Abstract
DNA double-strand breaks can result from closely opposed breaks induced directly in complementary strands. Alternatively, double-strand breaks could be generated during repair of clustered damage, where the repair of closely opposed lesions has to be well coordinated. Using single and multiple mutants of Saccharomyces cerevisiae (budding yeast) that impede the interaction of DNA polymerase δ and the 5′-flap endonuclease Rad27/Fen1 with the PCNA sliding clamp, we show that the lack of coordination between these components during long-patch base excision repair of alkylation damage can result in many double-strand breaks within the chromosomes of nondividing haploid cells. This contrasts with the efficient repair of nonclustered methyl methanesulfonate-induced lesions, as measured by quantitative PCR and S1 nuclease cleavage of single-strand break sites. We conclude that closely opposed single-strand lesions are a unique threat to the genome and that repair of closely opposed strand damage requires greater spatial and temporal coordination between the participating proteins than does widely spaced damage in order to prevent the development of double-strand breaks.
Endogenous metabolism or environmental factors such as oxidizing and alkylating agents can produce a wide variety of lesions in DNA. The genomes of mammalian cells experience from 10,000 to as many as 200,000 modifications per day (37, 44). Most lesions are repaired by a complex network of proteins that are part of an elaborate, multistep base excision repair (BER) system that generates single-strand break (SSB) intermediates. Importantly, defects in BER can lead to malignancies and can be associated with age-associated disease, especially neurodegeneration (60).
BER is initiated by specific DNA N-glycosylases that remove damaged bases, yielding apurinic/apyrimidinic (AP) sites. Subsequent incision by AP endonucleases results in SSBs, and excision results in a single base gap as a repair intermediate (33, 53). SSBs are expected to be frequent in the genome due to the abundance of base damage as well as intermediates of repair, recombination, replication, and other DNA transactions (15, 16). Because they are generally repaired efficiently by BER and SSB repair enzymes (16, 57), SSBs per se may not be a major source of genome instability. However, if lesions are clustered, the formation of two closely spaced SSBs on opposing strands (or a single SSB and a modified nucleotide or AP site) might pose a special risk in terms of the potential to generate mutations or the possibility of conversion to double-strand breaks (DSBs), which are potent genotoxic lesions. Clustered lesions can arise within cells by chance association of random DNA lesions in a small region or the induction of multiple events in a narrow region, as found for ionizing radiation and various chemicals, such as those used in cancer treatments (47, 58, 59). While efficient BER is important for genome integrity, the repair must be well coordinated to avoid the generation of closely opposed SSB intermediates at closely spaced lesions that could result in the secondary generation of DSBs, especially since cells have limited DSB repair capacity (<50 DSBs/cell in the case of Saccharomyces cerevisiae) (48). While the impact of clustered lesions on repair of DNA has been examined in vitro by use of purified enzymes or cell extracts (13, 14, 27, 39, 56), there has been little opportunity to address specifically the repair of clustered lesions, except for those arising from UV damage (49).
Whether formed directly from sugar damage or as BER intermediates, SSBs formed during the repair of base damage often possess 5′-deoxyribose phosphate (5′-dRP) ends that are not suitable for rejoining by DNA ligases (9, 15). In humans, removal and repair of 5′-dRP are accomplished by different combinations of proteins (3, 15) that result in short-patch repair, involving replacement of a single nucleotide (nt), or long-patch repair, involving 2 to 10 nt. The budding yeast Saccharomyces cerevisiae lacks a DNA polymerase β that provides AP lyase activity required for short-patch repair in mammalian cells. Instead, removal and repair of a 5′-dRP rely on the long-patch pathway, involving the successive actions of DNA polymerase δ (Pol δ) for strand displacement, the Rad27/Fen1 endonuclease to remove 5′ flaps, and DNA ligase (Cdc9) to rejoin the resulting nicks (9). The sliding clamp protein PCNA, which interacts with all three players, has been proposed to play a central role in coordinating these processes (18, 19, 34). The coupling between the strand displacement reaction by Pol δ and the flap cutting reaction by Fen1 is highly efficient, with over 90% of the products released by Fen1 being mononucleotides (17).
Although the coordination of Pol δ, PCNA, and Rad27/Fen1 provides efficient processing of individual lesions in DNA, closely opposed SSBs that arise during repair of base damage could manifest as DSBs, either directly or as a result of SSB processing. A DNA damaging agent that has been used frequently to characterize long- and short-patch BER is methyl methanesulfonate (MMS). Recently, we described the detection of closely opposed MMS-induced lesions in yeast (42). Since the closely opposed lesions might represent a special challenge to BER, we considered the possibility that they might specifically impact long-patch repair through Pol δ and/or coordination of events with Rad27/Fen1. Pol δ of S. cerevisiae is a heterotrimeric enzyme consisting of Pol3, Pol31, and Pol32 (23). The nonessential Pol32 subunit is involved in translesion DNA synthesis (TLS) (24, 30) and also break-induced replication (41). However, its role in other types of DNA repair remains unclear. Using our in vivo assay for specifically detecting closely spaced methylated DNA lesions (42) and SSBs, we examined the role of Pol32 as well as the cooperation between Pol δ, Rad27/Fen1, and PCNA in the repair of clustered DNA lesions induced by MMS in G1 stationary-phase haploid yeast. We found that Pol32 plays an important role in ensuring that clustered lesions are efficiently repaired and do not transition to DSBs.
MATERIALS AND METHODS
Yeast strains.
All strains used in this study are derivatives of two isogenic haploid yeast strains, MWJ49 and MWJ50 {MATα leu2-3,112 ade5-1 his7-2 ura3Δ trp1-289 [(chr II) lys2::Alu-DIR-LEU2-lys2Δ5′]}, which contain a circularized chromosome III (42). The strains have two copies of the LEU2 gene, one in chromosome III (original location) and the other in the vicinity of the LYS2 gene in chromosome II, which enables both chromosomes to be detected after induction of DNA damage and repair analysis by Southern hybridization analysis (42).
MMS treatment.
MMS treatment was done at the G1 stage of the cell cycle as previously described (42). Briefly, cells were grown in rich yeast extract-peptone-dextrose-agar medium for 48 h at 30°C until >95% of cells were in the G1 stage. Cells were resuspended in phosphate-buffered saline (PBS; 10 mM phosphate, 0.138 M NaCl, 0.0027 M KCl, pH 7.4) and incubated with 11.8 mM (0.1%) MMS for 15 or 30 min at 30°C with vigorous shaking and then neutralized by being mixed at a 1:1 (vol/vol) ratio with 10% Na2S2O3. After being washed with distilled H2O and resuspended in PBS, a portion of the MMS-treated cells was immediately processed for DNA preparation for either pulsed-field gel electrophoresis (PFGE) or quantitative PCR (QPCR) analysis. Another portion of the MMS-treated or control cells was kept in PBS buffer for 24 h at 30°C with constant shaking (referred to as liquid holding [LH]) and then processed for DNA sample preparation.
PFGE analysis to monitor chromosomal DNA breaks.
PFGE analysis to detect chromosomal DNA breaks (i.e., DSBs) as a result of closely spaced SSB formation on opposite DNA strands was performed as described previously (42). Briefly, control and MMS-treated cells before or after LH were embedded in 0.6% agarose (plug) to prepare chromosome-sized genomic DNA. The plug was first treated with 1 mg/ml Zymolyase (100 U/mg; MP Biochemicals, Solon, OH) for 2 h at 30°C in a “spheroplasting” solution (1 M sorbitol, 20 mM EDTA, 10 mM Tris, pH 7.5) to remove the cell wall. This was followed by digestion with proteinase K (10 mM Tris, pH 8.0, 100 mM EDTA, 1.0% N-lauroylsarcosine, 0.2% sodium deoxycholate, 1 mg/ml proteinase K) for 24 h at 30°C or 55°C. Electrophoresis was performed using a Bio-Rad Chef-Mapper XA PFGE system. Samples were resolved in a 1% agarose gel at 6 V/cm for 20 h, with a 60- to 120-s switch time ramp (14°C). After Southern blotting, hybridization was carried out with a probe for the LEU2 gene to detect both chromosomes II and III simultaneously. Autoradiographs were digitized and densitometric analysis was performed using Kodak 1D software (version 4.0). Quantification of chromosome breaks was based on the ratio between the band masses of chromosomes II and III by use of the following equation, as described previously (42): PChrIII(1)/PChrII(0) = Xe1.9X, where PChrIII(1) and PChrII(0) are the probabilities of a single break in chromosome III and no breaks in chromosome II, respectively, when the average number of DSBs in chromosome III is X. The ratio of PChrIII(1) to PChrII (0) was determined experimentally by the band intensities between the linearizing chromosome III and the intact chromosome III.
QPCR assay to measure total DNA lesions.
QPCR was performed and analyzed as described previously (1, 32). Briefly, genomic DNA was isolated from MMS-treated yeast cells before and after LH, and specific primers (sense, 5′-TATCATCCCGATTGCTGCCACTA-3′; and antisense, 5′-CGCTAAAATCCCGTGTATCCCTTG-3′) were used to amplify a 10-kb fragment of the genomic DNA. The presence of any DNA lesions (including base modifications, AP sites, and strand breaks) blocks the progression of the polymerase so that only undamaged templates can be amplified by PCR. Therefore, amplification is inversely proportional to the amount of DNA damage. Amplification of MMS-treated samples was compared with that of undamaged controls to calculate the relative amplification. Estimation of the average number of lesions per 10 kb of the genome, using a Poisson distribution, was described elsewhere (1, 51, 52).
S1 nuclease treatment to detect SSBs.
Plugs were treated with S1 nuclease (Sigma, St. Louis, MO) at a concentration of 4 U/ml (150 μl of reaction buffer for a 50-μl plug) for 20 min at room temperature in a buffer containing 33 mM sodium acetate, 50 mM NaCl, and 1 mM ZnCl2 (pH 4.6) as described previously (20, 46). The reaction was stopped using 50 mM EDTA, and the plugs were analyzed by PFGE (1% agarose gel at 6 V/cm for 24 h, 10- to 90-s switch time ramp, and 14°C). Single-stranded sites were cut by S1 nuclease, resulting in DSBs that could be monitored by PFGE analysis. Since S1 nuclease was used at a low concentration and for short treatment times to prevent degradation of intact DNA, it is likely that many SSBs were not converted into DSBs. Regardless, the assay provides a qualitative assay of random SSBs.
RESULTS
Monitoring intermediates created during repair of alkylation base damage.
We previously described an approach for assessing the engagement of BER proteins in the repair of alkylation base damage in G1 haploid yeast cells (42). This assay measures the induction and repair of methylation base damage by using PFGE to detect secondary DSBs resulting from closely spaced DNA lesions in full-size yeast chromosomes. The assay can detect the number of strand breaks arising during repair (in vivo), as well as other lesions, such as methylated bases and abasic (AP) sites (converted into DSBs in vitro). Using a circular chromosome, lesion frequency and repair kinetics can easily be determined as described in Materials and Methods. G1 haploid cells are treated with MMS (0.1% for 15 or 30 min in buffer), and repair is examined during a subsequent 24-h period of LH in buffer. This protocol enables us to focus specifically on BER, since other repair pathways, such as replication-associated damage bypass and homologous recombination, are not available during LH of G1 cells. As we and others have shown, MMS does not directly cause DSBs in vitro (40, 42). However, closely spaced lesions on opposite DNA strands (which can arise as a result of random damage) can be detected by PFGE as DSBs that are either formed in vivo or converted to DSBs in vitro (42).
The formation of DSBs following induction of MMS damage was proposed to be due to the accumulation of closely opposed SSBs that arise as repair intermediates during BER (42). Most MMS-induced closely opposed lesions can be repaired in the absence of the machinery required for DSB repair, such as through homologous recombination and nonhomologous end joining (42). As shown in Fig. 1A, lanes 1 to 3 (before LH) and lanes 13 to 15 (after LH), few closely opposed SSBs were detected in wild-type (WT) yeast, suggesting that if they are formed, at least one of the intermediate SSBs is repaired efficiently, thereby avoiding DSB formation. Closely opposed SSBs can accumulate in vivo when there is a defect in the BER pathway, such as both AP endonuclease genes APN1 and APN2. The closely opposed SSBs were previously referred to as heat-independent DSBs (42).
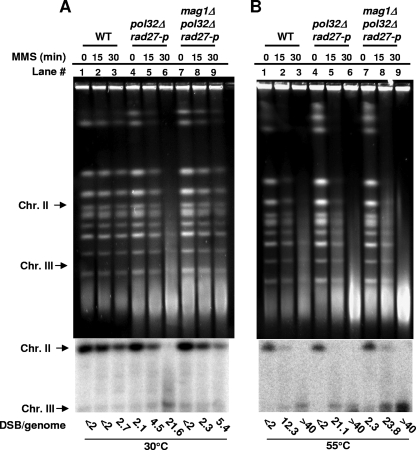
FIG. 1.
Accumulation of DSBs in pol32Δ rad27-p double mutants. G1 haploid yeast cells of different genotypes were treated with 0.1% (11.8 mM) MMS for 15 min (lanes 2, 5, 8, 11, 14, 17, 20, and 23) or 30 min (lanes 3, 6, 9, 12, 15, 18, 21, and 24), followed by immediate DNA purification and PFGE (lanes 1 to 12) or PFGE after LH for 24 h (lanes 13 to 24). Lanes 1, 4, 7, 10, 13, 16, 19, and 22 are mock-treated controls. Chromosomal DNA preparation with proteinase K digestion was performed either at 30°C (A) to measure preexisting closely opposed SSBs formed in vivo or at 55°C (B) (the order of lanes is the same as in panel A) to measure total closely opposed lesions. Chromosomes were visualized by ethidium bromide staining. Positions of some chromosome bands are indicated. A broken chromosome III that can be detected by Southern blotting is located between Chr I and Chr IX. The smear below the Chr I band (indicated by an asterisk) is nonspecific DNA material. A Southern blot detecting the two chromosome bands (Chr II and Chr III) is shown below the agarose gel (30°C).
MMS-induced base methylation damage (the direct adduct) and AP sites (repair intermediate) are heat labile (38, 45), and most of these lesions can be converted into strand breaks in vitro when DNA samples are incubated at 55°C during the 24-h proteinase K digestion step prior to electrophoresis. When closely opposed, these lesions yield DSBs in vitro and were previously referred to as heat-dependent DSBs. The heat-dependent DSBs provide an assessment of the overall number of MMS-induced lesions (42). As shown in Fig. 1B, the smearing of chromosome bands before LH (lanes 2 and 3) and their reconstitution after LH (lanes 14 and 15) in WT yeast indicate that repair is efficient for the bulk of MMS lesions. When DNA samples are processed at 30°C, the methylated bases and AP sites are relatively stable, so closely opposed lesions contribute little to DSB formation (40, 42, 50). Thus, if DSBs are detected in samples incubated at 30°C, they correspond to closely opposed strand breaks formed in vivo. Unless specifically stated, the current study mainly addresses DSBs formed in vivo at 30°C and detected by PFGE.
Deletion of POL32 combined with a defect in Rad27-PCNA interaction (rad27-p) leads to formation of DSBs.
As described in the Introduction, the actions of glycosylases and AP endonucleases at MMS-damaged nucleotides result in a single base gap with a 5′-dRP terminus that requires the successive action of Pol δ and Rad27/Fen1 to remove the 5′-dRP and generate a ligatable nick (4, 9, 61). While Pol δ may play an important role in displacing the DNA strands during repair synthesis to facilitate flap removal by Rad27/Fen1, the role of the nonessential subunit Pol32 is unknown. As shown in Fig. 1A, the incidence of DSBs due to closely opposed SSBs for samples treated at 30°C appeared to be somewhat greater for the pol32Δ mutant (lanes 5 and 6 [before LH] and lanes 17 and 18 [after LH]) than for WT cells (lanes 2 and 3 [before LH] and lanes 14 and 15 [after LH]), with or without incubation following MMS treatment. However, a severe repair defect was observed when the pol32Δ mutation was combined with a mutation in RAD27/FEN1 (rad27-p) that eliminates Rad27 interactions with PCNA (Fig. 1A, lanes 11 and 12 versus lanes 2 and 3 [before LH] and lanes 23 and 24 versus lanes 14 and 15 [after LH]). The rad27-p single mutant (Fig. 1A, lanes 8 and 9 [before LH] and lanes 20 and 21 [after LH]) was similar to the WT in the repair of MMS damage.
Presented in Table 1 are estimates (median numbers and ranges) of the numbers of DSBs resulting from closely opposed SSBs/haploid yeast genomes in various mutants before and after LH, based on Southern analysis of changes in chromosome II and circular chromosome III (42) (see Materials and Methods). Following a 30-min MMS treatment in the pol32Δ rad27-p mutant, there were ∼3 times more DSBs (24 DSBs/genome) than in the pol32Δ single mutant (7.3 DSBs/genome) and ∼4 times the number found in the rad27-p mutant (6.3 DSBs/genome) or the WT strain (5.0 DSBs/genome). The quantity of closely opposed SSBs giving rise to DSBs was further increased in pol32Δ rad27-p double mutants after LH incubation (Fig. 1A, lanes 23 and 24). As shown in Table 1, MMS treatment for 30 min resulted in ∼24 DSBs/genome before LH, which increased to >40 DSBs/genome after LH (six independent experiments). Complete deletion of RAD27 in the pol32Δ background led to an even higher accumulation of DSBs. The effect of the rad27Δ single mutant was less than that of the pol32Δ rad27-p double mutant, especially with the longer MMS treatment (30 min). The increased accumulation of DSBs after LH was observed only in the double pol32Δ mutants (pol32Δ rad27-p and pol32Δ rad27Δ mutants).
TABLE 1.
Median number of MMS-induced closely spaced SSBs in the genomea
| Strain (no. of independent expts) | Median (range) no. of closely spaced SSBs after treatment with MMS for indicated time (min)
|
|||||
|---|---|---|---|---|---|---|
| Before LH
|
After LH
|
|||||
| 0 | 15 | 30 | 0 | 15 | 30 | |
| WT (13) | <2 | 2.4 (<2-5.6) | 5.0 (2.7-9.5) | <2 (<2-3.7) | 2.2 (<2-4.6) | 3.0 (<2-5.9) |
| pol32Δ mutant (4) | 2.7 (<2-3.6) | 4.2 (2.3-9.1) | 7.3 (5.5-18.6) | 3.8 (<2-5.1) | 5.6 (<2-7.0) | 8.0 (6.1-14.6) |
| Rad27-p mutant (4) | <2 | 3.4 (<2-4.3) | 6.3 (<2-8.7) | 3.2 (<2-3.4) | 3.7 (<2-4.7) | 4.3 (<2-4.8) |
| rad27Δ mutant (7) | <2 (<2-2.2) | 5.9 (4.2-7.7) | 13.2 (10.4-19.6) | <2 (<2-2.2) | 6.0 (3.2-8.5) | 13.3 (7.9-20.9) |
| pol32Δ rad27-p mutant (6) | <2 (<2-3.6) | 6.1 (4.9-13.9) | 23.9 (16.4-28.2) | 2.4 (<2-2.7) | 9.2 (5.9-16.7) | >40 (27.7->40) |
| pol32Δ rad27Δ mutant (4) | <2 | 12.8 (10.5-15.2) | 32.1 (29.5-36.7) | <2 | 16.9 (14.5-20.6) | >40 |
The number of DSBs per haploid genome equivalent in chromosomes separated by PFGE was calculated for each experiment as described in Materials and Methods. The closely spaced SSBs were determined using chromosome samples prepared at 30°C. (Previously, these were referred to as heat-independent DSBs or HIBs [42].)
As noted above and previously described (42), the quantity of closely opposed MMS lesions and intermediates of repair, including closely opposed SSBs, can be estimated by incubating samples at 55°C during the 24-h proteinase K digestion step. As shown in Fig. 1B (lanes 1 to 12 [before LH] and lanes 13 to 24 [after LH]), repair was evident and comparable between the WT and the pol32Δ and rad27-p mutants. Importantly, the pol32Δ rad27-p double mutant exhibited some repair of closely opposed lesions that can be converted to DSBs at 55°C, although less than that in WT cells (Fig. 1B, lanes 11 and 12 [before LH] compared to lanes 23 and 24 [after LH]) (Southern blot data not shown). These results suggest that the pol32Δ rad27-p strain is inefficient in the repair of a subpopulation of MMS lesions that are detected as DSBs at 30°C. In line with the accumulation of DSBs, survival of the G1 haploid pol32Δ rad27-p cells treated with 11.8 mM MMS was dramatically decreased compared to that of WT cells and the pol32Δ or rad27-p single mutant, and the defect was even more severe than that of the rad27Δ mutant (Fig. 2).
FIG. 2.
Cell survival after MMS damage. (A) Sensitivity of yeast strains treated with 11.8 mM (0.1%) MMS for 15 or 30 min in PBS and plated, after 24 h of LH repair, on rich medium from serial 10-fold dilutions. (B) Survival of MMS-treated G1 yeast (after 24 h of LH). ○, WT; □, rad27Δ mutant; •, pol32Δ mutant; ▴, rad27-p mutant; ▪, pol32Δ rad27-p mutant.
Deletion of MAG1 in the pol32Δ rad27-p background prevents DSB formation.
In budding yeast, the Mag1 glycosylase is required for removal of the most abundant lesions caused by MMS (5), i.e., N7-methylguanine (80 to 85% of lesions) and N3-methyladenine (9 to 12% of lesions). Previously, we showed that in G1 yeast the MMS-induced methylated bases are prevented from forming AP sites if MAG1 is deleted (42). As shown in Fig. 3, the total amounts of MMS-induced lesions (as determined by the number of heat-dependent breaks at 55°C) (Fig. 3B) were comparable between pol32Δ rad27-p and pol32Δ rad27-p mag1Δ mutants, as well as the in vivo formation of DSBs (as determined by the number of heat-independent breaks at 30°C) (Fig. 3A). The ∼22 DSBs/genome observed in the pol32Δ rad27-p strain after 30 min of MMS treatment was reduced to 5.4 DSBs/genome if MAG1 was also inactivated (compare lanes 6 and 9), which is comparable to the WT level (3.4 to 7.6 DSBs/genome) (42). The prevention or slowing down of DSB formation by the MAG1 deletion demonstrates that the pol32Δ rad27-p mutant is defective in a repair process downstream of the generation of AP sites which is possibly related to strand displacement and/or flap removal.
FIG. 3.
A mag1 deletion in the pol32Δ rad27-p background prevents accumulation of DSBs. Genotypes are indicated. Each group of three lanes (from left to right) contains a mock-treated control and samples with 15 min and 30 min of MMS treatment. (A) DNA samples were processed at 30°C to detect closely opposed SSBs that were formed in vivo. (B) DNA samples were processed at 55°C to detect total closely opposed lesions (methylated lesions, AP sites, and SSBs). A Southern blot and the number of DSBs per haploid yeast genome, calculated as described in Materials and Methods, are shown under each lane.
Random SSBs induced by MMS treatment are repairable in a pol32Δ rad27-p strain.
Since the PFGE method employed in our study detects only opposing SSBs in close proximity, we next asked if the pol32Δ rad27-p strain was also defective in repairing random single SSBs. To identify the presence of random SSBs before and after LH, DNA samples prepared for PFGE (plugs) were subjected to S1 nuclease digestion to convert SSBs into DSBs (20, 21, 46). This single-strand-specific endonuclease can cut DNA regions containing nicks, gaps, and single-strand overhangs and thus will convert these lesions into DSBs which can be detected by PFGE (6, 21).
To test the utility of S1 nuclease in detecting SSBs, DNA plugs were treated with increasing doses of MMS. As previously shown (42), no DSBs were directly produced (Fig. 4, lanes 1 to 5). Following incubation of the MMS-treated plugs at 55°C to convert methylated bases and AP sites to SSBs, there was an appearance of up to 26 DSBs per genome equivalent (Fig. 4, lane 15). For MMS-treated DNA plugs that were heated at 55°C, the S1 nuclease greatly increased the number of DSBs (Fig. 4, lanes 16 to 20). For MMS-treated DNA plugs that were not heated, S1 nuclease treatment had, at most, a small but significant effect (Fig. 4, lanes 6 to 10). However, since there was no MMS dose-related increase in DSBs, these DSBs may have originated from preexisting SSBs in the genome.
FIG. 4.
Determination of SSBs by S1 nuclease treatment. MMS treatment of chromosomal DNA of WT yeast within plugs, along with the presence or absence of S1 nuclease, is indicated. DNA plugs were incubated in buffer either at 4°C (no heat treatment; lanes 1 to 10) or at 55°C (heat treatment) for 24 h to convert heat-labile sites into SSBs (lanes 11 to 20). Thereafter, S1 nuclease (4 U/ml for 20 min) treatment was applied to both mock heat-treated (lanes 6 to 10) and heat-treated (lanes 16 to 20) plugs.
The S1 nuclease approach was employed to assess SSBs in MMS-treated G1 cells before and following LH (Fig. 5). After 15 min of MMS treatment, few DSBs were produced in vivo, regardless of genotype (WT and pol32Δ rad27-p and rad27 mutants, corresponding to lanes 1, 2, and 3, respectively) (also see Fig. 1A). As shown in Table 2, there were comparable amounts of DSBs in the individual strains before and after LH (lanes 4 to 6 versus lanes 7 to 9), as follows: 2.9 versus <2 DSBs/genome equivalent for the WT, 6.6 versus 5.9 DSBs/genome for the rad27 mutant, and 5.1 versus 6.5 DSBs/genome for the pol32Δ rad27-p mutant. However, treatment of the DNA plugs with S1 nuclease revealed a large number of SSBs after MMS treatment (Fig. 5, lanes 13 to 15) that were reduced upon LH (Fig. 5, lanes 16 to 18) for all genotypes. The estimated number of S1-generated DSBs/genome equivalent (Table 2) decreased from 35 to 14 for the WT, from 35 to 23 for the rad27 mutant, and from 38 to 25 for the pol32Δ rad27-p mutant, corresponding to 75%, 38%, and >39% reductions, respectively, after accounting for the background of ∼7 DSBs/genome equivalent. The values (especially before LH) may correspond to transitional SSBs during repair instead of to total SSBs generated during BER. Therefore, the actual quantity of SSBs repaired during LH may be higher than that estimated. While there were more SSBs remaining in the rad27 and pol32Δ rad27-p mutants after LH than in the WT, the reductions in the level of S1 nuclease-converted DSBs clearly indicate that most random SSBs can be repaired in these two strains. This contrasts with the actual increase in DSBs due to closely opposed SSBs in the pol32Δ rad27-p strain after LH, further suggesting that DSBs are uniquely affected by the double mutation.
FIG. 5.
SSBs induced by MMS treatment are repairable in pol32Δ rad27-p mutants. Each group of three lanes (from left to right) represents WT, pol32Δ rad27-p, and rad27 genotypes. G1 haploid yeasts were treated with MMS as follows: mock-treated control (lanes 1 to 3 and 10 to 12), 15 min of MMS treatment (lanes 4 to 6 and 13 to 15), and 15 min of MMS treatment followed by LH (lanes 7 to 9 and 16 to 18). Chromosomal DNA samples were processed at 30°C in order to detect SSBs formed in vivo. S1 nuclease treatment was applied to DNA plugs (lanes 10 to 18).
TABLE 2.
S1 nuclease-detected MMS-induced SSBs in the genomea
| Strain (no. of independent expts) | Median (range) no. of SSBs
|
|||||
|---|---|---|---|---|---|---|
| No S1 treatment
|
S1 nuclease treatment
|
|||||
| No MMS | Before LH | After LH | No MMS | Before LH | After LH | |
| WT (5) | <2 (<2-2.1) | 2.9 (<2-4.5) | <2 (<2-2.39) | 7.2 (4-10.9) | 34.6 (25.7->40) | 14.4 (10.9-21.5) |
| rad27 mutant (5) | <2 | 6.6 (4.1-9.4) | 5.9 (3.4-9.4) | 8.5 (7.1-11.9) | 35.2 (32.1->40) | 23.6 (19.2-24.7) |
| pol32Δ rad27-p mutant (5) | <2 (<2-2.1) | 5.1 (3.2-8.4) | 6.5 (5.3-11.9) | 9.2 (7.4-13.1) | 37.8 (33.0->40) | 24.8 (20.2-27.6) |
The number of DSBs (converted from SSBs) per haploid genome in chromosomes separated by PFGE was calculated for each experiment as described in Materials and Methods.
Quantitation of total MMS-induced lesions in pol32Δ rad27-p mutant and WT.
Assuming that MMS-induced lesions are randomly distributed across the genome, there are hundreds more SSBs than closely opposed SSBs, with the actual numbers depending on the distance between two closely spaced SSBs required (42) to form a DSB. In order to further establish that the pol32Δ rad27-p mutant is specifically defective in repair of closely spaced lesions, total MMS damage was measured directly by QPCR, which detects all lesions that can block DNA amplification, as previously described (1, 12, 52, 54). The amount of MMS damage in a 10-kb window was assessed before and after LH (primers are described in Materials and Methods). As shown in Fig. 6, there were similar levels of MMS lesions in WT and pol32Δ rad27-p cells following MMS treatment, as follows: 1.1 to 1.3/10 kb (15-min MMS treatment) and ∼1.8/10 kb (30-min MMS treatment). Importantly, the extents of repair after LH were comparable between the WT and pol32Δ rad27-p strains for the two treatments, with 56% (15 min) and 38% (30 min) repair for the WT and 57% (15 min) and 35% (30 min) repair for the pol32Δ rad27-p mutant. Thus, the repair defect in the pol32Δ rad27-p mutant appears to be specific to closely spaced SSBs.
FIG. 6.
Comparable repair of total MMS-induced lesions in WT and pol32Δ rad27-p strains. Genomic DNAs were isolated from mock-treated and MMS-treated (15 and 30 min) G1 haploid cells before and after LH. QPCR was performed to amplify a 10-kb fragment and compared with controls to calculate the relative amplification. Estimation of the average number of lesions was done as described by Ayala-Torres et al. (1). Each bar represents the mean value and standard error for four independent MMS treatments (including six independent QPCR analyses for each sample).
Deletion of REV1 or REV3 does not affect the formation of MMS-induced DSBs in a rad27-p background.
Since Pol32 is involved in the bypass of DNA damage during replication (26, 29), we asked if TLS plays a role in preventing accumulation of closely opposed SSBs. The TLS pathway that involves Pol32 also requires the functions of Pol ζ (REV3) and Rev1 (26) but not that of Pol η (24, 25). If damage bypass is the underlying mechanism responsible for the accumulation of closely opposed SSBs in the pol32 rad27-p mutant, then deletion of REV1 or REV3 in the rad27-p background should have a similar effect. As shown in Fig. S1 in the supplemental material, rev1 rad27-p and rev1 rad27-p double mutants have a similar repair capacity to that of the rad27-p single mutant (only heat-dependent DSBs are shown so that repair can easily be visualized). Thus, it is likely that TLS plays only a minor role in the accumulation of closely opposed SSBs in the pol32 rad27-p strain.
DISCUSSION
Enzymatic processing of lesions during DNA repair must be coordinated to avoid the secondary creation of intermediates that in themselves would place the genome at risk. While single lesions are efficiently repaired through an SSB intermediate step by the many available redundant BER/SSB repair proteins, closely spaced lesions, if processed at the same time, would be expected to have a high risk of generating DSBs. The presence of nonligatable ends (i.e., “dirty” ends) at SSBs produced during BER might lead to DSBs that cause genome instability and cell death if not efficiently removed (48). In G1 haploid yeast cells, there is little opportunity for DSB repair through recombination or end joining involving nonligatable ends. In diploid and G2 cells, repair can occur via homologous recombination, although DSB repair capacity is limited in terms of the number of breaks repaired.
While there are extensive studies on the repair of clustered lesions in vitro, using oligonucleotides and purified enzymes or cell extracts, there are relatively few in vivo studies (for a recent review, see reference 22). Our recently developed in vivo assay has provided a unique opportunity to investigate the formation of DSBs arising from closely spaced lesions during repair of MMS-induced DNA damage in G1 stationary-phase haploid yeast. Since the assay measures clustered lesions in vivo as both SSBs and damaged bases (converted at a high temperature to DSBs in vitro), it is useful for addressing genetic interactions during BER of clustered lesions. The DSBs arising from MMS-induced closely spaced lesions are different from other types of DSBs induced by agents such as ionizing radiation or specific endonucleases, since the BER-associated DSBs are repaired well in G1 haploid yeast even in the absence of Rad52 (42), which is required for recombinational repair, or Ku70/80 (data not shown), which is required for end joining.
The generation of DSBs by closely spaced SSBs could occur if the overlapping complementary regions are short. Even if lesions are farther apart, repair processes that create SSBs could move SSBs closer to form DSBs, for example, during strand displacement repair synthesis (Fig. 7). This was proposed for SSBs produced on opposite DNA strands 80 to 90 bp apart, with an observed repair patch size of 40 nt (56). If a 5′ flap is not removed by Rad27/Fen1, strand displacement synthesis will continue without ligation, generating even longer flaps capable of binding replication protein A (RPA). Flaps longer than 20 to 30 nt are stably bound by RPA and are prevented from direct cleavage by Rad27/Fen1 (2, 8, 28); however, such flaps are subject to cutting by Dna2, which usually does not generate ligatable nicks. Strand displacement DNA synthesis would have the effect of bringing more distant ends closer together to generate DSBs. However, long flap formation appears unlikely when Rad27/Fen1 is fully functional during nick translation repair synthesis, since ∼90% of the products released by Fen1 are mononucleotides (17). Thus, the well-coordinated repair of SSBs in WT cells by Pol δ, PCNA, and Rad27/Fen1 would prevent the generation of DSBs via processing of widely (>30 nt) separated lesions.
FIG. 7.
Possible mechanisms for DSB formation from closely spaced lesions. When there is coordination of BER enzymes (WT), closely opposed base lesions can be repaired one by one. Once the first lesion is processed by glycosylases followed by AP endonucleases, strand displacement and 5′-flap removal are quickly finished to seal the nick generated during repair before starting nicking of the second lesion, thus avoiding the formation of DSBs. When there is a defect in the coordination of BER enzymes (pol32 rad27-p mutant), the strand displacement and/or 5′-dRP end flap removal might be slowed, resulting in increased time and space for the generation of a second nick 5′ of the closely opposed lesion, thereby leading to DSB formation. In this case, widely separated intermediate SSBs (e.g., >30 nt apart) might, in essence, be brought together to form DSBs. Alkylated bases are shown as filled red circles. 5′-dRP is shown as filled blue circles. Arrowheads on DNA strands correspond to 3′ ends. Red dashed lines represent repair-associated DNA synthesis, and red solid lines represent newly synthesized DNA. Note that this figure shows only one orientation of the two closely opposed SSBs, where DNA synthesis and strand displacement intermediates face each other. DSBs would not be generated if DNA synthesis and strand displacement at closely opposed SSBs proceeded in the opposite orientation.
Although the pol32Δ or rad27-p single mutant had little effect on the repair of MMS damage, not only was the pol32Δ rad27-p double mutant defective in repair of lesions that were closely spaced, resulting in DSBs, but there was also a further increase in these DSBs after LH (Fig. 1 and Table 1). Thus, the combination of Pol δ (with the Pol32 subunit) and Rad27 appears to prevent the generation of DSBs at closely spaced lesions. The model in Fig. 7 describes steps in the repair of closely spaced lesions that could lead to DSBs if there is a lack of coordination in BER components, as in the case of the pol32Δ rad27-p mutant. The DSBs could arise either directly after induction of SSBs or during subsequent processing of lesions that are not widely spaced.
Since deletion of MAG1 prevents the accumulation of closely opposed SSBs in the pol32Δ rad27-p double mutant (Fig. 3), the repair defect leading to DSBs is downstream of an SSB generated by an AP endonuclease. In vitro studies have demonstrated that for pairs of lesions as close as 1 to 3 nt apart, both can be incised by endonucleases/lysases to form a DSB (7, 39). Whether this can occur in vivo remains to be determined (22). Our present results showing that the number of closely opposed SSBs was increased during LH in the pol32Δ rad27-p mutant (Table 1) suggest that simultaneous processing of closely spaced lesions may normally be prohibited in vivo. Possibly, binding of the Polδ-PCNA-Rad27 complex obstructs access of glycosylase and AP endonuclease to nearby methylated bases, resulting in coordinated repair such that the first SSB is repaired before the second SSB is formed. Since Pol32 is required for optimum processivity of Pol δ and may affect strand displacement (10, 11, 31), the loss of Pol32 might slow the SSB repair process. Therefore, combined with the rad27-p mutation, the opportunity for generating a second SSB at a nearby lesion may be increased before finishing repair of the first SSB (Fig. 7). Also, there would be less of the Polδ-PCNA-Rad27 complex to physically obstruct processing of a nearby second lesion.
The synergy between the rad27-p mutation, which causes a defect in PCNA binding, and the pol32Δ mutation reveals the important role of PCNA in the repair of clustered lesions. PCNA interacts with Pol δ and DNA ligase and also forms a tight complex with Rad27/Fen1. This interaction provides efficient removal of 5′ nucleotides by means of “nick translation” to continuously maintain a nick that can be ligated (35, 36, 43). Displacement of a few nucleotides by Pol δ is enough to lead to removal of the 5′-dRP by Rad27/Fen1 (16). This may explain why the single pol32Δ mutation has only a mild effect, since a tight PCNA-Rad27 interaction could efficiently remove 5′-dRPs before a second incision at the nearby lesion or decrease the requirement for strand displacement and/or the length of a displaced flap. In the rad27-p mutant, a lack of PCNA-Rad27 interaction may lead to deficient recruitment of flap removal activity since the access of Rad27 protein to the site is most likely restricted. However, the reduced access of Rad27 protein to the repair site in the rad27-p mutant appeared to be compensated for by Pol32, since repair was comparable in the WT and rad27-p strains (Fig. 1 and Table 1). Only when both proteins were defective could the accumulation of DSBs occur.
Pol32 may play a role in helping to recruit the Rad27-p protein to a repair site, possibly by temporally releasing the Pol δ-PCNA complex from the repair site or by helping to generate a suitable flap. Since Pol32 can be involved in TLS (26, 29), Pol32 may aid in Pol δ strand displacement synthesis past a nearby opposing methylated base, resulting in the generation of a long flap that could be a better substrate for the Rad27-p protein. Pol32 is required in Pol ζ (REV3)- and Rev1-mediated (but not Pol η-dependent) TLS, at least for the initiation step, to bypass abasic sites (24-26). If TLS helps to prevent the accumulation of DSBs, deletion of other TLS proteins in the rad27-p background should result in MMS responses similar to those for the pol32Δ rad27-p mutant. However, as shown in Fig. S1 in the supplemental material, this is not the case for the rev1 rad27-p and rev1 rad27-p double mutants, suggesting that the synergistic effect between pol32Δ and rad27-p mutations is not due to a loss of TLS function.
While Pol32 is important for Pol δ processivity and strand displacement, the structural parameters in Pol δ that determine its strand displacement potential remain unknown (16). However, the Pol32 subunit has an extended structure which accounts for the unusual elongated shape of the entire three-subunit complex (31). Possibly, this structure helps to “open” the strands to enable bypass of an adjacent lesion without concomitant DNA synthesis. Recent in vitro results show that Pol δ in the absence of the Pol32 subunit has reduced activity for strand displacement (55).
Overall, this study and our previous report (42) reveal that closely spaced single-strand lesions can be converted into DSBs through BER processes in yeast. Closely spaced lesions could result from random lesions that are nearby by chance or from agents such as ionizing radiation, which creates clustered damage. The Pol32 component of Pol δ appears to play an important role in preventing the transition to DSBs by participating in the efficient coordinated repair of closely spaced lesions. It is interesting that Pol32 may also function elsewhere to prevent DSB formation, since the combination of a pol32Δ mutation with other homologous recombination repair pathway mutations results in slow growth or lethality (25).
Clustered lesions may possibly be generated during normal cellular metabolism by reactive oxygen or nitrogen species. The current study with G1 stationary-phase yeast cells has important implications for BER events in G1 human cells, especially postmitotic cells that cannot be replaced through cell division. Prevention of DSB formation by BER might be relevant to issues of neurodegeneration and aging.
Supplementary Material
Acknowledgments
We are grateful to Matthew Lau and Emilie Guerrent, who participated in the undergraduate NIEHS Summers of Discovery Program, for help in making yeast strains and to Julie Horton, Jeffrey D. Stumpf, and Vladimir Poltoratsky for critical readings of the manuscript and for suggestions.
This work was supported by the Intramural Research Program of the NIEHS (NIH, DHHS) under projects 1 Z01 ES065073 (M.A.R.) and 2 Z01 ES61062 (B.V.H.).
Footnotes
Published ahead of print on 15 December 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ayala-Torres, S., Y. Chen, T. Svoboda, J. Rosenblatt, and B. Van Houten. 2000. Analysis of gene-specific DNA damage and repair using quantitative polymerase chain reaction. Methods 22135-147. [DOI] [PubMed] [Google Scholar]
- 2.Bae, S. H., K. H. Bae, J. A. Kim, and Y. S. Seo. 2001. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412456-461. [DOI] [PubMed] [Google Scholar]
- 3.Barnes, D. E., and T. Lindahl. 2004. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 38445-476. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, R. A. 1999. The Saccharomyces cerevisiae ETH1 gene, an inducible homolog of exonuclease III that provides resistance to DNA-damaging agents and limits spontaneous mutagenesis. Mol. Cell. Biol. 191800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beranek, D. T. 1990. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 23111-30. [DOI] [PubMed] [Google Scholar]
- 6.Berk, A. J., and P. A. Sharp. 1978. Spliced early mRNAs of simian virus 40. Proc. Natl. Acad. Sci. USA 751274-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaisdell, J. O., L. Harrison, and S. S. Wallace. 2001. Base excision repair processing of radiation-induced clustered DNA lesions. Radiat. Prot. Dosimetry 9725-31. [DOI] [PubMed] [Google Scholar]
- 8.Bochkareva, E., S. Korolev, S. P. Lees-Miller, and A. Bochkarev. 2002. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 211855-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boiteux, S., and M. Guillet. 2004. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amsterdam) 31-12. [DOI] [PubMed] [Google Scholar]
- 10.Budd, M. E., C. C. Reis, S. Smith, K. Myung, and J. L. Campbell. 2006. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol. Cell. Biol. 262490-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgers, P. M., and K. J. Gerik. 1998. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 27319756-19762. [DOI] [PubMed] [Google Scholar]
- 12.Chen, K. H., F. M. Yakes, D. K. Srivastava, R. K. Singhal, R. W. Sobol, J. K. Horton, B. Van Houten, and S. H. Wilson. 1998. Up-regulation of base excision repair correlates with enhanced protection against a DNA damaging agent in mouse cell lines. Nucleic Acids Res. 262001-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.David-Cordonnier, M. H., J. Laval, and P. O'Neill. 2000. Clustered DNA damage, influence on damage excision by XRS5 nuclear extracts and Escherichia coli Nth and Fpg proteins. J. Biol. Chem. 27511865-11873. [DOI] [PubMed] [Google Scholar]
- 14.Eot-Houllier, G., S. Eon-Marchais, D. Gasparutto, and E. Sage. 2005. Processing of a complex multiply damaged DNA site by human cell extracts and purified repair proteins. Nucleic Acids Res. 33260-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fortini, P., and E. Dogliotti. 2007. Base damage and single-strand break repair: mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair (Amsterdam) 6398-409. [DOI] [PubMed] [Google Scholar]
- 16.Garg, P., and P. M. Burgers. 2005. How the cell deals with DNA nicks. Cell Cycle 4221-224. [PubMed] [Google Scholar]
- 17.Garg, P., C. M. Stith, N. Sabouri, E. Johansson, and P. M. Burgers. 2004. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 182764-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gary, R., K. Kim, H. L. Cornelius, M. S. Park, and Y. Matsumoto. 1999. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem. 2744354-4363. [DOI] [PubMed] [Google Scholar]
- 19.Gary, R., M. S. Park, J. P. Nolan, H. L. Cornelius, O. G. Kozyreva, H. T. Tran, K. S. Lobachev, M. A. Resnick, and D. A. Gordenin. 1999. A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Mol. Cell. Biol. 195373-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geigl, E. M., and F. Eckardt-Schupp. 1991. The repair of double-strand breaks and S1 nuclease-sensitive sites can be monitored chromosome-specifically in Saccharomyces cerevisiae using pulse-field gel electrophoresis. Mol. Microbiol. 51615-1620. [DOI] [PubMed] [Google Scholar]
- 21.Geigl, E. M., and F. Eckardt-Schupp. 1991. Repair of gamma ray-induced S1 nuclease hypersensitive sites in yeast depends on homologous mitotic recombination and a RAD18-dependent function. Curr. Genet. 2033-37. [DOI] [PubMed] [Google Scholar]
- 22.Georgakilas, A. G. 2008. Processing of DNA damage clusters in human cells: current status of knowledge. Mol. Biosyst. 430-35. [DOI] [PubMed] [Google Scholar]
- 23.Gerik, K. J., X. Li, A. Pautz, and P. M. Burgers. 1998. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 27319747-19755. [DOI] [PubMed] [Google Scholar]
- 24.Gibbs, P. E., J. McDonald, R. Woodgate, and C. W. Lawrence. 2005. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics 169575-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanna, M., L. G. Ball, A. H. Tong, C. Boone, and W. Xiao. 2007. Pol32 is required for Pol zeta-dependent translesion synthesis and prevents double-strand breaks at the replication fork. Mutat. Res. 625164-176. [DOI] [PubMed] [Google Scholar]
- 26.Haracska, L., I. Unk, R. E. Johnson, E. Johansson, P. M. Burgers, S. Prakash, and L. Prakash. 2001. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 15945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison, L., Z. Hatahet, A. A. Purmal, and S. S. Wallace. 1998. Multiply damaged sites in DNA: interactions with Escherichia coli endonucleases III and VIII. Nucleic Acids Res. 26932-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henricksen, L. A., C. B. Umbricht, and M. S. Wold. 1994. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 26911121-11132. [PubMed] [Google Scholar]
- 29.Huang, M. E., A. de Calignon, A. Nicolas, and F. Galibert. 2000. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase delta, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr. Genet. 38178-187. [DOI] [PubMed] [Google Scholar]
- 30.Huang, M. E., A. G. Rio, M. D. Galibert, and F. Galibert. 2002. Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase delta, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics 1601409-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson, E., P. Garg, and P. M. Burgers. 2004. The Pol32 subunit of DNA polymerase delta contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 2791907-1915. [DOI] [PubMed] [Google Scholar]
- 32.Karthikeyan, G., J. H. Santos, M. A. Graziewicz, W. C. Copeland, G. Isaya, B. Van Houten, and M. A. Resnick. 2003. Reduction in frataxin causes progressive accumulation of mitochondrial damage. Hum. Mol. Genet. 123331-3342. [DOI] [PubMed] [Google Scholar]
- 33.Krokan, H. E., R. Standal, and G. Slupphaug. 1997. DNA glycosylases in the base excision repair of DNA. Biochem. J. 3251-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levin, D. S., W. Bai, N. Yao, M. O'Donnell, and A. E. Tomkinson. 1997. An interaction between DNA ligase I and proliferating cell nuclear antigen: implications for Okazaki fragment synthesis and joining. Proc. Natl. Acad. Sci. USA 9412863-12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levin, D. S., A. E. McKenna, T. A. Motycka, Y. Matsumoto, and A. E. Tomkinson. 2000. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr. Biol. 10919-922. [DOI] [PubMed] [Google Scholar]
- 36.Li, X., J. Li, J. Harrington, M. R. Lieber, and P. M. Burgers. 1995. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J. Biol. Chem. 27022109-22112. [DOI] [PubMed] [Google Scholar]
- 37.Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature 362709-715. [DOI] [PubMed] [Google Scholar]
- 38.Lindahl, T., and A. Andersson. 1972. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry 113618-3623. [DOI] [PubMed] [Google Scholar]
- 39.Lomax, M. E., S. Cunniffe, and P. O'Neill. 2004. Efficiency of repair of an abasic site within DNA clustered damage sites by mammalian cell nuclear extracts. Biochemistry 4311017-11026. [DOI] [PubMed] [Google Scholar]
- 40.Lundin, C., M. North, K. Erixon, K. Walters, D. Jenssen, A. S. Goldman, and T. Helleday. 2005. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 333799-3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lydeard, J. R., S. Jain, M. Yamaguchi, and J. E. Haber. 2007. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448820-823. [DOI] [PubMed] [Google Scholar]
- 42.Ma, W., M. A. Resnick, and D. A. Gordenin. 2008. Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis. Nucleic Acids Res. 361836-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montecucco, A., R. Rossi, D. S. Levin, R. Gary, M. S. Park, T. A. Motycka, G. Ciarrocchi, A. Villa, G. Biamonti, and A. E. Tomkinson. 1998. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 173786-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura, J., and J. A. Swenberg. 1999. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 592522-2526. [PubMed] [Google Scholar]
- 45.Osborne, M. R., and D. H. Phillips. 2000. Preparation of a methylated DNA standard, and its stability on storage. Chem. Res. Toxicol. 13257-261. [DOI] [PubMed] [Google Scholar]
- 46.Petersen, S., G. Saretzki, and T. von Zglinicki. 1998. Preferential accumulation of single-stranded regions in telomeres of human fibroblasts. Exp. Cell Res. 239152-160. [DOI] [PubMed] [Google Scholar]
- 47.Regulus, P., B. Duroux, P. A. Bayle, A. Favier, J. Cadet, and J. L. Ravanat. 2007. Oxidation of the sugar moiety of DNA by ionizing radiation or bleomycin could induce the formation of a cluster DNA lesion. Proc. Natl. Acad. Sci. USA 10414032-14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Resnick, M. A., and P. Martin. 1976. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol. Gen. Genet. 143119-129. [DOI] [PubMed] [Google Scholar]
- 49.Reynolds, R. J. 1987. Induction and repair of closely opposed pyrimidine dimers in Saccharomyces cerevisiae. Mutat. Res. 184197-207. [DOI] [PubMed] [Google Scholar]
- 50.Rydberg, B. 2000. Radiation-induced heat-labile sites that convert into DNA double-strand breaks. Radiat. Res. 153805-812. [DOI] [PubMed] [Google Scholar]
- 51.Santos, J. H., L. Hunakova, Y. Chen, C. Bortner, and B. Van Houten. 2003. Cell sorting experiments link persistent mitochondrial DNA damage with loss of mitochondrial membrane potential and apoptotic cell death. J. Biol. Chem. 2781728-1734. [DOI] [PubMed] [Google Scholar]
- 52.Santos, J. H., J. N. Meyer, B. S. Mandavilli, and B. Van Houten. 2006. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 314183-199. [DOI] [PubMed] [Google Scholar]
- 53.Scharer, O. D., and J. Jiricny. 2001. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays 23270-281. [DOI] [PubMed] [Google Scholar]
- 54.Sobol, R. W., D. E. Watson, J. Nakamura, F. M. Yakes, E. Hou, J. K. Horton, J. Ladapo, B. Van Houten, J. A. Swenberg, K. R. Tindall, L. D. Samson, and S. H. Wilson. 2002. Mutations associated with base excision repair deficiency and methylation-induced genotoxic stress. Proc. Natl. Acad. Sci. USA 996860-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stith, C. M., J. Sterling, M. A. Resnick, D. A. Gordenin, and P. M. Burgers. 2008. Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J. Biol. Chem. 28334129-34140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vispe, S., E. L. Ho, T. M. Yung, and M. S. Satoh. 2003. Double-strand DNA break formation mediated by flap endonuclease-1. J. Biol. Chem. 27835279-35285. [DOI] [PubMed] [Google Scholar]
- 57.Wallace, S. S. 2002. Biological consequences of free radical-damaged DNA bases. Free Radic. Biol. Med. 331-14. [DOI] [PubMed] [Google Scholar]
- 58.Ward, J. F. 1986. Ionizing radiation induced DNA damage: identities and DNA repair. Basic Life. Sci. 38135-138. [DOI] [PubMed] [Google Scholar]
- 59.Ward, J. F. 1981. Some biochemical consequences of the spatial distribution of ionizing radiation-produced free radicals. Radiat. Res. 86185-195. [PubMed] [Google Scholar]
- 60.Wilson, D. M., III, and V. A. Bohr. 2007. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amsterdam) 6544-559. [DOI] [PubMed] [Google Scholar]
- 61.Wu, X., and Z. Wang. 1999. Relationships between yeast Rad27 and Apn1 in response to apurinic/apyrimidinic (AP) sites in DNA. Nucleic Acids Res. 27956-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.