Abstract
Under classical models for signal-dependent transcription in eukaryotes, DNA-binding activator proteins regulate the recruitment of RNA polymerase II (Pol II) to a set of target promoters. However, recent studies, as well as our results herein, show that Pol II is widely distributed (i.e., “preloaded”) at the promoters of many genes prior to specific signaling events. How Pol II recruitment and Pol II preloading fit within a unified model of gene regulation is unclear. In addition, the mechanisms through which cellular signals activate preloaded Pol II across mammalian genomes remain largely unknown. We show here that the predominant genomic outcome of estrogen signaling is the postrecruitment regulation of Pol II activity at target gene promoters, likely through specific changes in Pol II phosphorylation rather than through recruitment of Pol II to the promoters. Furthermore, we show that negative elongation factor binds to estrogen target promoters in conjunction with preloaded Pol II and represses gene expression until the appropriate signal is received. Finally, our studies reveal that the estrogen-dependent activation of preloaded Pol II facilitates rapid gene regulatory responses which play important physiological roles in regulating estrogen signaling itself. Our results reveal a broad use of postrecruitment Pol II regulation by the estrogen signaling pathway, a mode of regulation that is likely to apply to a wide variety of signal-regulated pathways.
The regulation of gene expression is an important means by which cells respond to physiological and environmental signals. Under classical models for signal-dependent transcription, DNA-binding activator proteins promote the recruitment of RNA polymerase II (Pol II) to a set of target promoters (27, 39, 46, 47). An alternative, and not mutually exclusive, mechanism controlling gene expression involves the regulation of Pol II activity, primarily through phosphorylation, at a step after recruitment to target genes (42, 54). In metazoans, gene-specific studies have provided examples of genes regulated by either Pol II recruitment or Pol II activation postrecruitment, although the predominant mechanism remains largely unknown (2, 33, 51). In addition, recent genome-wide studies have shown that Pol II localizes to the promoters of many unexpressed genes prior to specific signaling events (i.e., Pol II is “preloaded”) (6, 20, 23, 36, 45, 48, 65). Interestingly, preloaded or stalled Pol II has recently been suggested to play important roles in regulating promoter-proximal nucleosome assembly and gene expression (12, 18).
A current hypothesis about the role of preloaded Pol II across the genome suggests that it is poised for activation by physiological or developmental signals (34, 57). The validity of this hypothesis and the mechanisms through which specific signaling pathways activate preloaded Pol II have not been studied directly at a genome-wide scale. Estrogenic hormones, such as 17β-estradiol (E2), make a good model for signal-regulated transcription since they act through DNA-binding estrogen receptors (ERs) to control patterns of gene expression involved in reproduction, development, and metabolism (13, 38). The historical model for gene activation by E2 signaling, developed based on model E2-regulated genes, involves E2-dependent chromatin modifications followed by the recruitment of Pol II to target promoters (19, 35). However, the generality of this “Pol II recruitment” model has not been examined across the entire E2-regulated transcriptome.
The complexity of the transcription cycle provides many opportunities for exquisite regulatory control of Pol II-dependent transcriptional responses. Before transcription initiation, Pol II forms a preinitiation complex (PIC) with general transcription factors at gene promoters (9, 15). After initiation, Pol II is released from the promoter to enter productive elongation through the coding region of the gene (15, 54). Phosphorylation of specific residues within the heptapeptide repeat of the Pol II Rpb1 subunit carboxy-terminal domain (commonly referred to as the Pol II CTD) marks the transition from transcription initiation to transcription elongation (11). For example, phosphorylation at serine 5 (Ser5P) of the Pol II CTD generally occurs early in the transcription cycle. In contrast, phosphorylation at serine 2 (Ser2P), which is catalyzed primarily by the cyclin-dependent kinase 9 (Cdk9) of the positive transcription elongation factor-b (P-TEFb), generally occurs concomitantly with productive elongation and predominates toward the 3′ ends of genes (39, 42, 45, 54). Transacting factors, such as the negative-elongation factor (NELF) and the DRB-sensitivity-inducing factor (DSIF) complexes, cooperate to repress transcription elongation and their negative effects can be overcome by P-TEFb (41, 44, 60, 64). However, recent studies suggest that NELF may also play a positive role in transcriptional regulation (18). How Pol II phosphorylation and the functions of NELF and DSIF are regulated by cellular signals at a genomic scale remains largely unknown.
In the present study, we show that a significant portion of unexpressed genes (∼20%) in the human genome have preloaded Pol II prior to specific signaling events. Furthermore, our studies reveal that the predominant genomic outcome of estrogen signaling at target promoters is the postrecruitment activation of preloaded Pol II, likely through phosphorylation of the CTD, rather than recruitment of Pol II. This process involves the E2-dependent recruitment of Cdk9. Furthermore, our results suggest that transacting regulatory factors, such as NELF and DSIF, function to restrain transcription by preloaded Pol II until an appropriate signal, such as E2, is received. This mode of regulation by E2 facilitates rapid gene regulatory responses, which act in a feed-forward pathway to regulate the overall estrogen signaling program. The results described here, together with other recent findings, reveal a new mode of signal-dependent gene regulation that is likely to apply to a wide variety of signal-regulated pathways.
MATERIALS AND METHODS
Cells lines and RNA interference-mediated knockdown.
MCF-7 human breast cancer cells were kindly provided by Benita Katzenellenbogen (University of Illinois, Urbana-Champaign). The cells were maintained in minimal essential medium supplemented with 5% calf serum and plated for experiments in phenol red-free minimal essential medium supplemented with 5% charcoal-dextran treated calf serum. NELF-B-depleted MCF-7 cells were generated by retrovirus-mediated gene transfer of a short hairpin RNA (shRNA) sequence (5′-GACCTTCTGGAGAAGAGCT-3′) specifically targeting the NELF-B mRNA by using the pSUPER.retro system (Oligoengine). Control cells expressing a short hairpin RNA sequence directed against green fluorescent protein (GFP) were generated in parallel.
Antibodies.
The antibodies used in the present study were as follows: RNA Pol II (Santa Cruz N-20 and C-21 [SC-899 and SC-900, respectively]; the antibodies were combined in equal amounts before use), transcription factor IIB (TFIIB; Santa Cruz SI-1; SC-274X), Pol II Ser5-Phos CTD (Covance H14; MMS-134R), Pol II Ser2-Phos CTD (Covance H5; MMS-129R), NELF-A (Santa Cruz A20; SC-23599), NELF-B (mouse monoclonal) and NELF-E (rabbit polyclonal) (kindly provided by Hiroshi Handa, Tokyo Institute of Technology), Cdk9 (Santa Cruz H-169; SC-8338), and SPT5 (Santa Cruz H-300; SC-28678).
ChIP assays.
Chromatin immunoprecipitations (ChIPs) were performed as described previously (24). For the Ser5- and Ser2-Phos CTD ChIP assays, we used the protocol noted above with the following modifications: (i) immune complexes were captured using anti-mouse immunoglobulin M (μ-chain specific)-agarose beads (Sigma, A4540) after an overnight incubation with the antibodies at 4°C, and (ii) the agarose beads were washed once with each of the following buffers: (i) 1% Triton X-100, 0.1% sodium dodecyl sulfate, 2 mM EDTA, 20 mM Tris-HCl (pH 7.9), 150 mM NaCl, 2 mg of leupeptin/ml, and 2 mg of aprotinin/ml; (ii) 1% Triton X, 0.05% sodium dodecyl sulfate, 2 mM EDTA, 20 mM Tris-HCl (pH 7.9), 500 mM NaCl, 2 mg of leupeptin/ml, and 2 mg of aprotinin/ml; and (iii) 1% NP-40, 1 mM EDTA, 10 mM Tris-HCl (pH 7.9), 250 mM LiCl, 0.5% deoxycholate, 2 mg of leupeptin/ml, and 2 mg of aprotinin/ml; and (iv) TE. The beads were washed once again with Tris-EDTA and resuspended in elution buffer for analysis, as described previously (24). The enrichment of immunoprecipitated ChIP material relative to the input material was determined by using quantitative real-time PCR (qPCR) with gene-specific primer sets to specified genomic regions (see below). Each ChIP experiment was conduced a minimum of three times with independent chromatin isolates to ensure reproducibility.
Hybridization of ChIP material to DNA microarrays.
For microarray hybridization, purified ChIP DNA was blunted by using T4 DNA polymerase (New England Biolabs), ligated to linkers, and amplified by using ligation-mediated PCR, as described previously (24, 50). A total of 4 μg of immunoprecipitated (IP) and input DNA were sent to Nimblegen for labeling with Cy5 or Cy3 fluorophores, respectively, using 9-mer primers. The labeled IP and input DNA were cohybridized to the Nimblegen HG18 RefSeq promoter microarray, which contains ∼19,000 well-characterized RefSeq genes tiled from bp −2200 to +500 from the transcriptional start site (TSS) with 50- to 75-mer probes at ∼100-bp spacing. The raw ChIP-chip data can be accessed from the National Institutes of Health GEO website (http://www.ncbi.nlm.nih.gov/projects/geo/) using the series accession number GSE13051.
Controls to determine the IP efficiency, the average size of the sonicated DNA, and the efficiency of DNA amplification were performed to ensure that this protein-DNA interaction study was of high quality and the false-positive error rate was minimized. Gene-specific confirmation analyses showed that our ChIP-chip experiments had false-positive error rates of ∼15%, on average, which are similar to, if not lower, than those reported for other ChIP-chip analyses (26, 29, 31, 58).
ChIP-chip statistical analyses.
Statistical analysis was performed by using the statistical software R (GNU project, Free Software Foundation) (49), and all of the scripts that were used are available upon request. The signal ratio of IP-DNA over input-DNA was log2 transformed and Lowess normalized (55). For the experiments comparing −E2 and +E2 conditions, the data were scaled for total signal intensity to ensure that any differences observed were not due to variations in overall labeling or hybridization efficiencies. The corrected data were then analyzed by using moving average windows of 800 bp with a step of 150 bp, as described previously (8), and visualized by using the Treeview software (53). The Wilcoxon signed-rank test was used to ascertain whether a window average signal was significantly higher than the mean of all window signals.
To determine significant regions of factor binding, we applied an algorithm requiring: (i) the Wilcoxon signed-rank test P value for a certain window to be less than 0.05, (ii) each window to contain at least five probes on the microarray, and (iii) at least two consecutive windows to satisfy the criteria i and ii described above. For Ser5P, we also required that the significant regions should contain Pol II peaks, as described above. Based on gene-specific ChIP-qPCR experiments, our estimated confirmation rate using the thresholds described above was ∼85% for RNA Pol II, ∼93% for NELF-A, and ∼78% for Ser5P (genes tested = 33). Averaging analysis for several gene classes was performed by averaging the log2 (IP/input) signals of the appropriate windows across all genes within each class. The Student t test (two-tailed, unpaired homoschedastic) was applied to determine whether factor binding at a certain region was significantly different between two classes.
ChIP-qPCR.
For qPCR analyses of the ChIP material, 2 μl out of 50 μl of ChIP DNA, 1× SYBR green PCR master mix, and 250 nM concentrations of forward and reverse primers were used in 40 cycles of amplification (95°C for 15 s and 60°C for 1 min) using a DNA Engine Opticon detection system (MJ Research) after an initial 10-min incubation at 95°C. Melting-curve analysis was performed to ensure that only the targeted amplicon was amplified. Mock IP (i.e., nonspecific ChIP) values were subtracted from the IP values to generate the values shown in the graphs. The sequences of the primers used for the ChIP-qPCR are listed in the supplemental material.
Expression microarray analyses.
To determine the expression status of genes, we used previously published expression microarray data from MCF-7 cells treated with or without 100 nM E2 for 3 h (24) (GEO accession numbers GSM234903 through GSM234908). The raw data from three independent replicates were processed by Affymetrix GCOS software to obtain “present” or “absent” detection calls for each gene on the microarray. To account for the uncertainty due to variations within microarray data, we required a gene to have “present” or “absent” calls in all three replicates to be defined as unambiguously “expressed” or “unexpressed,” respectively. Genes with ambiguous expression status were eliminated from further analysis.
To define the E2-regulated transcriptome, the raw data were processed by Affymetrix GCOS software to obtain detection calls and signal values and then normalized by scaling. Only probe sets having “present” calls on at least two of the three arrays were included for further analysis; those signals were log2 transformed and median centered. The t test was applied to the normalized data matrix to identify differential genes between the E2-treated and untreated control samples. A significance level cutoff of 0.05 was applied to select each differential gene set and gene regulation was considered significant if the observed change was >1.5-fold.
Gene expression analyses by reverse transcription-qPCR.
MCF-7 cells were maintained in estrogen-free medium as described above and treated with ethanol or 100 nM E2 for 1, 3 and 6 h. For the 5,6-dichloro-1-d-ribofuranosyl-benzimidazole (DRB) gene expression analyses, MCF-7 cells were pretreated with vehicle (dimethyl sulfoxide) or 50 μM DRB (MP Biomedicals, catalog no. 157639) for 1 h prior to E2 treatment. Total RNA was isolated by using RNeasy columns and treatment with RNase-free DNase I (Qiagen) according to the manufacturer's specifications. First-strand cDNA synthesis was performed with 400 ng of total RNA, 1 μg oligo(dT), and 400 U of Moloney murine leukemia virus reverse transcriptase (Promega) in a 50-μl reaction, as described previously (1, 24). The resulting cDNA from each sample was then diluted 1:10 and analyzed by real-time PCR. Each real-time PCR consisted of 2 μl of cDNA, 1× SYBR green PCR master mix, and 0.25 μM concentrations each of the forward and reverse primers for a total reaction volume of 20 μl. Reactions were carried out in duplicates using a 96-well DNA Engine Opticon (MJ Research) or a 384-well Prism 7700 (ABI) real-time PCR thermocycler for 45 cycles (95°C for 15 s and 60°C for 1 min) after an initial 10-min incubation at 95°C. The change in the expression of each gene was calculated by using a standard curve of diluted cDNA from untreated samples (1:1, 1:10, 1:100) and normalized against the change of expression of the TBP gene, a housekeeping gene used as an internal control. Melting-curve analyses were performed to ensure that only the targeted amplicon was amplified. The sequences of the primers used for real-time PCR are listed in the supplemental material.
RESULTS
Pol II occupancy does not necessarily correspond with gene expression.
To explore the global mechanisms of signal-regulated transcription in human cells, we used ChIP coupled with genomic microarrays (i.e., ChIP-chip) to determine the localization of Pol II at ∼18,900 promoters in MCF-7 human breast cancer cells under basal, estrogen-free growth conditions. Using rigorous peak-finding criteria with a low false discovery rate, we found that Pol II was bound to and peaked at the TSSs of about half (i.e., 8,678) of the promoters on the array (see Fig. S1A in the supplemental material), in agreement with previous results (20, 23). Next, we matched the Pol II binding data with expression information for ∼14,000 of these promoters using microarray analyses in triplicate. By our criteria, genes having “present” calls in all three replicates were defined as unambiguously “expressed,” whereas those having “absent” calls in all three replicates were defined as unambiguously “unexpressed.” Genes with an ambiguous expression status in the microarray analyses were eliminated from further consideration. The expression status of several “expressed” and “unexpressed” genes was confirmed by quantitative reverse transcription-PCR (see Fig. S2 in the supplemental material). Our definition of unambiguously “unexpressed” does not, of course, exclude the possibility that low-level or unannotated transcripts, which escape detection by microarrays, are expressed from the promoters analyzed in our study. Nonetheless, it provides a useful working definition.
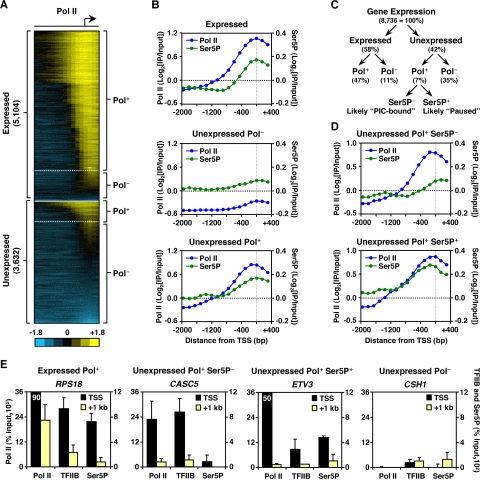
Using these stringent criteria, we identified 5,104 unambiguously “expressed” and 3,632 unambiguously “unexpressed” genes in MCF-7 cells under basal, estrogen-free growth conditions (see Fig. S1B in the supplemental material). As expected, the majority of the expressed genes contained Pol II at their promoters (expressed Pol+), whereas the majority of the unexpressed genes were devoid of Pol II (unexpressed Pol−) (Fig. 1A and B, top and middle). Surprisingly, however, a subset of the Pol II-bound genes was not expressed under the conditions tested (unexpressed Pol+; Fig. 1A and B, bottom). These findings indicate that the loading of Pol II at target promoters does not necessarily correlate with gene expression or Pol II activity.
FIG. 1.
The promoters of many unexpressed genes are preloaded with Pol II, which in many cases is prephosphorylated at Ser5 of the CTD. (A) Heat map of ChIP-chip binding analyses for Pol II at the promoters of expressed and unexpressed genes in MCF-7 cells. (B) Averaging of Pol II (blue) and Ser5P (green) promoter ChIP-chip signals for expressed, unexpressed Pol−, and unexpressed Pol+ genes. (C) Flowchart outlining the identification of promoters where Pol II is regulated at a postrecruitment level. (D) Averaging of Pol II (blue) and Ser5P (green) promoter ChIP-chip signals for unexpressed Pol+ genes with classification based on Ser5P status. (E) Gene-specific ChIP analyses of Pol II, TFIIB, and Ser5P binding at TSS (black) and downstream regions (+1 kb) (yellow) for a set of representative genes. Each bar represents the mean plus the standard error of the mean (SEM) (n ≥ 3).
Preloaded Pol II at unexpressed genes can undergo transcription initiation.
Pol II at the promoters of unexpressed genes may be associated with general transcription factors as part of a PIC. In this case, the CTD of the Pol II Rpb1 subunit should be largely unphosphorylated at both Ser2 and Ser5 of the heptapeptide repeat (15, 42). Alternatively, Pol II may be paused promoter-proximally at an early postinitiation/elongation stage, unable to progress into productive elongation (51, 54). In this case, the CTD of the Pol II Rpb1 subunit should be preferentially phosphorylated at Ser5 but not at Ser2 (7, 42, 52) (Fig. 1C). To explore these two possibilities in relation to our Pol II promoter binding results, we performed ChIP-chip using an antibody specifically recognizing the Ser5 phosphorylated form of the Pol II CTD (Ser5P) (Fig. 1B). As expected, Ser5P peaked at the TSS of expressed genes coincident with Pol II binding (Pol+), while little Ser5P was detected at the TSS of unexpressed Pol− genes (Fig. 1B, top and middle). The Ser5P status at the promoters of unexpressed Pol+ (i.e., “preloaded”) genes could be used to segregate the genes into two distinct groups. About two-thirds of the genes showed little Ser5P at the promoters (Ser5P−; perhaps representing PIC-bound promoters), while the remainder showed a peak of Ser5P at the TSS (Ser5P+; perhaps representing Pol II paused at an early elongation stage) (Fig. 1D). These results suggest that preloaded Pol II at the promoters of unexpressed genes may be poised at either the preinitiation or early elongation stages of transcription. Consistent with this observation, gene-specific ChIP analyses revealed the presence of Pol II and TFIIB at the TSS, but not the downstream (+1-kb) regions, of unexpressed Pol+ Ser5P− and unexpressed Pol+ Ser5P+ genes (Fig. 1E and see Fig. S3 in the supplemental material).
The majority of estrogen-regulated promoters contain Pol II prior to E2 treatment.
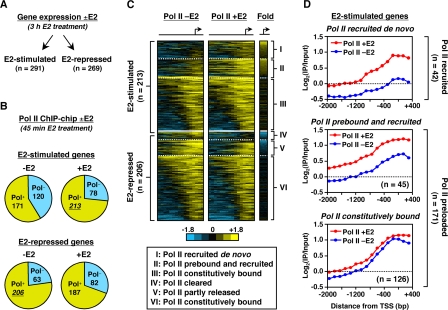
Although current models suggest a role for the postrecruitment activation of preloaded Pol II in signal-regulated gene expression (3, 7, 36, 48), the extent to which specific cellular signals can regulate Pol II activity postrecruitment has not been studied on a genome-wide scale. To explore this in higher eukaryotes, we examined the mechanisms through which estrogen, a developmentally and physiologically important signaling molecule, regulates Pol II occupancy and activity at target promoters. First, we defined the target genes for estrogen signaling by using expression microarray analyses from MCF-7 cells with or without a 3-h E2 treatment (24). This analysis revealed 560 genes whose expression changed significantly upon E2 treatment; 291 were stimulated and 269 were repressed, a finding consistent with previous reports (25) (Fig. 2A).
FIG. 2.
The majority of estrogen-regulated promoters are preloaded with Pol II prior to E2 treatment. (A) Gene expression microarray analysis in MCF-7 cells with (+) or without (−) E2 (3 h). (B) ChIP-chip analyses for Pol II at the promoters of all E2-regulated genes in MCF-7 cells ± E2 (45 min). (C) Heat maps of ChIP-chip for Pol II at the promoters of E2-stimulated and E2-repressed genes with (+) or without (−) E2 (45 min). (D) Averaging of Pol II promoter ChIP-chip signals for three classes of E2-stimulated genes with (+) or without (−) E2 (45 min).
Next, to determine the Pol II occupancy at the promoters of these E2-regulated genes, we performed ChIP-chip for Pol II in MCF-7 cells with or without a 45-min E2 treatment (Fig. 2B). We used a short E2 treatment to facilitate detection of direct, rather than secondary, effects. Surprisingly, in contrast to the historical “Pol II recruitment” model for gene activation by E2-liganded ERs (19, 35), the majority (171 of 291 [∼59%]) of E2-stimulated genes had elevated levels of Pol II at their promoters prior to E2 treatment (including two of the best studied mammalian “paused Pol II” genes, MYC and FOS [43, 56]; Fig. 2B, top). These levels either increased (i.e., Pol II prebound and further recruited) or stayed the same (i.e., Pol II prebound and unchanged) upon E2 treatment (collectively “Pol II preloaded” genes) (Fig. 2C and D). Similar trends were observed for the set of E2-repressed genes, with the majority of them containing Pol II both before and after E2 treatment (Fig. 2B, bottom, and C). We focused our remaining studies on the mechanisms regulating E2-dependent gene activation, which are better understood than E2-dependent gene repression. The presence of Pol II at the promoters of Pol II preloaded genes prior to E2 treatment was confirmed by nuclear run-on assays for selected target genes (see Fig. S4 in the supplemental material).
Together, our gene expression and Pol II ChIP-chip results suggest that for the majority of E2-regulated genes, changes in Pol II promoter occupancy are not sufficient to explain the changes in gene expression upon E2 treatment. In this regard, all of the “constitutively” Pol II preloaded genes (126 out of 171 total) show a significant increase in expression upon E2 treatment without a significant increase in Pol II occupancy at their promoters (Fig. 2D). Our present studies, which used an microarray platform containing all RefSeq promoters, have provided a higher estimate for the percentage of E2-regulated promoters containing preloaded Pol II than our previous study (24). This is likely due to the use of a smaller and more biased microarray platform used in our previous study.
Preloaded Pol II moves into the body of the gene in the presence of E2.
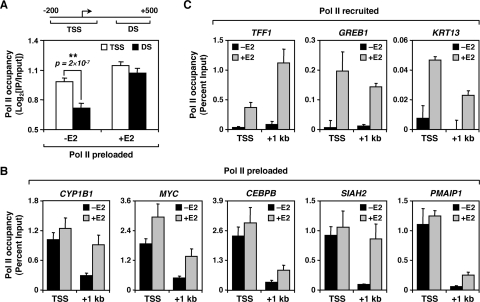
The presence of preloaded Pol II at the promoters of E2-stimulated genes prior to E2 exposure suggests a role for E2 signaling in regulating Pol II activity postrecruitment. For example, E2 may promote transcriptional elongation by Pol II into the body of the gene. To test this, we compared Pol II occupancy at the TSS to Pol II occupancy in the downstream region with or without E2 for all E2-stimulated Pol II preloaded genes identified in our ChIP-chip studies. Prior to E2 treatment, Pol II occupancy downstream was significantly lower than occupancy at the TSS (P = 2 × 10−7) (Fig. 3A, left). After E2 treatment, Pol II occupancy downstream increased to levels similar to those observed at the TSS (Fig. 3A, right), suggesting that E2 treatment promotes the movement of preloaded Pol II through the gene. To validate these results, we analyzed by ChIP-qPCR the occupancy of Pol II at the TSS and +1-kb regions of 12 E2-stimulated genes identified in our ChIP-chip experiments (Fig. 3B and C and data not shown). These experiments confirmed the ChIP-chip results and showed that E2-stimulated genes can be divided in two broad classes based on the regulation of Pol II binding at their promoters: (i) Pol II preloaded genes (n = 171), where Pol II is present primarily at the TSS prior to E2 treatment and moves into the body of the gene upon E2 treatment (e.g., CYP1B1, MYC, CEBPB, SIAH2, PMAIP1, FOS, MDM2, and STC2; Fig. 3B and data not shown), and (ii) Pol II recruited genes (n = 42), where Pol II is absent prior to E2 treatment and becomes recruited upon activation (e.g., TFF1, GREB1, KRT13, and HOXC5; Fig. 3C and data not shown). Together, our ChIP-chip and gene-specific studies indicate that in most cases, E2 controls target gene expression by regulating the activity, not the recruitment, of Pol II.
FIG. 3.
Preloaded Pol II localizes to TSS prior to E2 treatment and moves into the body of the gene upon activation. (A) Comparison of Pol II occupancy at the TSS (−200 to +100 bp) and downstream regions (DS; +300 to +500 bp) for all Pol II preloaded genes (n = 171), as determined by ChIP-chip with (+) or without (−) E2 (45 min). The associated t test P value is shown. (B and C) Gene-specific ChIP analyses of Pol II binding at the TSS and +1-kb regions of Pol II recruited (B) and Pol II preloaded (C) E2-stimulated genes with (+) or without (−) E2 (45 min). Each bar represents the mean plus the SEM (n ≥ 3).
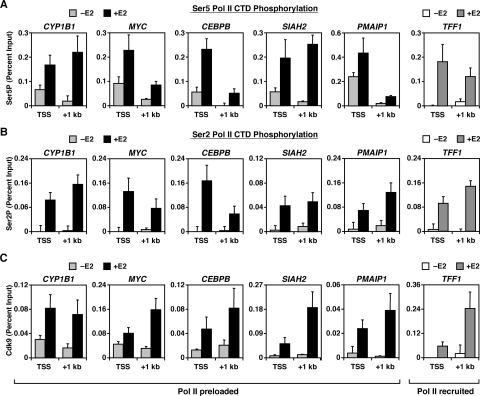
E2 stimulates the phosphorylation of preloaded Pol II.
To explore the mechanisms by which E2 signaling regulates Pol II activity, we determined the effects of E2 on phosphorylation of the Pol II CTD at E2 target genes by ChIP-qPCR. Prior to E2 treatment, preloaded Pol II at the TSS was phosphorylated at Ser5, but showed little phosphorylation of Ser2 (Fig. 4A and B). After E2 treatment, Ser5P and Ser2P increased at the TSS and downstream regions of both Pol II preloaded and Pol II recruited genes, indicating the movement of Ser5/Ser2-phoshorylated Pol II through the gene (Fig. 4A and B). These results, which are consistent with and extend the results of our previous study (24), suggest that preloaded Pol II may initiate transcription, but fails to enter productive elongation, before E2 treatment. Furthermore, they suggest that E2-induced Pol II phosphorylation, primarily Ser2P, is the trigger that stimulates the elongation activity of preloaded Pol II.
FIG. 4.
Preloaded Pol II is partially phosphorylated at Ser5, but not Ser2, prior to E2 treatment, and Pol II CTD phosphorylation increases in conjunction with Cdk9 recruitment upon E2 treatment. (A to C) Gene-specific ChIP analyses of Pol II Ser5P (A) and Pol II Ser2 (B), as well as Cdk9 occupancy (C), at the TSS and +1-kb regions of representative E2-stimulated genes with (+) or without (−) E2 (45 min). Each bar represents the mean plus the SEM (n ≥ 3).
To investigate further how E2 stimulates the phosphorylation and elongation activity of Pol II, we analyzed the localization of Cdk9 at the TSS and +1-kb regions of E2-stimulated genes. Cdk9, a subunit of P-TEFb, is the kinase primarily responsible for the phosphorylation of Ser2 in the Pol II CTD, as well as productive elongation (41, 44). Interestingly, Cdk9 was recruited to E2-stimulated genes in an E2-dependent manner and, for most of the genes, Cdk9 occupancy was higher in the +1-kb region, a finding consistent with its ability to travel with Pol II through the gene (7) (Fig. 4C). In this regard, inhibition of the activity of Pol II kinases (e.g., Cdk9) by DRB blocked the E2-dependent induction of representative Pol II preloaded and Pol II recruited genes (e.g., CYP1B1 and TFF1; see Fig. S5 in the supplemental material). Collectively, our findings present evidence connecting E2 signaling with Cdk9 recruitment, Pol II CTD phosphorylation, and transcriptional activation of E2-stimulated genes.
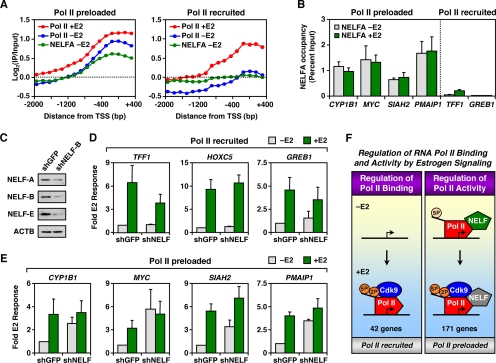
NELF binds to Pol II preloaded promoters and represses gene expression prior to E2.
In addition to the mechanisms regulating E2-induced Pol II activation, we also studied the mechanisms preventing preloaded Pol II from transcribing through the body of genes prior to E2 treatment. The NELF and DSIF complexes are two coregulators which cooperate to repress transcription elongation and establish paused Pol II at promoters (41, 60, 64). Thus, we examined a potential role of NELF and DSIF in the regulation of preloaded Pol II at E2-regulated promoters. We determined the binding of the NELF-A subunit to E2-stimulated promoters prior to E2 treatment by ChIP-chip, as described above for Pol II. Interestingly, we observed NELF-A binding to the majority of promoters with preloaded Pol II but little NELF-A binding to promoters with recruited Pol II (Fig. 5A), a finding consistent with that of our previous study (24). Gene-specific ChIP assays confirmed these results and revealed a similar binding pattern for the Spt5 subunit of DSIF (Fig. 5B and see Fig. S6 in the supplemental material). Both NELF-A and Spt5 remained associated with the promoters of these genes after E2 treatment, although E2-dependent Cdk9 recruitment and Pol II phosphorylation (Fig. 4) were sufficient to overcome the negative effects of NELF and DSIF and allow Pol II to transcribe through the gene. Together, our results implicate NELF and DSIF in the negative regulation of preloaded Pol II at the promoters of E2-stimulated genes prior to Pol II phosphorylation and activation by E2 signaling. They also suggest that NELF and DSIF, although still associated with the promoter, are unable to mediate their negative effects after E2 treatment.
FIG. 5.
NELF binds to promoters containing preloaded Pol II and prevents transcription through the gene prior to E2 treatment. (A) Averaging of Pol II and NELF-A ChIP-chip signals at 221 E2-stimulated promoters for the following conditions: Pol II -E2 (blue), Pol II +E2 (red), and NELF-A −E2 (green). (B) Gene-specific ChIP analysis of NELF-A binding at promoters of representative E2-stimulated genes with (+) or without (−) E2 (45 min). Each bar represents the mean plus the SEM (n ≥ 3). (C) Western blot showing the shRNA-mediated depletion of NELF-B (shNELF-B) in MCF-7 cells versus a GFP knockdown control (shGFP). NELF-B, as well as NELF-A and NELF-E, protein levels decrease in the shNELF-B cells. ACTB, loading control. (D and E) Gene-specific analysis of mRNA expression for representative Pol II recruited (D) and Pol II preloaded (E) genes in MCF-7 cells with or without NELF knockdown with (+) or without (−) E2 (3 h). Each bar represents the mean plus the SEM (n ≥ 3). (F) Summary of observations regarding the E2-dependent regulation of Pol II binding and activity. See the text for details.
To further understand the role of NELF in E2-dependent Pol II activation, we knocked down NELF-B, a NELF subunit that has been shown to interact with ERα (5), in MCF-7 cells. Interestingly, knockdown of NELF-B caused a reduction in NELF-A and NELF-E protein levels as well, possibly due to destabilization of the whole NELF complex (Fig. 5C). To study the effects of NELF depletion on E2-regulated gene expression, we assayed the mRNA levels of several Pol II recruited and Pol II preloaded genes with or without a 3-h E2 treatment (Fig. 5D and E). NELF depletion had no major effect on either the basal transcription or the E2 response of Pol II recruited genes (Fig. 5D) but did increase the levels of basal gene expression at Pol II preloaded genes, a finding consistent with a role of NELF in preventing preloaded Pol II from transcribing the genes prior to E2 treatment (Fig. 5E). Interestingly, NELF depletion had no major effect on gene expression after E2 treatment, confirming our hypothesis that NELF, although present, is unable mediate its negative effects after E2-dependent Pol II phosphorylation and Cdk9 recruitment (Fig. 5F).
Pol II preloaded genes are stimulated by E2 more rapidly than Pol II recruited genes.
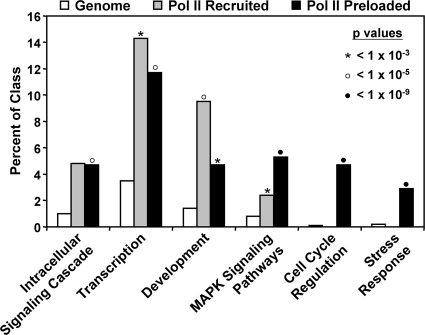
Current hypotheses suggest a biologically important role for the postrecruitment activation of preloaded Pol II in the rapid induction of genes in response to physiological and environmental signals (3, 7, 36, 48, 65). In this regard, we found that Pol II recruited and Pol II preloaded genes are enriched in different ontological categories (Fig. 6). For example, Pol II preloaded genes are enriched in ontological categories where rapid induction might be expected (e.g., cell cycle regulation and stress responses), whereas Pol II recruited genes are enriched in other ontological categories (e.g., development).
FIG. 6.
Pol II recruited and Pol II preloaded genes are enriched in different ontological categories. GO analysis was performed by using Genecodis and filtered for the universe of applicable GO terms (see Materials and Methods). Each gene set in each category was expressed as the percentage of genes in the class and assigned its own P value (chi-square analysis).
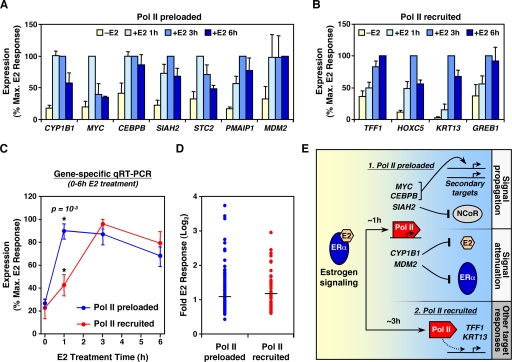
To explore this further, we examined whether there are differences in the kinetics of E2-dependent induction between Pol II preloaded and Pol II recruited genes. MCF-7 cells were treated with E2 for 0, 1, 3, and 6 h and the mRNA levels of a number E2-stimulated genes were measured by quantitative RT-PCR (Fig. 7A and B). On average, Pol II preloaded genes were stimulated by E2 significantly faster than Pol II recruited genes (P = 0.001), with the former peaking at 1 h and the latter peaking at 3 h after E2 treatment (Fig. 7C). Consistent with these gene expression results, preloaded Pol II moved more rapidly into the body of the CYP1B1 gene compared to Pol II recruited to the TFF1 gene (see Fig. S7 in the supplemental material). Although Pol II preloading and postrecruitment activation was associated with more rapid transcriptional responses, the absolute E2 induction after 3 h of treatment did not differ between Pol II preloaded and Pol II recruited genes (Fig. 7D). Interestingly, many of the Pol II preloaded genes that we examined play important roles in propagating (e.g., MYC, CEBPB, and SIAH2) or attenuating (e.g., CYP1B1 and MDM2) E2 signaling itself (see the Discussion) (Fig. 7E). Together, our results suggest that the regulation of Pol II by cellular signals at the postrecruitment level may facilitate immediate responses that contribute to the carefully coordinated regulation of the signaling response itself.
FIG. 7.
Pol II preloaded genes are induced by E2 more rapidly than Pol II recruited genes. (A and B) Gene-specific analysis of mRNA expression for representative Pol II preloaded (A) or Pol II recruited (B) genes in MCF-7 cells before or after E2 treatment for 1, 3, and 6 h. The E2 response is shown as the percentage of the maximum E2 response for each individual gene. Each bar represents the mean plus the SEM (n ≥ 3). (C) Averaging of E2 responses for all Pol II preloaded and Pol II recruited genes shown in panels A and B, respectively. Error bars represent the SEM. The associated t test P value for differences between the two points at 1 h is shown. (D) Scatterplot of absolute E2 responses for all Pol II preloaded and Pol II recruited genes, as determined by expression microarray analysis of MCF-7 cells treated with or without E2 for 3 h. The horizontal lines represent the mean for each class of genes. (E) Biological functions of E2-stimulated genes containing preloaded Pol II. Pol II preloaded genes, which are induced earlier than Pol II recruited genes, play key roles in propagating or attenuating estrogen signaling. See the text for details.
DISCUSSION
In this study, we have used genomic and gene-specific assays to examine the binding and regulation of Pol II to promoters across the genome under basal growth conditions, as well as in response to E2 signaling. Our results indicate that: (i) Pol II is widely distributed at the promoters of many unexpressed genes with Ser5-phosphorylated Pol II prior to E2 signaling, (ii) the predominant genomic outcome of E2 signaling is the postrecruitment regulation of Pol II activity through phosphorylation, (iii) NELF acts to repress transcription elongation by preloaded Pol II until the E2 signal is received, and (iv) E2-dependent activation of preloaded Pol II facilitates rapid gene regulatory responses which play important physiological roles in regulating estrogen signaling itself. These studies go beyond our previous work (24), which used a smaller microarray platform containing a limited number of promoters (∼1,000), including a biased set of E2-regulated genes identified in previous studies performed by other labs. Collectively, our results here establish a new mode of E2-dependent gene regulation that is likely to apply to a wide variety of signal-regulated pathways.
Many unexpressed genes have preloaded Pol II at their promoters.
Our results show that the promoters of a large fraction (∼20%) of unexpressed genes in the human genome have preloaded Pol II prior to specific signaling events (Fig. 1C). For about two-thirds of these genes, preloaded Pol II remains in the PIC, while for the remaining genes, preloaded Pol II has initiated transcription, as marked by Ser5P, but fails to proceed to productive elongation (Fig. 1D). Our results are consistent with other studies showing that Pol II is widely distributed at the promoters of many unexpressed genes prior to specific signaling events (6, 20, 23, 36, 48, 65), although the Ser5P status of these preloaded Pol II has not been previously studied. A current hypothesis suggests that preloaded Pol II at unexpressed genes becomes activated by certain physiological or developmental signals at appropriate times. Although this hypothesis has been tested in gene-specific studies (36, 54, 61), a genome-wide approach is needed to evaluate the generality and mechanisms of postrecruitment activation of Pol II by specific signaling pathways.
Estrogen signaling activates preloaded Pol II through phosphorylation.
Current models for signal-dependent transcription by E2 and other signaling pathways focus on the signal-dependent recruitment of Pol II to target promoters (19, 35, 46). This “Pol II recruitment” model was developed based on a limited subset of signal-regulated genes, and its generality has not been examined across the full E2-regulated transcriptome. Recent studies, as well as the results described here, have shown that Pol II is widely distributed at gene promoters prior to specific signaling events (6, 20, 23, 36, 48, 65), but the activation of preloaded Pol II by a specific cellular signal has not previously been studied on a global scale.
Here we show that the majority of E2-stimulated genes (∼59%) are preloaded with Pol II, which remains primarily at the TSS prior to E2 treatment and moves into the body of the gene after E2 treatment (Fig. 3). This movement of Pol II occurs concomitantly with E2-dependent Pol II phosphorylation, primarily Ser2P, as well as Cdk9 recruitment (Fig. 4). Accordingly, inhibition of Pol II kinase activity (e.g., Cdk9) by DRB abolished the induction of E2-stimulated genes (see Fig. S5 in the supplemental material). Cdk9 occupancy was higher in the coding region of the majority of E2-stimulated genes, a finding consistent with its ability to travel with Pol II through the gene (37). Together, our results are consistent with a direct transcriptional role for E2-stimulated Cdk9 recruitment and Pol II phosphorylation. Note, however, that DRB has also been shown to have nontranscriptional effects (e.g., interference with RNA processing) (37).
The E2-dependent recruitment of Cdk9 presents a novel means for regulatory control of Pol II-dependent transcription by E2 signaling. Cdk9 partners with cyclin T to form P-TEFb, which phosphorylates DSIF, NELF, and Ser2 of the Pol II CTD to allow paused Pol II to enter productive elongation (17, 44, 63). One interesting question is the mechanism through which Cdk9 becomes recruited to genes upon E2 treatment. Interestingly, cyclin T has been shown to interact with ERα and may be responsible for the Cdk9 recruitment to E2-stimulated genes (62). An alternative mechanism may involve the bromodomain containing protein BRD4, which mediates the association of P-TEFb to human genes through its interaction with acetylated histones (21). Collectively, our results expand the list of E2 signaling coregulators to include factors important for transcription elongation, in addition to the well-studied transcription initiation factors.
NELF mediates its negative elongation effects prior to, not after, E2 exposure.
Our studies show that NELF and DSIF bind to the majority of Pol II preloaded promoters prior to E2 treatment but show little binding to Pol II recruited promoters under the same conditions (Fig. 5A and B). These results are consistent with the role of NELF and DSIF in establishing paused Pol II at the 5′ end of genes (30, 36, 60, 64). Interestingly, though, NELF and DSIF remained associated with the promoters of Pol II preloaded genes after E2 treatment, suggesting that the E2-dependent activation of Pol II does not occur though the release of NELF and DSIF from the promoter. One possibility is that E2-dependent Pol II phosphorylation is sufficient to overcome the negative effects of NELF and DSIF by making the Pol II CTD accessible to Pol II CTD-binding elongation factors (42). In agreement with this hypothesis, our NELF knockdown studies showed that NELF represses the expression of Pol II preloaded genes prior to E2 treatment but is unable mediate its negative effects after the E2-dependent Pol II phosphorylation and Cdk9 recruitment, even though NELF is still present at the promoter (Fig. 5E). Another possibility is the inactivation of NELF through phosphorylation by Cdk9, as has been shown in vitro (17), although this has not been explored in vivo. A third possibility is that the persistent association of NELF and DSIF with the promoter is due to reassociation with reinitiated Pol II. It will be interesting to test these and other possibilities in future studies.
Aiyar et al. have previously examined the role of NELF in ligand-regulated activation of E2 target genes, focusing on genes with recruited Pol II (e.g., TFF1, also known as pS2) (4, 5). They found that both basal and E2-induced expression of TFF1 was increased upon depletion of NELF-B (also known as COBRA1) and concluded that COBRA1 causes Pol II to pause at the promoter-proximal region of TFF1 (5). This result is in apparent contradiction to our observation that NELF-B depletion does not have a major effect on the expression of TFF1 and other “Pol II recruited” genes. One possible explanation is that Aiyar et al. performed their experiments using T47D cells, whereas we used MCF-7 cells. The effect of NELF-B depletion on TFF1 expression may be cell-type specific, as are the estrogen-regulated gene expression patterns in different breast cancer cell lines (25). Interestingly, basal TFF1 expression in T47D cells is much lower than basal TFF1 expression in MCF-7 cells (data not shown), which is consistent with the hypothesis that NELF and other negative regulation complexes may play a larger role in TFF1 regulation in T47D cells than in MCF-7 cells.
Postrecruitment regulation of Pol II allows for rapid gene regulatory responses and possible autoregulatory control.
Previous studies have suggested a biologically important role for the postrecruitment activation of preloaded Pol II in the rapid induction of genes in response to physiological and environmental signals (3, 7, 48). Our kinetic analysis indicates that Pol II preloaded genes are stimulated by E2 significantly faster than Pol II recruited genes (Fig. 7C). These results, which are based on steady-state measurements of mRNA levels, are consistent with a direct transcriptional effect but could also include effects on mRNA stability and turnover. Interestingly, many of the E2-stimulated Pol II preloaded genes that we identified in our analyses play important roles in regulating the E2 signaling pathway. For example, MYC, CEBPB and FOS genes encode transcription factors that cooperate with ERα to regulate secondary targets (10, 28), thus propagating E2 signaling (Fig. 7E). In addition, SIAH2, which encodes a ubiquitin ligase, helps to propagate global estrogen signaling responses by targeting the nuclear corepressor NCoR for rapid degradation by the proteasome (16). Not all rapidly stimulated Pol II preloaded genes, however, regulate E2 signaling in a positive way. For example, CYP1B1 encodes a cytochrome P450 monooxygenase that metabolizes E2 and may be part of a rapid metabolic feedback response controlling E2 levels in target tissues (59). In addition, MDM2 encodes a ubiquitin ligase involved in the rapid degradation of ERα by the proteasome upon E2 treatment (14). Similar examples of positive and negative autoregulation by target genes have been previously described for a series of cellular signals, including mitogen-activated protein kinase-, NF-κB-, transforming growth factor β-, and STAT-regulated pathways (22, 32, 40, 66). Such autoregulatory functions were not readily apparent in the set of Pol II recruited genes that we examined. Collectively, our results, together with previous studies, suggest that the postrecruitment regulation of Pol II as an endpoint of cellular signaling pathways may facilitate rapid responses that contribute to the precise regulation of the cellular signal itself.
Supplementary Material
Acknowledgments
We thank John Lis and members of the Kraus and Lis labs for critical reading of the manuscript; Tong Zhang for performing the gene expression microarray analyses; and Matt Gamble, Raga Krishnakumar, Tong Zhang, and members of the Kraus lab for technical advice and helpful discussions. We also thank Benita Katzenellebogen for providing the MCF-7 cells and Hiroshi Handa for providing the NELF-B and NELF-E antibodies.
This study was supported by grants from the NIH/NIDDK (DK058110 and DK069710) and the Cornell Center of Vertebrate Genomics to W.L.K. and predoctoral fellowships from the Department of Defense Breast Cancer Research Program to M.K. (BC050755) and G.D.I. (BC050806).
Footnotes
Published ahead of print on 22 December 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Acevedo, M. L., K. C. Lee, J. D. Stender, B. S. Katzenellenbogen, and W. L. Kraus. 2004. Selective recognition of distinct classes of coactivators by a ligand-inducible activation domain. Mol. Cell 13725-738. [DOI] [PubMed] [Google Scholar]
- 2.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103667-678. [DOI] [PubMed] [Google Scholar]
- 3.Aida, M., Y. Chen, K. Nakajima, Y. Yamaguchi, T. Wada, and H. Handa. 2006. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol. Cell. Biol. 266094-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aiyar, S. E., A. L. Blair, D. A. Hopkinson, S. Bekiranov, and R. Li. 2007. Regulation of clustered gene expression by cofactor of BRCA1 (COBRA1) in breast cancer cells. Oncogene 262543-2553. [DOI] [PubMed] [Google Scholar]
- 5.Aiyar, S. E., J. L. Sun, A. L. Blair, C. A. Moskaluk, Y. Z. Lu, Q. N. Ye, Y. Yamaguchi, A. Mukherjee, D. M. Ren, H. Handa, and R. Li. 2004. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 182134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. [DOI] [PubMed] [Google Scholar]
- 7.Boehm, A. K., A. Saunders, J. Werner, and J. T. Lis. 2003. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 237628-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buck, M. J., and J. D. Lieb. 2004. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 83349-360. [DOI] [PubMed] [Google Scholar]
- 9.Cai, H., and D. S. Luse. 1987. Transcription initiation by RNA polymerase II in vitro: properties of preinitiation, initiation, and elongation complexes. J. Biol. Chem. 262298-304. [PubMed] [Google Scholar]
- 10.Cheng, A. S., V. X. Jin, M. Fan, L. T. Smith, S. Liyanarachchi, P. S. Yan, Y. W. Leu, M. W. Chan, C. Plass, K. P. Nephew, R. V. Davuluri, and T. H. Huang. 2006. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol. Cell 21393-404. [DOI] [PubMed] [Google Scholar]
- 11.Corden, J. L. 1990. Tails of RNA polymerase II. Trends Biochem. Sci. 15383-387. [DOI] [PubMed] [Google Scholar]
- 12.Core, L. J., and J. T. Lis. 2008. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 3191791-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couse, J. F., and K. S. Korach. 1999. Estrogen receptor null mice: what have we learned and where will they lead us? Endocrinol. Rev. 20358-417. [DOI] [PubMed] [Google Scholar]
- 14.Duong, V., N. Boulle, S. Daujat, J. Chauvet, S. Bonnet, H. Neel, and V. Cavailles. 2007. Differential regulation of estrogen receptor alpha turnover and transactivation by Mdm2 and stress-inducing agents. Cancer Res. 675513-5521. [DOI] [PubMed] [Google Scholar]
- 15.Dvir, A. 2002. Promoter escape by RNA polymerase II. Biochim. Biophys. Acta 1577208-223. [DOI] [PubMed] [Google Scholar]
- 16.Frasor, J., J. M. Danes, C. C. Funk, and B. S. Katzenellenbogen. 2005. Estrogen down-regulation of the corepressor N-CoR: mechanism and implications for estrogen derepression of N-CoR-regulated genes. Proc. Natl. Acad. Sci. USA 10213153-13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujinaga, K., D. Irwin, Y. Huang, R. Taube, T. Kurosu, and B. M. Peterlin. 2004. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 24787-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilchrist, D. A., S. Nechaev, C. Lee, S. K. Ghosh, J. B. Collins, L. Li, D. S. Gilmour, and K. Adelman. 2008. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 221921-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14121-141. [PubMed] [Google Scholar]
- 20.Guenther, M. G., S. S. Levine, L. A. Boyer, R. Jaenisch, and R. A. Young. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 13077-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang, M. K., K. Mochizuki, M. Zhou, H. S. Jeong, J. N. Brady, and K. Ozato. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19523-534. [DOI] [PubMed] [Google Scholar]
- 22.Javelaud, D., and A. Mauviel. 2005. Crosstalk mechanisms between the mitogen-activated protein kinase pathways and Smad signaling downstream of TGF-beta: implications for carcinogenesis. Oncogene 245742-5750. [DOI] [PubMed] [Google Scholar]
- 23.Kim, T. H., L. O. Barrera, M. Zheng, C. Qu, M. A. Singer, T. A. Richmond, Y. Wu, R. D. Green, and B. Ren. 2005. A high-resolution map of active promoters in the human genome. Nature 436876-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kininis, M., B. S. Chen, A. G. Diehl, G. D. Isaacs, T. Zhang, A. C. Siepel, A. G. Clark, and W. L. Kraus. 2007. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol. Cell. Biol. 275090-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kininis, M., and W. L. Kraus. 2008. A global view of transcriptional regulation by nuclear receptors: gene expression, factor localization, and DNA sequence analysis. Nuclear Recept. Signal 6e005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirmizis, A., S. M. Bartley, A. Kuzmichev, R. Margueron, D. Reinberg, R. Green, and P. J. Farnham. 2004. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 181592-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399609-613. [DOI] [PubMed] [Google Scholar]
- 28.Kushner, P. J., D. A. Agard, G. L. Greene, T. S. Scanlan, A. K. Shiau, R. M. Uht, and P. Webb. 2000. Estrogen receptor pathways to AP-1. J. Steroid Biochem. Mol. Biol. 74311-317. [DOI] [PubMed] [Google Scholar]
- 29.Laganiere, J., G. Deblois, C. Lefebvre, A. R. Bataille, F. Robert, and V. Giguere. 2005. From the cover: location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl. Acad. Sci. USA 10211651-11656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, C., X. Li, A. Hechmer, M. Eisen, M. D. Biggin, B. J. Venters, C. Jiang, J. Li, B. F. Pugh, and D. S. Gilmour. 2008. NELF and GAGA factor are linked to promoter proximal pausing at many genes in Drosophila. Mol. Cell. Biol. [DOI] [PMC free article] [PubMed]
- 31.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, D. B. Gordon, B. Ren, J. J. Wyrick, J. B. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298799-804. [DOI] [PubMed] [Google Scholar]
- 32.Lipniacki, T., and M. Kimmel. 2007. Deterministic and stochastic models of NFκB pathway. Cardiovasc. Toxicol. 7215-234. [DOI] [PubMed] [Google Scholar]
- 33.Lis, J. 1998. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harbor Symp. Quant. Biol. 63347-356. [DOI] [PubMed] [Google Scholar]
- 34.Margaritis, T., and F. C. Holstege. 2008. Poised RNA polymerase II gives pause for thought. Cell 133581-584. [DOI] [PubMed] [Google Scholar]
- 35.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocrinol. Rev. 20321-344. [DOI] [PubMed] [Google Scholar]
- 36.Muse, G. W., D. A. Gilchrist, S. Nechaev, R. Shah, J. S. Parker, S. F. Grissom, J. Zeitlinger, and K. Adelman. 2007. RNA polymerase is poised for activation across the genome. Nat. Genet. 391507-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ni, Z., B. E. Schwartz, J. Werner, J. R. Suarez, and J. T. Lis. 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 1355-65. [DOI] [PubMed] [Google Scholar]
- 38.Nilsson, S., S. Makela, E. Treuter, M. Tujague, J. Thomsen, G. Andersson, E. Enmark, K. Pettersson, M. Warner, and J. A. Gustafsson. 2001. Mechanisms of estrogen action. Physiol. Rev. 811535-1565. [DOI] [PubMed] [Google Scholar]
- 39.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108439-451. [DOI] [PubMed] [Google Scholar]
- 40.Paukku, K., and O. Silvennoinen. 2004. STATs as critical mediators of signal transduction and transcription: lessons learned from STAT5. Cytokine Growth Factor Rev. 15435-455. [DOI] [PubMed] [Google Scholar]
- 41.Peterlin, B. M., and D. H. Price. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23297-305. [DOI] [PubMed] [Google Scholar]
- 42.Phatnani, H. P., and A. L. Greenleaf. 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 202922-2936. [DOI] [PubMed] [Google Scholar]
- 43.Plet, A., D. Eick, and J. M. Blanchard. 1995. Elongation and premature termination of transcripts initiated from c-fos and c-myc promoters show dissimilar patterns. Oncogene 10319-328. [PubMed] [Google Scholar]
- 44.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 202629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price, D. H. 2008. Poised polymerases: on your mark … get set … go! Mol. Cell 307-10. [DOI] [PubMed] [Google Scholar]
- 46.Ptashne, M. 2005. Regulation of transcription: from lambda to eukaryotes. Trends Biochem. Sci. 30275-279. [DOI] [PubMed] [Google Scholar]
- 47.Ptashne, M., and A. Gann. 1997. Transcriptional activation by recruitment. Nature 386569-577. [DOI] [PubMed] [Google Scholar]
- 48.Radonjic, M., J. C. Andrau, P. Lijnzaad, P. Kemmeren, T. T. Kockelkorn, D. van Leenen, N. L. van Berkum, and F. C. Holstege. 2005. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell 18171-183. [DOI] [PubMed] [Google Scholar]
- 49.R Development Core Team. 2006. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org.
- 50.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rougvie, A. E., and J. T. Lis. 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of Drosophila melanogaster is transcriptionally engaged. Cell 54795-804. [DOI] [PubMed] [Google Scholar]
- 52.Ryser, S., T. Fujita, S. Tortola, I. Piuz, and W. Schlegel. 2007. The rate of c-fos transcription in vivo is continuously regulated at the level of elongation by dynamic stimulus-coupled recruitment of positive transcription elongation factor b. J. Biol. Chem. 2825075-5084. [DOI] [PubMed] [Google Scholar]
- 53.Saldanha, A. J. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 203246-3248. [DOI] [PubMed] [Google Scholar]
- 54.Saunders, A., L. J. Core, and J. T. Lis. 2006. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell. Biol. 7557-567. [DOI] [PubMed] [Google Scholar]
- 55.Smyth, G. K., and T. Speed. 2003. Normalization of cDNA microarray data. Methods 31265-273. [DOI] [PubMed] [Google Scholar]
- 56.Strobl, L. J., and D. Eick. 1992. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. EMBO J. 113307-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamkun, J. W. 2007. Stalled polymerases and transcriptional regulation. Nat. Genet. 391421-1422. [DOI] [PubMed] [Google Scholar]
- 58.Thibaud-Nissen, F., H. Wu, T. Richmond, J. C. Redman, C. Johnson, R. Green, J. Arias, and C. D. Town. 2006. Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J. 47152-162. [DOI] [PubMed] [Google Scholar]
- 59.Tsuchiya, Y., M. Nakajima, and T. Yokoi. 2005. Cytochrome P450-mediated metabolism of estrogens and its regulation in human. Cancer Lett. 227115-124. [DOI] [PubMed] [Google Scholar]
- 60.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, X., C. Lee, D. S. Gilmour, and J. P. Gergen. 2007. Transcription elongation controls cell fate specification in the Drosophila embryo. Genes Dev. 211031-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wittmann, B. M., K. Fujinaga, H. Deng, N. Ogba, and M. M. Montano. 2005. The breast cell growth inhibitor, estrogen down regulated gene 1, modulates a novel functional interaction between estrogen receptor alpha and transcriptional elongation factor cyclin T1. Oncogene 245576-5588. [DOI] [PubMed] [Google Scholar]
- 63.Yamada, T., Y. Yamaguchi, N. Inukai, S. Okamoto, T. Mura, and H. Handa. 2006. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21227-237. [DOI] [PubMed] [Google Scholar]
- 64.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 9741-51. [DOI] [PubMed] [Google Scholar]
- 65.Zeitlinger, J., A. Stark, M. Kellis, J. W. Hong, S. Nechaev, K. Adelman, M. Levine, and R. A. Young. 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 391512-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou, H., A. Lin, Z. Gu, S. Chen, N. H. Park, and R. Chiu. 2000. 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced c-Jun N-terminal kinase (JNK) phosphatase renders immortalized or transformed epithelial cells refractory to TPA-inducible JNK activity. J. Biol. Chem. 27522868-22875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.