Abstract
The molecular complexes involved in the nonhomologous end-joining process that resolves recombination-activating gene (RAG)-induced double-strand breaks and results in V(D)J gene rearrangements vary during mammalian ontogeny. In the mouse, the first immunoglobulin gene rearrangements emerge during midgestation periods, but their repertoires have not been analyzed in detail. We decided to study the postgastrulation DJH joints and compare them with those present in later life. The embryo DJH joints differed from those observed in perinatal life by the presence of short stretches of nontemplated (N) nucleotides. Whereas most adult N nucleotides are introduced by terminal deoxynucleotidyl transferase (TdT), the embryo N nucleotides were due to the activity of the homologous DNA polymerase μ (Polμ), which was widely expressed in the early ontogeny, as shown by analysis of Polμ−/− embryos. Based on its DNA-dependent polymerization ability, which TdT lacks, Polμ also filled in small sequence gaps at the coding ends and contributed to the ligation of highly processed ends, frequently found in the embryo, by pairing to internal microhomology sites. These findings show that Polμ participates in the repair of early-embryo, RAG-induced double-strand breaks and subsequently may contribute to preserve the genomic stability and cellular homeostasis of lymphohematopoietic precursors during development.
The adaptive immune system is characterized by the great diversity of its antigen receptors, which result from the activities of enzymatic complexes that cut and paste the genomic DNA of antigen receptor loci. The nonhomologous end-joining (NHEJ) machinery is then recruited to repair the double-strand DNA breaks (DSBs) inflicted by the products of the recombination-activating genes (RAGs) (45, 65). Within B cells, each immunoglobulin (Ig) receptor represents a singular shuffling of two heavy (H) and two light (L) chains, which are derived from the recombination of V, D, and J gene segments of the IgH locus and of V and J for IgL (71). Besides these combinatorial possibilities, most Ig variability derives from extensive processing of the coding ends, including exonucleolytic trimming of DNA ends, together with the addition of palindromic (P) nucleotides templated by the adjacent germ line sequence and of nontemplated (N) nucleotides secondary to the activity of the terminal deoxynucleotidyl transferase (TdT), a lymphoid-specific member of family X of DNA polymerases (reviewed in reference 56). During B-lineage differentiation, IgH rearrangements occur before those of the IgL locus, and D-to-JH rearrangements precede V-to-DJH rearrangements (62). DJH joints are formed in any of the three open reading frames (ORFs). ORF1 is predominantly used in mature Igs, ORF2 is transcribed as a Dμ protein that provides negative signals to the B-cell precursors, and ORF3 frequently leads to stop codons (32, 33, 37). Germ line V, D, and J gene segments display short stretches of mutually homologous nucleotides (SSH), which are frequently used in gene rearrangements during perinatal periods, when N additions are absent (27, 32, 55, 57). The actual Ig V-region repertoires represent both the results of the NHEJ process associated with genomic VDJ recombination and those of antigen-independent and -dependent selection events. Although the core NHEJ components (Ku-Artemis-DNA-PK and XLF-XRCC4-DNA ligase IV) are by themselves able to join RAG-induced, incompatible DNA ends, family X DNA polymerases can be recruited to fill gaps created by imprecise coding ends with 3′ overhangs (DNA polymerase μ [Polμ] and Polλ) and/or to promote diversity through the addition of N nucleotides (TdT) (34, 56).
The lymphoid differentiation pathways and clonotypic repertoires are developmentally regulated and differ between the embryo-fetal and adult periods (2, 44, 68). The perinatal B cells result from a wave of B lymphopoiesis occurring during the last third of mouse gestation (13, 14, 21, 70). Perinatal VH gene usage differs from that predominating in the adult (1, 69), and the former VDJ joints rarely display N additions, leading to V-region repertoires enriched in multi- and self-reactive specificities (36, 40). The program of B-cell differentiation starts at embryonic days 10 to 11 (E10 to E11) in embryo hematopoietic sites, after the emergence of multipotent progenitors (at E8.5 to E9.5) (18, 19, 23, 31, 51, 73). DJH rearrangements were detected in these early embryos, whereas full VDJH sequences were not observed before E14 (14, 18, 51, 66), when VJκ rearrangements were also found (63). The earliest mouse DJH/VDJH Ig sequences analyzed to date corresponded to late fetuses (E16) (14, 53). We reasoned that the true baseline of the Ig rearrangement process occurs in midgestation embryos, when the first DJHs are not yet transcribed and, consequently, not subjected to selection and are conditioned only for the evolutionarily established and developmentally regulated usage of distinct NHEJ machineries.
We report here the sequence profiles of the earliest embryo E10 to E12 DJH joints. Unexpected frequencies of embryonic DJH joints bearing N nucleotides, in the absence of detectable TdT expression, were found. Moreover, the embryo DJH joints lacking N nucleotides (N−) used fewer SSH to recombine than newborn DJHs, and these SSH were widely dispersed along the embryo D sequences, in contrast to the most joint-proximal ones, which predominated in newborn DJHs. Considering that Polμ is the closest relative of TdT (42% amino acid identity) (22), which is able to introduce N nucleotides in vitro (4, 22, 34, 39, 49) and to join DNA ends with minimal or even null complementarity (17, 58), and that it is expressed in early-embryo organs, we decided to investigate its putative contribution to the first embryo DJH joints. The DJH joints obtained from Polμ−/− embryos (48) showed a significant reduction of N nucleotides compared to wild-type (WT) embryos. Moreover, highly preserved DJH joints (with <3 deleted nucleotides) were selectively depleted in the Polμ−/− mouse embryos, while the remaining DJHs preferentially relied upon longer stretches of homology for end ligation. These findings support the idea that Polμ is active during early-embryo DJH rearrangements and that both its template-dependent and -independent ambivalent functions may be used to fill in small nucleotide gaps generated after asymmetric hairpin nicking and also to extend coding ends via a limited TdT-like activity.
MATERIALS AND METHODS
Embryo microsurgery and cell purifications.
BALB/c, BALB/c.RAG-2−/−, and BALB/c × 129/SV Polμ−/− (nine generations backcrossed to BALB/c) mice were maintained in the animal facilities of the Instituto de Salud Carlos III, Madrid, Spain, and the Centro de Biología Molecular Severo Ochoa, Madrid, Spain. The gestational age was determined by the vaginal plug after overnight mating (E0). Eggs containing the embryo attached to the placenta were washed by sequential transfers through 50-ml phosphate-buffered-saline tubes. After removal of the placenta, the embryo proper and yolk sac were separated and washed out again by passages through 20-ml petri dishes (51). All animal studies were performed in accordance with Spanish animal protection laws. The cells were incubated with anti-CD19-phycoerythrin (1D3), anti-CD45R/B220-allophycocyanin (RA3-6B2), anti-CD45-allophycocyanin (30F11), anti-cKit-fluorescein isothiocyanate (2B8), and anti-IgM-fluorescein isothiocyanate (331.13) antibodies (BD Pharmingen). Nonspecific background was eliminated by incubations with 10% mouse serum and Fc-Block (BD Pharmingen) and controlled with isotype-matched irrelevant antibodies. Cell debris and dead cells were discarded by adjusting light parameters and by propidium iodide staining. Labeled cells were purified in a DIVA cell sorter (BD Pharmingen), the degree of purity was controlled in a FACSCalibur with Cellquest software (BD Pharmingen), and only samples more than 95% pure were processed.
RT-PCR, genomic PCR, cloning, and sequencing.
Genomic DNA and total RNA were extracted, and oligo(dT)-primed cDNA samples were prepared with avian myeloblastosis virus reverse transcriptase (RT), as described previously (51). PCR amplifications were performed with 1 U of AmpliTaq Gold DNA polymerase (Roche Molecular Systems) in a PTC-200 DNA Engine cycler (Bio-Rad). DJH rearrangements were detected with a genomic amplification assay by using the Faststart PCR amplification kit (Roche Molecular Systems). DNA templates corresponding to 103 cells were amplified. For embryo-derived DJH rearrangements, 1 μl of the first PCR amplification reaction mixture was subjected to a nested PCR for 20 additional cycles. The primers and PCR conditions were as indicated in Table S1 in the supplemental material. In the case of newborn-derived samples and to facilitate cloning of the larger DJH structures, nested amplifications were performed by using JH intron-specific oligonucleotides as 3′ primers. The products of the primary (for JH4) or nested (JH1 to -3) PCRs obtained from adult and newborn samples, containing different DJH rearrangements in each of the JH-specific bands of the same size, were cloned using the pMBL-T vector kit (Dominion MBL) and transformed into JM109 Escherichia coli competent cells (Promega). After being plated, colonies were amplified, their products were separated electrophoretically on 2% agarose gels, and the bands were visualized with ethidium bromide and cleaned or gel purified with either PCR clean-up kits or gel spin kits (MoBio Laboratories), respectively. The purified bands were sequenced in an ABI7000 automatic sequencer with the BigDye sequencing mixture (Applied Biosystems).
The relative expression levels of TdT, Polμ, Polλ, and GαS genes were calculated by real-time PCR, performed on the LightCycler 2.0 system, by using the LightCycler FastStart DNA Master SYBR green I kit (Roche). The cycling steps were as follows: 1 cycle of 95°C for 10 min, followed by 45 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for either 15 s (TdT, Polμ, and Polλ) or 30 s (GαS), with a final cycle of 72°C for 5 min, before a melting curve was performed. LightCycler software 4.0 was used to calculate the threshold cycle (CT) of each reaction, and the relative amount of specific cDNA on each sample was determined using the 2−ΔΔCT method (46) by using GαS as the internal control gene. The values obtained for newborn samples were used as calibrators to determine the changes in the specific gene expression levels.
In vitro terminal-transferase assay.
Terminal-transferase-mediated incorporation of a single deoxynucleotide (ddTTP) was assayed as described previously (39) on double-stranded DNA molecules differing in the number of 3′ protruding nucleotides. The strand providing the 3′ overhang (5′-CGCAAGTCAGCGCTACGGG[T]0-5) was 5′ labeled with [γ-32P]ATP and T4 polynucleotide kinase and hybridized to the unlabeled complementary strand (5′-CCCGTAGCGCTGACTTGCG) in the presence of 50 mM Tris-HCl (pH 7.5) and 0.3 M NaCl. The reaction mixture contained 50 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, 1 mM MnCl2, 5 nM of labeled DNA, 200 nM of either human Polμ or TdT, 100 μM ddTTP, 4% glycerol, and 0.1 mg/ml bovine serum albumin in 12.5 μl. After incubation (37°C; 30 min), reactions were stopped by adding gel-loading buffer (95% formamide, 10 mM EDTA, 0.1% [wt/vol] xylene cyanol, and 0.1% [wt/vol] bromophenol blue). The +1 extension products were analyzed by 8 M urea-20% polyacrylamide gel electrophoresis, detected by autoradiography, and quantitated with a phosphorimager.
Statistical analyses.
GraphPad Prism 3.0 software was used. Means, standard deviations, and standard errors of the means (SEM) were calculated, and the normality of the distributions was determined with the Kolmogorov-Smirnov test. Comparisons of means were performed with the two-tailed unpaired t test with the Welch correction, from which P values were derived. For nonparametric samples, one-way analysis of variance with the Kruskal-Wallis test was performed. Contingency tables were used to compare frequencies among experimental groups, using chi-square and Fisher's exact tests to calculate P values. Deletion profiles were obtained by Gaussian kernel density estimation (The R Project for Statistical Computing [http://www.r-project.org]).
Nucleotide sequence accession numbers.
The sequences analyzed in this study are posted as supplemental material (see Fig. S1 to S5 in the supplemental material) and have been deposited in EMBL with the following accession numbers: FM162406 to FM162492 (WT embryos); AM998795 to AM998802, FM161938 to FM161957, and FM161968 to FM161985 (newborns); FM161991 to FM162014 and FM162405 (adults); FM162395 to FM162397 and FM162493 to FM162508 (RAG2+/− embryos); and FM162509 to FM162556 (Polμ−/− embryos).
RESULTS
The first DJH rearrangements of the mouse embryo.
A genomic PCR simultaneously amplified DJH1 to -4 gene rearrangements and revealed DJH-specific bands in hematopoietic organs of synchronized E10 to E12 BALB/c mouse embryos, as described previously (51). Fifty-eight percent of the PCRs performed in samples derived from a mean of 10 embryo explants per experiment were positive, and of these, two-thirds showed a unique DJH band, suggesting that they represented the earliest-appearing rearrangements during postgastrulation periods (Table 1; see Fig. S1 in the supplemental material). From E13 onward, the DJH rearrangements logarithmically expanded in the liver (14, 51). Control DJH groups recovered from 1-day-old livers and spleens and from 3-month-old spleens were included (see Fig. S2 and S3 in the supplemental material). DFL16.1 was the most utilized D gene, although this preference was more pronounced among newborn DJHs. All JH genes were similarly used in embryo and newborn samples, but most adult sequences contained the smallest DJH4 rearrangement, which was more easily cloned when the four DJH rearrangements were present in the PCR samples. Both adult and newborn DJHs preferentially utilized ORF1, whereas E10 to E12 embryo DJHs showed roughly similar frequencies of the three ORFs, indicating their unselected character.
TABLE 1.
Characteristics of DJH joint sequences in BALB/c mouse embryos, newborns, and adults
| Groupa | No. of sequences
|
||
|---|---|---|---|
| Embryo (n = 92) | Newborn (n = 57) | Adult (n = 25) | |
| P-Sp/AGM | 20 | ||
| Liver | 29 | 39 | |
| YS | 33 | ||
| Blood | 10 | ||
| Spleen | 18 | 25 | |
| E10/11/12 | 24/50/18 | ||
| DFL16b | 28 | 45 | 7 |
| DSP2 | 64 | 12 | 18 |
| JH1 | 10 | 15 | 0 |
| JH2 | 21 | 14 | 0 |
| JH3 | 28 | 12 | 2 |
| JH4 | 33 | 16 | 23 |
| ORF1 | 40 | 36 | 14 |
| ORF2 | 23 | 8 | 2 |
| ORF3 | 29 | 13 | 9 |
P-Sp/AGM, para-aortic splanchnopleura/aorta-gonad-mesonephros region; YS, yolk sac.
Two embryo DJH sequences and one adult DJH sequence used the DFL16.2 gene and were not included in the experimental analyses. The DFL16/DSP2 ratios were as follows: embryo, 0.36; newborn, 3.75; adult, 0.39.
E10 to E12 DJH coding joints include N nucleotides.
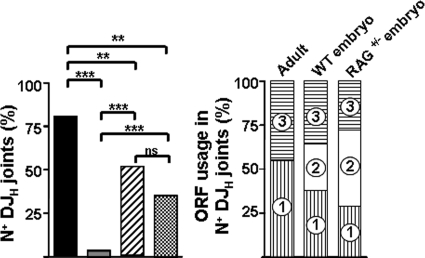
The processing of DJH joints is conditioned by at least four mechanisms: (i) usage of SSH in the recombination of two coding ends, (ii) exonucleolytic nibbling of germ line sequences, (iii) inclusion of templated P nucleotides, and (iv) random addition of N nucleotides by TdT. TdT is highly expressed in adult pro-B cells and hardly or not at all expressed in perinatal lymphoid progenitors (8, 44). In agreement with this, most of the adult spleen DJHs (80%) and only 2 out of 57 newborn DJHs (3.5%) showed N nucleotides. Unexpectedly, 51% of E10 to E12 DJHs included N nucleotides (47 out of 92; P < 0.001 and P < 0.01 with respect to newborn and adult DJHs, respectively) (Fig. 1, left). The lengths of embryo N stretches per N+ sequence were less than those of adult DJHs (3.2 ± 0.3 and 4.6 ± 0.7 for embryo and adult DJHs, respectively; mean ± SEM; P = 0.051). Considering that each exsanguinated embryo (and its dissected organs) was sequentially diluted in large buffer volumes (well over 106 times its own volume), the possibility of surgical contamination with B cells from the adult mice was low. Alternatively, DJH joints could represent mother-derived chimerism, although only a transient wave of maternal macrophages has been detected in E7.5 to E9.5 embryo yolk sacs and no other cell chimerism in fetuses up to E16 (6, 52). Additional support for the embryo origin of the N+ DJHs came from their unbiased ORF usage, while ORF2 was absent among adult N+ DJHs (P < 0.01 compared to embryo ORF2 usage) (Fig. 1, right); the slight preference for ORF1 in total embryo DJHs (Table 1) was actually accounted for by their N− sequences (data not shown). However, to definitively discard a maternal origin of the N+ DJHs, we sequenced DJHs from embryos derived from BALB/c.RAG2−/− females mated to BALB/c males. They also displayed an unselected distribution of ORFs, and 35% of these DJHs showed stretches of N nucleotides (Fig. 1, left; see Fig. S4 in the supplemental material), confirming that the E10 to E12 N+ DJH sequences were of embryonic origin.
FIG. 1.
N nucleotides in DJH joints derived from E10 to E12 BALB/c embryos. On the left, the frequencies of N+ DJH joints in adults (black), newborns (gray), WT embryos (hatched), and RAG2+/− embryos derived from BALB/c male × BALB/c.RAG2−/− female matings (crosshatched) are shown. The distribution of ORFs (circled numbers) among N+ DJH joints of adults and embryo DJH joints is displayed on the right. *, P < 0.05; **, P < 0.01; ***, P < 0.001, ns, not significant.
Developmental shifts of SSH utilization, D recombination sites, and nucleotide deletions in DJH joints.
It is thought that the SSH present in D and JH gene segments promote recombination whenever TdT is absent and N nucleotides are not introduced (28, 57). In agreement with this, 12% of adult and up to 78% of newborn DJHs used such microhomologies. In contrast, both total and N− embryo DJHs showed lower usage of SSH-related recombination sites than the equivalent DJHs of newborns (P < 0.001 and P < 0.01, respectively) (Fig. 2A). The analysis of D recombination sites also revealed distinctive patterns in both embryo and newborn DJHs. The most joint-proximal nucleotide was preferentially used to ligate both embryo and newborn DJHs, yet this recombination point was much more frequent in newborn than in embryo DJHs (30/57 and 35/108 DJH joints, respectively, ligating at position 1 versus total DJH joints [BALB/c and RAG+/− embryos are considered together hereafter]; P < 0.01). Compared to those of newborns, the embryos’ SSH-related recombination sites were distributed along the whole D segment sequence. When these SSH were classified between joint-proximal (first four D nucleotides) and joint-distal groups, the latter appeared more frequently in embryo than in newborn DJHs (P < 0.01) (Fig. 2B, left). The coding-end sequences may influence the DJH joint-processing events (3, 24, 29, 43, 74); however, the observed embryo/newborn differences applied equally to DJHs using the single DFL16.1 gene (P < 0.01) and to the DSP2 family members (P < 0.05) (Fig. 2B, right). The reduced numbers of DSP2 gene-specific newborn DJHs precluded a gene-by-gene comparison with the equivalent embryo DJHs. The increased utilization of joint-distal D recombination positions in embryo DJHs resulted in larger DJH nucleotide deletions: Both DFL16.1+ and DSP2+ newborn DJH coding ends were more “protected” than the equivalent embryo DJHs (P < 0.01) (Fig. 2C). We conclude that (i) embryo N− DJH joints used lower frequencies of SSH than newborn N− DJHs, (ii) embryo DJHs frequently rearranged to joint-distal SSH of D segments, and (iii) embryo DJHs supported larger nucleotide deletions than newborn DJHs.
FIG. 2.
Developmental shifts of SSH usage, of D recombination sites, and of nucleotide deletions in DJH joints. (A) Frequencies of usage of SSH-related recombination sites in total and N− embryo and newborn DJHs. (B) Embryo and newborn DJH joints recombine at SSH+ D sites. The DJH joints are arbitrarily located over the most joint-proximal SSH nucleotide; the progressively darker shades correspond to DJH joints using SSH 1, 2, 3, and >3 nt long; joint-proximal DJHs are delimited between the two vertical dotted lines (upper left). The two upper-right bar graphs represent the distributions of D recombination sites in DFL16.1+- and DSP2+-specific SSH+ DJHs (the nucleotide sequences are shown on the vertical axes, and for DSP2 genes, the numbers indicate variable nucleotide positions among the various gene family members). The bars below show the frequencies of joint-proximal DJHs in total, as well as in D family-specific, embryo and newborn SSH+ DJHs. (C) Degrees (means plus SEM) of nucleotide deletion in DFL16.1- and DSP2-specific embryo, newborn, and adult DJH joints. The symbols are as in Fig. 1, except for the shading of the upper bars in panel B.
High expression of Polμ in the early mouse embryo.
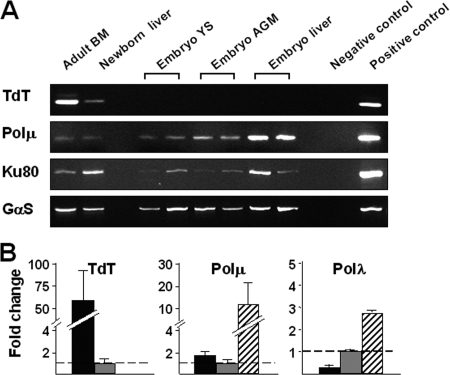
The expression of TdT, Polμ, and Ku80 genes in B-cell progenitors during mouse ontogeny was analyzed by conventional RT-PCRs in purified CD19+ IgM− cells from adult bone marrow (BM) and newborn liver, together with cKit+ CD45+ cells from embryo hematopoietic sites, which included a small subset of CD19+ B-cell precursors (5%) (19). TdT was present in adult BM, expressed at highly reduced levels in newborn pre-B cells, and undetected in embryo cells (even when a nested RT-PCR assay was used) (data not shown). In contrast, Polμ transcripts were more conspicuous in embryo cKit+ CD45+ cells than in both adult and newborn samples. The NHEJ core Ku gene was also well expressed in adult, embryo, and newborn samples (Fig. 3A). To better quantify the relative expression of X family DNA polymerase genes, we undertook real-time PCRs for TdT, Polμ, and Polλ genes in the above-mentioned populations of adult BM, newborn liver, and embryo liver cells. Compared to newborn values, the adult TdT expression was 50- to 100-fold increased, while it was completely absent in the embryo samples. In contrast, Polμ relative transcript levels were 5- to 10-fold higher in the embryo than in the newborn and adult cell samples. Polλ expression was also slightly increased in the embryo samples (Fig. 3B).
FIG. 3.
Differential expression of TdT, Polμ, Polλ, and Ku80 in lymphohematopoietic progenitors during mouse ontogeny. (A) CD19+ IgM− B-cell precursors were purified from adult BM and newborn liver, and cKit+ CD45+ cells were purified from the indicated embryo organs (YS, yolk sac; AGM, aorta-gonad-mesonephros region). Semiquantitative RT-PCRs were done as described in Materials and Methods. GαS gene expression was used as a reference for the total mRNA per sample. The negative and positive control samples were non-cDNA and adult thymus, respectively. (B) Normalized TdT, Polμ, and Polλ transcript levels relative to newborn 2−ΔΔCT. The expression of each specific gene was normalized to that of the control GαS gene, and the values obtained for both adult and embryo samples are relative to those obtained for newborn samples. The data shown are the means plus standard deviations of at least three different samples per experimental group. The symbols are as in Fig. 1.
Polμ and TdT polymerases show different size preferences for extending double-stranded DNA with 3′ overhangs.
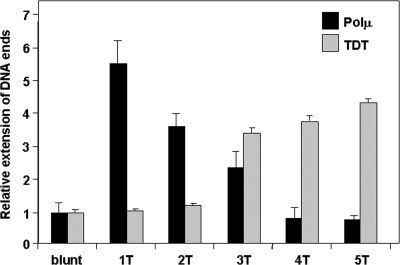
The finding of N additions in the embryo DJHs when TdT was absent but Polμ was highly expressed prompted us to analyze the differential efficiencies of both family X polymerases at introducing N nucleotides in vitro into different double-stranded DNA substrates, as they might be found in vivo in V(D)J rearrangements. As seen in Fig. 4, blunt ends were inefficient substrates for both enzymes. TdT also could not extend short protruding ends (1 or 2 nucleotide [nt] residues) but was active on >2-nt-long 3′ protruding ends. Polμ showed the highest efficiency on DNA ends with small 1- or 2-nt overhangs, later declining to its lowest level at 4- or 5-nt overhangs, those in which the TdT activity was maximal. In absolute terms, the terminal-transferase activity of Polμ on its optimal substrates was about threefold lower than that of TdT on ends protruding 4 nt. A mechanistic explanation is that the formation of a precatalytic ternary complex for terminal-transferase activity is a rate-limiting process in the case of Polμ, which would thus avoid excessive N additions and consequently favor templated insertions (P. Andrade, F. J. Lopez de Saro, and L. Blanco, unpublished data).
FIG. 4.
Differential in vitro terminal-transferase activities of Polμ and TdT. The assays were done on various double-stranded DNA substrates, either blunt ended or having increasing numbers of 3′ protruding T residues. The percentage of extended DNA of each type was related to the length of the blunt-ended molecule and is represented in a combined histogram (black and gray bars for Polμ and TdT, respectively; means plus SEM; n = 3 independent experiments).
Polμ contributes to the processing of mouse embryo DJH joints.
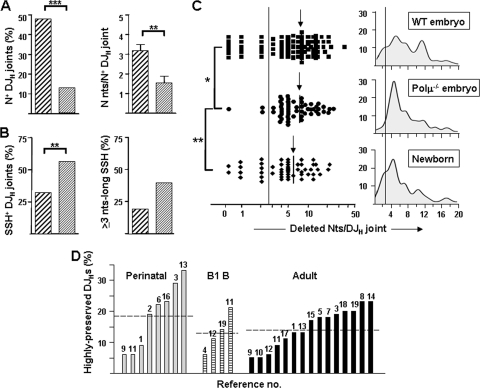
The lack of TdT, together with the potential of Polμ to add N nucleotides in DSBs (see above) (34, 39, 58) and its involvement in V(D)J rearrangements (4, 5), prompted us to investigate its putative activity in embryo DJH joints by analyzing Polμ−/− embryo mice. Fewer and late-appearing DJH rearrangements were detected in Polμ−/− compared to WT embryos (25 and 67% of E11 DJH+ samples, respectively), suggesting that the V(D)J rearrangement process is less efficient in the mutant mice. Polμ−/− embryo DJH joints were equally distributed between DFL16.1- and DSP2-bearing sequences, utilized all the JH gene segments, and showed an unbiased ORF distribution (see Fig. S5 in the supplemental material). They showed a significant decrease in N nucleotides relative to those of WT embryos, defined both as N+ DJH frequencies (13%; P < 0.001) and as the lengths of N stretches per N+ sequence (1.6 ± 0.4; P < 0.01) (Fig. 5A). Interestingly, the group of Polμ−/− DJHs included many embryos obtained from matings of mutant males with RAG2−/− females to eliminate the maternal contribution. Only 2 out of 26 of these DJHs showed N nucleotides (P < 0.05 with respect to the equivalent group of WT embryos derived from RAG2−/− females).
FIG. 5.
Differential features of Polμ−/− embryo DJH joints. (A) Frequencies of N+ DJH joints (left) and numbers of N nucleotides per N+ sequence (right) in WT (thickly hatched) and Polμ−/− (thinly hatched) embryos. (B) Frequencies of usage of SSH+ DJH joints (left) and of ≥3-nt-long SSH (right) in WT and Polμ−/− embryos. (C) Deleted nucleotides per DJH joint in WT and Polμ−/− embryos and in newborns (a logarithmic scale is used in the x axis on the left). The arrows indicate the mean values for each group. The distribution profiles of DJH joints related to the extents of nucleotide deletions in WT and Polμ−/− embryos and in WT newborns as defined by Gaussian kernel density estimations are shown in the right histograms (a linear scale is used in the x axis). The vertical lines delimit the highly preserved DJHs (≤2 deleted nucleotides). (D) Frequencies of highly preserved DJH joints detected in perinatal pre-B/B, adult B1, and adult pre-B/B cells from healthy mice in an extensive literature review (n = 1,572 DJH joints from 27 series in 19 papers). Each bar corresponds to an independent series of DJHs. The numbers above the bars refer to the original articles, which are listed in Table S2 in the supplemental material. The horizontal lines denote the mean of each group.
Embryo Polμ−/− DJHs relied more on the utilization of SSH for joining than those of WT embryos (P < 0.01), and the former SSH tended to be distinctively longer (39% and 19% of Polμ−/− and WT embryo SSH, respectively, were >3 nt long; P = 0.081) (Fig. 5B). Also, fewer joint-distal SSH were used among Polμ−/− than WT embryo DJHs, which therefore more closely resembled WT newborn DJHs (P = 0.09; data not shown). The average numbers of nucleotide deletions did not differ for WT and Polμ−/− embryo DJH joints. However, we observed that a subset of highly preserved DJH joints (≤2 deleted nucleotides), which accounted for 17% and 25% of WT embryo and newborn DJHs, respectively, were selectively depleted from Polμ−/− embryo DJHs (P < 0.05 and P < 0.01 compared to WT embryos and newborns, respectively) (Fig. 5C, left). The distribution profiles of DJH nucleotide deletions further revealed that the WT embryo DJHs showed a bimodal pattern, with peaks centered on 5- and 11-nt-long deletions, whereas newborn DJHs were predominantly limited to the less deleted subset. The Polμ−/− DJH profile resembled that of newborn DJHs in the rarity of highly deleted joints, although it differed from the other two in the selective absence of highly preserved joints (Fig. 5C, right). To elucidate whether the latter sequence subset was a consistent feature of every DJH repertoire, we undertook an extensive review of published DJH/VDJH joints from WT adult and perinatal mice (n = 1,572 DJH joints). All the analyzed series included a fraction of highly preserved DJH joints (15.2% ± 1.5%) (Fig. 5D) (the original references are shown in Table S2 in the supplemental material), suggesting that highly preserved DJHs represent an intrinsic result of the V(D)J rearrangement process and that the activity of Polμ is essential for their generation. Taken together, the above-mentioned findings show that Polμ participates in DJH coding-end ligation in the mouse embryo by (i) introducing a limited number of N nucleotides, (ii) filling in small gaps in a template-dependent manner, and (iii) contributing to recombination with end-distal SSH sites of the D sequence.
DISCUSSION
The Ig V region repertoires differ between the perinatal and adult periods of life, with the former preferentially expressing D-proximal VH genes (1, 69), showing distinct recombination sites (41), and, more importantly, lacking the diversifying potential of the N-nucleotide-producing TdT polymerase (8, 27, 32, 44, 55, 57). In the absence of TdT activity, the usage of germ line SSH in the recombination between most coding gene partners is predominant. B-lymphoid differentiation, however, starts much earlier, in the midgestation mouse embryo (14, 19, 51, 73). We have shown here that the embryo DJH joints have unusual characteristics, such as (i) short stretches of N nucleotides in the absence of detectable TdT expression, (ii) reduced utilization of SSH in embryo N− DJHs compared to those of newborn N− DJHs, and (iii) frequent DJHs rearranging with joint-distal D-gene SSH, which results in extensive DJH nucleotide deletions. The embryo N+ DJHs were also found among those recombined in ORF2 (which are strongly counterselected for in the adult) and, more importantly, in DJH RAG2+/− embryos derived from RAG2−/− mothers, demonstrating that they were of embryonic origin. Most adult-derived N nucleotides are introduced by TdT (30, 42), although a few N nucleotides remained in TdT−/− mice and low levels of N+ V(D)J joints were also detected in non-TdT-expressing perinatal life periods (21, 63). In agreement with others, we could not detect any TdT transcript signal in embryo hematopoietic cells, which might account for the N nucleotides found. Also, when a limited pool of TdT−/− embryo and adult DJHs (n = 23 and 13, respectively) were sequenced, N nucleotides were not found in the adult, whereas a subset of N+ DJHs were detected in the embryos, supporting the TdT independence of the embryo DJH N additions (M. A. R. Marcos and M. L. Gaspar, unpublished data). There is evidence of selection against the utilization of N nucleotides in neonatal B and T cells (7, 11, 12), as well as of positive selection for N− IgHs among marginal-zone B cells (10) and for N− public (26) and highly shared T-cell receptor β chains (38), suggesting that the levels of N+ V(D)J joints under TdT− conditions (as is the case in the perinatal periods) might even be higher in the absence of antigen selection and that there is room for the activities of other polymerases (30).
The embryo N− DJH joints did not show a predominant usage of SSH-related recombination sites (as happened in the newborn DJHs), suggesting that SSH were initially used stochastically and that only afterwards were SSH+ DJH joints preferentially expanded, because SSH increased the efficiency of DJH rearrangements and/or because SSH+ DJHs were positively selected for, as previously proposed (59). Whereas most SSH+ newborn DJHs used joint-proximal D nucleotides, thus preserving the genomic sequence, a significant fraction of embryo DJHs ligated to joint-distal SSH, resulting in large germ line nucleotide deletions. These highly deleted DJH joints were infrequent in later ontogenic periods, but they have been described in Bcl-XL transgenic mice, suggesting that B-cell precursors bearing “inefficient” V(D)J products were normally eliminated by apoptosis (25). Interestingly, the few V(D)J rearrangements rescued in DNA-PK-deficient mice also displayed significant nucleotide deletions that were joined in embryo but not in adult V-Jλ1 joints, implying that the NHEJ machineries acting in the two stages of life are different (43, 66). Both embryo and perinatal DJH+ B-cell precursors might represent independent compartments in which distinctive NHEJ complexes act. Alternatively, we are tempted to suggest that neonatal DJHs are a fraction selected from those emerging in the postgastrulation mouse embryo, after negative selection of most ORF2-expressing N+ and highly deleted DJH joints and/or promotion for ORF1+ SSH+ DJHs. As the embryo DJH patterns (particularly the presence of N nucleotides) were already undetectable in E16 DJHs when IgH and the λ5/V preB-encoded surrogate L chain were present (51, 63), it is possible that differential pairing of the emerging IgHs with the surrogate L chain is involved in filtering out the embryos’ “inadequate” IgH chains during late fetal periods (54, 72) and subsequently gives rise to a select perinatal IgH repertoire.
The processing of V(D)J coding joints can be sustained by the core NHEJ components (Ku-Artemis-DNA-PK and XLF-XRCC4-DNA ligase IV) (34), but the family X polymerases (Polμ, Polλ, and TdT) further contribute, especially when the nick leaves 3′ overhangs, which are good substrates for them (45, 49, 58). There is a gradient of template dependence in the activities of X polymerases (56, 58), as Polλ requires an extensive alignment of the ends to exclusively catalyze template-directed gap filling (60, 61), whereas the lymphoid-specific TdT activity is incompatible with the template strand due to interference with its rigid loop 1 domain (20) and so is restricted to catalyzing N additions in V(D)J reactions. In contrast, Polμ has a more flexible loop 1, which allows ambivalent strategies (39): (i) it preferentially inserts templated nucleotides at small gaps created from overhangs with minimal or even null complementarities and (ii) it may also add nucleotides in a terminal-transferase-like fashion to create a “connector” that provides de novo microhomology. The latter N nucleotides would then be initially masked but, if elongated with a second N addition or with a templated insertion or used for promiscuous ligation, would be detected as bona fide N nucleotides. Polμ is biased to insert pyrimidines versus purines, but it is important to consider that any untemplated insertion of Cs and Gs would be favorably selected for upon end joining, due to the greater strength of G·C than A·T pairs. By analyzing Polμ−/− embryos, Polμ was found to be responsible for most N nucleotides detected in the embryonic WT DJHs, although a few N nucleotides still remained, which might be accounted for by either Polλ (also expressed in the embryo) or other activities. Some in vitro insights into the Polμ potential to catalyze N nucleotides (including the experiments shown here) have been produced; the results obtained in the embryo DJH joints represent (to our knowledge) the first in vivo evidence of the TdT-like Polμ activity. Polμ was also required for the formation of highly preserved DJH sequences (<3 deleted nucleotides), which are always present in every DJH repertoire observed in healthy mice. Another feature of Polμ−/− embryo DJHs was the relative reduction in sequences with large nucleotide deletions, which were observed in WT embryos. These simultaneous decreases in highly deleted and in highly preserved DJHs (plus reduced N additions) in Polμ−/− embryos explained why the mean deletion rates were similar in WT and Polμ−/− embryo DJHs, despite the fact that the latter became a more homogeneous group. Finally, more and larger recombination-related SSH were used in the Polμ−/− than in the WT embryo DJHs, likely representing a backup mechanism, as is observed when the classical NHEJ process is altered (15). It has been reported that the VJκ joints of adult Polμ−/− mice showed increased nucleotide deletions (4). The authors did not observe any change in V(D)JH processing, presumably due to insufficient Polμ expression in IgH-rearranging adult BM pro-B cells. The apparent discrepancy with our work may be accounted for by the different ontogenic origins of the samples and, in particular, by the higher expression of Polμ in embryo DJH-rearranging precursors, which could thus reach the threshold level required to participate in DJH joining.
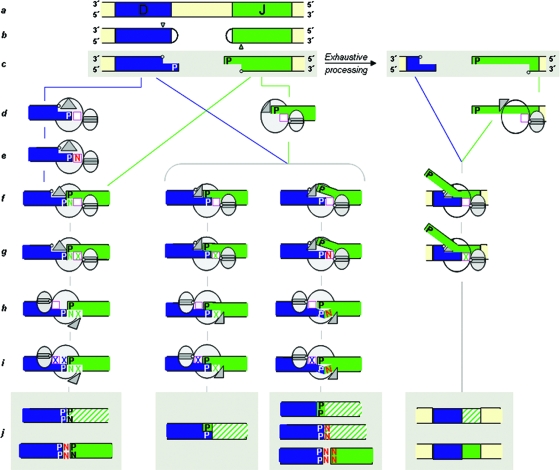
A scenario for the variations in the V(D)J rearrangement process during ontogeny might be proposed as follows. RAG activity is switched on in the postgastrulation mouse embryo when it induces the first DSBs at immune-specific loci. During early embryo ontogeny, somatic cells proliferate very actively and are notably sensitive to genotoxic stresses. Subsequently, robust machineries are required to ligate imprecise and highly processed coding joints in order to preserve cell viability and genome stability (35, 64). The evolutionarily ancient X polymerase Polμ, which is part of the cellular response to DSBs (9, 50), is well equipped to connect these embryo DSBs and to avoid collateral genomic damage (Fig. 6). First, Polμ can use templated nucleotide insertion to allow complete or almost complete conservation of the germ line sequence (Fig. 6, middle left) (17, 58). In good agreement with this, we observed that highly preserved DJH joints disappeared from Polμ−/− embryos. Secondly, Polμ's terminal-transferase activity can selectively target short DSB overhangs arising after hairpin nicking and opening (2-nt-long overhangs are the shortest nonblunt ends) (47), where it can introduce a few Ns, which may contribute to the pairing of DNA ends (Fig. 6, left) (17), particularly in the cases of C and G additions. Alternatively, after end bridging by Polμ, N nucleotides could also be inserted due to an imprecise orientation of the templating base that leads to an insertion error/point mutation, ready for ligation (Fig. 6, middle right). Finally, exhaustive processing of DNA ends, resulting in deletion and extensive 3′ overhangs, can still be handled by Polμ through its primer-realigning capacity (67, 75) and avidity for internal 5′ P, subsequently promoting pairing to internal SSH sites (16) at the cost of germ line sequence losses (Fig. 6, right). Challenged by strong evolutionary pressures to resolve cell-stressing DSBs in the early embryo, Polμ thus may use all its skills to repair the coding ends, even if many of the resulting products are “useless” and will be counterselected for later on. During the last third of gestation, N− RF1+ DJH/VDJH rearrangements, which preferentially used genome-preserving joint-proximal SSHs, selectively expanded, a process that may be secondary to changes in the NHEJ complexes used and/or to selection for pairing to surrogate light-chain and/or (self) antigens. Both in the newborn and in the adult, Polμ will concentrate on the preservation of germ line coding ends by filling up small nucleotide gaps at those DSBs where it is recruited (IgL chain-encoding genes in the adult BM) (67, 75). The most recently evolved TdT will then be upregulated in adult pro-B cells, and its exclusive template-independent activity will be devoted to the generation of the extensive junctional diversity of IgH antibody chains. Finally, Polμ and TdT are not only compartmentalized during ontogeny and in different lymphoid differentiation stages, they also target distinct DSB DNA ends related to the extent of the 3′ overhangs produced by the nicking step, an event that might also influence the V(D)J repertoires during ontogeny.
FIG. 6.
A model for nucleotide insertions catalyzed by Polμ during embryonic DJH rearrangements. The Polμ structure (in gray) is depicted as two ellipses, the large one representing the polymerization core and the small one indicating the 8-kDa domain, which has a single-stranded DNA binding cleft for the 5′ P binding site. The triangle represents mobile loop 1 of Polμ (39, 56). The deoxynucleotide triphosphate (dNTP) binding site is represented by a square (magenta). The hatched areas are “out-of-frame” J segments. (a) Germ line genomic region with D and J segments of the IgH chain. (b) RAG-induced hairpinned coding ends at both D and J segments. A nicking site is indicated by a small triangle at each end. (c) After nicking by Artemis-DNA-PK, a 3′ overhang (2 nt in this case), which contains a P nucleotide, is generated at both ends. An internal 5′ phosphate is indicated by a small circle. (d) Binding of Polμ to DNA ends can occur in two different ways: an enzyme-DNA complex can be formed in which the 3′ protruding end is oriented as a primer strand whose primer terminus (P nucleotide) is close to the dNTP binding site (left); alternatively, the 5′ P of one DNA end is bound by the Polμ 8-kDa domain next to the dNTP binding site (center and right). (e to j, left) Terminal-transferase N addition occurs before end joining (e). The inserted N nucleotide can serve as a connector for end joining if it is complementary to the other DNA end and the synapsis is stabilized by loop 1 (f). The N nucleotide, which is now inapparent as a template-independent insertion, is further extended by insertion of a templated dXTP (g). Once this strand is ligated, Polμ is adjusted to the remaining gap, coupled to a conformational change of loop 1 (h). After gap filling and ligation (i), the end joining is completed and produces an “out-of-frame” J segment (hatched area) due to the two extra P nucleotides added at the junction. An “in-frame” product (shown below) could be obtained by the addition of two N nucleotides before the end-joining step. In this case, one of the Ns would be evident (j). (f to j, middle left) Polμ's loop 1-mediated synapsis without requiring N nucleotides. If the two P nucleotides are complementary, their extension (X) will be template directed at both gaps, and the resulting product will not include N nucleotides (an “out-of-frame” J segment; one P nucleotide added). (f to j, middle right) If the two P nucleotides are not complementary and the templating base (located in front of the dNTP binding site) is not properly adjusted, Polμ can add N nucleotides. One or more N nucleotides could be immediately ligated and detected in the final recombination product. (c to j, right) When the ends are heavily processed and have large single-stranded overhangs, Polμ can bind the internal 5′ P and attract a second 3′ overhang to its vicinity, based on its capacity to accept microhomologies. After gap filling and further processing of the flapped strand, a largely deleted product is obtained that could contain either “out-of-frame” or “in-frame” J segments.
Supplementary Material
Acknowledgments
This work was supported by grants SAF2007-65265, BFU2006-14390/BMC, and CSD2007-00015 from the Ministry of Education of Spain; by the Fundación Mutua Madrileña; and by S-SAL-0304-2006 from the Community of Madrid. The Centro de Biología Molecular Severo Ochoa receives institutional funding from Fundación Ramón Areces. B.G.-L. and N.S. are recipients of fellowships from Instituto de Salud Carlos III, and G.T. has a fellowship from the Spanish Ministry of Science and Technology.
We thank Fernando Martínez and Mario Alía for expert technical assistance, David Abia for statistical support, and Susana Morales and Daniel Lucas for contributions in the early stages of the work.
Footnotes
Published ahead of print on 22 December 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alt, F. W., T. K. Blackwell, and G. D. Yancopoulos. 1987. Development of the primary antibody repertoire. Science 2381079-1087. [DOI] [PubMed] [Google Scholar]
- 2.Baba, Y., R. Pelayo, and P. W. Kincade. 2004. Relationships between hematopoietic stem cells and lymphocyte progenitors. Trends Immunol. 25645-649. [DOI] [PubMed] [Google Scholar]
- 3.Bentolila, L. A., S. Olson, A. Marshall, F. Rougeon, C. J. Paige, N. Doyen, and G. E. Wu. 1999. Extensive junctional diversity in Ig light chain genes from early B cell progenitors of mu MT mice. J. Immunol. 1622123-2128. [PubMed] [Google Scholar]
- 4.Bertocci, B., A. De Smet, C. Berek, J. C. Weill, and C. A. Reynaud. 2003. Immunoglobulin kappa light chain gene rearrangement is impaired in mice deficient for DNA polymerase mu. Immunity 19203-211. [DOI] [PubMed] [Google Scholar]
- 5.Bertocci, B., A. De Smet, J. C. Weill, and C. A. Reynaud. 2006. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J. recombination in vivo. Immunity 2531-41. [DOI] [PubMed] [Google Scholar]
- 6.Bertrand, J. Y., A. Jalil, M. Klaine, S. Jung, A. Cumano, and I. Godin. 2005. Three pathways to mature macrophages in the early mouse yolk sac. Blood 1063004-3011. [DOI] [PubMed] [Google Scholar]
- 7.Bogue, M., S. Candeias, C. Benoist, and D. Mathis. 1991. A special repertoire of alpha:beta T cells in neonatal mice. EMBO J. 103647-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogue, M., S. Gilfillan, C. Benoist, and D. Mathis. 1992. Regulation of N-region diversity in antigen receptors through thymocyte differentiation and thymus ontogeny. Proc. Natl. Acad. Sci. USA 8911011-11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capp, J. P., F. Boudsocq, A. G. Besnard, B. S. Lopez, C. Cazaux, J. S. Hoffmann, and Y. Canitrot. 2007. Involvement of DNA polymerase mu in the repair of a specific subset of DNA double-strand breaks in mammalian cells. Nucleic Acids Res. 353551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carey, J. B., C. S. Moffatt-Blue, L. C. Watson, A. L. Gavin, and A. J. Feeney. 2008. Repertoire-based selection into the marginal zone compartment during B cell development. J. Exp. Med. 2052043-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlsson, L., C. Overmo, and D. Holmberg. 1992. Selection against N-region diversity in immunoglobulin heavy chain variable regions during the development of pre-immune B cell repertoires. Int. Immunol. 4549-553. [DOI] [PubMed] [Google Scholar]
- 12.Cassady-Cain, R. L., and A. K. Kaushik. 2006. Increased negative selection impairs neonatal B cell repertoire but does not directly lead to generation of disease-associated IgM auto-antibodies. Int. Immunol. 18661-669. [DOI] [PubMed] [Google Scholar]
- 13.Ceredig, R., E. ten Boekel, A. Rolink, F. Melchers, and J. Andersson. 1998. Fetal liver organ cultures allow the proliferative expansion of pre-B receptor-expressing pre-B-II cells and the differentiation of immature and mature B cells in vitro. Int. Immunol. 1049-59. [DOI] [PubMed] [Google Scholar]
- 14.Chang, Y., C. J. Paige, and G. E. Wu. 1992. Enumeration and characterization of DJH structures in mouse fetal liver. EMBO J. 111891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corneo, B., R. L. Wendland, L. Deriano, X. Cui, I. A. Klein, S. Y. Wong, S. Arnal, A. J. Holub, G. R. Weller, B. A. Pancake, S. Shah, V. L. Brandt, K. Meek, and D. B. Roth. 2007. Rag mutations reveal robust alternative end joining. Nature 449483-486. [DOI] [PubMed] [Google Scholar]
- 16.Covo, S., L. Blanco, and Z. Livneh. 2004. Lesion bypass by human DNA polymerase mu reveals a template-dependent, sequence-independent nucleotidyl transferase activity. J. Biol. Chem. 279859-865. [DOI] [PubMed] [Google Scholar]
- 17.Davis, B. J., J. M. Havener, and D. A. Ramsden. 2008. End-bridging is required for polμ to efficiently promote repair of noncomplementary ends by nonhomologous end joining. Nucleic Acids Res. 363085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Andres, B., I. Cortegano, N. Serrano, B. del Rio, P. Martin, P. Gonzalo, M. A. Marcos, and M. L. Gaspar. 2007. A population of CD19highCD45R−/lowCD21low B lymphocytes poised for spontaneous secretion of IgG and IgA antibodies. J. Immunol. 1795326-5334. [DOI] [PubMed] [Google Scholar]
- 19.de Andres, B., P. Gonzalo, S. Minguet, J. A. Martinez-Marin, P. G. Soro, M. A. Marcos, and M. L. Gaspar. 2002. The first 3 days of B-cell development in the mouse embryo. Blood 1004074-4081. [DOI] [PubMed] [Google Scholar]
- 20.Delarue, M., J. B. Boule, J. Lescar, N. Expert-Bezancon, N. Jourdan, N. Sukumar, F. Rougeon, and C. Papanicolaou. 2002. Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase. EMBO J. 21427-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delassus, S., S. Darche, P. Kourilsky, and A. Cumano. 1998. Ontogeny of the heavy chain immunoglobulin repertoire in fetal liver and bone marrow. J. Immunol. 1603274-3280. [PubMed] [Google Scholar]
- 22.Dominguez, O., J. F. Ruiz, T. Lain de Lera, M. Garcia-Diaz, M. A. Gonzalez, T. Kirchhoff, A. C. Martinez, A. Bernad, and L. Blanco. 2000. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 191731-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egawa, T., K. Kawabata, H. Kawamoto, K. Amada, R. Okamoto, N. Fujii, T. Kishimoto, Y. Katsura, and T. Nagasawa. 2001. The earliest stages of B cell development require a chemokine stromal cell-derived factor/pre-B cell growth-stimulating factor. Immunity 15323-334. [DOI] [PubMed] [Google Scholar]
- 24.Ezekiel, U. R., T. Sun, G. Bozek, and U. Storb. 1997. The composition of coding joints formed in V(D)J recombination is strongly affected by the nucleotide sequence of the coding ends and their relationship to the recombination signal sequences. Mol. Cell. Biol. 174191-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang, W., D. L. Mueller, C. A. Pennell, J. J. Rivard, Y. S. Li, R. R. Hardy, M. S. Schlissel, and T. W. Behrens. 1996. Frequent aberrant immunoglobulin gene rearrangements in pro-B cells revealed by a bcl-xL transgene. Immunity 4291-299. [DOI] [PubMed] [Google Scholar]
- 26.Fazilleau, N., J. P. Cabaniols, F. Lemaitre, I. Motta, P. Kourilsky, and J. M. Kanellopoulos. 2005. Vα and Vβ public repertoires are highly conserved in terminal deoxynucleotidyl transferase-deficient mice. J. Immunol. 174345-355. [DOI] [PubMed] [Google Scholar]
- 27.Feeney, A. J. 1990. Lack of N regions in fetal and neonatal mouse immunoglobulin V-D-J. junctional sequences. J. Exp. Med. 1721377-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feeney, A. J. 1992. Predominance of VH-D-JH junctions occurring at sites of short sequence homology results in limited junctional diversity in neonatal antibodies. J. Immunol. 149222-229. [PubMed] [Google Scholar]
- 29.Gerstein, R. M., and M. R. Lieber. 1993. Coding end sequence can markedly affect the initiation of V(D)J. recombination. Genes Dev. 71459-1469. [DOI] [PubMed] [Google Scholar]
- 30.Gilfillan, S., A. Dierich, M. Lemeur, C. Benoist, and D. Mathis. 1993. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science 2611175-1178. [DOI] [PubMed] [Google Scholar]
- 31.Godin, I. E., J. A. Garcia-Porrero, A. Coutinho, F. Dieterlen-Lievre, and M. A. Marcos. 1993. Para-aortic splanchnopleura from early mouse embryos contains B1a cell progenitors. Nature 36467-70. [DOI] [PubMed] [Google Scholar]
- 32.Gu, H., I. Forster, and K. Rajewsky. 1990. Sequence homologies, N sequence insertion and JH gene utilization in VHDJH joining: implications for the joining mechanism and the ontogenetic timing of Ly1 B cell and B-CLL progenitor generation. EMBO J. 92133-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu, H., D. Kitamura, and K. Rajewsky. 1991. B cell development regulated by gene rearrangement: arrest of maturation by membrane-bound D mu protein and selection of DH element reading frames. Cell 6547-54. [DOI] [PubMed] [Google Scholar]
- 34.Gu, J., H. Lu, B. Tippin, N. Shimazaki, M. F. Goodman, and M. R. Lieber. 2007. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 261010-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heyer, B. S., A. MacAuley, O. Behrendtsen, and Z. Werb. 2000. Hypersensitivity to DNA damage leads to increased apoptosis during early mouse development. Genes Dev. 142072-2084. [PMC free article] [PubMed] [Google Scholar]
- 36.Holmberg, D., A. A. Freitas, D. Portnoi, F. Jacquemart, S. Avrameas, and A. Coutinho. 1986. Antibody repertoires of normal BALB/c mice: B lymphocyte populations defined by state of activation. Immunol. Rev. 93147-169. [DOI] [PubMed] [Google Scholar]
- 37.Ichihara, Y., H. Hayashida, S. Miyazawa, and Y. Kurosawa. 1989. Only DFL16, DSP2, and DQ52 gene families exist in mouse immunoglobulin heavy chain diversity gene loci, of which DFL16 and DSP2 originate from the same primordial DH gene. Eur. J. Immunol. 191849-1854. [DOI] [PubMed] [Google Scholar]
- 38.Johnson, M. E., Z. Cheng, V. A. Morrison, S. Scherer, M. Ventura, R. A. Gibbs, E. D. Green, and E. E. Eichler. 2006. Recurrent duplication-driven transposition of DNA during hominoid evolution. Proc. Natl. Acad. Sci. USA 10317626-17631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Juarez, R., J. F. Ruiz, S. A. Nick McElhinny, D. Ramsden, and L. Blanco. 2006. A specific loop in human DNA polymerase mu allows switching between creative and DNA-instructed synthesis. Nucleic Acids Res. 344572-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kearney, J. F., and M. Vakil. 1986. Idiotype-directed interactions during ontogeny play a major role in the establishment of the adult B cell repertoire. Immunol. Rev. 9439-50. [DOI] [PubMed] [Google Scholar]
- 41.Kepler, T. B., M. Borrero, B. Rugerio, S. K. McCray, and S. H. Clarke. 1996. Interdependence of N nucleotide addition and recombination site choice in V(D)J. rearrangement. J. Immunol. 1574451-4457. [PubMed] [Google Scholar]
- 42.Komori, T., A. Okada, V. Stewart, and F. W. Alt. 1993. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science 2611171-1175. [DOI] [PubMed] [Google Scholar]
- 43.Lewis, S. M. 1994. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv. Immunol. 5627-150. [DOI] [PubMed] [Google Scholar]
- 44.Li, Y. S., K. Hayakawa, and R. R. Hardy. 1993. The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J. Exp. Med. 178951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lieber, M. R., H. Lu, J. Gu, and K. Schwarz. 2008. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 18125-133. [DOI] [PubMed] [Google Scholar]
- 46.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 47.Lu, C. P., J. E. Posey, and D. B. Roth. 2008. Understanding how the V(D)J. recombinase catalyzes transesterification: distinctions between DNA cleavage and transposition. Nucleic Acids Res. 362864-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lucas, D., T. Lain de Lera, M. A. Gonzalez, J. F. Ruiz, O. Dominguez, J. C. Casanova, A. C. Martinez, L. Blanco, and A. Bernad. 2005. Polymerase mu is up-regulated during the T cell-dependent immune response and its deficiency alters developmental dynamics of spleen centroblasts. Eur. J. Immunol. 351601-1611. [DOI] [PubMed] [Google Scholar]
- 49.Ma, Y., H. Lu, B. Tippin, M. F. Goodman, N. Shimazaki, O. Koiwai, C. L. Hsieh, K. Schwarz, and M. R. Lieber. 2004. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell 16701-713. [DOI] [PubMed] [Google Scholar]
- 50.Mahajan, K. N., S. A. Nick McElhinny, B. S. Mitchell, and D. A. Ramsden. 2002. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol. Cell. Biol. 225194-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marcos, M. A., S. Morales-Alcelay, I. E. Godin, F. Dieterlen-Lievre, S. G. Copin, and M. L. Gaspar. 1997. Antigenic phenotype and gene expression pattern of lymphohemopoietic progenitors during early mouse ontogeny. J. Immunol. 1582627-2637. [PubMed] [Google Scholar]
- 52.Marleau, A. M., J. D. Greenwood, Q. Wei, B. Singh, and B. A. Croy. 2003. Chimerism of murine fetal bone marrow by maternal cells occurs in late gestation and persists into adulthood. Lab. Investig. 83673-681. [DOI] [PubMed] [Google Scholar]
- 53.Marshall, A. J., N. Doyen, L. A. Bentolila, C. J. Paige, and G. E. Wu. 1998. Terminal deoxynucleotidyl transferase expression during neonatal life alters D(H) reading frame usage and Ig-receptor-dependent selection of V regions. J. Immunol. 1616657-6663. [PubMed] [Google Scholar]
- 54.Martin, F., X. Chen, and J. F. Kearney. 1997. Development of VH81X transgene-bearing B cells in fetus and adult: sites for expansion and deletion in conventional and CD5/B1 cells. Int. Immunol. 9493-505. [DOI] [PubMed] [Google Scholar]
- 55.Meek, K. 1990. Analysis of junctional diversity during B lymphocyte development. Science 250820-823. [DOI] [PubMed] [Google Scholar]
- 56.Moon, A. F., M. Garcia-Diaz, V. K. Batra, W. A. Beard, K. Bebenek, T. A. Kunkel, S. H. Wilson, and L. C. Pedersen. 2007. The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair 61709-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nadel, B., S. Tehranchi, and A. J. Feeney. 1995. Coding end processing is similar throughout ontogeny. J. Immunol. 1546430-6436. [PubMed] [Google Scholar]
- 58.Nick McElhinny, S. A., J. M. Havener, M. Garcia-Diaz, R. Juarez, K. Bebenek, B. L. Kee, L. Blanco, T. A. Kunkel, and D. A. Ramsden. 2005. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell 19357-366. [DOI] [PubMed] [Google Scholar]
- 59.Pandey, A., L. W. Tjoelker, and C. B. Thompson. 1993. Restricted immunoglobulin junctional diversity in neonatal B cells results from developmental selection rather than homology-based V(D)J joining. J. Exp. Med. 177329-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Picher, A. J., and L. Blanco. 2007. Human DNA polymerase lambda is a proficient extender of primer ends paired to 7,8-dihydro-8-oxoguanine. DNA Repair 61749-1756. [DOI] [PubMed] [Google Scholar]
- 61.Picher, A. J., M. Garcia-Diaz, K. Bebenek, L. C. Pedersen, T. A. Kunkel, and L. Blanco. 2006. Promiscuous mismatch extension by human DNA polymerase lambda. Nucleic Acids Res. 343259-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajewsky, K. 1996. Clonal selection and learning in the antibody system. Nature 381751-758. [DOI] [PubMed] [Google Scholar]
- 63.Ramsden, D. A., C. J. Paige, and G. E. Wu. 1994. Kappa light chain rearrangement in mouse fetal liver. J. Immunol. 1531150-1160. [PubMed] [Google Scholar]
- 64.Reddy, Y. V., E. J. Perkins, and D. A. Ramsden. 2006. Genomic instability due to V(D)J recombination-associated transposition. Genes Dev. 201575-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rooney, S., J. Chaudhuri, and F. W. Alt. 2004. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol. Rev. 200115-131. [DOI] [PubMed] [Google Scholar]
- 66.Ruetsch, N. R., G. C. Bosma, and M. J. Bosma. 2000. Unexpected rearrangement and expression of the immunoglobulin λ1 locus in scid mice. J. Exp. Med. 1911933-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ruiz, J. F., D. Lucas, E. Garcia-Palomero, A. I. Saez, M. A. Gonzalez, M. A. Piris, A. Bernad, and L. Blanco. 2004. Overexpression of human DNA polymerase mu (Pol mu) in a Burkitt's lymphoma cell line affects the somatic hypermutation rate. Nucleic Acids Res. 325861-5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schroeder, H. W., Jr. 2006. Similarity and divergence in the development and expression of the mouse and human antibody repertoires. Dev. Comp. Immunol. 30119-135. [DOI] [PubMed] [Google Scholar]
- 69.Schroeder, H. W., Jr., J. L. Hillson, and R. M. Perlmutter. 1987. Early restriction of the human antibody repertoire. Science 238791-793. [DOI] [PubMed] [Google Scholar]
- 70.Strasser, A., A. Rolink, and F. Melchers. 1989. One synchronous wave of B cell development in mouse fetal liver changes at day 16 of gestation from dependence to independence of a stromal cell environment. J. Exp. Med. 1701973-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tonegawa, S. 1983. Somatic generation of antibody diversity. Nature 302575-581. [DOI] [PubMed] [Google Scholar]
- 72.Wasserman, R., Y. S. Li, S. A. Shinton, C. E. Carmack, T. Manser, D. L. Wiest, K. Hayakawa, and R. R. Hardy. 1998. A novel mechanism for B cell repertoire maturation based on response by B cell precursors to pre-B receptor assembly. J. Exp. Med. 187259-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yokota, T., J. Huang, M. Tavian, Y. Nagai, J. Hirose, J. C. Zuniga-Pflucker, B. Peault, and P. W. Kincade. 2006. Tracing the first waves of lymphopoiesis in mice. Development 1332041-2051. [DOI] [PubMed] [Google Scholar]
- 74.Yu, K., and M. R. Lieber. 1999. Mechanistic basis for coding end sequence effects in the initiation of V(D)J. recombination. Mol. Cell. Biol. 198094-8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, Y., X. Wu, F. Yuan, Z. Xie, and Z. Wang. 2001. Highly frequent frameshift DNA synthesis by human DNA polymerase mu. Mol. Cell. Biol. 217995-8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.