Abstract
Patients carrying mutations in the XPB helicase subunit of the basal transcription and nucleotide excision repair (NER) factor TFIIH display the combined cancer and developmental-progeroid disorder xeroderma pigmentosum/Cockayne syndrome (XPCS). Due to the dual transcription repair role of XPB and the absence of animal models, the underlying molecular mechanisms of XPBXPCS are largely uncharacterized. Here we show that severe alterations in Xpb cause embryonic lethality and that knock-in mice closely mimicking an XPCS patient-derived XPB mutation recapitulate the UV sensitivity typical for XP but fail to show overt CS features unless the DNA repair capacity is further challenged by crossings to the NER-deficient Xpa background. Interestingly, the XpbXPCS Xpa double mutants display a remarkable interanimal variance, which points to stochastic DNA damage accumulation as an important determinant of clinical diversity in NER syndromes. Furthermore, mice carrying the XpbXPCS mutation together with a point mutation in the second TFIIH helicase Xpd are healthy at birth but display neonatal lethality, indicating that transcription efficiency is sufficient to permit embryonal development even when both TFIIH helicases are crippled. The double-mutant cells exhibit sensitivity to oxidative stress, suggesting a role for endogenous DNA damage in the onset of XPB-associated CS.
Defects in XPD and XPB, the two helicase subunits of transcription/repair factor TFIIH can lead to a surprising clinical heterogeneity. They cause the cancer-prone disorder xeroderma pigmentosum (XP) or a combination of XP and the neurodevelopmental progeroid condition Cockayne syndrome (XPCS). The latter displays striking clinical variance, with a life expectancy ranging from 7 months to over 30 years (38). In addition, defects in both helicase subunits can give rise to trichothiodystrophy (TTD), a condition similar to CS but with characteristic brittle hair and nails and scaly skin (7, 36), or a combined form of XPTTD (12). In addition, although mutations are overall disease specific, extensive phenotypic variance among patients carrying even the same causative mutation has been observed (16), suggesting that variables other than differences in the genetic background, such as the stochastic nature of DNA damage accumulation, may be involved. The participation of TFIIH in multiple cellular processes, including basal and activated transcription as well as the two modes of nucleotide excision repair (NER), further complicates the genotype-phenotype correlation. The above complexity highlights the need for genetically defined model organisms for resolving composite phenotypes, such as developmental delay and/or accelerated aging.
NER is one of the most versatile DNA repair systems, responsible for removing a wide variety of helix-distorting DNA lesions, including UV-induced photoproducts and several forms of oxidative lesions (7, 10, 22, 26, 36, 56, 59). NER consists of two subpathways: global genome NER (GG-NER), which surveys the genome overall, and transcription-coupled NER (TC-NER), which focuses on NER-type lesions in the transcribed strand of active genes. Next to TC-NER, a broader transcription-coupled repair (TCR) process likely exists that also eliminates non-NER-type transcription-blocking lesions. In GG-NER, the hHR23B-XPC complex functions as a damage sensor and primary initiator of the NER reaction. In TC-NER, lesion-induced blockage of transcribing RNA-polymerase II is believed to be the trigger of repair, requiring the CSA and CSB proteins (29, 33, 36, 55) that may also be involved in the broader TCR. After DNA damage recognition, the DNA around the lesion is unwound by TFIIH helicase components XPD and XPB and the lesion is removed in a multistep “cut and patch”-type reaction that involves ∼25 proteins and is similar in both pathways. The requirement for either TC-NER or GG-NER may vary with cell type, further complicating the extension of in vitro findings to complex phenotypes in vivo (23, 26, 44, 47).
Complete inactivation of NER by deletion of XPA, a central NER component, does not cause CS- and TTD-like neurodevelopmental progeroid features but instead leads to extreme UV sensitivity and skin cancer predisposition, as in XP. Therefore, it has been proposed that some of the CS and TTD features may be the outcome of defects in the additional transcription function of TFIIH; hampered basal and activated transcription have been reported in a XpdR722W mouse model for TTD and in in vitro studies with a panel of TTD patient-derived mutations in a basal transcription assay (18, 28, 35). On the other hand, the neurodevelopmental and accelerated aging component in CS and TTD could also be associated with defective repair and/or damage processing of oxidative DNA lesions, a feature also not seen in totally NER-deficient XPA mutants (13, 22, 30, 39, 56, 59). Indeed, cancer-prone Xpa knockout (KO) mice display only a very mild aging phenotype, whereas mouse models for NER-related neurodevelopmental accelerated ageing syndromes CS, XPCS, and TTD exhibit many of the basic features of the human disorders, albeit in a milder fashion (3, 24, 27, 61).
Despite the fact that developmental and progeroid features in TTD and CS are attributed mostly to transcriptional deficits and defective repair of oxidative DNA lesions, respectively, complete inactivation of NER by removing Xpa, which has no role in transcription, in TTD, CS, or XPCS mice displaying mild premature aging features leads to a very similar and severe aggravation of the premature aging phenotype. The phenotype has a postnatal onset, showing that it is not developmental per se, and includes cachexia (loss of weight; failure to thrive), kyphosis (hunchback), neurodegeneration, early cessation of growth, and premature death at 2 to 4 weeks, all consistent with a severe form of each syndrome (3, 19, 41, 62, 63). The phenotypic overlap described above suggests that the DNA repair capacity modulates CS, XPCS, and TTD phenotypes alike.
Notably, the currently available mouse models for TTD, CS, and XPCS genocopy “severe” disease-causing mutations, which in humans result in early juvenile lethality (Csa and Csb truncation mutants modeling CS; XpdG602D for XpdXPCS and XpdR722W for genocopying XpdTTD). Mouse models mimicking mutations that cause a milder phenotype in humans for CS, TTD, or XPCS are currently not available.
Unlike XPD and CS proteins, for which mouse models have been described, much less is known about the etiology of XPB-related developmental and progeroid disease. Patients carrying mutations in XPB helicase are extremely rare, with only six affected families reported (48). The lack of individuals carrying defects in XPB has been attributed to the essential function of XPB in basal transcription (73), suggesting that most mutations in XPB cause very early embryonic lethality (48). Here we report the generation and characterization of the first mouse model carrying a “mild” XPCS type of splice site mutation in Xpb. The new mouse model enabled us to shed light onto the etiology of Xpb-associated XPCS and highlight the potential of stochastic DNA damage accumulation in determining phenotypic diversity in NER disorders.
MATERIALS AND METHODS
Targeting vectors.
The pXpbfs-targeting construct was generated as follows: from the 7-kb SalI-NsiI fragment containing the murine Xpb exon 15, a 4-kb EcoRI fragment containing exon 15 was subcloned into pTZ18R (Pharmacia). The 4-bp insertion (GATC) and a diagnostic BglII site was introduced after the first codon of exon 15 using the mutagenesis kit (Stratagene). The XpbXPCS mutation was introduced in a similar fashion (see Fig. 1). Mutations were confirmed by double-stranded sequence analysis. Further details about targeting constructs will be provided upon request. 129/Ola embryonic stem (ES) cells were targeted according to standard procedures.
FIG. 1.
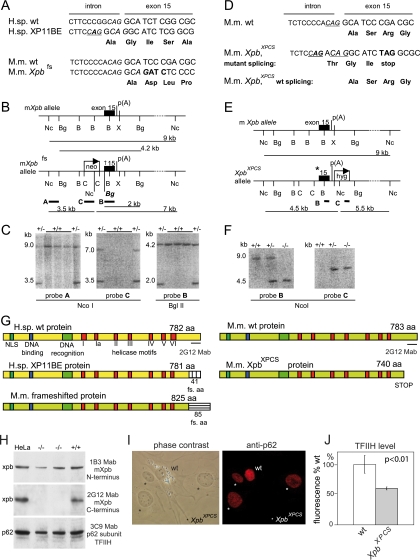
Targeting of the mouse gene Xpb. (A) Sequence of the last intron-exon border of the human XPB and XP11BE and the corresponding mouse Xpb alleles. C-to-A transversion (bold) creates a new splice acceptor site (CAG; underlined italics), resulting in a frame-shifted transcript and 40 novel amino acids at the C terminus derived from the frameshift (first five frameshifted amino acids are indicated). A mouse 4-bp insertion in mXpb creates a similar frameshift (Xpbfs; bold). Due to differences in the nucleotide sequence between mice and humans, in mice the 4-bp insertion leads to substitution of the last 41 amino acids to 85 frame-shifted amino acids (G). (B) Schematic representation of the genomic structure and partial restriction map of the WT and targeted mouse Xpb loci. Black box, exon 15; p(A), polyadenylation signal; †, Xpbfs 4-bp insertion. Probes A, B, and C are indicated with thick black lines. Restriction sites are abbreviated as follows: Nc, NcoI; Bg, BglII; B, BamHI; X, XbaI; C, ClaI. The diagnostic BglII site (Bg) introduced in the sequence is indicated in bold italics. (C) Southern blot analysis of NcoI- and BglII-digested genomic DNA from WT and Xpbfs recombinant ES clones hybridized with probes A, C, and B as indicated. (D) Sequence of the last intron-exon border of mouse WT and XpbXPCS alleles. The WT splice acceptor site is underlined.Changed nucleotides in the mutated allele (bold) create an additional 5′ splice acceptor site (underlined) and stop codon (TAG; bold) in the altered reading frame only. Amino acids encoded by both WT and XpbXPCS reading frames are indicated. (E) Genomic structure of the Xpb WT and XpbXPCS alleles. The asterisk indicates the introduced 5′ splice acceptor site. (F) Southern blot analysis of NcoI-digested genomic DNA from WT ES cells (+/+), XpbXPCS recombinant ES clones (+/−), and homozygous XpbXPCS/XPCS mutant mice (−/−) hybridized with probes B and C. (G) Schematic representation of human (NP_000113.1) and mouse (NP_598419.1) XPB proteins derived from alleles depicted in panels A and D. NLS, nuclear localization signal; fs. aa, novel amino acids derived from the frameshift; the C-terminal recognition epitope of 2G12 MAb is indicated with a line. (H) Immunoblot analysis of whole-cell extracts from HeLa cells, two independent homozygous mutant XpbXPCS MEF lines (−/−), and WT MEFs (+/+). Note that the αXPB MAbs 1B3 and 2G12 recognize conserved epitopes within the N and C termini, respectively, and that the C-terminal epitope is absent in the truncated protein (G). The p62 subunit of TFIIH (stained with MAb 3C9; lower panel) served as qualitative control for loading. (I) Reduction of TFIIH protein levels in homozygous XpbXPCS primary MEFs visualized by comparative immunofluorescence assay. WT cells were labeled with 0.79-μm latex beads; XpbXPCS cells were unlabeled (asterisk). Left panel, phase contrast image; right panel, corresponding immunofluorescent image of the p62 subunit of TFIIH. Note the reduced signal in the XpbXPCS cells. (J) Quantification of the TFIIH level in XpbXPCS MEFs. The immunofluorescence signal from XpbXPCS cells was determined (at least 50 nuclei per genotype, two separate experiments with two independent XpbXPCS and WT cell lines) and expressed as the average percentage of the level in WT cells analyzed on the same microscopic slide. Error bars indicate the SEM between experiments. H.sp., Homo sapiens; M.m., Mus musculus.
RT-PCR and dot blot analysis.
Xpb cDNA generated by reverse transcriptase PCR (RT-PCR) was subcloned into a Bluescript TA cloning vector. Individual clones were analyzed by dot blot analysis using 32P-labeled primers hybridizing either to the wild-type (WT) or to a 4-bp insertion, splice-acceptor, or stop codon mutation.
Generation of Xpb mutant mice and histology.
Chimeric mice were obtained by using standard procedures and backcrossed with WT C57BL/6 mice for more than eight generations. Genotyping was performed either by Southern blotting (see Fig. 1) or for XpbXPCS mice using primers hybridizing Xpb exon 15, the PGK promoter, and sequence 3′ of the Xpb gene. Histology was performed using standard procedures and hematoxylin and eosin (H&E) staining.
DNA damage sensitivity assays.
UV-induced unscheduled DNA synthesis (UDS) and recovery of RNA synthesis (RRS) assays as well as cellular UV survival were established using primary mouse embryonic fibroblasts (MEFs). For gamma ray and paraquat survival assays, spontaneously transformed MEF lines were utilized. For UDS, cells were irradiated with UVC at a dose of 16 J/m2. Thereafter, cells were incubated for 2.5 h in culture medium containing [3H]thymidine and were subsequently fixed and subjected to autoradiography. Numbers of grains in nuclei per fixed square were counted in at least 25 non-S-phase nuclei per genotype in three independent experiments. A representative experiment is shown in Fig. 2A. Protocols can be found in reference 67, and a detailed protocol can be found from references therein. For RRS, cells were irradiated with 10 J/m2 UVC and labeled 16 h later with [3H]uridine for 1 h. Grains were counted in at least 25 nuclei per genotype in three independent experiments. Methods are described in reference 26; see references therein for precise details. Paraquat and gamma ray cellular survival assays were performed with a [3H]thymidine incorporation assay or a colony assay as described in references 19 and 26.
FIG. 2.
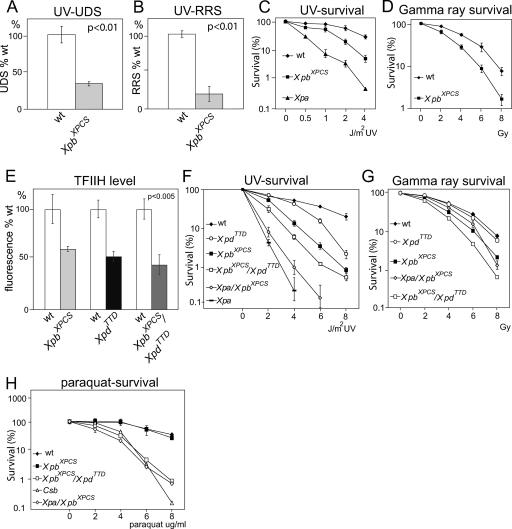
Cellular DNA repair characteristics. (A) UV-induced (15 J/m2 UVC) unscheduled DNA repair synthesis capacity (UDS) of primary homozygous XpbXPCS MEFs. A representative experiment (three experiments total) is depicted. The P value indicates the significance of the difference between WT and XpbXPCS within the representative experiment. Error bars indicate the SEM. (B) Recovery of RNA synthesis after UV (10 J/m2 UVC) irradiation (RRS). A representative experiment (three experiments total) is depicted. The P value indicates the significance of the difference between WT and XpbXPCS within the representative experiment. Error bars indicate the SEM. (C) UV survival curves averaged from four independent experiments. At least 2 cell lines per genotype were included. Error bars indicate SEM between experiments. (D) Gamma ray survival curves averaged from five independent experiments with two cell lines per genotype. Error bars indicate the SEM between experiments. (E) Reduction of TFIIH protein levels in XpbXPCS, XpdTTD single-mutant, and XpbXPCS XpdTTD double-mutant primary MEFs by comparative immunofluorescence analysis of p62 subunit of TFIIH. Quantification of the immunofluorescence signal is based on analysis of at least 50 nuclei per genotype in two separate experiments with two independent cell lines per genotype. WT cells are labeled with latex beads, mixed with the mutant cells, and cultured and immunostained on the same microscopic slide. Bars representing cell lines analyzed on the same microscopic slide are depicted side by side. The P value indicates the minimum significant difference between WT versus mutant cell lines analyzed on the same microscopic slide within one experiment. (F) UV survival curves averaged from four independent experiments. At least two cell lines per genotype were included. Error bars indicate the SEM between experiments. (G) Hypersensitivity of the indicated cells to acute oxidative damage. Gamma ray survival curves averaged from two independent experiments with two cell lines per genotype. Error bars indicate the SEM between experiments and lie within the symbol size. (H) Hypersensitivity of XpbXPCS XpdTTD and Xpa XpbXPCS cells to chronic oxidative injury. MEF cells of the indicated genotype were cultured in the continuous presence of the indicated concentration of paraquat for 3 days. Two cell lines per genotype were tested. For reasons of simplicity, on the depicted representative survival experiment, results from two independent cell lines for XpbXPCS and Xpa XpbXPCS cells were averaged and error bars depict the SEM between two independent cell lines within the given experiment. For the other genotypes in the given experiment, one cell line per genotype was used and error bars depict the SEM within the experiment.
Immunoblotting.
Anti-XPB (1B3) monoclonal antibody (MAb) was described earlier (52). MAb2G12 is a MAb raised toward the human WT 42 C-terminal amino acids of XPB. Western blotting was performed by using standard procedures.
Immunofluorescence analysis of p62 subunit of TFIIH.
Latex bead labeling and comparative immunofluorescence analysis of the p62 subunit of the TFIIH were performed essentially as described earlier in similar studies (2, 8, 65, 66, 68). Briefly, cells were either labeled or not labeled with beads for 2 days, mixed, plated on coverslips, and immunostained. For TFIIH level quantification, 25 to 50 microscopic fields in each experiment were photographed in a “blind” fashion. Nuclear red fluorescence intensity was measured using Adobe Photoshop. A standard Student's t test using the two-tailed two-sample equal variance setting was used to verify the statistical significance of the data. Standard karyotyping was performed to verify that the observed differences in signal intensity are not due to the altered karyotype of the primary MEF cells. In all the cell lines used in this study, ∼80% of the nuclei were found to be diploid while ∼20% had a tetraploid karyotype. Two or more cell lines per genotype were used in this study.
RESULTS
Xpb is essential for mouse development.
Three XPBXPCS patients carry a germ line C-to-A transversion at the last intron-exon border of the gene. This mutation generates a novel splice-acceptor sequence 4 bp upstream of the normal 3′ splice site. The abnormal splicing from this site results in a frameshift at codon 741 and leads to the replacement of the last 42 amino acids by 41 frameshifted nonsense residues (Fig. 1A and G). No WT XPB mRNA is detected in XPBXPCS patient cells, suggesting that the disease-causing 5′ proximal splice acceptor site is used in most (if not all) mutant XPB mRNAs (70). The second XPB allele in all three patients harbors different nonsense mutations, leading to an early stop in the open reading frame and rendering it unlikely that this allele contributes to the phenotype.
To generate a mouse model mimicking XPBXPCS, 4 bp were inserted at the last intron-exon border of the mouse gene Xpb (Xpbframeshifted, designated as Xpbfs) (Fig. 1A). This modification leads to an Xpb protein in which the last 43 amino acids are substituted by frameshifted nonsense amino acids, quite similar to the incorrectly spliced mutant gene in the patients. Due to the sequence differences, in mice this results in the addition of 85 instead of 41 frameshifted residues (Fig. 1G). The Xpb locus was targeted, and ES cells were screened with conventional methods (Fig. 1A to C). RT-PCR and dot blot analysis from heterozygous ES cells revealed that steady-state mRNA levels from the Xpbfs-targeted allele are substantially (∼10-fold) lower than those of the WT untargeted allele (data not shown).
Mice heterozygous for the Xpbfs allele appeared phenotypically normal, consistent with a recessive mode of inheritance. However, intercrosses of heterozygous Xpbfs animals yielded 43 WT and 103 heterozygous pups but failed to generate homozygous mutants, and an analysis of embryos showed that homozygous mutants were missing as early as embryonic day 8.5 (E8.5). The absence of extraembryonic tissue suggested that embryonic lethality occurred during the preimplantation stage. Our results indicate that the engineered Xpbfs mutation causes early embryonic lethality and lend support to the notion that scarcity of XPB-related disease is due to the essential function of this helicase.
Generation of a viable XpbXPCS mouse model carrying a deletion of the last 43 amino acids of the Xpb (XpbΔ43).
As the completely frameshifted homozygous Xpbfs mutation led to embryonic lethality, probably resulting from the low mRNA expression and/or the presence of the different nonsense C terminus of the mutant protein in the case of mice, we reengineered the targeting construct. Since it is not excluded that in cells of the patients a low quantity of correctly spliced mRNA encoding the WT Xpb protein is generated due to occasional usage of the proper 3′ splice acceptor site, we closely mimicked the human proximal splice acceptor mutation in the murine genomic Xpb locus (Fig. 1D). Furthermore, a stop codon was introduced to prevent translation of the altered amino acid tail in the incorrectly spliced reading frame while leaving the WT frame unaltered. Thus, usage of the new splice acceptor site would result in a frame-shifted mRNA sequence encoding a truncated protein lacking the last 43 amino acids (XpbΔ43, hereafter referred to as XpbXPCS), whereas occasional usage of the WT splice site would lead to an unaltered (WT) Xpb protein (Fig. 1D and G).
The targeted ES clones were screened by Southern blotting (Fig. 1F and data not shown). DNA sequencing of independent RT-PCR clones revealed that ∼90% of the mRNA derived from the targeted allele was obtained by utilizing the proximal splice site encoding the truncated Xpb protein, while the remaining ∼10% of splicing had occurred via the WT splice acceptor site. Thus, in ES cells, the introduced proximal splice site was used preferentially but not exclusively.
To obtain isogenic XpbXPCS animals, heterozygous mice were back-crossed to WT C57BL/6 mice for more than eight generations. Consistent with the recessive nature of XPB mutations in the human population, heterozygous XpbXPCS/wt mice were indistinguishable from WT animals in all aspects studied. Intercrosses between heterozygous XpbXPCS/wt animals yielded XpbXPCS/XPCS homozygous offspring (hereafter designated as XpbXPCS) at Mendelian frequency (Fig. 1F and data not shown).
Analysis of Xpb expression and TFIIH levels in XpbXPCS mice.
To determine the frequency of the mutant splice acceptor usage in homozygous XpbXPCS mice, total RNA was isolated from the liver, amplified by RT-PCR, and subcloned. Sequencing of independent cDNA clones revealed that 26 of 28 clones contained the expected 4-bp intron-derived insertion encoding the C-terminally truncated mutant Xpb protein; the remaining 2 clones were products of normal splicing. Thus, in ∼5% to 10% of splicing events, the WT splice acceptor was used. Analysis of the mRNA derived from primary MEFs, testes, kidneys, spleens, and brains of XpbXPCS mice yielded similar results (data not shown). Thus, as in heterozygous ES cells, the WT 3′ splice acceptor was used in all cells and tissues at a low but detectable frequency in XpbXPCS mice, leading to the low-level presence of WT Xpb-encoding transcripts.
The expression of Xpb protein levels in XpbXPCS cells was analyzed by immunoblot analysis. Two MAbs were used; the N-terminal MAb (1B3) recognized both mutant and WT Xpb, while the C-terminal MAb (2G12) recognized only the WT protein, as the epitope is absent in the truncated Xpb protein (Fig. 1G). The mutant protein was detected in immunoblots, but despite the low-level presence of WT mRNA in XpbXPCS cells, WT Xpb protein appeared below the level of detection (Fig. 1H).
Because mutations in the XPB or XPD genes frequently lead to a reduced content of the entire 10-subunit TFIIH complex, which is known to impact its function in NER both in humans and mice (2, 8, 66), we determined TFIIH levels in the XpbXPCS primary fibroblasts by measuring the immunofluorescence intensity of the TFIIH p62 subunit. An approximately 40% reduction of TFIIH levels was observed compared to that of the WT cells (Fig. 1I and J).
XpbXPCS mice do not display overt developmental or accelerated ageing features.
To investigate whether XpbXPCS mice develop CS-related accelerated ageing features, animals were screened for developmental delay, neuromotor deficiencies, fecundity, kyphosis, and cachexia. Measuring body weight, starting from 12 days of age, did not reveal significant developmental delay in XpbXPCS mice (data not shown). Neuromotor function, as tested by tail suspension and accelerating rotarod tests, was indistinguishable from that of the WT. Both male and female XpbXPCS mice were fertile up to at least 7 months of age, with litter sizes comparable to those of WT littermates. Compared to gender-matched littermate controls, XpbXPCS mice (n = 6) did not display overtly accelerated aging features typical of CS, such as kyphosis or cachexia, until 24 months of age when surveillance was concluded.
Histological examination at the age of 1.5 years revealed no obvious signs of CS or any other type of pathological abnormalities in the brain, sciatic nerve, eyes, heart, lungs, lymph nodes, kidneys, liver, colon, intestines, spleen, thymus, pancreas, gall bladder, urinary bladder, testes, ovaries, bones (femur/joint), skin, and skeletal muscles. Compared to results from the littermate controls (n = 8), no significant increase or decrease in the number of tumors was noted (a total of nine XpbXPCS mice analyzed). In conclusion, XpbXPCS mice failed to show detectable CS-like developmental or accelerated segmental ageing features and did not display phenotypic variance compared to WT animals with any of the parameters tested.
XpbXPCS cells are sensitive to UV and acute oxidative stress.
Fibroblasts from XPBXPCS patients are largely defective in TC-NER and GG-NER and are thus hypersensitive to UV-induced cell killing (48, 70). The absence of a CS-like phenotype suggested that NER in XpbXPCS mice might also be unaffected. To assess the repair status of the mutant mice, GG-NER and TC-NER activities of primary MEFs derived from XpbXPCS embryos were measured using UDS and RRS assays, respectively. XpbXPCS cells retained ∼30% of WT UDS and ∼20% of WT RRS (Fig. 2A and B), which appears slightly less severe than in cells of XPBXPCS patients (50, 70). In UV-induced survival experiments, XpbXPCS cells showed intermediate hypersensitivity compared to Xpa mutant cells (Fig. 2C). Xpa mutants lack NER activity of UV-induced DNA lesions entirely and were used as a positive control in the assay, showing obvious hypersensitivity to UV. Ionizing (gamma) radiation (IR) was used to test the sensitivity of the cells to acute oxidative stress. In line with data reported for human and mouse cells from CS individuals (3, 22, 25, 26, 56, 59), XpbXPCS cells were slightly but significantly hypersensitive to IR (Fig. 2D). Thus, despite the lack of CS-related ageing features in the XpbXPCS animals, XpbXPCS MEFs exhibited typical CS defects, showing clear but not complete sensitivity to UV light and acute oxidative stress.
XpbXPCS mice are hypersensitive to UV irradiation.
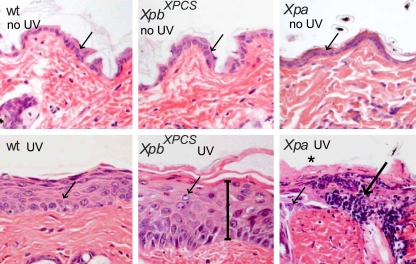
To examine whether the partial NER defect detected in cultured XpbXPCS cells is also detectable in vivo, photosensitivity of XpbXPCS mice was tested by exposing the shaven dorsal skin of mice to UVB light at a dose of 500 J/m2/day for four consecutive days. Xpa mice were included as a positive control for UV hypersensitivity. One week after the initiation of the treatment, WT and heterozygous animals appeared normal, whereas Xpa mice exhibited pronounced redness of the skin, indicative of erythema and edema (generally known as sunburn), and acanthosis (thickening of the epidermis), accompanied with recurrent loss of the whole epidermis (Fig. 3). XpbXPCS mice displayed moderately enhanced acanthosis (Fig. 3), an intermediate phenotype consistent with the partial NER defect in cultured fibroblasts. Hence, the XpbXPCS mice reconstitute the hallmark XP feature, sensitivity to UV light, both in vitro and in vivo.
FIG. 3.
Acute effects of UV-B on the skin of XpbXPCS mice. H&E staining of dorsal skin sections from shaven mice exposed to daily doses of 500 J/m2 UVB for four consecutive days and sacrificed 1 week after the start of the treatment. Note the moderate epidermal hyperplasia, consisting of an increased number of cell layers (acanthosis, indicated with a bar) in the XpbXPCS skin and the much more severe effect in Xpa mice as evident from hyperemia (dilated capillaries filled with blood, indicated with a long arrow) and the absence of keratinized and epidermal layers (asterisk), reflecting severe “scaling of the skin.” Note the “swelling” of the nuclei after UV treatment in all genotypes compared to untreated controls (short arrows). Magnification, ×400.
Xpa XpbXPCS double-mutant mice display CS-like features and large phenotypic variation.
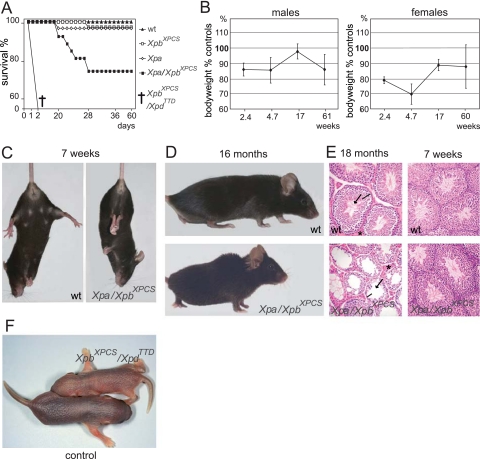
XpbXPCS mice exhibit a partial NER deficiency and XP features. In addition, they show a slight sensitivity to IR, which is observed with CS-type mutants, yet they fail to display overt CS symptoms. To find out whether the substantial residual NER activity in XpbXPCS mice has a suppressive effect on the CS phenotype, we crossed the mice into an Xpa-deficient background. Mice lacking Xpa do not display overt developmental or progeroid (CS-like) features, but are completely NER deficient and highly susceptible to UV-induced skin cancer (24, 27, 42). Double-mutant Xpa XpbXPCS mice were born healthy and at expected Mendelian frequencies and initially developed normally. However, at about 10 days after birth, most Xpa XpbXPCS mice failed to gain weight at a comparable rate to WT, single-mutant, and double-heterozygous, gender-matched littermates. As a consequence, in the third and fourth week, the most severely affected individuals even died, concomitant with failure to thrive and progressive cachexia. Within this group of juvenile lethality (7/30; ∼23% of animals) the average life span was only 23.3 days (standard error of the mean [SEM], 1.1 days) (Fig. 4A). During the last week of their lives, these animals progressively developed severe kyphosis and gait anomalies, including balance problems (data not shown), very similar to what has previously been reported for mouse models for CS, XPCS, and TTD (Csa and Csb truncation mutants; XpdG602D [XpdXPCS], XpdR722W [XpdTTD], and Xpg [with a deleted exon 15]) in an Xpa-deficient background (3, 19, 41, 54, 62). Like XPB and XPD, XPG is involved in GG-NER as well as TC-NER and may also be implicated in the broader TCR process. Notably, while juvenile lethality affected ∼23% of Xpa XpbXPCS mice, it has been observed to be nearly 100% penetrant in Xpa-deficient CSA, CSB, XPDXPCS, XPDTTD, and XpgDexon15 mouse models. However, exceptions have been noted. Two of 12 Xpa XpdTTD mice which survived weaning lived for 4 and 12 months, respectively, and 2 out of 25 XpgDexon15 mice lacking Xpa lived for 5 to 5.5 months (19, 54). Remarkably, by ∼3 months of age, most of the surviving Xpa XpbXPCS mice (77%; n = 23) were able to “catch up” (close) to the range of the body weight of the WT littermate controls (Fig. 4B).
FIG. 4.
Effects of additional NER mutations in XpbXPCS mice. (A) Kaplan-Meier survival curve of Xpa XpbXPCS mice. Shown are results for the WT (n = 36), Xpa (n = 34), XpbXPCS (n = 41), Xpa XpbXPCS (n = 30), and XpbXPCS XpdTTD (n = 5). Please note that for XpbXPCS XpdTTD mice, the exact time of death is unknown; it likely occurred within 1 to 2 days after birth (see Table 2 and above). (B) Body weight of Xpa XpbXPCS mice, plotted as a percentage of age and gender-matched littermate controls. Note the increase in body weight between 4.7 and 17 weeks. Error bars indicate the SEM. (C) Tail suspension test of 7-week-old male mice. The WT mouse depicted displays normal spreading of the hind limbs. Xpa XpbXPCS mice displayed heterogeneous behavior, ranging from normal to severe cramp-like seizures, spastic movements, and tremors of hind limbs as depicted here. Note also the smaller size of the Xpa XpbXPCS mouse. (D) Severe kyphosis in a 16-month-old male Xpa XpbXPCS mouse. (E) Premature testicular tubular atrophy of Xpa XpbXPCS mice. H&E-stained sections of the testis of 18-month and 7-week-old WT and Xpa XpbXPCS mice. Note the reduced thickness or absence of the germinal epithelium (bar), reduced occurrence of mature spermatids (arrow), and the increase in interstitial cells (asterisk) in Xpa XpbXPCS males at 18 months but not at 7 weeks. Magnification, ×100. (F) Photograph of a ∼24-h-old XpbXPCS XpdTTD double-mutant mouse and a double-heterozygote littermate. After apparent normal embryogenesis, double-mutant mice failed to grow and died within ∼36 h.
To analyze whether Xpa XpbXPCS mice display the neurological features typical for CS, we performed visual examination and tail suspension tests. In about 30% of the 23 analyzed animals, this revealed occasional tremors and ataxia, spasticity, and abnormal coordination of hind limbs up to ∼2 months of age (Fig. 4C). Interestingly, later in life, these features became less pronounced and gradually disappeared among most of the double-mutant mice, although some of the animals remained mildly affected. Visual observation occasionally revealed abnormal hyperactivity, excitability, and “nervous” behavior in 5 out of 23 double-mutant mice, yet only 1 mouse sustained these features beyond ∼2 months of age. About half of the Xpa XpbXPCS mice, which showed anomalies during the tail suspension test, also displayed developmental delay; for nervous behavior, no such correlation was observed (Table 1).
TABLE 1.
Individual phenotypes of XpbXPCS Xpa mice (n = 23) over timea
| ID no., gender | % Reduction in wt at postnatal days 10-21 | Postnatal day 21-2 mo
|
2-7 mo
|
8-12 mo
|
13-18 mo
|
18 mo
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % Reduction in wt | TS | K | N | % Reduction in wt | TS | K | N | % Reduction in wt | TS | K | N | % Reduction in wt | TS | K | N | Sc | Te | ||
| 1, f | −25 | −30, −15 | ++ | − | − | −15 | ± | ± | − | −30 | ++ | +++ | − | −20, 30 | ++ | +++ | − | Yes | NA |
| 2, m | −30 | −35, −25 | + | − | + | −25 | ++ | ++ | − | −40 | ± | ++ | − | −45 | ± | +++ | − | Yes | Yes |
| 3, f | −30, −40 | −60, −65 | ++ | ++ | − | Diedb | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 4, f | −25 | −25, −10 | − | − | − | 0 | − | + | − | −20 | − | − | − | ND | ND | ND | ND | ND | NA |
| 5, f | −15 | ND | − | − | − | 0 | − | − | − | −25 | ± | − | − | −40 | − | +++ | − | Yes | NA |
| 6, f | −15 | ND | − | − | − | −10, 0 | − | − | − | ND | ND | − | ND | ND | ND | ND | ND | ND | NA |
| 7, m | −10, 0 | 0 | − | − | − | 0 | − | − | − | 10, 0 | − | − | − | 0 | − | − | − | − | Yes |
| 8, m | −10, −30 | −30, −15 | + | + | + | −10, 0 | − | − | − | −10, 0 | ++ | − | − | −10, 0 | − | − | − | − | Yes |
| 9, f | −10, 0 | 0 | − | − | − | 0 | − | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | NA |
| 10, f | −10, −20 | −20 | − | − | − | −15 | ± | − | − | 0 | − | − | −25, −30 | − | + | − | − | NA | |
| 11, m | ND | −15 | + | − | + | −15 | ± | − | − | −30 | − | − | − | −40 | − | ++ | − | − | Yes |
| 12, f | ND | ND | − | ND | ND | − | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | NA | |
| 13, m | 0 | 0 | − | − | + | +15 | − | − | ++ | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 14, m | −10, 0 | −25, −10 | + | − | + | 0 | ± | − | − | 0 | − | − | − | −10 | − | − | − | − | ND |
| 15, m | −10, 0 | −20 | − | − | − | −10, 0 | − | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 16, m | −25, −10 | −10, 0 | − | − | + | 0 | ± | − | − | 0 | − | − | − | 0 | − | − | − | − | ND |
| 17, f | −15 | −10, 0 | − | − | − | 0 | + | − | − | +10 | − | − | − | 0 | − | − | − | − | NA |
| 18, m | −10, 0 | 0 | − | − | − | +10, 0 | − | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 19, m | −20 | −10, 0 | − | − | − | 0 | − | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 20, m | −20 | −10, 0 | − | − | − | ND | − | − | − | −10, 0 | − | ND | − | −10 | − | − | − | − | ND |
| 21, m | ND | ND | ND | ND | ND | −15 | − | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 22, f | −10, 0 | ND | ND | ND | ND | ND | − | − | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | NA |
| 23, m | ND | ND | ND | ND | ND | ND | − | − | − | 0 | − | ND | ND | +10 | − | − | − | − | Yes |
Body weight is expressed as the percentage of littermate gender-matched control mice. When measurements were performed more than once during the given period of time and the parameter changed during that time, results from both measurements are noted. For example, a body weight of −30, −10 means that the first measurement was a reduction of 30% and the second measurement was a reduction of 10% compared to that of the gender-matched littermate controls. f, female; m, male; TS, tail suspension test; K, kyphosis; N, hyperactivity/nervous behavior; Sc, skull deformation; Te, testicular atrophy; NA, not applicable; ND, not detected; −, feature absent; ±, mild symptoms compared to background; +, moderate feature; ++, severe feature; +++, very severe feature.
Animal no. 3 is exceptional because it survived weaning but died at about 2 months of age with severe cachexia.
Both male and female Xpa XpbXPCS mice were fertile until at least 7 months of age, with litter sizes comparable to those of WT animals. Between 8 and 12 months of age, Xpa XpbXPCS mice displayed premature kyphosis (Fig. 4D) with heterogeneous penetrance (5/23). Mild gait abnormalities were observed during both the juvenile developmental delay period (from ∼12 days old up to ∼1 to 2 months old) among the most affected individuals, as well as among the ageing mice with the most pronounced kyphosis.
At 7 to 8 weeks and 1.5 years of age, Xpa XpbXPCS and age-matched controls (Xpa, XpbXPCS, and WT littermates) were sacrificed for histological analysis. Macroscopical examination revealed that the skulls of 1.5-year-old double-mutant mice with the most pronounced kyphosis (n = 3) showed deformation and thickening, mainly in the occipital region, which most likely relates to the severe kyphosis of the vertebral column. All 1.5-year-old male Xpa XpbXPCS mice investigated (five total; two with pronounced kyphosis and three without clear kyphosis) revealed tubular testicular atrophy. This feature is associated with normal ageing in mice but at later age, and was not noted in 1.5-year-old control mice in this study (Fig. 4E). Compared to the controls, histological analysis of the brain, sciatic nerve, heart, lungs, lymph nodes, kidneys, liver, colon, intestines, spleen, thymus, pancreas, gall bladder, urinary bladder, ovaries, skin, and skeletal muscles appeared grossly normal (data not shown). Except for the kyphosis, no further overt abnormalities, such as early demineralization, indicative of osteoporosis, were observed in X-ray images of Xpa XpbXPCS and control skeletons (oldest individual studied at 23 months old). Histopathological analysis of the trabecular and cortical bones at 1.5 years old did not reveal evident premature bone aging as was reported in some CS patients (43) and TTD mice (19; data not shown). Finally, we did not observe significant differences in spontaneous tumor incidence between Xpa XpbXPCS mice (n = 7) and controls (n = 10) at 1.5 years of age.
Taken together, a significant proportion of Xpa XpbXPCS mice initially showed both developmental and accelerated ageing features, which were similar to the typical symptoms of CS patients (43), including prominent progressive cachexia, early cessation of growth, neurological abnormalities, and severely reduced life span. Although during the first weeks, most mice exhibited the features described above to a variable degree, a remarkable phenotypic variance also became apparent after the period of weaning (Table 1) (Fig. 4). Strong heterogeneity in the severity of clinical features has occasionally been observed in NER patients as well, even with identical predisposing mutations (16). Notably, the dramatic phenotypic variation of the Xpa XpbXPCS mice occurred in spite of the uniform C57BL/6 genetic background, ruling out the possibility that the heterogeneity in clinical manifestations would be due to intrinsic differences in genetic makeup.
XpdTTD XpbXPCS double-mutant mice die within 2 days of age.
In addition to a NER and a TCR defect, patient-derived mutations in TFIIH helicases are thought to result in a transcriptional defect. XPB and XPD helicases carrying either XPBXPCS patient-derived XPBfs42(XPCS) or XPDTTD patient-derived XPDR722W(TTD) mutations show defects in a basal transcription initiation assay in vitro (17, 28). Furthermore, XPDTTD (XPDR722W) alongside with XPDXP-type causative mutations appear to impair transcriptional activation of retinoic acid receptor α1 target genes (35), and TFIIH-dependent thyroid hormone gene regulation was reported to be defective in XpdTTD (XpdR722W) mice (18). On the organismal level, XpdTTD (XpdR722W) single-mutant animals display a variety of both developmental and progeroid features, including developmental delay, osteoporosis, kyphosis, sclerosis of the scull, cachexia, and reduced life span (19, 27).
In view of the vital function of TFIIH in transcription and the critical TFIIH levels in TTD patients and mice due to mutations in the XPD helicase, we wondered whether double-mutant animals carrying mutations in both Xpb and Xpd helicases would be viable. When XpbXPCS (XpbΔ43) mice were crossed with XpdTTD (XpdR722W) animals, the double-homozygous XpbXPCS XpdTTD mice were absent at the age of genotyping (10 to 12 days after birth), suggesting that simultaneous defects in both helicases of TFIIH are lethal indeed. However, surprisingly, observation of litters immediately after birth revealed that double-mutant mice were born quite healthy and showed normal activity and movement but failed to grow and died within 1 to 2 days (Fig. 4A). The oldest double-mutant mouse observed alive was ∼36 h old. Absence of any signs of violence and the presence of milk in the stomachs suggested that XpbXPCS XpdTTD mice were nursed normally but died due to some internal complication. All newborn double-mutant animals analyzed (Table 2) appeared slightly smaller than their WT littermates (Fig. 4F), but their body weights were still within the normal newborn range (data not shown). Gross anatomical and histological examination of major organ systems and blood in three double-mutant animals found alive after birth did not disclose any overt pathological or developmental anomalies (data not shown). Analysis of E13.5 and E18.5 embryos revealed no obvious difference in size and body weight from that of littermate controls, and the proportion of XpbXPCS XpdTTD mice found in utero suggests Mendelian inheritance (Table 2). Neither terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick-end labeling staining nor histological analysis of the liver, lungs, heart, and kidneys isolated at E18.5 and ∼12 h after birth revealed differences in the amount of apoptotic cells, leaving the reason of death in the double mutants unknown. In conclusion, normal in utero development of XpbXPCS XpdTTD mice suggests that at least the basal transcription function of TFIIH carrying defects in both helicase subunits is still largely intact.
TABLE 2.
Analysis of XpbXPCS XpdTTD embryos and mice
| Age | No. of pups/embryos analyzed | No. of XpbXPCS XpdTTD mice
|
|
|---|---|---|---|
| Expecteda | Found | ||
| Postnatal days 10-12 | 155 | 14.25 | 0 |
| Postnatal days 0.5-1.5 | 61 | 10.375 | 5b |
| E18.5 | 4 | 1 | 2 |
| E13.5 | 14 | 1.75 | 2 |
If Mendelian inheritance occurred.
Two mice were dead and three were alive.
TFIIH levels and DNA repair properties of the double-mutant cell lines.
To examine whether TFIIH defects can be registered at the cellular level, we established MEFs of various single- and double-mutant combinations. In human and mouse XpdTTD cells, TFIIH levels were found to be diminished due to reduced TFIIH stability (2, 8, 66). Therefore, we first assessed the TFIIH levels of XpbXPCS XpdTTD MEFs. Although TFIIH levels were reduced in both single mutants, they did not appear significantly further reduced in double-mutant MEFs (Fig. 1I and J and 2E), suggesting that the observed double-mutant phenotypes resulted from qualitative rather than quantitative properties of TFIIH. As expected based on the essential role of Xpa in NER of UV-induced DNA damage, the ability of cells to endure increasing doses of UV was the most reduced in Xpa and Xpa XpbXPCS MEFs (Fig. 2F). GG-NER and TC-NER activities as measured with UV-based UDS and RRS assays, respectively, are essentially absent in Xpa KO cells and therefore cannot be further reduced with additional NER mutations (3, 19, 24, 62). Double-mutant XpbXPCS XpdTTD MEFs were slightly more UV sensitive than XpbXPCS or XpdTTD single-mutant cells, indicating an additive effect of the two mutations within the same TFIIH complex (Fig. 2F). XpdTTD cells showed only mild UV sensitivity (Fig. 2F), which correlates with the relatively well-preserved GG-NER and TC-NER in these cells (∼50% and ∼40% of that of the WT, respectively) (2). GG-NER capacity in XpbXPCS XpdTTD MEFs, assessed with UDS assays, was reduced to ∼8%, while the TC-NER activity in RRS assays remained in the ∼20% range of the WT levels (similar to the XpbXPCS single mutant) (Fig. 2A and data not shown). This low-residual-NER activity explains the higher UV survival of XpbXPCS XpdTTD cells compared to entirely NER-deficient Xpa and Xpa XpbXPCS cells.
Analysis of the IR-induced acute oxidative stress tolerance of double-mutant XpbXPCS XpdTTD cells revealed a mild increase in sensitivity compared to XpbXPCS MEFs. XpdTTD single-mutant cells showed behavior close to that of the WT (Fig. 2G). Xpa XpbXPCS double-mutant cells showed no increase in gamma ray sensitivity compared to XpbXPCS single mutants in this assay, consistent with the finding that Xpa KO cells are insensitive to gamma irradiation (19, 26). Importantly, these data suggest that certain TFIIH mutations compromise a repair activity of oxidative DNA lesions, induced by IR, which is independent of Xpa and, therefore, independent of the classical NER. Because accumulation of oxidative DNA damage in vivo is likely to be a chronic, time-dependent process, poorly modeled by acute oxidative stress sensitivity assessed by a single dose of IR, we also mimicked chronic low-dose oxidative injury by culturing MEFs for 3 days in the presence of paraquat, a compound which delivers oxidative stress slowly over time. XpdTTD and Xpa single-mutant cells are known to be insensitive, but cells lacking Csb are sensitive to paraquat, and therefore, Csb KO cells were used as a positive control in the assay (19, 26). Unlike XpbXPCS single-mutant cells, which did not show increased sensitivity compared to WT cells, both Xpa XpbXPCS and XpbXPCS XpdTTD double-mutant MEFs were hypersensitive in this assay (Fig. 2H).
Finally, because some of the NER proteins such as the ERCC1-XPF complex are, in addition to NER, also involved in interstrand cross-link and recombinational repair where defects can lead to severe progeria (45), we tested the sensitivity of all the mutant cell lines described above to a cross-linking agent mitomycin C. None of the single mutants and combinations of Xpa, XpbXPCS, and XpdTTD appeared significantly sensitive to this compound, consistent with the cross-link repair connection being specific to ERCC1-XPF (data not shown).
DISCUSSION
An Xpb mouse model for XPCS.
The first attempt to construct an XpbXPCS mouse model unintentionally led to early embryonic lethality, reflecting the essential function of the Xpb helicase in basal transcription initiation. Similar results have previously been reported for animals lacking Xpd, the other helicase subunit of TFIIH (21). A second targeting attempt yielded a viable Xpb mutant, which lacked the last 43 amino acids of Xpb. In the human XPBXPCS (XP11BE) patient, the corresponding last 42 amino acids are substituted by 41 novel, frameshifted amino acids. XpbΔ43 mutant mice recapitulated the UV sensitivity in vivo and in vitro and were therefore designated XpbXPCS. However, overt CS-like developmental/early progeroid features were absent in XpbXPCS mice. This is consistent with previous findings with NER-deficient mice. To date, mouse models for CS, XPCS, and TTD have been designed to genocopy severe mutations, which in humans result in death before puberty. In mice, these mutations lead to relatively milder symptoms. Here, we genocopied a “mild” XPBXPCS causative mutation, which in humans associates with a life span of over 30 years. Consequently, XpbXPCS mice, unless further challenged, do not display clear developmental or progeroid features typical of CS. A similar generally weaker phenotype in mice is also observed for other models for human progeroid syndromes, such as Werner and Hutchison-Gilford progeria syndromes (32). It appears that translation of progeria from humans to mice involves a reduction factor in terms of phenotype, which may be due to the fact that the time scales for phenotypes to develop in humans are an order of magnitude longer than in mice.
Lessons from mouse models for CS, XPCS, and TTD: how do Xpa XpbXPCS mice fit in?
In humans, TTD-specific brittle hair and nails and scaling skin aside, CS and TTD share a principal set of severe progeroid-like symptoms, including early cessation of growth, cachexia, progressive sensorineuronal loss, demyelination, calcification of basal ganglia, and features of osteoporosis (1, 11). As for the corresponding mouse models, particularly XpdTTD mice exhibit a multitude of bona fide progeroid symptoms that develop in a time frame of 1.5 to 2 years, and in fact this mouse model revealed for the first time that the human syndrome is characterized by many aspects of premature aging (19, 71), although TTD mice also do not recapitulate the strong reduction in life span observed with TTD patients carrying the same XPDR722W point mutation (∼20% shortening of life span in mice versus >90% in humans). Within the perspective of NER defects, it is provocative that Xpa KO mice, which totally lack GG-NER/TC-NER activity, display XP-like cancer predisposition (24, 42) but age normally, whereas mouse mutants for CS, XPCS, or TTD, which harbor only a partial NER defect, exhibit developmental/early progeroid features and, in the case of TTD, also exhibit a reduced life span. The most plausible explanation is that the genes involved in the latter diseases have an additional function outside of the classical GG-NER/TC-NER pathway, defects which cause XP. Indeed, XPD and XPB proteins implicated in TTD, XPCS, and XP are helicase components of TFIIH and function in basal and activated transcription (18, 28, 35). In fact, the role in basal transcription explains the best features which are specific for TTD and never seen in CS or XPCS, i.e., brittle hair and scaling skin. TTD-type mutations in XPD, XPB, and TTDA subunits of TFIIH cause instability of the whole complex, which in turn causes transcription to extinguish before the final stage of terminal differentiation of hair, nail, and skin keratinocytes, interfering with the last cross-linking step of the keratin filaments. Consequently, hair and nails are brittle and skin is scaly (8, 66).
Similarly, several additional functions have been reported for CS proteins CSA and CSB. Besides TC-NER of helix-distorting, transcription-blocking lesions for RNA polymerase II, these additional activities include chromatin remodeling, repair of some oxidative DNA lesions, transcription elongation itself, processing of lesion-blocked RNA-polymerase I transcription, and E3 ubiquitin ligase pathway (9, 13-15, 22, 30, 39, 59). Each of these functions separately or in combination has been put forward to explain the progeroid CS symptoms and the CS component in TTD. Some of the additional functions described above do not correlate in all cases with the clinical phenotype. For example, defects in transcription activation of various hormonal promoters by different XPD mutations fail to show a strict correlation with the CS/TTD phenotype, as they have also been reported for XP without CS (HD2 cell line, described in reference 35) and for some but not all TTD cell lines (35). Thus, this renders such a function invalid as an explanation for the progeroid CS/TTD symptoms.
A scenario that correlates with the phenotype and in which most of the functions described above can be accommodated involves the repair of DNA lesions that interfere with transcription. This process is the common denominator of all these disorders. The spectrum of lesions blocking transcription covers NER-type distorting damage that requires TC-NER and consequently also XPA for their repair. However, different indications support the notion that also some types of oxidative, non-NER DNA damage may arrest transcription and require the CS/TFIIH/XPG (but not the other) components of the NER machinery for a broader TCR reaction (3, 19, 22, 26, 56, 59). This implies that the CS phenotype is largely the consequence of an overall TCR defect, i.e., the inability to rescue transcription arrested by NER- and non-NER-type (oxidative) DNA damage. In the case of CSA and CSB mutations, TCR is completely impaired but GG-NER is unaffected. In the case of TTD- or XPCS-type XPD or XPB mutations, the partial TCR defect is coupled with impaired GG-NER activity. Because of the GG-NER defect, DNA damage accumulates, enhancing the transcriptional repair problems, which may give rise to the severe phenotype in some of the patients. This model also provides a plausible explanation for the dramatic aggravation of the early progeroid phenotype when any of the TCR mutants (Csa, Csb, XpdTTD, XpdXPCS, and XpbXPCS, described here) are crossed with Xpa or Xpc GG-NER KO mice (Table 3). The complete GG-NER defect in double-mutant animals causes persistence of much higher levels of DNA lesions anywhere in the genome, such as a distorting type of cyclopurines induced by oxidative agents (10), which dramatically complicates matters for the defective TCR machinery, causing a very severe early premature aging syndrome, which we term “NER progeria.” This dramatic phenotype is characterized by healthy in utero development, indicating that the condition is not a developmental disorder per se. The healthy birth is followed by postnatal cessation of growth, cachexia, disproportionally big head and limbs, kyphosis, ataxia and other features of neurodegeneration, death around weaning, and, where examined, loss or atrophy of Purkinje neurons in the cerebellum (3, 19, 31, 41, 62). Importantly, as mentioned, the effect is not Xpa specific, as the same set of symptoms is observed in Csa and Csb mice lacking the Xpc gene, which abolishes only the global genome repair capacity of the NER pathway (31, 37). This implies that persisting DNA damage must be the driver behind the acceleration of the aging phenotype of double mutants. Notably, when endonucleases responsible for incising DNA around the lesion in NER Xpg, Xpf, or Ercc1 genes are knocked out, again NER progeria is observed (34, 40, 58, 69). When a milder mutation is engineered in Xpg, namely, a deletion of exon 15 (Xpg DelEx15), the animals grow normally, whereas additional ablation of Xpa leads yet again to the NER progeria (54). The phenotypes of the published mouse models are summarized in Table 3. In all of the cases described above, endogenous DNA damage is the root cause of increased cell death and/or impaired proliferation triggered by unrepaired DNA damage, which induces the arrest of transcription (in the case of a TCR defect) or interstrand DNA cross-links blocking transcription as well as replication (in the case of a defect in the NER/cross-link repair proteins Ercc1/Xpf). Enhanced cell death in turn contributes to accelerated aging (and may protect from cancer, as is observed in CS and TTD). The finding that the bona fide premature aging symptoms in XpdTTD mice that arise in a time frame of months to >1 year are basically the same in XpdTTD Xpa double mutants but only in a accelerated fashion indicates that these features are not developmental defects but dramatically accelerated progeroid features that now interfere indirectly with normal development.
TABLE 3.
Summary of mouse models exhibiting NER progeriaa
| Mouse mutant (reference) | Life span and impact of mutation on life span (reference) | Common histological findings |
|---|---|---|
| Without NER progeria | ||
| Xpa KO (24, 42) | ≥2 yr, mild reduction (27) | |
| Xpc KO (51) | ≥2 yr, mild reduction (72) | |
| Csa KO (60) | ≥2 yr, ND (60) | |
| Csb KO (61) | Normal (60) | |
| XpdTTD (20) | ≥2 yr, modest reduction (27) | |
| XpdXPCS (3) | ≥2 yr, mild reduction (3) | |
| XpdXPCS/TTDb (2) | ≥2 yr, NDb (2) | |
| Xpg-DelEx15 (53) | ≥2 yr, ND (53) | |
| With NER progeria | ||
| Csb Xpa (41, 62) | ∼3 wk | Purkinje neuron degeneration or atrophy |
| Csb Xpc (37) | ∼3 wk | Purkinje neuron degeneration or atrophy |
| Csa Xpc (31) | ∼3 wk | ND |
| XpdTTD Xpa (19) | ∼3 wk | ND |
| XpdXPCS Xpa (3) | ∼3 wk | Purkinje neuron degeneration or atrophy |
| Xpg-DelEx15 Xpa (54) | ∼3 wk | ND |
| Xpg KO (34, 57) | ∼3 wk | Purkinje neuron degeneration or atrophy |
| Xpf KO (58) | ∼3 wk | ND |
| Ercc1 KO (40, 69) | ∼3 wk | ND |
| XpgD811stop (53) | A few days past 3 wk | ND |
| XpdXPCS/TTD Xpa (64) | Up to 6 mob | No defectb |
Note the emergence of NER progeria in Xpa-deficient background independent of the involvement of the second affected gene in CS, XPCS, or TTD. Some of the mouse models described above display gene-specific extra features which for reasons of simplicity are not included in the table. These include polyploidy in liver and kidney in Ercc1 and Xpf mice, intestinal anomalies in Xpg mice, and scaling skin and brittle hair in XpdTTD mice. NER progeria is defined as normal in utero development followed by failure to thrive, cachexia, disproportionally big head and limbs, kyphosis, death around weaning, absence of acute pathology in any organ system except for the cases specified below and, where examined, loss of Purkinje neurons in the cerebellum. ND, not detected.
In XpdXPCS/TTD and XpdXPCS/TTD Xpa mice, tissue-specific usage of different Xpd allele products and/or interallelic complementation alleviates the phenotype compared to that of each Xpd homozygous mutant.
On top of the phenotypic overlap, there are also gene expression data, which point to common abnormalities in NER progeria. In Csb Xpa, XpdTTD Xpa, XpdXPCS Xpa, and compound heterozygous XpdTTD/XPCS Xpa mice, the growth hormone (GH)/insulin-like growth factor 1 (IGF-1) axis, which controls growth and metabolism, is clearly dampened, indicating that at least in mouse models for CS, XPCS, and TTD, the genetic addition of DNA damage by depleting NER leads to a similar organismal, metabolic, and molecular phenotype (62, 63, 64). Suppression of the GH/IGF-1 somatotrophic axis conveniently explains the severe growth retardation observed in these double mutants after birth. Moreover, the expression profiles of several NER progeria mouse mutants displayed significant similarities with the changes in gene expression observed with natural aging, stressing the relevance of accelerated aging for the normal process of aging (45, 62).
Animals that escape from severe NER progeria.
There are also remarkable exceptions to the early death observed for many of the double TCR/GG-NER mutants. After the critical initial postnatal period of ∼2 to 3 weeks, some animals survive much longer. Two of 24 Xpg D811stop mice (C-terminal truncation of Xpg) survived over 2 months, and 2 of 25 Xpg DelEx15 Xpa double mutants survived for nearly half a year. Similarly, 2 of 12 XpdTTD Xpa mice survived for 4 and 12 months (19, 53, 54). This suggests the existence of a “developmental bottleneck” that occurs around weaning; when successfully passed by, it enables life for much longer. Data from compound heterozygous XpdTTD/XPCS Xpa mice indeed support the case. In those animals, tissue-specific usage of specific Xpd allele products and/or interallelic complementation “broadens” the developmental bottleneck, enabling most animals to survive the critical weaning point, and by 10 weeks of age, the dampening of the GH/IGF-1 axis is also released, enabling animals to catch up in growth at least to some extent (2, 64). Unlike the majority of mice displaying NER progeria (Table 3), most of the Xpa XpbXPCS animals survive weaning and catch up with littermates to a large extent.
What can be the explanation for the various survival rates of different double mutants beyond weaning? Previously, we have demonstrated that chronic exposure of WT mice to nontoxic doses of the prooxidant peroxisome proliferator DHEP or the DNA cross-linking agent MMC also triggers transient suppression of the GH/IGF-1 axis concomitant with upregulation of the antioxidant response (45, 62). Thus, accumulation of DNA damage is the likely cause of the somatotropic growth suppressive response in the NER progeria animals, which in turn leads to the runted phenotype. We observed that the more severe the growth retardation, the lower the chance that a NER progeroid animal will survive the “bottleneck” period. Assuming that the degree of growth delay reflects the level of accumulated DNA damage, a correlation is expected between the extent of the combined GG-NER/TCR defect and the fraction of surviving animals. This seems grosso modo to be the case indeed. The Csa or Csb double mutants with Xpc or Xpa KOs are completely devoid of TCR and GG-NER and belong to the most severe variants of the NER progeria phenotype, with hardly any animal surviving weaning. In contrast, a high proportion of Xpa XpbXPCS animals pass the bottleneck correlating with the “milder” nature of the TCR repair defect. Importantly, the developmental “catch up” that can occur in the TCR-defective mice opens perspectives for the development of therapeutic options for human CS, XPCS, and TTD patients.
What is the rationale of the damage-induced growth suppression?
The evolutionary raison d'être of the suppression of the GH/IGF-1 system is apparent from mice carrying a mutation in the GH receptor gene or many other mutants in which the GH/IGF1 signaling cascade is suppressed (5). Such mice generally exhibit dwarfism and a lower metabolic rate, but they have a longer life span and less aging-related pathology, including reduced cancer susceptibility. Apparently, these mice invest less in growth in favor of maintenance and defenses. A similar response of decreased GH/IGF1 signaling is triggered by caloric restriction, the only intervention currently known to extend life span in many species (6). The rationale of this reaction is to promote survival past the critical period of, e.g., food shortage. The most logical interpretation for the same “survival” response in the short-lived double-mutant mice is that they similarly attempt to invest more in maintenance and repair at the expense of growth and in this manner attempt to extend their life span. In view of the inherited DNA repair defect, the response is futile. The more severe the combined TCR/GG-NER repair defect, the stronger the response and the more pronounced the cachexia and runted phenotype, which correlates with the inability to overcome the critical period up to weaning. Animals with the most severe DNA repair defect will accumulate the highest DNA damage load and consequently display the strongest growth retardation. Below a certain threshold, the animal will die.
Why do animals surviving the weaning period live so much longer? At present, the answer to this question is a matter of speculation. Assuming that the level of DNA damage induction is not grossly different between the nursing and the postweaning period, the sensitivity of the organism to respond to accumulating DNA damage by transiently arresting growth is apparently reduced after development is largely completed. Perhaps this is merely an adaptation to no longer postpone growth and resume development even though conditions are not very favorable. Indeed, it seems to make sense to be more conservative early in development to prevent mutations and too much cell death as this will have a major impact on proper further development. Later in life, the growth suppressive effect may be less important as the animal has already developed to become independent and the penalty for increased cell death and mutations is smaller when generation of progeny is close.
Phenotypic variance in XpbXPCS Xpa mice—a potential role of stochastic DNA damage accumulation?
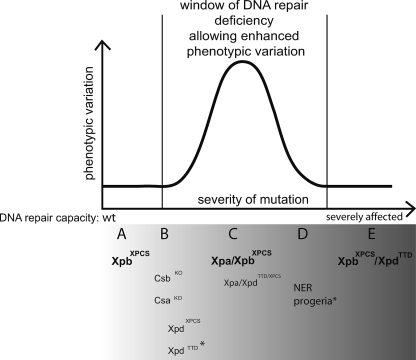
Stochastic DNA damage accumulation has been set forth to explain various phenotypes both in DNA repair mutants and upon normal aging. Data supporting this hypothesis include the observation that monozygotic Fanconi anemia twins with an inborn defect in interstrand cross-link repair can display profound interindividual differences in congenital malformations, which is likely caused by spontaneous stochastic chromosome aberrations occurring in progenitor cells during early embryonic development (46). Also, among NER-defective patients with the same predisposing mutations, remarkable phenotypic variation has been noted (16). In the laboratory, cells from aging mice and cells exposed to oxidative DNA-damaging agents display an increase in cell-to-cell variation in gene transcription, a process directly affected by DNA damage (4). Here, using an isogenic mammalian model system under controlled environmental conditions, we found that when DNA repair (NER) capacity was reduced, interindividual phenotypic variation both in juvenile development and upon aging increased (Table 1) (Fig. 4). These new findings with XpbXPCS Xpa mice are consistent with the idea that stochastic DNA damage next to epigenetic diversity may explain at least in part the phenotypic variation between individual mice in an isogenic mutant background in terms of life span and premature aging features. Figure 5 groups the different GG-NER/TCR and NER/cross-link repair-deficient mice according to the degree of the repair deficiency and depicts the postulated relationship with the extent of phenotypic variation; diversity is smallest when the repair defect is either very small or when it is too severe.
FIG. 5.
Schematic representation of the relation between the extent of the DNA repair deficiency and the phenotypic variation. With a mild DNA repair defect, limited phenotypic variation is noted, whereas mice with a severe DNA repair defect die too fast to allow stochastic effects to manifest. The biggest phenotypic variation is expected in animals with an intermediate DNA repair defect. Lower panel: the existing mouse models are grouped from A to E based on phenotypic variation and presumed DNA repair (or other) defects. (A) XpbXPCS animals with a “mild” causative allele exhibit no detectable phenotypic deviation from the WT. (B) In mice mimicking the human “severe” CS, XPCS, or TTD causative mutations, phenotypes are relatively mild, suggesting that the DNA repair capacity is still proficient enough to battle most of the endogenous DNA damage load. (C) Xpa XpbXPCS mice display the widest phenotypic variation. On top of complete NER deficiency, these animals are also compromised in the removal of non-NER-type oxidative DNA lesions. Some symptoms in Xpa XpbXPCS mice appeared less penetrant (23% juvenile lethality; kyphosis and/or cachexia in 30% of animals) than others (tubular testicular atrophy was seen in all tested ageing males). Presumably, a random build-up of genome damage in the critical progenitor cells of individual mice may lead to diverse phenotypic consequences both during development and upon aging, depending on the number and tissue context of the affected cells. Xpa XpdTTD/XPCS mice were also placed within or near group C. Here the effect of the mutation is made “milder” by tissue-specific usage of each Xpd allele product and/or interallelic complementation. (D) Animals with NER progeria (Table 3) die nearly uniformly around weaning, leaving little time for stochastically accumulating lesions to manifest in phenotype. (E) XpbXPCS XpdTTD mice have defects in the repair of oxidative DNA damage (Fig. 2G and H) but may, in addition, have defects in activated transcription (18, 35), explaining their failure to survive beyond 1 to 2 days after birth (Table 3). Mouse models generated in this study are indicated (bold).
Normal embryonal development and postnatal death of XpbXPCS XpdTTD mice.
One remarkable observation is that the XpbXPCS XpdTTD mice develop quite normally up to birth and the second striking finding is that they die during the first ∼36 h after birth. The first observation implies that TFIIH with both helicases crippled by mutations, which individually causes severe human syndromes, is still able to function in basal transcription initiation to the extent that normal embryonal development is permitted. This may be consistent with the idea that the DNA opening capacity of the complex for transcription initiation is less critical than for GG-NER and TCR. Also, the instability of TFIIH caused by each of the mutations is apparently not dramatically enhanced in the double-mutant cells.
What is the cause of the uniform death of the XpbXPCS XpdTTD newborns? Inspection of dead pups revealed normal feeding and failed to disclose a clear cause. Birth is a stressful event, and also for other GG-NER/TCR mutants we observed increased perinatal lethality (62). It is possible that increased oxidative stress, associated with the sudden transition from the very low oxygen tension in the embryo (∼1%) to atmospheric oxygen when the pup starts to breathe causes transient high levels of oxidative damage (49), which may cause a specific problem for GG-NER/TCR-deficient mice. Although it is difficult to extend in vitro findings to in vivo, here we find that cells from the most severely affected XpbXPCS XpdTTD mice were hypersensitive to two sources of oxidative stress, acute and chronic.
In vitro sensitivity to oxidative stress in single and double-mutant cell lines.
It was of interest to systematically compare various single and double GG-NER/TCR mutants at the cellular level for different repair endpoints to see whether a link with the phenotype could be made. UV sensitivity and UDS and RRS indicated the NER status. In this respect, XpbXPCS cells and mice display partial deficiency, which appears slightly enhanced in XpbXPCS XpdTTD cells. As a mimic for oxidative DNA damage, we used gamma rays and paraquat, which induce different types of lesions and act with different kinetics. Our data are consistent with the idea that an inability to remove oxidative lesions may be involved in XpbXPCS XpdTTD neonatal death. Cells from Xpa XpbXPCS mice, which displayed moderate developmental and progeroid features, were hypersensitive to paraquat but not to gamma ray-induced oxidative stress compared to single-mutant controls (Fig. 2). Importantly, Xpa KO cells do not display sensitivity to either type of oxidative DNA damage, indicating that the repair of oxidative lesions occurs through pathways outside the classically defined NER, where Xpa plays an indispensable role (24). XpbXPCS single-mutant cells were mildly hypersensitive to gamma rays but not significantly to paraquat. As mentioned, cells from the very-short-lived XpbXPCS XpdTTD mutants exhibit sensitivity to both gamma irradiation and paraquat. Taken together, these observations are in line with previously published data demonstrating a repair defect of oxidative DNA lesions in CS and XPCS (3, 22, 25, 26, 56, 59). However, which lesions and mechanism trigger overlapping premature aging symptoms in CS, XPCS, and TTD and in mouse models exhibiting NER progeria awaits to be discovered.
Summary.
The current study describes the first mouse model for XpbXPCS and sheds light on the disease etiology of XPB-linked disease. Based on the in vivo and in vitro data, the developmental and progeroid features linked to XPB-XPCS are likely to at least in part result from the reduction of DNA repair capacity, and this reduction includes non-NER-type TCR of oxidative lesions. We suggest that in NER-related early progeroid conditions, stochastic DNA damage accumulation may be an important phenotypic determinant responsible for interindividual variation in clinical symptoms.
Acknowledgments
We are very grateful to Mart Saarma, Urmas Arumäe, and Maria Lindahl for critical reading, to Saara Ollila for essential editing of the manuscript, and to Ruud Koppenol and Tom de Vries for photography.
This research was supported by the Netherlands Organization for Scientific Research (NWO), through the foundation of the Research Institute for Diseases of the Elderly, The Netherlands Genomic Initiative, as well as grants from the National Institutes of Health (1PO1 AG17242-02), the National Institute of Environmental Health Sciences (1UO1 ES011044), the European Commission (QRTL-1999-02002), and the Dutch Cancer Society (EUR 99-2004). J.R.M. was a fellow of the Damon Runyon Cancer Research Fund (DRG 1677). J.O.A. is the fellow of the Academy of Finland, postdoctoral research grant no. 108973 and 214526. J.H.H. is CSO of Pharming.
Footnotes
Published ahead of print on 29 December 2008.
REFERENCES
- 1.Andressoo, J. O., and J. H. Hoeijmakers. 2005. Transcription-coupled repair and premature ageing. Mutat. Res. 577179-194. [DOI] [PubMed] [Google Scholar]
- 2.Andressoo, J. O., J. Jans, J. de Wit, F. Coin, D. Hoogstraten, M. van de Ven, W. Toussaint, J. Huijmans, H. B. Thio, W. J. van Leeuwen, J. de Boer, J. M. Egly, J. H. Hoeijmakers, G. T. van der Horst, and J. R. Mitchell. 2006. Rescue of progeria in trichothiodystrophy by homozygous lethal Xpd alleles. PLoS Biol. 4e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andressoo, J. O., J. R. Mitchell, J. de Wit, D. Hoogstraten, M. Volker, W. Toussaint, E. Speksnijder, R. B. Beems, H. van Steeg, J. Jans, C. I. de Zeeuw, N. G. Jaspers, A. Raams, A. R. Lehmann, W. Vermeulen, J. H. Hoeijmakers, and G. T. van der Horst. 2006. An Xpd mouse model for the combined xeroderma pigmentosum/Cockayne syndrome exhibiting both cancer predisposition and segmental progeria. Cancer Cell 10121-132. [DOI] [PubMed] [Google Scholar]
- 4.Bahar, R., C. H. Hartmann, K. A. Rodriguez, A. D. Denny, R. A. Busuttil, M. E. Dolle, R. B. Calder, G. B. Chisholm, B. H. Pollock, C. A. Klein, and J. Vijg. 2006. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature 4411011-1014. [DOI] [PubMed] [Google Scholar]
- 5.Bartke, A. 2005. Minireview: role of the growth hormone/insulin-like growth factor system in mammalian aging. Endocrinology 1463718-3723. [DOI] [PubMed] [Google Scholar]
- 6.Bauer, M., A. C. Hamm, M. Bonaus, A. Jacob, J. Jaekel, H. Schorle, M. J. Pankratz, and J. D. Katzenberger. 2004. Starvation response in mouse liver shows strong correlation with life-span-prolonging processes. Physiol. Genomics 17230-244. [DOI] [PubMed] [Google Scholar]
- 7.Bootsma, D., K. H. Kraemer, J. E. Cleaver, and J. H. Hoeijmakers. 2002. The genetic basis of human cancer. McGraw-Hill Medical Publishing Division, New York, NY.
- 8.Botta, E., T. Nardo, A. R. Lehmann, J. M. Egly, A. M. Pedrini, and M. Stefanini. 2002. Reduced level of the repair/transcription factor TFIIH in trichothiodystrophy. Hum. Mol. Genet. 112919-2928. [DOI] [PubMed] [Google Scholar]
- 9.Bradsher, J., J. Auriol, L. Proietti de Santis, S. Iben, J. L. Vonesch, I. Grummt, and J. M. Egly. 2002. CSB is a component of RNA pol I transcription. Mol. Cell 10819-829. [DOI] [PubMed] [Google Scholar]
- 10.Brooks, P. J. 2007. The case for 8,5′-cyclopurine-2′-deoxynucleosides as endogenous DNA lesions that cause neurodegeneration in xeroderma pigmentosum. Neuroscience 1451407-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brooks, P. J., T. F. Cheng, and L. Cooper. 2008. Do all of the neurologic diseases in patients with DNA repair gene mutations result from the accumulation of DNA damage? DNA Repair 7834-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broughton, B. C., M. Berneburg, H. Fawcett, E. M. Taylor, C. F. Arlett, T. Nardo, M. Stefanini, E. Menefee, V. H. Price, S. Queille, A. Sarasin, E. Bohnert, J. Krutmann, R. Davidson, K. H. Kraemer, and A. R. Lehmann. 2001. Two individuals with features of both xeroderma pigmentosum and trichothiodystrophy highlight the complexity of the clinical outcomes of mutations in the XPD gene. Hum. Mol. Genet. 102539-2547. [DOI] [PubMed] [Google Scholar]
- 13.Charlet-Berguerand, N., S. Feuerhahn, S. E. Kong, H. Ziserman, J. W. Conaway, R. Conaway, and J. M. Egly. 2006. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 255481-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Citterio, E., V. Van Den Boom, G. Schnitzler, R. Kanaar, E. Bonte, R. E. Kingston, J. H. Hoeijmakers, and W. Vermeulen. 2000. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 207643-7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cleaver, J. E., E. Hefner, R. R. Laposa, D. Karentz, and T. Marti. 2007. Cockayne syndrome exhibits dysregulation of p21 and other gene products that may be independent of transcription-coupled repair. Neuroscience 1451300-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleaver, J. E., L. H. Thompson, A. S. Richardson, and J. C. States. 1999. A summary of mutations in the UV-sensitive disorders: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. Hum. Mutat. 149-22. [DOI] [PubMed] [Google Scholar]
- 17.Coin, F., E. Bergmann, A. Tremeau-Bravard, and J. M. Egly. 1999. Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J. 181357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Compe, E., M. Malerba, L. Soler, J. Marescaux, E. Borrelli, and J. M. Egly. 2007. Neurological defects in trichothiodystrophy reveal a coactivator function of TFIIH. Nat. Neurosci. 101414-1422. [DOI] [PubMed] [Google Scholar]
- 19.de Boer, J., J. O. Andressoo, J. de Wit, J. Huijmans, R. B. Beems, H. van Steeg, G. Weeda, G. T. van der Horst, W. van Leeuwen, A. P. Themmen, M. Meradji, and J. H. Hoeijmakers. 2002. Premature aging in mice deficient in DNA repair and transcription. Science 2961276-1279. [DOI] [PubMed] [Google Scholar]
- 20.de Boer, J., J. de Wit, H. van Steeg, R. J. W. Berg, M. Morreau, P. Visser, A. R. Lehmann, M. Duran, J. H. J. Hoeijmakers, and G. Weeda. 1998. A mouse model for the basal transcription/DNA repair syndrome trichothiodystrophy. Mol. Cell 1981-990. [DOI] [PubMed] [Google Scholar]
- 21.de Boer, J., I. Donker, J. de Wit, J. H. J. Hoeijmakers, and G. Weeda. 1998. Disruption of the mouse xeroderma pigmentosum group D DNA repair/basal transcription gene results in preimplantation lethality. Cancer Res. 5889-94. [PubMed] [Google Scholar]
- 22.D'Errico, M., E. Parlanti, M. Teson, P. Degan, T. Lemma, A. Calcagnile, I. Iavarone, P. Jaruga, M. Ropolo, A. M. Pedrini, D. Orioli, G. Frosina, G. Zambruno, M. Dizdaroglu, M. Stefanini, and E. Dogliotti. 2007. The role of CSA in the response to oxidative DNA damage in human cells. Oncogene 264336-4343. [DOI] [PubMed] [Google Scholar]
- 23.D'Errico, M., M. Teson, A. Calcagnile, T. Nardo, N. De Luca, C. Lazzari, S. Soddu, G. Zambruno, M. Stefanini, and E. Dogliotti. 2005. Differential role of transcription-coupled repair in UVB-induced response of human fibroblasts and keratinocytes. Cancer Res. 65432-438. [PubMed] [Google Scholar]
- 24.de Vries, A., C. T. M. van Oostrom, F. M. A. Hofhuis, P. M. Dortant, R. J. W. Berg, F. R. de Gruijl, P. W. Wester, C. F. van Kreijl, P. J. A. Capel, H. van Steeg, and S. J. Verbeek. 1995. Increased susceptibility to ultraviolet-B and carcinogens of mice lacking the DNA excision repair gene XPA. Nature 377169-173. [DOI] [PubMed] [Google Scholar]
- 25.de Waard, H., J. de Wit, J. O. Andressoo, C. T. van Oostrom, B. Riis, A. Weimann, H. E. Poulsen, H. van Steeg, J. H. Hoeijmakers, and G. T. van der Horst. 2004. Different effects of CSA and CSB deficiency on sensitivity to oxidative DNA damage. Mol. Cell. Biol. 247941-7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Waard, H., J. de Wit, T. G. Gorgels, G. van den Aardweg, J. O. Andressoo, M. Vermeij, H. van Steeg, J. H. Hoeijmakers, and G. T. van der Horst. 2003. Cell type-specific hypersensitivity to oxidative damage in CSB and XPA mice. DNA Repair 213-25. [DOI] [PubMed] [Google Scholar]
- 27.Dollé, M. E., R. A. Busuttil, A. M. Garcia, S. Wijnhoven, E. van Drunen, L. J. Niedernhofer, G. van der Horst, J. H. Hoeijmakers, H. van Steeg, and J. Vijg. 2006. Increased genomic instability is not a prerequisite for shortened lifespan in DNA repair deficient mice. Mutat. Res. 59622-35. [DOI] [PubMed] [Google Scholar]
- 28.Dubaele, S., L. Proietti De Santis, R. J. Bienstock, A. Keriel, M. Stefanini, B. Van Houten, and J. M. Egly. 2003. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol. Cell 111635-1646. [DOI] [PubMed] [Google Scholar]
- 29.Fousteri, M., and L. H. Mullenders. 2008. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 1873-84. [DOI] [PubMed] [Google Scholar]
- 30.Fousteri, M., W. Vermeulen, A. A. van Zeeland, and L. H. Mullenders. 2006. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell 23471-482. [DOI] [PubMed] [Google Scholar]
- 31.Friedberg, E. C., and L. B. Meira. 2004. Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage (Version 6). DNA Repair 31617-1638. [DOI] [PubMed] [Google Scholar]
- 32.Garinis, G. A., G. T. van der Horst, J. Vijg, and J. H. Hoeijmakers. 2008. DNA damage and ageing: new-age ideas for an age-old problem. Nat. Cell Biol. 101241-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanawalt, P. C. 2002. Subpathways of nucleotide excision repair and their regulation. Oncogene 218949-8956. [DOI] [PubMed] [Google Scholar]
- 34.Harada, Y. N., N. Shiomi, M. Koike, M. Ikawa, M. Okabe, S. Hirota, Y. Kitamura, M. Kitagawa, T. Matsunaga, O. Nikaido, and T. Shiomi. 1999. Postnatal growth failure, short life span, and early onset of cellular senescence and subsequent immortalization in mice lacking the xeroderma pigmentosum group G gene. Mol. Cell. Biol. 192366-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keriel, A., A. Stary, A. Sarasin, C. Rochette-Egly, and J. M. Egly. 2002. XPD mutations prevent TFIIH-dependent transactivation by nuclear receptors and phosphorylation of RARα. Cell 109125-135. [DOI] [PubMed] [Google Scholar]
- 36.Kraemer, K. H., N. J. Patronas, R. Schiffmann, B. P. Brooks, D. Tamura, and J. J. DiGiovanna. 2007. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship. Neuroscience 1451388-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laposa, R. R., E. J. Huang, and J. E. Cleaver. 2007. Increased apoptosis, p53 up-regulation, and cerebellar neuronal degeneration in repair-deficient Cockayne syndrome mice. Proc. Natl. Acad. Sci. USA 1041389-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindenbaum, Y., D. Dickson, P. Rosenbaum, K. Kraemer, I. Robbins, and I. Rapin. 2001. Xeroderma pigmentosum/cockayne syndrome complex: first neuropathological study and review of eight other cases. Eur. J. Paediatr. Neurol. 5225-242. [DOI] [PubMed] [Google Scholar]
- 39.Marietta, C., and P. J. Brooks. 2007. Transcriptional bypass of bulky DNA lesions causes new mutant RNA transcripts in human cells. EMBO Rep. 8388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McWhir, J., J. Seldridge, D. J. Harrison, S. Squires, and D. W. Melton. 1993. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat. Gen. 5217-223. [DOI] [PubMed] [Google Scholar]
- 41.Murai, M., Y. Enokido, N. Inamura, M. Yoshino, Y. Nakatsu, G. T. van der Horst, J. H. Hoeijmakers, K. Tanaka, and H. Hatanaka. 2001. Early postnatal ataxia and abnormal cerebellar development in mice lacking xeroderma pigmentosum Group A and Cockayne syndrome Group B DNA repair genes. Proc. Natl. Acad. Sci. USA 9813379-13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakane, H., S. Takeuchi, S. Yuba, M. Saijo, Y. Nakatsu, H. Murai, Y. Nakatsuru, T. Ishikawa, S. Hirota, Y. Kitamura, et al. 1995. High incidence of ultraviolet-B- or chemical-carcinogen-induced skin tumours in mice lacking the xeroderma pigmentosum group A gene. Nature 377165-168. [DOI] [PubMed] [Google Scholar]
- 43.Nance, M. A., and S. A. Berry. 1992. Cockayne syndrome: review of 140 cases. Am. J. Med. Genet. 4268-84. [DOI] [PubMed] [Google Scholar]
- 44.Niedernhofer, L. J. 2008. Tissue-specific accelerated aging in nucleotide excision repair deficiency. Mech. Ageing Dev. 129408-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niedernhofer, L. J., G. A. Garinis, A. Raams, A. S. Lalai, A. R. Robinson, E. Appeldoorn, H. Odijk, R. Oostendorp, A. Ahmad, W. van Leeuwen, A. F. Theil, W. Vermeulen, G. T. van der Horst, P. Meinecke, W. J. Kleijer, J. Vijg, N. G. Jaspers, and J. H. Hoeijmakers. 2006. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature 4441038-1043. [DOI] [PubMed] [Google Scholar]
- 46.Niedernhofer, L. J., A. S. Lalai, and J. H. Hoeijmakers. 2005. Fanconi anemia (cross)linked to DNA repair. Cell 1231191-1198. [DOI] [PubMed] [Google Scholar]
- 47.Nouspikel, T. 2008. Nucleotide excision repair and neurological diseases. DNA Repair 71155-1167. [DOI] [PubMed] [Google Scholar]
- 48.Oh, K. S., S. G. Khan, N. G. Jaspers, A. Raams, T. Ueda, A. Lehmann, P. S. Friedmann, S. Emmert, A. Gratchev, K. Lachlan, A. Lucassan, C. C. Baker, and K. H. Kraemer. 2006. Phenotypic heterogeneity in the XPB DNA helicase gene (ERCC3): xeroderma pigmentosum without and with Cockayne syndrome. Hum. Mutat. 271092-1103. [DOI] [PubMed] [Google Scholar]
- 49.Randerath, E., G. D. Zhou, and K. Randerath. 1997. Organ-specific oxidative DNA damage associated with normal birth in rats. Carcinogenesis 18859-866. [DOI] [PubMed] [Google Scholar]
- 50.Robbins, J. H., K. H. Kraemer, M. A. Lutzner, B. W. Festoff, and H. G. Coon. 1974. Xeroderma pigmentosum. An inherited disease with sun sensitivity, multiple cutaneous neoplasms and abnormal repair. Ann. Intern. Med. 80221-248. [DOI] [PubMed] [Google Scholar]
- 51.Sands, A. T., A. Abuin, A. Sanchez, C. J. Conti, and A. Bradley. 1995. High susceptibility to ultraviolet-induced carcinogenesis in mice lacking XPC. Nature 377162-165. [DOI] [PubMed] [Google Scholar]
- 52.Schaeffer, L., R. Roy, S. Humbert, V. Moncollin, W. Vermeulen, J. H. J. Hoeijmakers, P. Chambon, and J. Egly. 1993. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science 26058-63. [DOI] [PubMed] [Google Scholar]
- 53.Shiomi, N., S. Kito, M. Oyama, T. Matsunaga, Y. N. Harada, M. Ikawa, M. Okabe, and T. Shiomi. 2004. Identification of the XPG region that causes the onset of Cockayne syndrome by using Xpg mutant mice generated by the cDNA-mediated knock-in method. Mol. Cell. Biol. 243712-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiomi, N., M. Mori, S. Kito, Y. N. Harada, K. Tanaka, and T. Shiomi. 2005. Severe growth retardation and short life span of double-mutant mice lacking Xpa and exon 15 of Xpg. DNA Repair 4351-357. [DOI] [PubMed] [Google Scholar]
- 55.Shuck, S. C., E. A. Short, and J. J. Turchi. 2008. Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res. 1864-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spivak, G., and P. C. Hanawalt. 2006. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA Repair 513-22. [DOI] [PubMed] [Google Scholar]
- 57.Sun, X. Z., Y. N. Harada, S. Takahashi, N. Shiomi, and T. Shiomi. 2001. Purkinje cell degeneration in mice lacking the xeroderma pigmentosum group G gene. J. Neurosci. Res. 64348-354. [DOI] [PubMed] [Google Scholar]
- 58.Tian, M., R. Shinkura, N. Shinkura, and F. W. Alt. 2004. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol. Cell. Biol. 241200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tuo, J., P. Jaruga, H. Rodriguez, V. A. Bohr, and M. Dizdaroglu. 2003. Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. FASEB J. 17668-674. [DOI] [PubMed] [Google Scholar]
- 60.van der Horst, G. T., L. Meira, T. G. Gorgels, J. de Wit, S. Velasco-Miguel, J. A. Richardson, Y. Kamp, M. P. Vreeswijk, B. Smit, D. Bootsma, J. H. Hoeijmakers, and E. C. Friedberg. 2002. UVB radiation-induced cancer predisposition in Cockayne syndrome group A (Csa) mutant mice. DNA Repair 1143-157. [DOI] [PubMed] [Google Scholar]
- 61.van der Horst, G. T. J., H. van Steeg, R. J. W. Berg, A. van Gool, J. de Wit, G. Weeda, H. Morreau, R. B. Beems, C. F. van Kreijl, F. R. de Gruijl, D. Bootsma, and J. H. J. Hoeijmakers. 1997. Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell 89425-435. [DOI] [PubMed] [Google Scholar]
- 62.van der Pluijm, I., G. A. Garinis, R. M. Brandt, T. G. Gorgels, S. W. Wijnhoven, K. E. Diderich, J. de Wit, J. R. Mitchell, C. van Oostrom, R. Beems, L. J. Niedernhofer, S. Velasco, E. C. Friedberg, K. Tanaka, H. van Steeg, J. H. Hoeijmakers, and G. T. van der Horst. 2007. Impaired genome maintenance suppresses the growth hormone—insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 5e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van de Ven, M., J. O. Andressoo, V. B. Holcomb, P. Hasty, Y. Suh, H. van Steeg, G. A. Garinis, J. H. Hoeijmakers, and J. R. Mitchell. 2006. Extended longevity mechanisms in short-lived progeroid mice: identification of a preservative stress response associated with successful aging. Mech. Ageing Dev. 21858-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van de Ven, M., J. O. Andressoo, V. B. Holcomb, M. von Lindern, W. M. Jong, C. I. Zeeuw, Y. Suh, P. Hasty, J. H. Hoeijmakers, G. T. van der Horst, and J. R. Mitchell. 2006. Adaptive stress response in segmental progeria resembles long-lived dwarfism and calorie restriction in mice. PLoS Genet. 2e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vermeulen, W., E. Bergmann, J. Auriol, S. Rademakers, P. Frit, E. Appeldoorn, J. H. Hoeijmakers, and J. M. Egly. 2000. Sublimiting concentration of TFIIH transcription/DNA repair factor causes TTD-A trichothiodystrophy disorder. Nat. Genet. 26307-313. [DOI] [PubMed] [Google Scholar]
- 66.Vermeulen, W., S. Rademakers, N. G. Jaspers, E. Appeldoorn, A. Raams, B. Klein, W. J. Kleijer, L. K. Hansen, and J. H. Hoeijmakers. 2001. A temperature-sensitive disorder in basal transcription and DNA repair in humans. Nat. Genet. 27299-303. [DOI] [PubMed] [Google Scholar]
- 67.Vermeulen, W., R. J. Scott, S. Potger, H. J. Muller, J. Cole, C. F. Arlett, W. J. Kleijer, D. Bootsma, J. H. J. Hoeijmakers, and G. Weeda. 1994. Clinical heterogeneity within xeroderma pigmentosum associated with mutations in the DNA repair and transcription gene ERCC3. Am. J. Hum. Gen. 54191-200. [PMC free article] [PubMed] [Google Scholar]
- 68.Volker, M., M. J. Mone, P. Karmakar, A. van Hoffen, W. Schul, W. Vermeulen, J. H. Hoeijmakers, R. van Driel, A. A. van Zeeland, and L. H. Mullenders. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8213-224. [DOI] [PubMed] [Google Scholar]
- 69.Weeda, G., I. Donker, J. de Wit, H. Morreau, R. Janssens, C. J. Vissers, A. Nigg, H. van Steeg, D. Bootsma, and J. H. J. Hoeijmakers. 1997. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr. Biol. 7427-439. [DOI] [PubMed] [Google Scholar]
- 70.Weeda, G., R. C. A. Van Ham, W. Vermeulen, D. Bootsma, A. J. Van der Eb, and J. H. J. Hoeijmakers. 1990. A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne's syndrome. Cell 62777-791. [DOI] [PubMed] [Google Scholar]
- 71.Wijnhoven, S. W., R. B. Beems, M. Roodbergen, J. van den Berg, P. H. Lohman, K. Diderich, G. T. van der Horst, J. Vijg, J. H. Hoeijmakers, and H. van Steeg. 2005. Accelerated aging pathology in ad libitum fed Xpd(TTD) mice is accompanied by features suggestive of caloric restriction. DNA Repair 41314-1324. [DOI] [PubMed] [Google Scholar]
- 72.Wijnhoven, S. W., E. M. Hoogervorst, H. de Waard, G. T. van der Horst, and H. van Steeg. 2007. Tissue specific mutagenic and carcinogenic responses in NER defective mouse models. Mutat. Res. 61477-94. [DOI] [PubMed] [Google Scholar]
- 73.Winkler, G. S., S. J. Araujo, U. Fiedler, W. Vermeulen, F. Coin, J. M. Egly, J. H. Hoeijmakers, R. D. Wood, H. T. Timmers, and G. Weeda. 2000. TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. J. Biol. Chem. 2754258-4266. [DOI] [PubMed] [Google Scholar]