microtubules associate with the Ci complex, and as a result, proteasomal processing of this complex occurs (6, 22, 38, 40, 41, 43, 48, 53). Shh activation results in disassociation of the complex and inhibition of Ci processing. Consistent with this notion, we found that inhibition of microtubule assembly mimics Shh activation in murine osteoblasts, i.e., it disrupts the microtubule filament structure and increases Gli2 protein concentrations in cells (Fig. 5A and B). Unlike the case with the processing of Gli2, which is apparently complete, Gli3 is cleaved to release a C′-terminally truncated form (Gli3rep) (55). We previously reported that Gli3rep represses BMP-2 transcription and bone formation (12). Therefore, it is tempting to speculate that microtubule inhibition not only reduces degradation of full-length Gli2 but may also reduce generation of repressor Gli3 by effectively inhibiting Gli3 processing, which could be also anticipated to enhance BMP-2 transcription. This requires further investigation.
Anchoring of the Ci complex to microtubules initiates the proteolytic processing of the Ci protein through the β-TrCP-proteasome pathway (38, 43, 48). In this study, we have demonstrated that microtubules also associate with the Gli2 complex and facilitate Gli2 degradation in osteoblasts via the same pathway. Herein, we described the role of β-TrCP in mediating Gli2 degradation and inhibiting BMP-2 promoter activity in osteoblasts (Fig. 6A to D). We consistently found that microtubule inhibition blocked such β-TrCP function (Fig. 6F). In other cell systems, it has been shown that β-TrCP-mediated Gli2 degradation requires Gli2 phosphorylation by PKA or other kinases (40, 41, 55). We demonstrated that IBMX, a PKA activator, is a negative regulator of the Gli2 protein concentration in osteoblasts (Fig. 6F). However, we unexpectedly found that IBMX has dual effects on BMP-2 promoter activity; that is, treatment with IBMX for 24 h increases BMP-2 promoter activity (data not shown), but treatment for 48 h inhibits BMP-2 transcription (Fig. 6E). The following explanations are possible reasons for this phenomenon. IBMX induces PKA activity, which activates the cAMP response element binding protein (CREB). We have found that CREB is a powerful enhancer of BMP-2 transcription in bone cells (data not shown). Phosphorylation-induced activation of CREB activity is a rapid response within 24 h. However, if the IBMX treatment is extended, it will trigger Gli2 degradation and in turn inhibit BMP-2 expression. Taking these things together, we propose that the molecular basis for enhancement of Gli2 protein concentrations by inhibition of microtubule assembly is dissociation of the microtubule-Gli complex, which impairs β-TrCP function in targeting the complex to the proteasome. These findings suggest an important role of β-TrCP in osteoblast differentiation and bone formation by Gli2 and BMP-2. However, a number of questions remain, including how microtubules, Gli2, and β-TrCP associate and how drugs that promote microtubule disassembly affect such an association.
Our data with BMP-2 conditional knockout mice (Fig. 8F) suggest that BMP-2 is a major player in mediating the anabolic effects of microtubule inhibitors on bone, and we did find that microtubule inhibition has an effect on the canonical Wnt/β-catenin signaling activity in osteoblasts. However, we do not exclude the possibility that multiple signaling pathways may also be involved in the effects of microtubule drugs on bone. For example, Gli2 has also recently been shown to upregulate expression and activity of Runx2 (46). Runx2 controls osteoblast maturation and bone formation (7, 26, 39, 60). Therefore, it is possible that microtubule activity may also regulate Runx2 expression through Gli2 and that part of the effect of microtubule inhibitors on osteoblast differentiation is mediated through Runx2. The relationship between Gli2, BMP-2, and Runx2 requires further investigation.
Recently Chandler et al. identified a specific DNA sequence of 600 bp located more than +156 kb 3′ from the mouse BMP-2 promoter and demonstrated this short sequence is responsible for controlling BMP-2 transcription specifically in osteoblasts (5). This regulatory region contains a Gli binding site. We are presently investigating the involvement of this remote sequence in BMP-2 regulation by Gli proteins and microtubules.
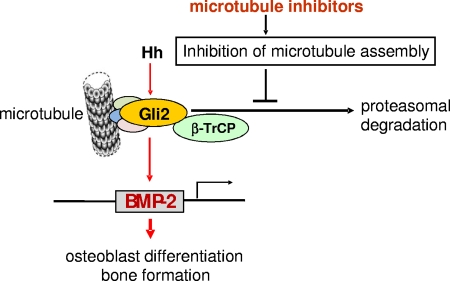
In summary, we have identified inhibitors of microtubule assembly as powerful stimulators of bone formation and have investigated the potential molecular mechanisms responsible for this effect. Based on our results and those of others, we propose a model (Fig. 9) in which microtubule inhibition causes disassociation of the microtubule-Gli complex in osteoblasts and that in turn inhibits β-TrCP-dependent proteasomal degradation of Gli2. This results in enhancement of Gli2 protein concentrations within cells and increased BMP-2 transcription. Through this mechanism, microtubule inhibition results in stimulation of osteoblast differentiation and new bone formation.
FIG. 9.
Proposed mechanism by which inhibition of microtubule assembly regulates Gli2 and BMP-2 in osteoblasts. Microtubule inhibitors cause inhibition of microtubule assembly and disassociation of the microtubule-Gli complex in osteoblastic cells. This in turn impairs the E3 ligase β-TrCP-dependent proteolytic processing of Gli2 in the proteasome, resulting in accumulation of Gli2 within cells. In turn, increased concentrations of Gli2 lead to enhanced transactivation of the BMP-2 gene and consequently increased osteoblast differentiation and new bone formation.
Acknowledgments
We thank Alexandra Joyner for kindly providing Gli2+/− null mice, Hiroshi Sasaki for kindly providing Gli expression vectors and Gli reporter constructs, and Irina Kaverina for suggestions for microtubule experiments. We also thank Gloria Gutierrez, I. Ross Garrett, and Gianni Rossini for their help in the early experiments and Andrew Hart for proofreading.
This study was supported by NIH grants AG024637, AR051165, and AR050605 and grants from the Department of Veterans Affairs.
Footnotes
Published ahead of print on 22 December 2008.
REFERENCES
- 1.Arai, T. 1983. Inhibition of microtubule assembly in vitro by TN-16, a synthetic antitumor drug. FEBS Lett. 155273-276. [DOI] [PubMed] [Google Scholar]
- 2.Baron, R., G. Rawadi, and S. Roman-Roman. 2006. Wnt signaling: a key regulator of bone mass. Curr. Top. Dev. Biol. 76103-127. [DOI] [PubMed] [Google Scholar]
- 3.Boden, S. D. 2005. The ABCs of BMPs. Orthop. Nurs. 2449-52. [DOI] [PubMed] [Google Scholar]
- 4.Boyden, L. M., J. Mao, J. Belsky, L. Mitzner, A. Farhi, M. A. Mitnick, D. Wu, K. Insogna, and R. P. Lifton. 2002. High bone density due to a mutation in LDL-receptor-related protein 5. N. Engl. J. Med. 3461513-1521. [DOI] [PubMed] [Google Scholar]
- 5.Chandler, R. L., K. J. Chandler, K. A. McFarland, and D. P. Mortlock. 2007. Bmp2 transcription in osteoblast progenitors is regulated by a distant 3′ enhancer located 156.3 kilobases from the promoter. Mol. Cell. Biol. 272934-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, M. M., Jr. 2003. The Hedgehog signaling network. Am. J. Med. Genet. 1235-28. [DOI] [PubMed] [Google Scholar]
- 7.Ducy, P., R. Zhang, V. Geoffroy, A. L. Ridall, and G. Karsenty. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89747-754. [DOI] [PubMed] [Google Scholar]
- 8.Feng, J. Q., L. P. Xing, J. H. Zhang, M. Zhao, D. Horn, J. Chan, B. F. Boyce, S. E. Harris, G. R. Mundy, and D. Chen. 2003. NF-κB specifically activates BMP-2 gene expression in growth plate chondrocytes in vivo and in a chondrocyte cell line in vitro. J. Biol. Chem. 27829130-29135. [DOI] [PubMed] [Google Scholar]
- 9.Feng, J. Q., D. Chen, N. Ghosh-Choudhury, J. Esparza, G. R. Mundy, and S. E. Harris. 1997. Bone morph genetic protein2 transcripts in rapidly developing deer antler tissue contain an extended 5′ non-coding region arising from a distal promoter. Biochim. Biophys. Acta 135047-52. [DOI] [PubMed] [Google Scholar]
- 10.Feng, J. Q., M. A. Harris, N. Ghosh-Choudhury, M. Feng, G. R. Mundy, and S. E. Harris. 1994. Structure and sequence of mouse bone morphogenetic protein-2 gene (BMP-2): comparison of the structures and promoter regions of BMP-2 and BMP-4 genes. Biochim. Biophys. Acta 1218221-224. [DOI] [PubMed] [Google Scholar]
- 11.Fleet, J. C., K. Cashman, K. Cox, and V. Rosen. 1996. The effects of aging on the bone inductive activity of recombinant human bone morphogenetic protein-2. Endocrinology 1374605-4610. [DOI] [PubMed] [Google Scholar]
- 12.Garrett, I. R., D. Chen, G. Gutierrez, M. Zhao, A. Escobedo, G. Rossini, S. E. Harris, W. Gallwitz, K. B. Kim, S. Hu, C. M. Crews, and G. R. Mundy. 2003. Selective inhibitors of the chymotrypsin-like activity of the osteoblast proteasome are potent stimulators of bone formation in vivo and in vitro. J. Clin. Investig. 1111771-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh-Choudhury, N., J. J. Windle, B. A. Koop, M. A. Harris, D. L. Guerrero, J. M. Wozney, G. R. Mundy, and S. E. Harris. 1996. Immortalized murine osteoblasts derived from BMP-2T antigen expressing transgenic mice. Endocrinology 137331-339. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh-Choudhury, N., G. Ghosh-Choudhury, M. A. Harris, J. M. Wozney, G. R. Mundy, S. L. Abboud, and S. E. Harris. 2001. Autoregulation of mouse BMP-2 gene transcription is directed by the proximal promoter element. Biochem. Biophys. Res. Commun. 286101-108. [DOI] [PubMed] [Google Scholar]
- 15.Govender, S., C. Csimma, H. K. Genant, A. Valentin-Opran, Y. Amit, R. Arbel, H. Aro, D. Atar, M. Bishay, M. G. Borner, P. Chiron, P. Choong, J. Cinats, B. Courtenay, R. Feibel, B. Geulette, C. Gravel, N. Haas, M. Raschke, E. Hammacher, D. van der Velde, P. Hardy, M. Holt, C. Josten, R. L. Ketterl, B. Lindeque, G. Lob, H. Mathevon, G. McCoy, D. Marsh, R. Miller, E. Munting, S. Oeyre, L. Nordsletten, A. Patel, A. Pohl, W. Rennie, P. Revnders, P. M. Rommens, J. Rondia, W. C. Rossouw, P. J. Daneel, S. Ruff, A. Ruter, S. Santavirta, T. A. Schildhauer, C. Gekle, R. Schnettler, D. Segal, H. Seiler, R. B. Snowdowne, J. Stapert, G. Taglang, R. Verdonk, L. Vogels, A. Weckbach, A. Wentzensen, T. Wisniewski, and the BMP-2 Evaluation in Surgery for Tibial Trauma (BESTT) Study Group. 2002. Recombinant human bone morphogenetic protein-2 for treatment of open tibial fractures: a prospective, controlled, randomized study of four hundred and fifty patients. J. Bone Joint Surg. Am. 842123-2134. [DOI] [PubMed] [Google Scholar]
- 16.Gundersen, G. G., and T. A. Cook. 1999. Microtubules and signal transduction. Curr. Opin. Cell Biol. 1181-94. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton, D. W., K. S. Wong, and D. M. Brunette. 2006. Microfabricated discontinuous-edge surface topographies influence osteoblast adhesion, migration, cytoskeletal organization, and proliferation and enhance matrix and mineral deposition in vitro. Calcif. Tissue Int. 78314-325. [DOI] [PubMed] [Google Scholar]
- 18.Hande, K. R., A. Hagey, J. Berlin, Y. Cai, K. Meek, H. Kobayashi, A. C. Lockhart, D. Medina, J. Sosman, G. B. Gordon, and M. L. Rothenberg. 2006. The pharmacokinetics and safety of ABT-751, a novel, orally bioavailable sulfonamide antimitotic agent: results of a phase 1 study. Clin. Cancer Res. 122834-2840. [DOI] [PubMed] [Google Scholar]
- 19.Harris, S. E., J. Q. Feng, M. A. Harris, N. Ghosh-Choudhury, M. R. Dallas, J. M. Wozney, and G. R. Mundy. 1995. Recombinant bone morphogenetic protein 2 accelerates bone cell differentiation and stimulates BMP-2 mRNA expression and BMP-2 promoter activity in primary fetal rat calvarial osteoblast cultures. Mol. Cell. Diff. 3138-155. [Google Scholar]
- 20.Harris, S. E., M. Sabatini, M. A. Harris, J. Q. Feng, J. M. Wozney, and G. R. Mundy. 1994. Expression of bone morphogenetic protein messenger RNA in prolonged cultures of fetal rat calvarial cells. J. Bone Miner. Res. 9389-394. [DOI] [PubMed] [Google Scholar]
- 21.Holmen, S. L., T. A. Giambernardi, C. R. Zylstra, B. D. Buckner-Berghuis, J. H. Resau, J. F. Hess, V. Glatt, M. L. Bouxsein, M. Ai, M. L. Warman, and B. O. Williams. 2004. Decreased BMD and limb deformities in mice carrying mutations in both Lrp5 and Lrp6. J. Bone Miner. Res. 192033-2040. [DOI] [PubMed] [Google Scholar]
- 22.Ingham, P. W. 1998. Transducing Hedgehog. The story so far. EMBO J. 173505-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordan, M. A., and L. Wilson. 2004. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 4253-265. [DOI] [PubMed] [Google Scholar]
- 24.Katoh, Y., and M. Katoh. 2004. KIF27 is one of orthologs for Drosophila Costal-2. Int. J. Oncol. 251875-1880. [PubMed] [Google Scholar]
- 25.Katoh, Y., and M. Katoh. 2005. Comparative genomics on Sonic hedgehog orthologs. Oncol. Rep. 141087-1090. [PubMed] [Google Scholar]
- 26.Komori, T., H. Yagi, S. Nomura, A. Yamaguchi, K. Sasaki, K. Deguchi, Y. Shimizu, R. T. Bronson, Y. H. Gao, M. Inada, M. Sato, R. Okamoto, Y. Kitamura, S. Yoshiki, and T. Kishimoto. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89755-764. [DOI] [PubMed] [Google Scholar]
- 27.Lipinski, R. J., J. J. Gipp, J. Zhang, J. D. Doles, and W. Bushman. 2006. Unique and complimentary activities of the Gli transcription factors in Hedgehog signaling. Exp. Cell. Res. 3121925-1938. [DOI] [PubMed] [Google Scholar]
- 28.Liu, J. H., M. H. Yang, F. S. Fan, C. C. Yen, W. S. Wang, Y. H. Chang, K. K. Chen, and P. M. Chen. 2001. Tamoxifen and colchicine-modulated vinblastine followed by 5-fluorouracil in advanced renal cell carcinoma: a phase II study. Urology 57650-654. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto, A., K. Yamaji, M. Kawanami, and H. Kato. 2001. Effect of aging on bone formation induced by recombinant human bone morphogenetic protein-2 combined with fibrous collagen membranes at subperiosteal sites. J. Periodontal Res. 36175-182. [DOI] [PubMed] [Google Scholar]
- 30.McGarry, J. G., J. Klein-Nulend, and P. J. Prendergast. 2005. The effect of cytoskeletal disruption on pulsatile fluid flow-induced nitric oxide and prostaglandin E2 release in osteocytes and osteoblasts. Biochem. Biophys. Res. Commun. 330341-348. [DOI] [PubMed] [Google Scholar]
- 31.Meyer, R. A., M. H. Meyer, M. Tenholder, W. R. Wondracel, and P. Garges. 2003. Gene expression in older rats with delayed union of femoral fractures. J. Bone Joint Surg. Am. 851243-1254. [DOI] [PubMed] [Google Scholar]
- 32.Meyers, C. A., A. P. Kudelka, C. A. Conrad, C. K. Gelke, W. Grove, and R. Pazdur. 1997. Neurotoxicity of CI-980, a novel mitotic inhibitor. Clin. Cancer Res. 3419-422. [PubMed] [Google Scholar]
- 33.Miao, D., H. L. Liu, P. Plut, M. J. Niu, R. J. Huo, D. Goltzman, and J. E. Henderson. 2004. Impaired endochondral bone development and osteopenia in Gli2-deficient mice. Exp. Cell Res. 294210-222. [DOI] [PubMed] [Google Scholar]
- 34.Mo, R., A. M. Freer, D. L. Zinyk, M. A. Crackower, J. Michaud, H.H.-Q. Heng, K. W. Chik, X.-M. Shi, L.-C. Tsui, S. H. Cheng, A. L. Joyner, and C.-C. Hui. 1997. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development 124113-123. [DOI] [PubMed] [Google Scholar]
- 35.Mundy, G. R., I. R. Garrett, S. E. Harris, J. Chan, D. Chen, G. Rossini, B. Boyce, M. Zhao, and G. Gutierrez. 1999. Stimulation of bone formation in vitro and in rodents by statins. Science 2861946-1949. [DOI] [PubMed] [Google Scholar]
- 36.Nogales, E., and H. W. Wang. 2006. Structural intermediates in microtubule assembly and disassembly: how and why? Curr. Opin. Cell Biol. 18179-184. [DOI] [PubMed] [Google Scholar]
- 37.Nogales, E., and H. W. Wang. 2006. Structural mechanisms underlying nucleotide-dependent self-assembly of tubulin and its relatives. Curr. Opin. Struct. Biol. 16221-229. [DOI] [PubMed] [Google Scholar]
- 38.Ogden, S. K., M. Ascano, Jr., M. A. Stegman, L. M. Suber, J. E. Hooper, and D. J. Robbins. 2003. Identification of a functional interaction between the transmembrane protein Smoothened and the kinesin-related protein Costal2. Curr. Biol. 131998-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otto, F., A. P. Thornell, T. Crompton, A. Denzel, K. C. Gilmour, I. R. Rosewell, G. W. Stamp, R. S. Beddington, S. Mundlos, B. R. Olsen, P. B. Selby, and M. J. Owen. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89765-771. [DOI] [PubMed] [Google Scholar]
- 40.Pan, Y., and B. Wang. 2007. A novel protein-processing domain in Gli2 and Gli3 differentially blocks complete protein degradation by the proteasome. J. Biol. Chem. 28210846-10852. [DOI] [PubMed] [Google Scholar]
- 41.Pan, Y., C. B. Bai, A. L. Joyner, and B. Wang. 2006. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol. Cell. Biol. 263365-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rahimov, F., L. A. Ribeiro, E. de Miranda, A. Richieri-Costa, and J. C. Murray. 2006. GLI2 mutations in four Brazilian patients: how wide is the phenotypic spectrum? Am. J. Med. Genet. A 1402571-2576. [DOI] [PubMed] [Google Scholar]
- 43.Robbins, D. J., K. E. Nybakken, R. Kobayashi, J. C. Sisson, J. M. Bishop, and P. P. Therond. 1997. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell 90225-234. [DOI] [PubMed] [Google Scholar]
- 44.Sasaki, H., C. C. Hui, M. Nakafuku, and H. Kondoh. 1997. A binding site for Gli proteins is essential for HNF-3b floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development 1241313-1322. [DOI] [PubMed] [Google Scholar]
- 45.Sasaki, H., Y. Nishizaki, C. C. Hui, M. Nakafuku, and H. Kondoh. 1999. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 1263915-3924. [DOI] [PubMed] [Google Scholar]
- 46.Shimoyama, A., M. Wada, F. Ikeda, K. Hata, T. Matsubara, A. Nifuji, M. Noda, K. Amano, A. Yamaguchi, R. Nishimura, and T. Yoneda. 2007. Ihh/Gli2 signaling promotes osteoblast differentiation by regulating Runx2 expression and function. Mol. Cell. Biol. 182411-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slosar, P. J., R. Josey, and J. Reynolds. 2006. Accelerating lumbar fusions by combining rhBMP-2 with allograft bone: a prospective analysis of interbody fusion rates and clinical outcomes. Spine J. 7301-307. [DOI] [PubMed] [Google Scholar]
- 48.Stegman, M. A., J. E. Vallance, G. Elangovan, J. Sosinski, Y. Cheng, and D. J. Robbins. 2000. Identification of a tetrameric hedgehog signaling complex. J. Biol. Chem. 27521809-21812. [DOI] [PubMed] [Google Scholar]
- 49.Styrkarsdottir, U., J. B. Cazier, A. Kong, O. Rolfsson, H. Larsen, E. Bjarnadottir, V. D. Johannsdottir, M. S. Sigurdardottir, Y. Bagger, C. Christiansen, I. Reynisdottir, S. F. A. Grant, K. Jonasson, M. L. Frigge, J. R. Gulcher, G. Sigurdsson, and K. Stefansson. 2003. Linkage of osteoporosis to chromosome 20p12 and association to BMP-2. Public Libr. Sci. Biol. 11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takae, R., S. Matsunaga, N. Origuchi, T. Yamamoto, N. Morimoto, S. Suzuki, and T. Sakou. 1999. Immunolocalization of bone morphogenetic protein and its receptors in degeneration of intervertebral disc. Spine 241397-1401. [DOI] [PubMed] [Google Scholar]
- 51.Tsuji, K., A. Bandyopadhyay, B. D. Harf, K. Cox, S. Kakar, L. Gerstenfeld, T. Einhorn, C. J. Tabin, and V. Rosen. 2006. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat. Genet. 381424-1429. [DOI] [PubMed] [Google Scholar]
- 52.Urist, M. R. 1965. Bone formation by autoinduction. Science 150893-899. [DOI] [PubMed] [Google Scholar]
- 53.van den Heuvel, M. 2003. Hedgehog signalling: off the shelf modulation. Curr. Biol. 13R686-R688. [DOI] [PubMed] [Google Scholar]
- 54.Vuong, B. Q., T. L. Arenzana, B. M. Showalter, J. Losman, X. P. Chen, J. Mostecki, A. S. Banks, A. Limnander, N. Fernandez, and P. B. Rothman. 2004. SOCS-1 localizes to the microtubule organizing complex-associated 20S proteasome. Mol. Cell. Biol. 249092-9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, B., J. F. Fallon, and P. A. Beachy. 2000. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100423-434. [DOI] [PubMed] [Google Scholar]
- 56.Wozney, J. M., V. Rosen, A. J. Celeste, L. M. Mitsock, M. J. Whitters, R. Kriz, R. Hewick, and E. A. Wang. 1988. Novel regulators of bone formation: molecular clones and activities. Science 2421528-1534. [DOI] [PubMed] [Google Scholar]
- 57.Yang, W. C., D. Guo, J. Gluhak, M. A. Harris, A. Lichtler, B. Kream, J. Edwards, G. R. Mundy, Y. Mishina, and S. E. Harris. 2007. Role of BMP-2 in postnatal bone biology: conditional knockout (cKO) of BMP-2 using 3.6 collagen type 1a1-Cre model. J. Bone Miner. Res. 22(suppl. 1)S31. [Google Scholar]
- 58.Yazaki, Y., S. Matsunaga, T. Onishi, T. Nagamine, N. Origuchi, T. Yamamoto, Y. Ishidou, T. Imamura, and T. Sakou. 1998. Immunohistochemical localization of bone morphogenetic proteins and the receptors in epiphyseal growth plate. Anticancer Res. 182339-2344. [PubMed] [Google Scholar]
- 59.Zhao, M., H. X. Wang, and T. C. Zhou. 1994. Expression of recombinant mature peptide of human bone morphogenetic protein-2 in Escherichia coli and its activity of bone formation. Chin. Biochem. J. 10319-324. [Google Scholar]
- 60.Zhao, M., M. Qiao, B. O. Oyajobi, G. R. Mundy, and D. Chen. 2003. E3 ubiquitin ligase Smurf1 mediates Cbfa1 degradation and plays a specific role in osteoblast differentiation. J. Biol. Chem. 27827939-27944. [DOI] [PubMed] [Google Scholar]
- 61.Zhao, M., M. Qiao, S. E. Harris, D. Chen, B. O. Oyajobi, and G. R. Mundy. 2006. The zinc finger transcription factor Gli2 mediates bone morphogenetic protein 2 expression in osteoblasts in response to hedgehog signaling. Mol. Cell. Biol. 266197-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]