Abstract
Eukaryotic translation initiation factor 4E (eIF4E) is encoded by a potent oncogene which is highly elevated in many human cancers. Few studies have investigated how the level, and thus activity, of eIF4E is regulated in healthy (noncancerous) cells and how they become elevated in malignant cells. Here, our studies reveal a novel mechanism by which eIF4E levels are regulated at the level of mRNA stability. Two factors known to modulate transcript stability, HuR and the p42 isoform of AUF1, compete for binding to the 3′ untranslated regions (3′UTRs) of eIF4E mRNAs. We identified a distinct AU-rich element in the 3′UTR of eIF4E which is responsible for HuR-mediated binding and stabilization. Our studies show that HuR is upregulated in malignant cancer specimens characterized by high eIF4E levels and that its depletion leads to reduction in eIF4E levels. Further, HuR and eIF4E regulate a common set of transcripts involved in cellular proliferation (cyclin D1 and c-myc) and neoangiogenesis (vascular endothelial growth factor), which suggests a functional connection between HuR and eIF4E in the regulation of these important processes. In summary, we present a novel model for the regulation of eIF4E expression and show that this model is relevant to elevation of eIF4E levels in malignant cells.
Eukaryotic translation initiation factor 4E (eIF4E) is encoded by a potent oncogene (23). Its overexpression leads to malignant transformation in cell culture and to tumorigenesis in animal models (6). Accordingly, elevated levels of eIF4E are observed in a wide variety of human cancers where higher levels correlate with poor prognosis (6). eIF4E is a potent posttranscriptional regulator of gene expression. Particularly, it is a central node in an RNA regulon governing proliferation and cell survival (3). Underlying this, eIF4E acts in cap-dependent translation, and it also promotes the nuclear export of specific growth-promoting transcripts (5, 10). For mRNAs to be regulated by eIF4E, they must contain specific elements in their 5′ and 3′ untranslated regions (5′UTRs and 3′UTRs, respectively), referred to as USER codes. The 5′ USER codes are long and highly structured (14). The 3′ USER code is a 50-nucleotide element referred to as an eIF4E sensitivity element (4). Importantly, eIF4E must bind the m7G caps on the 5′ ends of mRNAs for its activities in translation, export, transformation, and cell survival (3). Targeting eIF4E by impairing its cap binding activity is a novel therapeutic strategy being tested in refractory acute myeloid leukemia patients (in a phase I/II clinical trial in Canada [www.ribatrial.com]).
Although the mechanisms and physiological effects of eIF4E activity were studied extensively, few studies have focused on the control of eIF4E expression. Early studies suggested that eIF4E is a myc target gene as it has an E-box in its promoter (12). However, eIF4E is still produced in myc−/− cells, indicating that eIF4E can be controlled by alternative mechanisms (2a, 34). The importance of understanding cellular mechanisms and factors controlling the level of eIF4E level is highlighted by the observation that eIF4E RNA and protein levels are frequently elevated in cancer. In breast cancer and head and neck squamous cell carcinoma (HNSCC), eIF4E is thought to be elevated due to gene amplification (27, 28). eIF4E is highly elevated in a subset (M4 or M5) of acute myeloid leukemias (AMLs) (31). In these patient specimens, eIF4E RNA levels are substantially reduced by the introduction of a dominant-negative inhibitor of NF-κB (31). Clearly, there is little understanding of how eIF4E is controlled in healthy (noncancerous) cells or how it becomes elevated in cancer cells.
To better understand regulation of eIF4E expression, we examined its 5′UTR and 3′UTR to assess whether eIF4E could be a target of posttranscriptional control. We identified three conserved AU-rich elements (AREs) in eIF4E. The HuR protein frequently regulates the stability of ARE-containing transcripts. Here, we examine the role of HuR in the regulation of eIF4E mRNA stability. Further, we identified a competing protein which generally destabilizes transcripts, AUF1, as another regulator of eIF4E mRNA stability. These studies are the first to demonstrate that eIF4E is regulated at the posttranscriptional level, specifically at the level of mRNA stability. We also show that eIF4E stability is upregulated in cancer cells as a function of elevated HuR levels. Primary leukemia specimens with elevated eIF4E protein levels consistently overexpress HuR. Together, these data indicate that HuR likely plays a major role in the elevation of eIF4E levels in cancer. Our studies also reveal another level of complexity: we find that HuR upregulates the eIF4E regulon and that eIF4E is required for at least a subset of HuR's effects on gene expression.
MATERIALS AND METHODS
Cell culture.
All cells lines used in this study were obtained from the ATCC. U2Os and HEK293 cells were maintained in Dulbecco modified Eagle medium (Invitrogen). FaDu and Detroit 551 cells were maintained in ATCC-formulated Eagle's minimum essential medium (ATCC). In both cases, media were supplemented with 10% fetal bovine serum (FBS) and penicillin or streptomycin (both obtained from Invitrogen). CD34+ M1 and M4 AML specimens were obtained from the Banque de Cellules Leucémiques du Québec (BCLQ). The specimens were stripped of all identifiers, and the experiments were done with the approval of the ethics board.
Plasmids, RNA interference (RNAi), and transfections.
pcDNA3HuR, pcDNA2FeIF4E, and pcDNA2FW56AeIF4E were described previously (4, 8). The pcDNA2FHuR construct was generated by cloning full-length HuR cDNA under the EcoRI/XhoI restriction site of pcDNA2F backbone. To generate pcDNA3.1HisLacZ/4E3′UTR constructs, we amplified the 3′UTR of eIF4E (gi|54873625; bases 1506 to 2476) from the DNase-treated RNA isolated from HEK293 or U2Os cells using primers 4EUTRHMF (5′-TTAAGAAGACACCTTCTGAGTATTCT) and 4EUTRHMR (5′-AAGACAATTCACTGTACACATTTTATT-3′) and Titan reverse transcription-PCR (RT-PCR) kit (Boehringer). PCR product was reamplified using the same primers with EcoRI and XhoI restriction sites and pfuTurbo (Stratagene) and subsequently cloned distal to the LacZ open reading frame in the pcDNA3.1HisLacZ backbone (Invitrogen) (using the EcoRI/XhoI restriction site). Notably, 3′UTR of eIF4E obtained from both cell lines was missing bases 2058 to 2094 compared to the eIF4E sequence submitted under gi|54873625. Detailed information about other plasmids used in this study is provided in the supplemental material.
Plasmid transfections were carried out using Fugene 6 (Roche) according to the manufacturer's instructions. We routinely achieve transfection efficiencies of >80% using Fugene 6 in Detroit 551, U2Os, and HEK293 cells. For transient transfection, cells were harvested after 48 h. Stably transfected cells were generated through selection with 1 mg/ml G418 24 h posttransfection.
For small interfering RNA (siRNA) analysis, siRNA duplexes and double-scrambled negative control (DS) (all obtained from IDT) were transfected with Lipofectamine 2000 (Invitrogen) at the final concentration of 10 nM, unless otherwise indicated. The siRNA duplexes used in this study were HuR siRNA (HSC.RNAI.N001419.4.2; IDT), eIF4E siRNA (sense, 5′-CCCAAAUCUCGAUUGCUUGACGCAGUC-3′; antisense, 5′-CUGCGUCAAGCAAUCGAGAUUUGGG-3′), and AUF1 siRNA (HSC.RNAI.N001003810.4.2; IDT). In each experiment, transfection efficiency was >90% as quantified using dsRED control (IDT), except in Detroit 551 cells; we repeatedly failed to efficiently transfect Detroit 551 cells with either dsRED control (IDT) or HuR siRNA. For the rescue experiments, U2Os/HuR cells were transfected with human-specific eIF4E siRNA duplexes. After 24 h, cells were washed three times with Dulbecco modified Eagle medium supplemented with 10% FBS and subsequently transfected with flag-tagged, murine eIF4E constructs (wild-type and W56A eIF4E) using Fugene 6. The eIF4E siRNA is complementary to the segment of the 3′UTR of eIF4E mRNA which is specific for human eIF4E transcripts, and therefore, it does not affect the expression of the murine eIF4E. Importantly, murine eIF4E shares 98% identity with its human homologue and can efficiently substitute for its function (20). Twenty-four hours posttransfection, G418 (1 mg/ml; Invitrogen) was added to the medium. Cells were lysed in radioimmunoprecipitation assay buffer 48 h after the second round of transfection.
Western and Northern blots.
Western and Northern blots were carried out as described previously (30). Antibodies (Ab) used for Western blots are mouse monoclonal HuR Ab (3A2; 1:500), rabbit polyclonal anti-eIF4E Ab (Cell Signaling), mouse monoclonal anti-eIF4E Ab (1:2,000; BD Transduction Laboratories), mouse monoclonal anti-cyclin D1 Ab (1:500; BD PharMingen), rabbit polyclonal anti-AUF1 Ab (Upstate Biotechnologies), mouse monoclonal anti-c-myc Ab (9E10; Covance), mouse monoclonal anti-FLAG-M2 Ab (1:2,000), mouse monoclonal anti-β-actin Ab (1:10,000), and mouse monoclonal anti-α-tubulin Ab (1:2,000) (all from Sigma), mouse monoclonal anti-Xpress Ab (1:5,000; Invitrogen), mouse monoclonal anti-hnRNP C1/C2 Ab (4F4; 1:1,000), mouse monoclonal anti-hnRNP A1 Ab (4B10; 1:1,000), rabbit polyclonal anti-RNA polymerase II (1:500), rabbit polyclonal anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) Ab (1:1,000), rabbit polyclonal c-fos Ab (1:500), rabbit polyclonal anti-vascular endothelial growth factor (anti-VEGF) Ab (1:500), rabbit polyclonal anti-cyclin D1 Ab (1:500), and mouse monoclonal anti-p53 Ab (1:1,000) (all from Santa Cruz).
Northern blots were carried out using the NorthernMax kit (Ambion) according to the manufacturer's instruction. The probes used in the Northern blots were eIF4E cDNA probe (5 pM), GAPDH cDNA probe (5 pM; Ambion), LacZ cDNA probe (5 pM), tRNALys antisense oligoprobe (30 pM), and U6 snRNA antisense oligoprobe (30 pM) (5, 30).
Immunoprecipitation and RIP.
To minimize the possibility of messenger RNP (mRNP) reassortment in cell extracts (22), initial immunoprecipitations were carried out using two different protocols. We have used the first protocol extensively. Briefly, U2Os (or where indicated HEK293) cells were lysed in ice-cold NET-2 buffer (50 mM Tris-HCl [pH 7.4], 300 mM NaCl, 0.5% [vol/vol] Nonidet P-40, 1× complete protease inhibitors [Roche], 200 U/ml RNase OUT [Invitrogen]), and the immunoprecipitations were carried out as described previously (4, 5, 30). The second protocol, designed for RNA immunoprecipitation (RIP) on CHIP analysis, is described in detail elsewhere (13).
Notably, both immunoprecipitation protocols yielded the same results, which suggests that the immunoprecipitated material represents the composition of endogenous mRNPs and that the protein-RNA interactions were not due to the reassortment of mRNPs in the lysate.
The antibodies used for immunoprecipitations were mouse monoclonal anti-eIF4E Ab agarose conjugate and mouse immunoglobulin G (IgG) agarose conjugate (Santa Cruz), rabbit polyclonal anti-eIF4E Ab (Cell Signaling), mouse monoclonal anti-HuR Ab (3A2), mouse IgG (Calbiochem), rabbit polyclonal anti-AUF1 Ab (Cell Signaling), and rabbit IgG (Calbiochem). Immunoprecipitated material was eluted using nonreducing sample buffer (Pierce).
Synthesis of biotinylated transcripts and RNA pulldown assays.
Templates for in vitro synthesis of biotinylated transcripts were generated by PCR from pcDNA3.1HisLacZ/4E3′UTR, GAPDH and h4E plasmid using primers listed in the supplemental material. All 5′ primers contained the T7 polymerase sequence. In vitro transcriptions were carried out using the Megascript kit (Ambion) according to the manufacturer's instructions, using a 1:10 ratio of CTP and 11-biotin CTP (NEB). In vitro-transcribed, biotinylated probes were purified using phenol-chloroform extraction and MegaClean columns (Ambion), and the efficiency of biotinylation was verified by Northern Max (Ambion).
Biotin pulldown assays were carried out as described previously (29). Briefly, U2Os cells (109) were lysed in ice-cold buffer X (10 mM HEPES [pH 7.5], 100 mM potassium acetate, 1.5 mM magnesium acetate). Lysates were cleared by centrifugation (13,200 rpm, 10 min, 4°C), and the resulting supernatants were supplemented with yeast tRNA (15 μg/ml; Sigma) and RNase OUT (100 U/ml; Invitrogen) and incubated with 50 μl of equilibrated affigel-heparin beads (Bio-Rad) for 10 min at 4°C. The supernatants were transferred to fresh tubes and incubated with 100 μl of equilibrated streptavidin-Sepharose beads (Sigma) for 1 h at 4°C, the beads were then spun down, and the resulting supernatant was supplemented with 10 μg of in vitro-transcribed, biotinylated RNA probe. The incubations were carried out for 1 h at 4°C after which 30 μl of equilibrated streptavidin-Sepharose beads (Sigma) was added to each reaction mixture and incubated for an additional 2 h at 4°C. Beads were collected by centrifugation and washed five times with buffer X, and the pulldown material was eluted by boiling in Laemmli buffer. Protein content in the eluted material was monitored by Western blotting. RNA assays with the recombinant glutathione S-transferase (GST) and GST-HuR protein (prepared as described elsewhere [18]) were carried out under the same conditions as described above, except that the incubation with affigel-heparin beads (Bio-Rad) was omitted. In each reaction mixture, 5 nM of recombinant protein was incubated with 10-fold molar excess of in vitro-transcribed, biotinylated RNA probe (50 nM).
Immunofluorescence and laser-scanning confocal microscopy.
Immunofluorescence was carried out as described previously (9). Briefly, FaDu and Detroit 551 cells were grown on coverslips, fixed in 3% paraformaldehyde for 20 min at room temperature, washed two times with phosphate-buffered saline (PBS), and permeabilized in blocking solution (1× PBS [pH 7.4], 1% FBS) supplemented with 0.5% (vol/vol) Triton X-100 for 15 min at room temperature. Upon permeabilization, cells were washed three times with blocking solution and incubated with mouse monoclonal anti-HuR Ab (3A2; diluted 1:100 in blocking solution) for 1 h at room temperature, followed by two washes in blocking solution. Subsequently, cells were incubated with secondary donkey anti-mouse IgG fluorescein isothiocyanate Ab (Jackson Immunolaboratories; diluted 1:150 in blocking solution), washed three times with 1× PBS (pH 7.4), rinsed in double-distilled H2O, mounted in Vectashield with 4′,6′-diamidino-2-phenylindole (DAPI) (Vector Laboratories), and sealed with nail polish. Analysis was carried out using a laser-scanning confocal microscope (LSM510; Carl Zeiss, Inc.), exciting at 405 and 543 nm using a 100× objective.
Actinomycin D treatments, cell fractionation, semiquantitative RT-PCR (sqRT-PCR), and RT-qPCR.
When indicated, cells were treated with 5 μg/ml of actinomycin D (Sigma) or with the appropriate amount of carrier (dimethyl sulfoxide). At each time point, RNA was isolated using Trizol (Invitrogen), treated with Turbo DNase (Ambion), and quantified by RT-quantitative PCR (RT-qPCR). The values obtained for target genes were normalized against 18S values and plotted on a semilogarithmic graph. The values obtained for the dimethyl sulfoxide-treated cells were set at 100%. Results obtained by RT-qPCR were verified by Northern blotting.
Fractionation and RNA isolation were performed as described previously (30). sqRT-PCR and RT-qPCR were performed using OneStep RT-PCR kit (Qiagen) and Sybr Green PCR Master Mix (Applied Biosystems), respectively. The primers and the conditions under which the PCRs were carried out are provided elsewhere (4, 5).
Computational analysis.
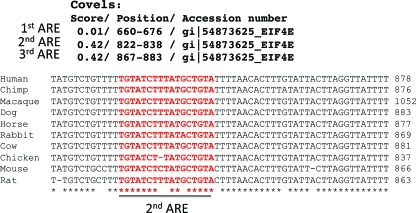
To identify the putative HuR binding motifs in eIF4E 3′UTR of human origin, we deployed the stochastic context-free grammar model, capturing both the primary and secondary features of the RNA motif using the program COVELs as described previously (17). Accession numbers and COVELs scores are provided in Fig. 1.
FIG. 1.
eIF4E mRNA contains AU-rich elements in its 3′UTR. (A) (Top) COVELs scores and the positions of putative HuR binding sites in the 3′UTR of human eIF4E (gi|54873625). (Bottom) Sequence alignments of the 3′UTR of eIF4E surrounding the putative HuR binding site designated 2ndARE (shown in red) from the indicated animals suggest that this region is evolutionarily conserved.
RESULTS
The 3′UTR of eIF4E mRNA contains AU-rich elements.
To determine whether eIF4E was likely to be regulated posttranscriptionally, we searched for putative RNA regulatory elements in the human eIF4E transcript. The COVELs model algorithm (17) revealed three putative HuR binding sites resembling AREs in the 3′UTR of eIF4E mRNA. Considering the fact that HuR binding sites frequently overlap with AREs, hitherto these sites will be referred to as the 1stARE, 2ndARE, and 3rdARE. Notably, the 2ndARE was the most evolutionarily conserved among the sequences examined (Fig. 1). AREs and similar elements in the UTRs sensitize transcripts to regulation by HuR as well as other ARE-binding proteins, such as AUF1 (1). With this in mind, we investigated whether HuR regulates the fate of eIF4E transcripts, and thereby eIF4E expression.
HuR overexpression increases eIF4E protein levels.
We first determined whether eIF4E levels changed as a function of HuR expression. In U2Os cells, HuR overexpression led to a dose-dependent increase in eIF4E protein levels (Fig. 2A). The increase in eIF4E levels was comparable to the increase in the levels of c-fos protein, which is an established target of HuR (24) (Fig. 2A). Experiments using different cell types and constructs yielded the same results, which suggests that these findings were not cell type or construct specific (see Fig. S1A in the supplemental material). Conversely, there were no apparent changes in HuR protein levels as a function of eIF4E overexpression (see Fig. S1B in the supplemental material). As expected, eIF4E-overexpressing cells showed a marked increase in the protein levels of the well-established eIF4E target, cyclin D1 (see Fig. S1B in the supplemental material) (26). These findings suggest that HuR regulates eIF4E expression, while eIF4E does not affect HuR expression.
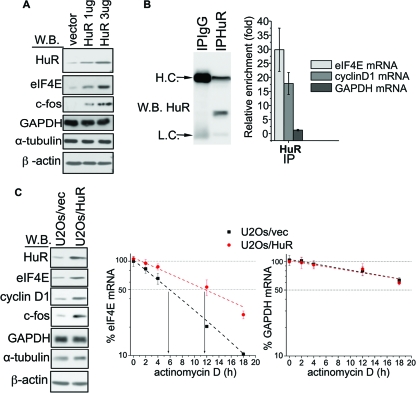
FIG. 2.
HuR binds and stabilizes eIF4E mRNA, which leads to the elevation of eIF4E protein levels. (A) U2Os cells were transfected with an empty vector or with the indicated amounts (1 or 3 μg) of HuR construct. The expression of HuR, eIF4E, c-fos, β-actin, α-tubulin, and GAPDH (the last two proteins were used as a loading control) was analyzed by Western blotting (W.B.). (B) RIP reactions carried out with the anti-HuR antibody (IPHuR) or with the control IgG (IPIgG). Western blotting indicates that HuR specifically immunoprecipitated with anti-HuR antibody. The positions of heavy chain (H.C.) and light chain (L.C.) are shown to the left of the blot. The amount of eIF4E, cyclin D1, and GAPDH mRNA in each immunoprecipitation (IP) was determined by RT-qPCR. Results are presented as the relative enrichment in HuR RIP compared to IgG RIP. Values are means ± standard deviations (SD) (error bars) (n = 3). (C) Overexpression of HuR leads to stabilization of eIF4E mRNA. (Left) Western blot showing the expression of HuR, eIF4E, cyclin D1, c-fos, β-actin, α-tubulin, and GAPDH (the last two proteins were used as a loading control) in U2Os/HuR and U2Os/vec cell lines. (Right) The same cells were treated with actinomycin D. (D) The amounts of eIF4E and GAPDH mRNA for each time point were determined by RT-qPCR and normalized (see Materials and Methods). Values are means ± SD (error bars) (n = 3).
HuR associates with eIF4E transcripts and increases their half-lives.
In order to understand the molecular mechanism(s) that underlie the apparent HuR-dependent regulation of eIF4E expression, we first tested whether HuR associates with eIF4E transcripts by RNA immunoprecipitation. RIPs were carried out in U2Os cells, and the amount of a given transcript in each reaction mixture was determined by RT-qPCR. This analysis revealed that eIF4E mRNA was significantly enriched in the HuR immunoprecipitation relative to the IgG control (Fig. 2B). Comparable enrichment was detected for cyclin D1 mRNA, which is a well-established HuR target (32), and was used here as a positive control. Notably, GAPDH transcripts, used here as a negative control (15), were not enriched in the HuR immunoprecipitation (Fig. 2B). These results were confirmed by sqRT-PCR (see Fig. S1C in the supplemental material).
Given that HuR affects various aspects of mRNA metabolism, we tested its effects on the nuclear export, translation, and stability of eIF4E mRNA. For these studies, we generated U2Os cell lines that stably overexpress HuR (U2Os/HuR) or that were stably transfected with the empty vector (U2Os/vec). There was no apparent difference in the nucleocytoplasmic ratios of eIF4E transcripts between U2Os/HuR and U2Os/vec cells (see Fig. S2A in the supplemental material). Similarly, there was no major alteration in the polysomal loading of eIF4E transcripts in HuR-overexpressing cells (see Fig. S2B in the supplemental material). These findings indicate that HuR does not affect the nuclear export rate or translational efficiency of eIF4E mRNA.
Next, we examined the effects of HuR on the stability of endogenous eIF4E mRNA. Here, we treated U2Os/HuR and U2Os/vec cells with the transcriptional inhibitor actinomycin D and measured the half-lives of eIF4E transcripts using RT-qPCR. In these experiments, the half-life of eIF4E mRNA was increased from ∼5.5 h in U2Os/vec cells to ∼12 h in U2Os/HuR cells, with no apparent change in the stability of HuR-insensitive GAPDH transcripts between the two cell lines (Fig. 2C). The RNAi knockdown of HuR levels consistently decreased the half-life of eIF4E mRNA, without affecting GAPDH transcripts (see Fig. S3A in the supplemental material). In both cases, changes in the eIF4E transcript stability were paralleled by appropriate changes in eIF4E protein levels (Fig. 2C) (see Fig. S3A in the supplemental material). Results obtained by RT-qPCR were confirmed by Northern blotting (see Fig. S3B in the supplemental material). Thus, we conclude that HuR binds to and stabilizes endogenous eIF4E mRNA transcripts, which leads to elevated eIF4E protein levels.
HuR stabilizes eIF4E mRNA through binding to its 3′UTR.
Next, we set out to identify whether elements in the 3′UTR of eIF4E were used for HuR binding and whether these were necessary and sufficient for HuR-induced stabilization of eIF4E. Here we used a RNA pulldown assay where biotinylated RNA probes corresponding to the 5′UTR, coding region, or 3′UTR of eIF4E were incubated with U2Os cell extracts. For a negative control, we used the GAPDH 3′UTR probe. In these experiments, HuR associated exclusively with the eIF4E 3′UTR probe. Notably, all of the probes associated with hnRNP A1, indicating that they interacted with the RNA-binding proteins in the conditions we used in the pulldown assay (Fig. 3A). RT-qPCR analysis revealed a marked enrichment of LacZ/4E3′UTR transcripts in the HuR immunoprecipitation compared to the IgG control, while there was no apparent difference in the enrichment of LacZ RNA between IgG and HuR immunoprecipitations (Fig. 3C). These findings were confirmed by sqRT-PCR (see Fig. S4B in the supplemental material).
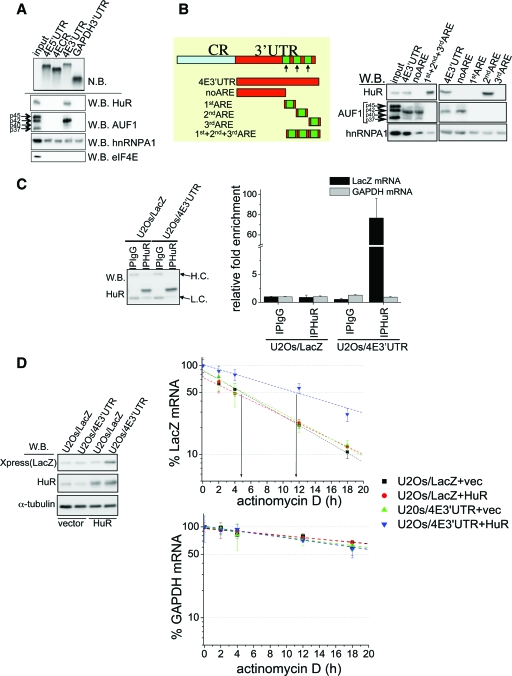
FIG. 3.
The 3′UTR of eIF4E is sufficient for both HuR binding and HuR-mediated stabilization of eIF4E transcripts. (A) Proteins pulled down with biotinylated RNA probes corresponding to the 5′UTR (4E5′UTR), coding region (4ECR), 3′UTR of eIF4E (4E3′UTR), or 3′UTR of GAPDH (GAPDH3′UTR) were visualized by Western blotting (W.B.) using the indicated antibodies. Biotinylated probes were visualized by Northern blotting (N.B.). (B) (Left) Schematic representations of the RNA probes used in the RNA pulldown assay. Green boxes show the positions of putative HuR-AREs. CR, coding region. (Right) Pulled down proteins were visualized by Western blotting using the indicated antibodies. The positions of individual AUF1 isoforms are indicated by the arrows to the left of the blots. (C) RIP reactions carried out with the anti-HuR antibody (IPHuR) or with the control IgG (IPIgG). Western blotting indicates the same efficiency of HuR immunoprecipitation in U2Os/LacZ and U2Os/4E3′UTR cells. The positions of heavy chain (H.C.) and light chain (L.C.) are indicated by arrows to the right of the blot. The amount of GAPDH and LacZ mRNA in each immunoprecipitation was determined by RT-qPCR. Results are presented as changes in enrichment in HuR RIPs compared to IgG RIPs. Values are means ± standard deviations (SD) (error bars) (n = 3). (D) Stability of LacZ/4E3′UTR transcripts is affected in an HuR-dependent manner. (Left) Expression of LacZ, HuR, and α-tubulin in U2Os/LacZ and U2Os/4E3′UTR cells transfected with HuR or the empty vector (vec) were determined by Western blotting. (Right) The same cells were treated with actinomycin D, and the amount of LacZ and GAPDH mRNA at each time point was determined by RT-qPCR (see Materials and Methods). Values are means ± SD (error bars) (n = 3).
To map the HuR binding site in the 3′UTR of eIF4E mRNA, we generated biotinylated RNA probes corresponding to the fragments schematically depicted in Fig. 3B. In this assay, HuR was exclusively pulled down with the probes containing the 2ndARE (i.e., 1st+2nd+3rd ARE and 2ndARE) in amounts comparable to those of the full-length 3′UTR (4E3′UTR) (Fig. 3B). These results were corroborated in RNA pulldown assays where U2Os cell extracts were replaced with a recombinant, purified GST-HuR. GST-HuR specifically associated with the 2ndARE, whereas noARE and the 1stARE and 3rdARE probes failed to pull it down (see Fig. S4A in the supplemental material). Thus, HuR directly binds to a 73-nucleotide stretch comprising the 2ndARE. In summary, the 2ndARE within the 3′UTR of eIF4E is necessary and sufficient for association with HuR.
To determine whether this 73-nucleotide element was sufficient for enhancing stabilization, we examined the stability of LacZ chimeric constructs with the eIF4E 3′UTRs cloned downstream of LacZ (U2Os/4E3′UTR) or with the various ARE constructs generated above. We first carried out RIP with either an anti-HuR antibody or a control IgG to ensure that LacZ chimeric constructs bound HuR (Fig. 4A) as expected from our RNA pulldown results (Fig. 3B). Comparable amounts of HuR immunoprecipitated itself in all experiments, enabling us to directly compare the results obtained from these two cell lines (see Fig. S6A in the supplemental material).
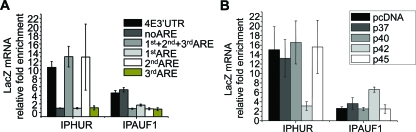
FIG. 4.
HuR binds to a distinct region within the 3′UTR of eIF4E, and the p42AUF1 AUF1 isoform disrupts this interaction through association with the 3′UTR of eIF4E. (A) RIP analysis of the lysates obtained from U20s cells transfected with the chimeric LacZ construct harboring the indicated eIF4E 3′UTR fragments. IPHUR, RIP with an anti-HuR antibody; IPAUF1, RIP with a panisoform-specific anti-AUF1 antibody. The amount of LacZ mRNA in each RIP was determined by RT-qPCR and represented as change in enrichment relative to IgG. Values are means ± standard deviations (SD) (error bars) (n = 3). (B) Amount of LacZ mRNA in the material immunoprecipitated with anti-HuR (IPHUR) or with panisoform-specific anti-AUF1 antibody (IPAUF1) from U2Os/4E3′UTR cells transiently overexpressing individual AUF1 isoforms was determined by RT-qPCR. For each RIP, the values obtained were normalized against the corresponding inputs and presented as changes in enrichment compared to the appropriate IgG RIP. Values are means ± SD (error bars) (n = 3).
Next, we examined mRNA stability of these constructs as a function of HuR overexpression or knockdown as described above. There was no apparent change in the half-life of LacZ mRNA in control and HuR-overexpressing cells (Fig. 3D). In contrast, HuR increased the half-life of LacZ/4E3′UTR mRNA compared to vector controls (Fig. 3D). The knockdown of HuR levels by siRNA consistently resulted in shortening of the half-life of LacZ/4E3′UTR, without affecting the stability of LacZ mRNA (see Fig. S4C in the supplemental material). The alterations in the stability of LacZ/4E3′UTR mRNA were paralleled by the expected changes in LacZ protein levels (Fig. 3D) (see Fig. S4C in the supplemental material). Thus, the 3′UTR is sufficient for enabling HuR binding and HuR-mediated mRNA stabilization.
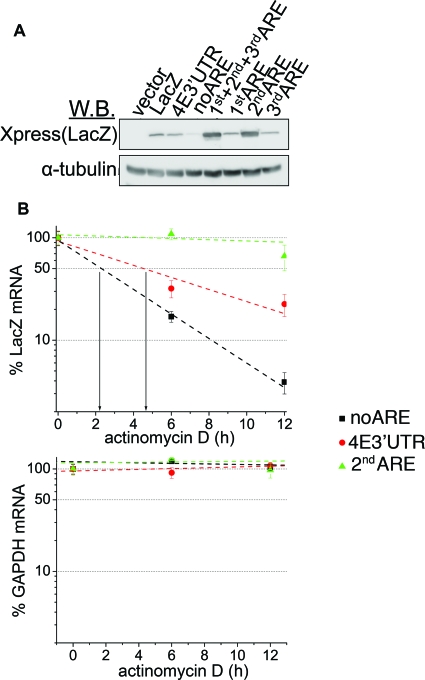
We investigated whether the 2ndARE is sufficient to act as an RNA-stabilizing element. The levels of LacZ protein in chimeras carrying the noARE fragment were markedly decreased compared to the levels in cells expressing LacZ/4E3′UTR (Fig. 5A). Further, expression of chimeras carrying the 2ndARE (i.e., 1st+2nd+3rd ARE and 2ndARE) led to the upregulation of LacZ protein levels compared to cells expressing LacZ/4E3′UTR (Fig. 5A). To ascertain whether these effects were due to the alteration of transcript stability, we determined the half-lives of LacZ/4E3′UTR, LacZ/noARE, and LacZ/2ndARE transcripts in U2Os cells (Fig. 5B). As expected, LacZ/2ndARE has a substantially longer half-life than LacZ/noARE does. However, it is interesting that it also has a longer half-life than lacZ/4E3′UTR does, which suggests that the eIF4E 3′UTR also contains a destabilizing mRNA element(s) (Fig. 5B).
FIG. 5.
The 2ndARE is an RNA-stabilizing element. (A) U2Os cells were transfected with vector, LacZ, or with the indicated LacZ chimera. The levels of LacZ (Xpress) and α-tubulin (used as a loading control) were monitored by Western blotting (W.B.). (B) U2Os cells transfected with LacZ/4E3′UTR, LacZ/noARE, and LacZ/2ndARE were treated with actinomycin D for the indicated time period. The amount of LacZ and GAPDH mRNA for each time period was determined by RT-qPCR (see Materials and Methods). Values are means ± standard deviations (error bars) (n = 3).
p42AUF1 interacts with the 3′UTR of eIF4E and decreases the stability of eIF4E mRNA.
Two key findings led us to investigate whether eIF4E mRNA is regulated by not only enhancing but also reducing transcript stability. First, in the absence of HuR overexpression, LacZ and LacZ/4E3′UTR mRNAs have comparable half-lives (Fig. 3D). Hence, the 3′UTR itself does not affect the stability of eIF4E mRNA. This can be explained by a model where a destabilizing RBP(s) antagonizes the effects of HuR. Further, LacZ/2ndARE constructs are more stable than LacZ/4E3′UTR constructs are (Fig. 5B). Together, these findings suggest that the posttranscriptional regulation of eIF4E, in its totality, is likely mediated by competition between stabilizing and destabilizing interactions. AUF1 is known to destabilize transcripts containing AREs. Thus, we investigated its association here. AUF1 is expressed as four different isoforms (i.e., p37AUF1, p40AUF1, p42AUF1, and p45AUF1) (33). Using an RNA pulldown assay, we found that AUF1 (mainly p42AUF1) interacts with the 3′UTR of eIF4E (Fig. 3A). Furthermore, eIF4E mRNA was markedly enriched in AUF1 immunoprecipitations (see Fig. S5A in the supplemental material). Finally, AUF1 immunoprecipitated with LacZ/4E3′UTR, but not with LacZ mRNA (see Fig. S5B in the supplemental material).
To test whether AUF1 affects the expression of eIF4E, we first depleted the levels of all AUF1 isoforms using a panisoform-specific siRNA. Surprisingly, reduction of AUF1 expression failed to affect eIF4E protein levels (see Fig. S5C in the supplemental material). For a positive control, we show that the AUF1 RNAi treatment elevated the levels of one of its established targets, cyclin D1 (15) (see Fig. S5C in the supplemental material).
Previous studies indicate that there are AUF1 isoform-specific effects. Thus, we transfected U2Os cells with the individual AUF1 isoforms in order to assess their effects on the expression of eIF4E. Importantly, all of the AUF1 isoforms were expressed at similar levels, thereby enabling us to directly compare results between different transfectants (Fig. 6A). Overexpression of p42AUF1 decreased eIF4E protein levels relative to cells transfected with other AUF1 isoforms or vector (Fig. 6A). Notably, p42AUF1 was the isoform which most prominently associated with the 3′UTR of eIF4E in the RNA pulldown assay (Fig. 3A and B). Overexpression of the p37AUF1 isoform resulted in upregulation of eIF4E protein expression (Fig. 6A), thereby providing an explanation as to why the panisoform-specific AUF1 siRNA did not affect eIF4E levels (see Fig. S5C in the supplemental material). This is consistent with previous studies suggesting that individual AUF1 isoforms can have different and even opposing roles (25).
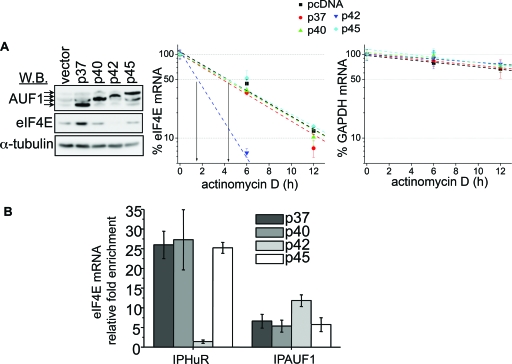
FIG. 6.
p42AUF1 disrupts HuR-eIF4E mRNA interaction and decreases the half-life of eIF4E mRNA. (A) (Left) Western blot (W.B.) showing the expression of AUF1, eIF4E, and α-tubulin (used as a loading control) in U2Os cell lines transfected with the empty vector or with the individual AUF1 isoforms (p37, p40, p42, and p45; indicated by arrows). (Right) These cells were treated with actinomycin D for the indicated time. The amount of eIF4E and GAPDH mRNA was determined by RT-qPCR (see Materials and Methods). Values are means ± standard deviations (SD) (error bars) (n = 3). (B) RIPs were carried out in the lysates obtained from U20s cells transfected with individual AUF1 isoforms (p37, p40, p42, and p45) using panisoform-specific AUF1 antibody (IPAUF1), anti-HuR antibody (IPHuR), or appropriate IgG controls. The amount of eIF4E mRNA in each RIP was determined by RT-qPCR and represented as change in enrichment relative to the corresponding IgG. Values are means ± SD (error bars) (n = 3).
Since two isoforms of AUF1 affected the levels of eIF4E protein, we examined the effects of specific isoforms on eIF4E mRNA stability. For these studies, U2Os cells were transfected with the individual AUF1 isoforms and subsequently treated with actinomycin D. Overexpression of p42AUF1 markedly decreased the half-life of eIF4E mRNA compared to the overexpression of all other isoforms. As expected, none of the AUF1 isoforms affected the stability of GAPDH transcripts (15) (Fig. 6A) (see Fig. S5D in the supplemental material).
Finally, we examined whether p42AUF1-mediated destabilization of eIF4E transcripts required the 3′UTR of eIF4E by overexpressing individual AUF1 isoforms in U2Os/LacZ and U2Os/4E3′UTR cells. Overexpression of p42AUF1 substantially decreased the protein levels of LacZ/4E3′UTR compared to LacZ cells or to overexpression of other AUF1 isoforms (see Fig. S5E in the supplemental material). p42AUF1 overexpression consistently prominently decreased the stability of LacZ/4E3′UTR transcripts (see Fig. S5E in the supplemental material). Thus, we conclude that p42AUF1 binds to the 3′UTR of eIF4E mRNA and in contrast to HuR, decreases the stability of eIF4E transcripts. Of note, p37AUF1 failed to affect the stability of eIF4E mRNA (Fig. 6A).
Overexpression of the p42AUF1 isoform disrupts the HuR-eIF4E mRNA complex.
At this point, we hypothesized that HuR and p42AUF1 alter the stability of eIF4E transcripts by competing for binding to its 3′UTR. In order to test this, we overexpressed the individual AUF1 isoforms in U2Os cells and assessed their effect on the HuR-eIF4E mRNA interaction by RIP (Fig. 6B) (see Fig. S6B in the supplemental material). Overexpression of p42AUF1 markedly decreased the amount of eIF4E mRNA immunoprecipitating with HuR relative to overexpression of other AUF1 isoforms (Fig. 6B). More eIF4E transcripts were consistently found in the immunoprecipitated fraction when a panisoform-specific anti-AUF1 antibody was used in p42AUF1-overexpressing cells relative to other isoforms (Fig. 6B). Moreover, p42AUF1 overexpression reduced the levels of LacZ/4E3′UTR mRNA in HuR immunoprecipitation and increased the levels of these transcripts in AUF1 immunoprecipitation (Fig. 4B) These findings indicate that p42AUF1 binds to the 3′UTR of eIF4E and disrupts its association with HuR.
Given that p42AUF1 competes for HuR binding of eIF4E transcripts, we examined whether these proteins bound overlapping sequences in the 3′UTR. Interestingly, p42AUF1 was pulled down with the probe lacking the three AREs (i.e., noARE) and failed to associate with any of the probes derived from the region that contains the HuR-binding element or the other AREs (Fig. 3B). Therefore, despite the fact that the p42AUF1 isoform abrogates the HuR-eIF4E mRNA interactions, these proteins bind to distinct regions within the primary sequence of the 3′UTR of eIF4E. RIP assays with LacZ chimeras corresponding to the RNA probes used in the pulldown assays consistently indicated that the p42AUF1 isoform does not bind the ARE-containing region, whereas HuR does (Fig. 4A) (see Fig. S6A in the supplemental material). Thus, HuR and p42AUF1 modulate eIF4E mRNA stability through distinct elements in the 3′UTR of eIF4E.
eIF4E and HuR collaboratively regulate gene expression.
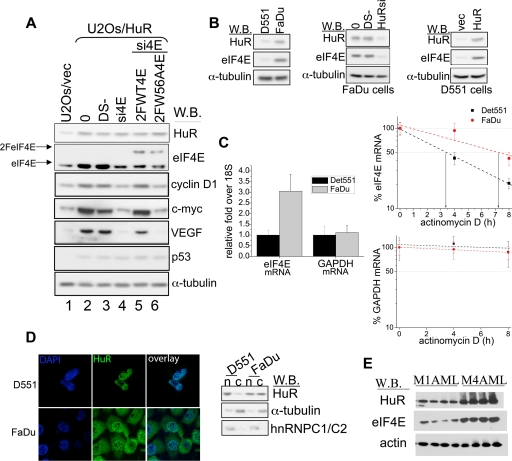
Given that HuR stabilizes eIF4E and that HuR and eIF4E target common transcripts, we decided to investigate whether these two important posttranscriptional regulators collaboratively regulate the expression of common target genes. We further investigated whether some of the effects of HuR on gene expression could be mediated through eIF4E. To test this possibility, we depleted the levels of eIF4E by RNAi in U2Os/HuR cells and monitored the expression of cyclin D1, VEGF, and c-myc by Western blotting. Depletion of eIF4E levels resulted in a prominent decrease in the levels of these proteins compared to double-scrambled controls (DS) (Fig. 7A, compare lanes 3 and 4). Importantly, knockdown of eIF4E levels did not affect the expression of p53, which is a well-established translational target of HuR (21) but not a target of eIF4E. In order to test the specificity of eIF4E siRNA, we transfected siRNA-treated cells with eIF4E siRNA-insensitive constructs. Overexpression of wild-type eIF4E rescued the levels of cyclin D1, c-myc, and VEGF protein expression to levels detected in DS-treated cells. As expected, the inactive form of eIF4E (W56A) failed to rescue the expression of all three proteins (Fig. 7A, lanes 5 and 6). These data indicate that the HuR-induced upregulation of cyclin D1, VEGF, and c-myc depends, at least in part, on eIF4E levels.
FIG. 7.
eIF4E and HuR are functionally linked. (A) Western blot (W.B.) analysis of the expression of indicated proteins in U2Os/vec (lane 1) or U2Os/HuR cells (lanes 2 to 6). U2Os/HuR cells were treated with Lipofectamine (0), double-scrambled negative control (DS−), or with eIF4E siRNA (si4E). Upon si4E treatment, cells were transfected with RNAi-insensitive flag-tagged eIF4E constructs (2FeIF4E) coding for wild-type (2FWT4E; lane 5) or inactive mutant of eIF4E (2FW56A4E; lane 6). (B) Western blot analysis of HuR, eIF4E, and α-tubulin (used as a loading control) in Detroit 551 (D551) and FaDu cells (left panel), untreated (0), DS- or HuR siRNA (HuRsi)-treated FaDu cells (middle panel), and Detroit 551 (D551) cells transfected with an empty vector (vec) or with HuR (right panel). (C) (Left) Total steady-state levels of eIF4E and GAPDH mRNA in D551 and FaDu cells were determined by RT-qPCR and normalized against 18S values. Results are presented as changes in the difference between D551 (set at 1) and FaDu cells. (Right) FaDu and Detroit 551 (D551) cells were treated with actinomycin D. The amount of eIF4E and GAPDH mRNA for each time point was determined by RT-qPCR (see Materials and Methods). Values are means ± standard deviations (error bars) (n = 3). (D) (Left) Representative confocal micrographs of Detroit 551 (D551) and FaDu cells stained with the HuR antibody (green). Nuclei were counterstained with DAPI (blue). (Right) Western blot showing the nuclear (n) and cytoplasmic (c) levels of HuR protein in FaDu and D551 cells. hnRNP C1/C2 and α-tubulin were used as the nuclear and cytoplasmic marker, respectively. (E) Levels of HuR, eIF4E, and actin (served as a loading control) in CD34+ cells isolated from the bone marrow of M1 and M4 AML patients were monitored by Western blotting (W.B.).
Elevated eIF4E levels in human cancers is functionally correlated with elevated HuR levels.
In order to investigate whether HuR-mediated upregulation of eIF4E levels is relevant to elevated levels of eIF4E in cancer, we investigated whether tumors characterized by elevated eIF4E expression show increased HuR protein levels. FaDu cells, derived from a hypopharyngeal squamous cell carcinoma, are characterized by elevated levels of eIF4E, compared to a healthy (noncancerous) skin fibroblast cell line, Detroit 551 (7). We observe that HuR levels were markedly elevated in FaDu cells relative to Detroit 551 cells (Fig. 7B, left panel). Increased HuR expression in FaDu cells correlated with increased steady-state levels of eIF4E mRNA and with elevated levels of eIF4E protein (Fig. 7A and C). As expected, overexpression of HuR in Detroit 551 cells led to a marked increase in eIF4E protein levels relative to vector controls (Fig. 7B, right panel), consistent with the above studies. siRNA-mediated knockdown of HuR in FaDu cells consistently led to a downregulation of eIF4E protein levels (Fig. 7B, middle panel).
Upon activation, HuR shifts from the nucleus to the cytoplasm (2). The cytoplasmic accumulation of HuR leads to stabilization of its target mRNAs. To test the potential differences in the subcellular distribution of HuR in FaDu and Detroit 551 cells, we fractionated the cells and determined the amount of HuR in the nuclear and cytoplasmic fractions by Western blot analysis. FaDu cells had comparable levels of HuR in both compartments, while in Detroit 551 cells, HuR was mostly found in the nucleus. These findings were confirmed by immunofluorescence (Fig. 7D). Importantly, elevated levels and cytoplasmic redistribution of HuR in FaDu cells correlated with the increased stability of eIF4E transcripts relative to Detroit 551 cells (Fig. 7C). Hence, in FaDu cells, high levels and cytoplasmic redistribution of HuR correlate with the increased stability of eIF4E transcripts and with elevated eIF4E protein levels. These findings strongly suggest that eIF4E overexpression in tumors could be mediated by HuR.
M4 and M5 subtypes of AML are characterized by elevated eIF4E levels (31). Thus, we investigated the expression of HuR and eIF4E proteins in these primary AML specimens. In CD34+ M4 AML cells, high levels of eIF4E correlated with the overexpression of HuR, while the CD34+ M1 AML cells, with normal levels of eIF4E (31), had levels of HuR similar to those of healthy controls (Fig. 7E). Taken together, these data suggest that HuR regulates the levels of eIF4E in histologically distinct malignancies (i.e., AML and HNSCC) characterized by elevated eIF4E levels. In this way, HuR-induced upregulation of eIF4E expression could be a global phenomenon in the etiology of eIF4E-dependent oncogenesis.
DISCUSSION
Little is known about the regulation of eIF4E expression. Cellular factors such as c-myc and p53 have been implicated as transcriptional regulators of eIF4E expression (16, 34). Our present study reveals that eIF4E expression is also modulated at the level of mRNA stability, which is achieved through the competition between HuR and p42AUF1 for binding to the 3′UTR of eIF4E mRNA. Interestingly, p42AUF1 and HuR associate with eIF4E transcripts through distinct, nonoverlapping regions, indicating that they do not bind to the same site with comparable affinity. This is consistent with previous findings, showing that AUF1 and HuR could simultaneously, as well as competitively, bind to p21 and cyclin D1 mRNA (15).
A large body of data indicates that the increase in eIF4E activity in cancer stems from its overexpression (11, 19). However, the factors that lead to upregulation of eIF4E expression in cancer remain largely elusive. Here, we show that HuR levels are increased in two unrelated malignancies characterized by elevated eIF4E levels (i.e., HNSCC and AML). Furthermore, in an HNSCC cell line, high levels of HuR correlated with increased stability of eIF4E transcripts, suggesting that HuR-induced upregulation of eIF4E is a plausible mechanism that underlies overexpression of eIF4E in human tumors. This is in addition to the role that gene amplification may play in HNSCC. Thus, several mechanisms may simultaneously contribute to the elevation of eIF4E in cancer cells. Moreover, our data indicate that eIF4E and HuR are positioned to collaboratively regulate the expression of several factors that play an important role in oncogenesis (i.e., cyclin D1 and c-myc) and neoangiogeneisis (i.e., VEGF). Taken together, these findings suggest that HuR plays an important role in eIF4E-mediated oncogenesis and vice versa. Accordingly, the aberrant activation of the posttranscriptional network comprised of eIF4E and HuR could provide the changes in the proteome that favor proliferation and survival of malignant cells.
In summary, we demonstrate a novel mode for the control of eIF4E RNA levels that depends on the interplay between HuR and p42AUF1. Elevation of HuR levels appears to contribute to the elevation of eIF4E observed in cancer cells. Further, we provide evidence that HuR and eIF4E coregulate the expression of transcripts, indicating that their mutual dysregulation will have a profound impact on the proteome of associated cancer cells.
Supplementary Material
Acknowledgments
We are grateful to Serafin Piñol-Roma (anti-hnRNP A1 [4B10] and anti-hnRNP C1/C2 [4F4] antibodies), Jack Keene (3A2 anti-HuR antibody and pcDNA/HuR construct), and Gary Brewer (pcDNA/AUF1 constructs). We thank Christian Charbonneau for help with microscopy, Matthew Friedersdorf for help with the COVELs algorithm, and Jack D. Keene for helpful discussions.
This work is supported by funding from NIH (RO1 R56 98571) to K.L.B.B. She holds a Canada Research Chair in Molecular Biology of the Cell Nucleus. I.T. is a Special Fellow of the Leukemia and Lymphoma Society of the United States. T.H. is supported by grants from the Canadian Institute for Health Research (CIHR) and from the National Cancer Institute of Canada. She holds a Canada Research Chair in Cell Differentiation and the Genetics of Acute Leukemias. M.T. is supported by a studentship award from the Fonds de Recherche en Santé du Québec and the CIHR.
Footnotes
Published ahead of print on 29 December 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Barreau, C., L. Paillard, and H. B. Osborne. 2005. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 337138-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Bush, A., M. Mateyak, K. Dugan, A. Obaya, S. Adachi, J. Sedivy, and M. Cole. 1998. c-myc null cells misregulate cad and gadd45 but not other proposed c-Myc targets. Genes Dev. 123797-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Culjkovic, B., I. Topisirovic, and K. L. Borden. 2007. Controlling gene expression through RNA regulons: the role of the eukaryotic translation initiation factor eIF4E. Cell Cycle 665-69. [DOI] [PubMed] [Google Scholar]
- 4.Culjkovic, B., I. Topisirovic, L. Skrabanek, M. Ruiz-Gutierrez, and K. L. Borden. 2006. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J. Cell Biol. 175415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Culjkovic, B., I. Topisirovic, L. Skrabanek, M. Ruiz-Gutierrez, and K. L. Borden. 2005. eIF4E promotes nuclear export of cyclin D1 mRNAs via an element in the 3′UTR. J. Cell Biol. 169245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Benedetti, A., and J. R. Graff. 2004. eIF-4E expression and its role in malignancies and metastases. Oncogene 233189-3199. [DOI] [PubMed] [Google Scholar]
- 7.DeFatta, R. J., C. O. Nathan, and A. De Benedetti. 2000. Antisense RNA to eIF4E suppresses oncogenic properties of a head and neck squamous cell carcinoma cell line. Laryngoscope 110928-933. [DOI] [PubMed] [Google Scholar]
- 8.Fan, X. C., and J. A. Steitz. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 173448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallouzi, I. E., C. M. Brennan, M. G. Stenberg, M. S. Swanson, A. Eversole, N. Maizels, and J. A. Steitz. 2000. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. USA 973073-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68913-963. [DOI] [PubMed] [Google Scholar]
- 11.Graff, J. R., and S. G. Zimmer. 2003. Translational control and metastatic progression: enhanced activity of the mRNA cap-binding protein eIF-4E selectively enhances translation of metastasis-related mRNAs. Clin. Exp. Metastasis 20265-273. [DOI] [PubMed] [Google Scholar]
- 12.Jones, R. M., J. Branda, K. A. Johnston, M. Polymenis, M. Gadd, A. Rustgi, L. Callanan, and E. V. Schmidt. 1996. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol. Cell. Biol. 164754-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keene, J. D., J. M. Komisarow, and M. B. Friedersdorf. 2006. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 1302-307. [DOI] [PubMed] [Google Scholar]
- 14.Koromilas, A. E., A. Lazaris-Karatzas, and N. Sonenberg. 1992. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 114153-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lal, A., K. Mazan-Mamczarz, T. Kawai, X. Yang, J. L. Martindale, and M. Gorospe. 2004. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 233092-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, C. J., R. Cencic, J. R. Mills, F. Robert, and J. Pelletier. 2008. c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res. 685326-5334. [DOI] [PubMed] [Google Scholar]
- 17.Lopez de Silanes, I., M. Zhan, A. Lal, X. Yang, and M. Gorospe. 2004. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. USA 1012987-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma, W. J., S. Cheng, C. Campbell, A. Wright, and H. Furneaux. 1996. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 2718144-8151. [DOI] [PubMed] [Google Scholar]
- 19.Mamane, Y., E. Petroulakis, L. Rong, K. Yoshida, L. W. Ler, and N. Sonenberg. 2004. eIF4E-from translation to transformation. Oncogene 233172-3179. [DOI] [PubMed] [Google Scholar]
- 20.Marcotrigiano, J., A. C. Gingras, N. Sonenberg, and S. K. Burley. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89951-961. [DOI] [PubMed] [Google Scholar]
- 21.Mazan-Mamczarz, K., S. Galban, I. Lopez de Silanes, J. L. Martindale, U. Atasoy, J. D. Keene, and M. Gorospe. 2003. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. USA 1008354-8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mili, S., and J. A. Steitz. 2004. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 101692-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montanaro, L., and P. P. Pandolfi. 2004. Initiation of mRNA translation in oncogenesis: the role of eIF4E. Cell Cycle 31387-1389. [DOI] [PubMed] [Google Scholar]
- 24.Peng, S. S., C. Y. Chen, N. Xu, and A. B. Shyu. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 173461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raineri, I., D. Wegmueller, B. Gross, U. Certa, and C. Moroni. 2004. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 321279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenwald, I. B., A. Lazaris-Karatzas, N. Sonenberg, and E. V. Schmidt. 1993. Elevated levels of cyclin D1 protein in response to increased expression of eukaryotic initiation factor 4E. Mol. Cell. Biol. 137358-7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorrells, D. L., D. R. Black, C. Meschonat, R. Rhoads, A. De Benedetti, M. Gao, B. J. Williams, and B. D. Li. 1998. Detection of eIF4E gene amplification in breast cancer by competitive PCR. Ann. Surg. Oncol. 5232-237. [DOI] [PubMed] [Google Scholar]
- 28.Sorrells, D. L., Jr., G. E. Ghali, A. De Benedetti, C. A. Nathan, and B. D. Li. 1999. Progressive amplification and overexpression of the eukaryotic initiation factor 4E gene in different zones of head and neck cancers. J. Oral Maxillofac. Surg. 57294-299. [DOI] [PubMed] [Google Scholar]
- 29.Takagi, M., M. J. Absalon, K. G. McLure, and M. B. Kastan. 2005. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 12349-63. [DOI] [PubMed] [Google Scholar]
- 30.Topisirovic, I., A. D. Capili, and K. L. Borden. 2002. Gamma interferon and cadmium treatments modulate eukaryotic initiation factor 4E-dependent mRNA transport of cyclin D1 in a PML-dependent manner. Mol. Cell. Biol. 226183-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Topisirovic, I., M. L. Guzman, M. J. McConnell, J. D. Licht, B. Culjkovic, S. J. Neering, C. T. Jordan, and K. L. Borden. 2003. Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes hematopoietic differentiation and contributes to leukemogenesis. Mol. Cell. Biol. 238992-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, W., M. C. Caldwell, S. Lin, H. Furneaux, and M. Gorospe. 2000. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 192340-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson, G. M., and G. Brewer. 1999. The search for trans-acting factors controlling messenger RNA decay. Prog. Nucleic Acid Res. Mol. Biol. 62257-291. [DOI] [PubMed] [Google Scholar]
- 34.Zhu, N., L. Gu, H. W. Findley, and M. Zhou. 2005. Transcriptional repression of the eukaryotic initiation factor 4E gene by wild type p53. Biochem. Biophys. Res. Commun. 3351272-1279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.