Abstract
Kinetoplastid flagellates attach a 39-nucleotide spliced leader (SL) upstream of protein-coding regions in polycistronic RNA precursors through trans splicing. SL modifications include cap 2′-O-ribose methylation of the first four nucleotides and pseudouridine (ψ) formation at uracil 28. In Trypanosoma brucei, TbMTr1 performs 2′-O-ribose methylation of the first transcribed nucleotide, or cap 1. We report the characterization of an SL RNA processing complex with TbMTr1 and the SLA1 H/ACA small nucleolar ribonucleoprotein (snoRNP) particle that guides SL ψ28 formation. TbMTr1 is in a high-molecular-weight complex containing the four conserved core proteins of H/ACA snoRNPs, a kinetoplastid-specific protein designated methyltransferase-associated protein (TbMTAP), and the SLA1 snoRNA. TbMTAP-null lines are viable but have decreased SL RNA processing efficiency in cap methylation, 3′-end maturation, and ψ28 formation. TbMTAP is required for association between TbMTr1 and the SLA1 snoRNP but does not affect U1 small nuclear RNA methylation. A complex methylation profile in the mRNA population of TbMTAP-null lines indicates an additional effect on cap 4 methylations. The TbMTr1 complex specializes the SLA1 H/ACA snoRNP for efficient processing of multiple modifications on the SL RNA substrate.
Mature RNA molecules contain extensive posttranscriptional nucleotide modifications of both base and ribose moieties. The isomerization of uracil to pseudouridine (5-ribosyluracil) and 2′-O-ribose methylation occurs by either site-specific (31, 50) or small nucleolar RNA (snoRNA)-guided enzymes (13). Conserved snoRNA-guided rRNA nucleotide modifications occur in functionally relevant regions (14), with their absence resulting in disease states due to defective catalytic RNA activity (76). Cap ribose methylations of RNA polymerase II transcripts by cap-specific methyltransferases (MTases) are conserved in higher eukaryotes and implicated in the enhancement of translational efficiency (28a).
Kinetoplastid protozoa, including human-pathogenic Leishmania major, Trypanosoma brucei, and Trypanosoma cruzi, transcribe all protein-encoding genes polycistronically (28, 40). Maturation to translatable monocistronic units requires resolution of each coding region by trans splicing of a 39-nucleotide (nt) spliced leader (SL) exon and 3′-end polyadenylation. The SL RNA is involved in the maturation of each and every nuclear mRNA, accounting for approximately 7% of total RNA synthesis (8, 23). Rapid substrate SL consumption supports the argument for a dynamic processing mechanism. Substrate SL RNA is modified by eight methylations of the 5-nt cap structure and pseudouridylation at nt 28 (ψ28). Along with those of the m7G (cap 0), the methylations of the kinetoplastid cap 4 are the most extensive, with 2′-O-ribose methylation of the first four nucleotides and additional base methylations on the first (m26A) and fourth (m3U) positions (5, 21, 49). The SL cap 4 and/or exon primary sequence and pseudouridylation have been implicated in kinetoplastid trans splicing (6, 39, 62, 67) and polysome association (82).
Among higher eukaryotes, cotranscriptional cap 0 formation on mRNA and small nuclear RNA (snRNA) is accompanied often by conserved cap 2′-O-ribose methylation of the first and second transcribed nucleotides, known as cap 1 and cap 2 (22). TbMTr1 is the sequence-specific cap 1 2′-O-ribose MTase acting on the SL RNA and U1 snRNA in T. brucei (44, 77). Two additional T. brucei cap 2′-O-ribose MTases are named for the nucleotide positions modified: TbMTr2 and TbMTr3 (1, 2, 24, 78). Cap 4 formation occurs sequentially from the 5′ end (66), with cotranscriptional (37) and intracellular trafficking-dependent (81) cap formation proposed. TbMTr2 and TbMTr3 activities are dependent on SL RNA association with the heptameric Sm protein complex in vivo (38, 61, 79). Knockout lines for each of the cap 2′-O-ribose MTases are viable (1, 2, 77).
The majority of snoRNAs direct nucleotide modifications of snRNA and rRNA by base-pairing with substrate RNA molecules (41). Box C/D snoRNAs guide predominantly 2′-O-ribose methylation, while box H/ACA snoRNAs specify pseudouridine formation; both classes may direct the cleavage of pre-rRNA. The C/D and H/ACA snoRNAs are each associated with four distinct subsets of core proteins, conserved from archaea to eukaryota (41). Dual-function small Cajal-body-specific RNAs guide methylation and pseudouridylation on snRNAs (11, 27). The H/ACA ribonucleoprotein (RNP) pseudouridylation core proteins are Cbf5p (also known as Dyskerin), Nop10p, Nhp2p, and Gar1p (72). Cbf5p is the pseudouridine synthase that is essential in Saccharomyces cerevisiae (29) and T. brucei (6). Mutations in the mammalian H/ACA snoRNP core protein Dyskerin, Nop10, or Nhp2 cause dyskeratosis congenita (56, 64, 70). snoRNA complexes have other functions, such as the five C/D RNPs directing rRNA cleavage and the mammalian H/ACA telomerase RNA that templates telomere extension (9). The cleavage snoRNPs are associated specifically with additional proteins to tailor their functions (17, 35, 57); however, none of the pseudouridylation snoRNPs is known to complex with factors beyond the four core components.
The T. brucei H/ACA snoRNAs form a single stem-loop structure with an AGA motif (32), in contrast to the two stem-loop structures with ACA motif observed with other eukaryotes. Thirty-four H/ACA RNAs are predicted to guide modification of 32 rRNA nucleotides in T. brucei (34). The SLA1 H/ACA snoRNA guides ψ28 formation (33) and resides primarily in the nucleolus (53) but is also detected in the nucleoplasm (33). Cbf5 knockdown in T. brucei destabilizes SLA1 snoRNA, with defects seen in SL RNA cap 4 formation and decreased trans splicing (6). Pseudouridine formation is not required for trans splicing (39, 62).
We report the in vivo purification of a specialized SLA1 snoRNP complex containing TbMTr1 for enhanced SL RNA processing. TbMTr1 tandem affinity purification followed by protein identification using multidimensional protein identification technology (MudPIT) (20, 36) and the T. brucei genome database (7, 25) revealed association with the four conserved core proteins of H/ACA snoRNPs and with a kinetoplastid-specific protein designated MTase-associated protein (TbMTAP). The SLA1 snoRNA is specifically associated with the TbMTr1 complex. The TbMTr1 complex establishes an SL RNA processing center for Sm protein-independent modifications and demonstrates specialization of a pseudouridylation snoRNP for enhanced processing of a specific RNA.
MATERIALS AND METHODS
DNA cloning.
The pMOT3H-TbMTr1 vector for in situ C-terminal hemagglutinin (HA) fusion protein expression was cloned using pMOT3H (45). The last 724 nt of the TbMTr1 coding region was released from a pTOPO-TA (Invitrogen) vector containing the full 1,110-bp coding region (77) using an internal ApaI and an XhoI site engineered into the reverse primer for amplification. The TbMTr1 3′ UTR was amplified with TbMT370-3′UTR-R-NotI (5′-ATC TAG ACA AGT CAT ACC TAG TCC) and BamHI370-3′UTR-F (5′-AGG ATC CCA AGT CAT AGG TAG TCC) and inserted using XbaI and BamHI sites. The pMOT3H-TbMTr1 vector was digested with ApaI and BamHI for transfection. For PTP tagging, the TbMTr1 sequence was amplified from the pTOPO-TA-TbMTr1 vector using MT370 Xba-F (GGT CTA GAA TGC CTG CCG TTG CAG AC) and pC-PTP-Neo370R (TGC GGC CGC AAT GAC TTG ACC TC). The last 724 nt of the TbMTr1 open reading frame (ORF) was then excised with ApaI and NotI and ligated to the pC-PTP-NEO vector (58). The pC-PTP-NEO-TbMTr1 vector was linearized with BbsI for transfection.
The full 1,230-nt genomic coding sequence of TbMTAP (GeneDB no. Tb11.01.8210) was amplified using primers TbSLA1_ApaI_F (AGG GCC CAT GCC AGC AAA AC) and TbSLA1_NotI_R (GCG GCC GCG ACA TTG AAA ATA G). The TbMTAP knockout vectors were made using the pKO vector (30) where the drug marker was replaced with phleomycin or puromycin. The 648-nt 5′ UTR directly upstream of the ORF was cloned using TbSLA1_5′UTR_HindIII_F (AAA GCT TGA GAA TTT AGC ACA C) and TbSLA1_5′-UTR_EcoRI_R (TGA ATT CCT CTG ATT CCT GAT G). The 496-nt 3′ UTR was amplified with TbSLA1_3′-UTR_Xba_F (ATC TAG AAA TAA TTT TTA AAG AAC AC) and TbSLA1_3′-UTR_NotI_R (GCG GCC GCT ACT CAC AAA AC). Both UTRs were cloned into pKOpuro and pKOphleo, sequenced, and digested with HindIII/NotI for transfection.
T. brucei strains and cell culture.
YTAT procyclic T. brucei was grown at 27°C in SM medium (10) supplemented with 10% fetal bovine serum. Transfection was performed as described previously (71), and cells were selected with hygromycin (20 μg/ml), puromycin (10 μg/ml), phleomycin (2.5 μg/ml), or G418 (15 μg/ml).
DNA and RNA analyses.
Total cell RNA was isolated with TRIzol reagent (Invitrogen), and high-resolution acrylamide RNA blotting, RNA primer extension using Moloney murine leukemia virus reverse transcriptase (RT), and DNA sequencing reactions were performed as described previously (80). The [γ-32P]-labeled oligonucleotides are exon-specific TbWTexon (CAA TAT AGT ACA GAA ACT G), intron-specific SL RNA TbSL40 (CTA CTG GGA GCT TCT CAT AC), U1 snRNA TbU1-40 (CCC ACT CAA AGT TTA CTG), TbSLA1 snoRNA TbSLA (GTC TCG CTC TCC AGT TCG TG), Tb9cs2H1 snoRNA Tb9cs2H1R (AAT TCT CGG ACC ACG TGA), Tb6CS1H2 snoRNA Tb6CS1H2R (CGC GGG TCC GAT TGA G), and U3 snoRNA TbU3 (CCG GGC GGA GCC AGC AAC CTT C). Poly(A)+ RNA was isolated using the MicroPoly(A)Purist kit (Ambion). All blots were visualized on PhosphorImager screens (Amersham).
Total RNA from 5 × 108 T. brucei cells was used for pseudouridine detection by N-cyclohexyl-N′-(2-morpholinoethyl)-carbodiimide metho-p-toluolsulfonate (CMC) (Sigma) or hydrazine-aniline treatment as described previously (3). The RNA pellet was treated with 35% hydrazine solution (Sigma) and resuspended in 1.0 M aniline (Sigma).
Sucrose gradient fractionation.
Nuclear extracts were prepared essentially as described previously (15) from a 1-liter culture of T. brucei grown to a density of 107 cells/ml. Recombinant TbMTr1 was expressed and purified from E. coli as described previously (44) and was subjected to sucrose gradient sedimentation simultaneously. Twenty serial fractions were collected from the top of the gradients using a piston gradient fractionator (BioComp Instruments), trichloroacetic acid (TCA) precipitated, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then immunoblot analysis. Protein standards (Amersham Bioscience), soybean trypsin inhibitor (20.1 kDa), bovine serum albumin (67 kDa), lactate dehydrogenase (140 kDa), catalase (232 kDa), and ferritin (440 kDa) were loaded on sucrose gradients under similar conditions and detected by Coomassie staining after SDS-PAGE.
Tandem affinity purification and mass spectrometry.
Purification was performed with 2.5 liters of 1 × 107 cell/ml culture. For tandem affinity purification, the PTP tag with the protein A and human protein C epitope was utilized and purification performed as described previously (58). Total elution from the protein C column was resolved by SDS-PAGE and stained with GelCode blue stain reagent (Thermo Scientific) or TCA precipitated. For analysis by mass spectrometry, TCA precipitates were resolubilized in digestion buffer (8 M urea, 100 mM Tris-HCl, pH 8.5) and digested by the sequential addition of Lys-C and trypsin proteases as described previously (42). Peptide digests were analyzed using a two-dimensional liquid chromatography-tandem mass spectrometry approach in which they were fractionated using a seven-step multidimensional separation strategy and eluted directly into a Thermo Fisher LTQ-Orbitrap mass spectrometer, where tandem mass spectra were collected in a data-dependent fashion. Additional technical details regarding this method can be found in the studies of MacCoss et al. and Florens et al. (20, 36). The proteomic data were analyzed using the SEQUEST and DTASelect2 algorithms (18, 63) against the T. brucei genome database (7, 25). The final filtering criteria used for this analysis was a peptide level false-positive rate of 5%, as estimated using a decoy database and a minimum of two peptides per protein (48).
Immunoprecipitation and Western blotting.
Cells from 150 ml of culture at 1 × 107 cells/ml were harvested and lysed as described for the PTP purification, and 5 μg of HA.11 monoclonal antibody (Covance) was added. After mixing gently for 1 h at 4°C, 50 μl of protein G-Sepharose suspension (G.E. Healthcare) was added and mixed for 1 h. The samples were spun at 12,000 × g at 4°C, and the pellet was washed four times in buffer C (100 mM NaCl; 20 mM Tris-HCl, pH 8.0; 3 mM MgCl2; 0.1% Tween 20) and resuspended directly into SDS-PAGE loading buffer, or the RNA was extracted with TRIzol reagent. Protein blots were probed with a 1:1,000 dilution of HA.11 anti-HA monoclonal antibody (Covance), a 1:1,000 dilution of anti-HIS polyclonal antibody (Santa Cruz), a 1:1,000 dilution of polyclonal rabbit anti-HA antibody (Santa Cruz), or a 1:1,000 dilution of peroxidase antiperoxidase agent (Fisher).
RESULTS
In vivo TbMTr1 is in a higher molecular weight complex.
TbMTr1 is the cap 1 2′-O-ribose MTase modifying the SL RNA and U1 snRNA in T. brucei (77). To search for additional proteins involved in snRNA biogenesis, TbMTr1 protein interactions were queried.
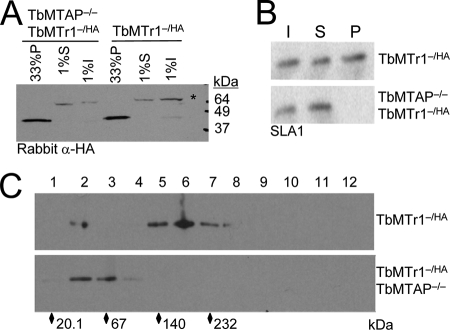
Procyclic T. brucei cell line TbMTr1−/HA expressing a functional C-terminal HA-tagged TbMTr1 (TbMTr1-HA) was established. TbMTr1-green fluorescent protein (GFP) fusion protein localized to the nucleus (77); thus, nuclear extracts from TbMTr1−/HA cells were fractionated on a 2-to-20% sucrose gradient and compared to the sedimentation of purified recombinant His-tagged TbMTr1 (rTbMTr1-His) expressed in E. coli. Fractions collected from each gradient were assessed for the presence of TbMTr1 by use of antibodies recognizing the appropriate epitope tags. Purified rTbMTr1-His peaked in fractions 1 and 2 between protein standards of 20.1 kDa and 67 kDa (Fig. 1A), consistent with a monomeric 41.9-kDa form. The TbMTr1-HA protein from T. brucei nuclear extracts sedimented faster, peaking in fraction 6, near the 232-kDa protein standard. A detectable amount of monomeric protein was observed in some experiments (see Fig. 4C) and may reflect either small amounts of monomeric TbMTr1 within T. brucei cells or disassociation of the complex under fractionation conditions. The absence of higher-molecular-weight rTbMTr1-His forms indicated that the in vivo complexes were not multimeric oligomerization products, implying that in vivo TbMTr1-HA was associated with additional proteins to form the faster-sedimenting complex.
FIG. 1.
Identification and purification of the TbMTr1 complex. (A) Western blot probing alternate fractions of a 2-to-20% sucrose gradient loaded with either purified His-tagged, E. coli-expressed TbMTr1 (rTbMTr1-His) with anti-His antibody (top) or the TbMTr1-HA fusion protein (TbMTr1−/HA) from T. brucei nuclear extracts with monoclonal anti-HA antibody (bottom). Diamonds indicate the following protein standards: soybean trypsin inhibitor (20.1 kDa), bovine serum albumin (67 kDa), lactate dehydrogenase (140 kDa), and catalase (232 kDa). (B) Western blot using peroxidase anti-peroxidase reagent detecting the TbMTr1-PTP fusion protein in whole-cell extracts resolved with a Benchmark prestained molecular mass marker. (C) Colloidal Coomassie stain of SDS-PAGE gel from final purification step of TbMTr1-PTP purification (right lane). Bands are labeled with apparent molecular masses compared to that of the Benchmark protein ladder (left and center lanes) (Invitrogen).
FIG. 4.
TbMTAP is required for SL processing complex formation. (A) Western blot probing for TbMTr1-HA fusion protein using polyclonal rabbit HA antibody in HA.11 monoclonal antibody immunoprecipitation input (I), supernatant (S), and pellet (P) from TbMTr1−/HA and TbMTr1−/HA TbMTAP−/− cells. The asterisk indicates the nonspecific band detected by the polyclonal antibody. (B) Primer extension using oligonucleotide complementary to the SLA1 snoRNA from 2% input (I), 2% supernatant (S), and 10% pellet (P) RNA isolated by TbMTr1-HA immunoprecipitation from TbMTr1−/HA and TbMTr1−/HA TbMTAP−/− cells. (C) Western blot with monoclonal anti-HA antibody probing alternate fractions of a 2-to-20% sucrose gradient loaded with nuclear extracts from either TbMTr1−/HA (top) or TbMTr1−/HA TbMTAP−/− cells (bottom). Diamonds indicate the following protein standards: soybean trypsin inhibitor (20.1 kDa), bovine serum albumin (67 kDa), lactate dehydrogenase (140 kDa), and catalase (232 kDa).
To identify the interacting components, the TbMTr1 complex was isolated by tandem affinity purification.
Purification of the TbMTr1 complex.
Procyclic T. brucei cell line TbMTr1−/PTP was engineered to express a functional TbMTr1 C-terminal PTP tag fusion, which provides a higher yield and less contaminants than the original TAP epitopes (58). A slightly faster-migrating single band corresponding to the predicted 59.7-kDa TbMTr1-PTP protein was confirmed in whole-cell extracts by detection of the protein A epitope with peroxidase anti-peroxidase reagent (Fig. 1B). Following purification, SDS-PAGE revealed five protein bands with apparent molecular masses of 48, 46, 45, 28, and <20 kDa in approximately stoichiometric ratios (Fig. 1C).
To identify the proteins in the final elution of the purification protocol, analysis of the complex peptide mixture was performed in solution using MudPIT and the T. brucei genomic database.
TbMTr1 associates with core H/ACA snoRNA proteins.
Applying a two-peptide hit per protein criterion for MudPIT analysis, the 105 total peptide sequences obtained corresponded to six T. brucei proteins, summarized in Table 1. In addition to the TbMTr1-PTP bait protein, four conserved core protein components of H/ACA snoRNP Cbf5p, Gar1p, Nhp2p, and Nop10p were identified, along with a hypothetical T. brucei ORF.
TABLE 1.
Protein components of TbMTr1 RNP complex and peptides identified by MudPIT
| Protein | Mol wt (in thousands) from SDS gelb | Predicted mol wt (in thousands) | No. of peptides from TbMTr1-PTP | No. of peptides from TbMTAP-PTPc | T. brucei GeneDB no. | Homology to human protein I (%)/S (%)a |
|---|---|---|---|---|---|---|
| TbMTr1 | 45 | 40.8 | 32 | 11 | Tb10.6k15.2610 | 32/51 |
| TbCbf5 | 48 | 48.2 | 25 | 6 | Tb10.100.0060 | 59/77 |
| TbNhp2 | <20 | 16.3 | 7 | ND | Tb927.4.750 | 41/66 |
| TbGar1 | 28 | 21.7 | 5 | ND | Tb927.4.470 | 41/56 |
| TbNop10 | NA | 7.4 | 2 | ND | Tb10.70.2465 | 65/79 |
| TbMTAP | 46 | 44.0 | 34 | 28 | Tb11.01.8210 | NA |
I, sequence identity; S, sequence similarity.
ND, not detected.
NA, not applicable.
Proteins identified by MudPIT had predicted sizes that were consistent with the results from the SDS-PAGE of the purified TbMTr1-PTP complex. The largest protein detected was the 48-kDa homolog of the H/ACA snoRNA-guided pseudouridine synthase, Cbf5p. The 28-kDa and <20-kDa bands matched homologs to known Cbf5p-associated proteins Gar1p and Nhp2p. The final core component of H/ACA snoRNPs, Nop10p, with a predicted molecular mass of 7.4 kDa, was run off the bottom of the SDS-PAGE gel but was captured in the MudPIT analysis.
The association of TbMTr1 with the H/ACA snoRNP core components at stoichiometric levels defines a complex for cap 1 ribose methylation and pseudouridylation of T. brucei RNAs. The copurification of TbCbf5 and TbMTr1 in a complex may explain the deficient cap 4 phenotype in TbCbf5 knockdowns (6).
Specialized TbMTr1-SLA1 snoRNP complex for SL RNA processing.
To determine if the TbMTr1 complex is specialized for SL RNA biogenesis, TbMTr1-HA immunoprecipitations were performed on T. brucei whole-cell extracts from TbMTr1−/HA cells and then any RNA component in the immunoprecipitate was extracted. The coprecipitated RNA was used as a template for detection of a variety of small RNAs, including TbMTr1 substrates SL RNA and U1 snRNA, and four snoRNAs (Fig. 2). The SLA1 snoRNA alone yielded a signal in the pellet and appeared enriched relative to the supernatant. As SLA1 snoRNA guides ψ28 formation of the SL RNA (33), the association of TbMTr1 with the SLA1 snoRNP establishes an SL RNA processing complex for Sm protein-independent modifications (38, 79, 81). To test for the exclusion of other H/ACA snoRNAs in the TbMTr1 complex, TbMTr1-HA-coprecipitated RNA was assayed for the presence of two verified RNA-modifying H/ACA snoRNAs, Tb9Cs2H1 and Tb6Cs1H2 (6). Neither was detected in the pellet, indicating an SL specificity for the TbMTr1 complex. Neither TbMTr1 substrate SL RNA nor U1 snRNA was detected in the pellet, suggesting their interactions are transient. Previous immunoprecipitations of the H/ACA core protein TbNhp2 likewise reveal stable association with SLA1 snoRNA, but not SL RNA (6). The nucleolar box C/D U3 snoRNA served as a negative control for TbMTr1-HA-coprecipitated RNA and remained in the supernatant.
FIG. 2.
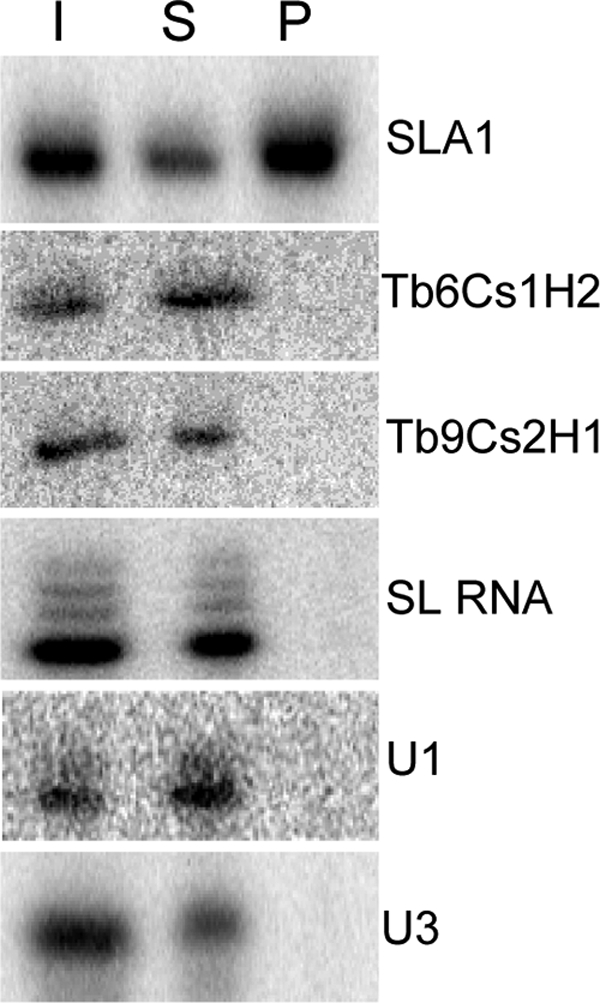
SLA1 snoRNA specifically associates with TbMTrl complex. Primer extension analysis of RNA species in TbMTr1-HA immunoprecipitated samples from 3% input (I), 3% supernatant (S), and 10% pellet (P) performed with HA.11 monoclonal antibody. Oligonucleotides targeting the SLA1, Tb6Cs1H2, and Tb9Cs2H1 H/ACA snoRNAs, TbMTr1 substrates SL RNA and U1 snRNA, and the C/D box U3 snoRNA were used.
The TbMTr1 complex is specific for processing SL RNA, as only SLA1 snoRNA copurified with this snoRNP. The determinants linking SL cap 1 methylation and pseudouridylation may be within the SLA1 RNA, the only distinguishing component of the pseudouridylation machinery. Whether TbMTr1 acts on the U1 snRNA within this complex, as a monomeric protein or as part of a distinct lower abundance snoRNP complex guiding U1 snRNA pseudouridylation, remains to be determined.
Kinetoplastid-specific TbMTAP and the TbMTr1 complex.
A hypothetical protein of a predicted molecular mass of 44.0 kDa was the only uncharacterized component that copurified with TbMTr1 and was designated MTase-associated protein or TbMTAP. TbMTAP is conserved throughout the kinetoplastid family (see Fig. S1 in the supplemental material), with a predicted WD40 domain often found in scaffold proteins coordinating multiprotein complex assemblies (60).
To validate TbMTAP participation in the TbMTr1 complex, a TbMTAP−/PTP line expressing an N-terminal PTP-TbMTAP protein was established for reciprocal complex purification. T. brucei expression of C-terminal-tagged TbMTAP with PTP, HA, and c-Myc epitopes was unsuccessful, likely due to destabilization of the protein. N-terminal-tagged TbMTAP−/PTP cells expressed 30% of the predicted full-length protein product, with the remainder containing C-terminal truncations visible by Western analysis (data not shown). TbCbf5 and TbMTr1 were identified by MudPIT analysis of the purification, confirming their stable association (Table 1). Although TbNhp2, TbGar1, and TbNop10 were not detected, their presence in the complex cannot be eliminated. The more likely explanation is that smaller amounts of the intact complex were purified from the TbMTAP-PTP-expressing strain than from the TbMTr1-PTP-expressing strain, and the other proteins were not abundant enough to be identified reliably. Several common contaminants, such as HSP70 and β-tubulin, also were present, possibly associating with the truncated forms of the PTP-TbMTAP fusion.
The identification of TbMTAP and TbMTr1 with the SLA1 RNA snoRNP reflects the specific association of unique protein components to an individual pseudouridylation complex.
TbMTAP is not essential.
The nucleotide modifications of the SL exon found on every mature kinetoplastid mRNA (Fig. 3A) are implicated in trans splicing and translation. Individual knockouts of SL RNA cap ribose MTases do not affect viability in procyclic cell culture (1, 2, 77), and pseudouridine formation is not required for trans splicing (39, 62).
FIG. 3.
SL RNA is underprocessed in TbMTAP−/− cells. (A) The SL RNA-modified nucleotides implicated in kinetoplastid trans splicing and translation are shown within the exon sequence. The nomenclature for cap intermediates is listed above the modified nucleotides. An m above the sequence indicates base methylation; an m below the sequence indicates 2′-O-ribose methylation. (B) SL RNA cap phenotype in the wild type (WT) and TbMTAP knockout (MTAP KO) (center) determined using γ-32P-labeled oligonucleotide specifically targeting substrate SL RNA in primer extensions (left). Quantification of cap intermediates was performed with QuantOne software from Amersham (right) and are listed as percentages of the total. (C) Primer extension with SL RNA intron-specific oligonucleotide detecting CMC attachment to ψ28 in wild-type (WT), TbMTr1 knockout (MTr1 KO), and TbMTAP knockout (MTAP KO) RNA samples. (D) Primer extension analysis of hydrazine-aniline-treated RNA samples as described above for detection of unmodified uridine nucleotides at position 28 of the SL RNA. (E) High-resolution RNA blot probed for substrate SL RNA in wild-type and TbMTAP−/− cells with radiolabeled oligonucleotide complementary to the intron sequence. 3′ ext., 3′-extended forms. (F) Mature mRNA cap structures are undermethylated in TbMTAP-null mutants. Primer extension assay on poly(A)-purified RNA with [γ-32P]-labeled oligonucleotide complementary to SL exon sequence for wild type (WT) and TbMTAP knockout (MTAP KO).
To determine if TbMTAP is essential for T. brucei cell viability, RNA interference experiments targeting TbMTAP transcripts were performed. No effect on cellular growth was observed (data not shown); thus, TbMTAP-null cell lines were created. Knockout of both TbMTAP alleles was confirmed by Southern blot analysis (see Fig. S2 in the supplemental material). Similar to those of TbMTr1−/− lines, TbMTAP−/− cells showed no apparent defects in growth under continuous log-phase growth conditions (data not shown).
The viability of TbMTAP−/− cells further supports the specialization of TbMTr1 and TbMTAP for the SL RNA processing complex, since global essential H/ACA snoRNP function was not compromised by the absence of TbMTAP. Potential roles for TbMTAP in the TbMTr1 complex include catalysis, regulation, and scaffolding. Elucidation of TbMTAP function continued with examination of SL RNA processing phenotypes in TbMTAP−/− lines.
TbMTAP absence retards SL capping.
Primer extension analysis was performed on total RNA isolated from wild-type and TbMTAP−/− cells to assess cap 4 formation. Most of the intermediates of cap 4 formation are detected by primer extension using a radiolabeled oligonucleotide complementary to the intron portion of substrate SL RNA, as methylation inhibits RT extension.
The wild-type SL RNA cap 4 formation profile showed intermediates corresponding to cap 0, cap 1, cap 2, and cap 4 forms, with cap 4 comprising the majority of substrate SL RNA (Fig. 3B). The cap 3 termination product is not visualized by this assay, attributed to a short-lived intermediate consistent with a hypothesized dual methylation by TbMTr3 (78). Comparison of the TbMTAP−/− substrate SL RNA cap profile revealed an accumulation of cap 0 as the majority form. Detection of the cap 1 intermediate represents 2′-O-ribose methylation of the first transcribed nucleotide (77); thus, TbMTr1 remains active in the absence of TbMTAP, although base dimethylation cannot be excluded. The U1 snRNA cap structure in TbMTAP knockouts showed no detectable differences in cap formations (data not shown).
Since TbMTr1−/− and TbMTAP−/− cells show similar defects in SL cap 4 formation with substantial accumulation of cap 0 substrate, loss of TbMTAP may affect TbMTr1 activity directly or interaction with the SL RNA substrate. Normal U1 snRNA cap formation in TbMTAP−/− cells is consistent with impairment of a specialized complex for SL RNA. A distinct complex for U1 snRNA processing cannot be excluded. The T. brucei Tgs1 protein for guanosine cap trimethylation of spliceosomal snRNAs is nucleoplasmic (55). As TbMTr1 does not recognize the trimethyl cap as substrate (44), TbMTr1 must act before U1 snRNA trimethylation.
Reduced SL pseudouridylation in TbMTAP−/−.
TbMTAP−/− SL pseudouridine formation was assayed by two methods: CMC treatment and hydrazine-aniline cleavage.
The ψ28 on SL RNA can be visualized by the irreversible attachment of CMC to the isomerized nucleotide, as CMC impedes RT extension (3). Using an oligonucleotide complementary to the SL RNA intron, the wild-type CMC-treated RNA sample yielded a termination product at position 29 dependent on CMC treatment (Fig. 3C). The RNA from TbMTr1−/− cells showed little effect on the RT stop at this position, indicating minimal effect on ψ28 formation. In contrast, TbMTAP−/− SL RNA showed decreased amounts of the ψ28 termination product compared to that of the wild type. Steady-state levels of SLA1 snoRNA were unchanged in TbMTAP−/− cells (data not shown), indicating that availability was not limiting. To verify defects in pseudouridine formation, hydrazine-aniline treatment was employed (3), in which pseudouridine resistance to hydrazine-aniline treatment is exploited, while unmodified uridine nucleotides are cleaved, providing different termination points for RT. In the hydrazine-aniline-treated wild-type sample, no termination products for uridine at position 28 were detected, suggesting near-complete ψ28 modification (Fig. 3D). TbMTAP−/− RNA samples had uridines at position 28, confirming CMC treatment results. Examination of TbMTr1−/− SL RNA ψ28 by hydrazine-aniline treatment also showed uridine 28, but to a lesser extent than in TbMTAP−/− cells.
The absence of TbMTAP resulted in decreased processing of cap methylation and ψ28 formation, both activities associated with the SL RNA processing complex. The greater decrease in SL RNA ψ28 in TbMTAP−/− cells compared to TbMTr1−/− cells suggests that TbMTAP plays a significant functional role in coordinating SLA1 snoRNP activity.
TbMTAP−/− SL RNA is underprocessed at the 3′ end.
The 3′-end maturation of SL RNA residual nucleotides from attenuated transcriptional termination requires multiple enzymatic activities, including removal of the final uridine nucleotide by TbSNIP (80). TbMTr1−/− displayed defects in 3′-end maturation (77). To determine if TbMTAP−/− lines had similar delays in 3′ processing, high-resolution RNA blots were employed to assay for 3′-extended forms. TbMTAP−/− cells accumulated higher-molecular-weight substrate SL RNA, consistent with reduced 3′-end maturation (Fig. 3E).
SL RNA processing phenotypes between TbMTr1-, TbCbf5-, and TbMTAP-compromised cells show multiple similarities, consistent with the association of these components in a single functional complex. To determine the downstream consequences of SL RNA function in TbMTAP−/− lines, cap structure in the mRNA population was assessed.
TbMTAP−/− mRNAs are undermethylated.
TbMTr1−/− mRNA cap structure displays a homogeneous profile with complete RT termination at position 4, but lacking cap 1 ribose methylation (77). To test for defects in mRNA cap structure in TbMTAP−/− cells, poly(A)+ RNA was isolated and assayed by primer extension using an oligonucleotide complementary to the exon portion of the SL RNA.
Wild-type mRNA showed a single extension product corresponding to position 4-modified SL. In TbMTAP−/−, additional undermethylated cap structures corresponding to cap 1 and cap 2 modifications were detected in addition to the mature cap 4 (Fig. 3F). The quality of the poly(A)+ RNA was controlled by primer extension using the intron-specific oligonucleotide TbSL40, which showed no products from contaminating substrate SL (data not shown). The ψ28 mRNA levels could not be assessed broadly, as it is too close to the splice junction.
Undermethylated cap structures were not detected in TbMTr1−/− mRNA; thus, the defect in TbMTAP−/− SL RNA processing was distinctive, disrupting further cap 4 progression and forcing the use of underprocessed SL RNA in trans splicing. In addition to delays in cap 4 formation, the extent of decreased ψ28 in TbMTAP−/− cells compared to that in TbMTr1−/− cells may also influence subsequent processing. Thus, complete cap 4 formation and 3′-end maturation may be affected directly by the absence of TbMTAP or indirectly by decreased efficiency in acquiring the Sm-independent modifications.
TbMTAP is required for TbMTr1 complex formation.
To test the hypothesis that TbMTAP provides a structural linkage between the modifying enzymes, the composition of the TbMTr1 complex was analyzed in TbMTAP−/− cells.
A TbMTr1−/HA TbMTAP−/− cell line was established. The detection of the fusion protein at similar levels to that of the wild type demonstrated that TbMTr1 protein stability was uninfluenced by the absence of TbMTAP (data not shown). Anti-HA immunoprecipitation studies were performed with detection of the SLA1 snoRNA as a marker for the snoRNP, isolating similar amounts of TbMTr1-HA protein in both TbMTr1−/HA and TbMTr1−/HA TbMTAP−/− cells (Fig. 4A). However, the SLA1 snoRNA was not detected by primer extension in the HA-immunoprecipitated RNA from the TbMTr1−/HA TbMTAP−/− cells (Fig. 4B), indicating that TbMTAP is required for stable TbMTr1 association with the SLA1 snoRNP. Since SLA1 snoRNA levels are unchanged in TbMTAP knockouts and association with TbCbf5 is required for SLA1 snoRNA stability (6), the SLA1 core snoRNP is likely to be intact. To confirm disassociation of TbMTr1 from the other protein components of the complex, sucrose gradient fractionation of nuclear extracts was performed on the TbMTr1−/HA TbMTAP−/− cell lines. In the absence of TbMTAP, TbMTr1-HA sediments much slower (peaks at fraction 2 or 3) compared to the presence of TbMTAP in wild-type cells, consistent with a monomeric protein (Fig. 4C).
TbMTAP is thus required for the association between TbMTr1 and the SLA1 snoRNP complex (Fig. 5). TbMTr1 is lost from the SLA1 snoRNP, while reduced ψ28 is still detected on substrate SL RNA in TbMTAP−/−, indicating that the functional SLA1 snoRNP particle is still present in these cells. TbMTr1 may be linked to the snoRNP by TbMTAP, possibly added to the snoRNP as a TbMTAP-TbMTr1 subcomplex.
FIG. 5.
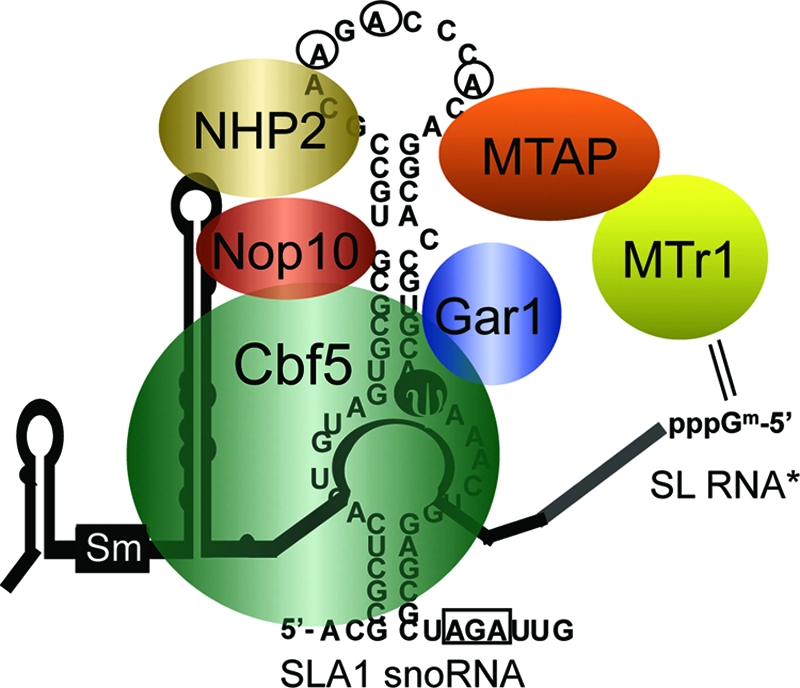
Members of the TbMTr1 complex for SL RNA processing. Six polypeptides and the SLA1 snoRNA coordinate cap 1 2′-O-ribose methylation and pseudouridine formation on SL RNA in T. brucei. Spatial organization is speculative. *, SL RNA is not found in stable association with the complex.
DISCUSSION
We describe an SL RNA processing complex consisting of TbMTr1 cap 1 2′-O-ribose MTase and an H/ACA pseudouridylation snoRNP complex that efficiently processes the most abundant cellular snRNA in kinetoplastids. The TbMTr1 complex contains the four core protein components of H/ACA snoRNPs, the SLA1 snoRNA responsible for SL RNA ψ28 formation, and a WD40 domain protein named TbMTAP that is unique to kinetoplastids. TbMTAP−/− cells are delayed in substrate SL cap 4 formation, 3′-end maturation, and ψ28 formation, while mRNAs are undermethylated, suggesting that TbMTAP has a larger role in SL RNA biogenesis beyond TbMTr1 complex association (see model in Fig. 6). The complex represents a center for Sm-independent modifications early in SL RNA biogenesis. The association of TbMTr1 and pseudouridine synthase activities in one complex explains the unexpected SL RNA cap 4 phenotype observed in TbCbf5 knockdown cells where the complex was disrupted (6).
FIG. 6.
Working model for SL RNA biogenesis. The enzymatic reactions required for maturation of the SL RNA are shown. Gray backgrounds indicate enzymes that have not been assigned. Assembly of the heptameric Sm ring defines early and late steps in the process. RNA synthesis (16, 26, 68) and cap 0 formation (54) occur at a discrete site in the nucleoplasm. Cap 1 modifications and ψ28 formation are early modifications; however, their relative order is not known. TbNhp2 and TbNop10 localize to the nucleolus (6, 55); thus, TbMTr1 complex modifications are placed in that location. The two-step removals of the poly(U) tail and the remaining four cap methylations are late Sm-dependent events. The relative timing of 5′ and 3′ processing is not known.
The stoichiometric association of TbMTr1 and TbMTAP with the SLA1 snoRNP suggests stable association of these unique components with the SLA1 snoRNP. Based on a structural model of an assembled H/ACA snoRNP (51), a bipartite protein-RNA structure accommodates the association of different substrates with their cognate H/ACA snoRNA and the four core proteins (43). The structure of the core H/ACA snoRNP must allow for binding of additional specializing protein components, most likely by direct access to the RNA but possibly through slight shifts in the protein assembly on the RNA scaffold. The most probable mechanism for specificity of TbMTAP/TbMTr1 association with the SLA1 snoRNP is through the SLA1 snoRNA itself, as it is the only distinguishing component relative to the other H/ACA snoRNPs. Direct protein-protein interactions with core protein components are likely to stabilize the mature complex. Since TbMTAP is required for TbMTr1 association with the SLA1 snoRNP, TbMTAP may serve as the link between the two enzymatic components. The identification of TbMTAP and TbMTr1 domains for RNA binding and protein-protein interactions will reveal the structure of this SL processing complex.
Assembly of H/ACA snoRNA-associated proteins in S. cerevisiae begins with the cotranscriptional recruitment of Cbf5p and the RNA binding protein Naf1p (4, 74). Naf1p is complexed to Shq1p and is required for H/ACA snoRNP biogenesis (75). T. brucei has a homolog to Shq1p (Tb11.01.8060). H/ACA snoRNP core Gar1p displaces Naf1p in the nucleolus to form the mature H/ACA snoRNP (12); thus, its presence in the TbMTr1 complex argues that the SLA1 snoRNP component is in a functional form. TbMTAP and/or TbMTr1 may be recruited to the SLA1 snoRNA genomic locus as part of a cotranscriptional assembly pathway or join the preformed SLA1 snoRNP. T. brucei Nop10-GFP fusion protein (55) and TbNhp2-TAP tag immunolocalization (65) show accumulation in the nucleolus, consistent with H/ACA snoRNP proteins in other eukaryotic systems. The purification of the TbMTr1 complex in association with nucleolar RNP components required reexamination of TbMTr1 localization. The TbMTr1-GFP fusion protein localized to the nucleoplasm (77). Preliminary immunolocalization studies suggest that TbMTr1 resides within a distinct area in the nucleus that is deficient in 4′,6-diamidino-2-phenylindole staining (data not shown).
The TbMTr1 complex composition sets a likely nucleolar stage for early SL RNA processing. Localization of RNA polymerase II and the SL RNA genes at different nonnucleolar sites in a variety of kinetoplastids (16, 26, 68) indicates intranuclear trafficking from the site of synthesis to the nucleolus for early modifications of the SL RNA. Trafficking of SL RNA to the nucleolus may be accomplished by specific sequences within the transcript itself or by preassociation with individual components of the TbMTr1 complex. SL RNA was not associated with the TbMTr1 complex; thus, the interaction with the SLA1 snoRNP is likely to be transient. SLA1 snoRNA and SL RNA copurify with Sm proteins (46), and the SLA1 snoRNP may be a chaperone for SL RNA biogenesis (6). As SLA1 RNA lacks an Sm-binding site, stable interaction of the two small RNAs may occur. The purification of the TbMTr1/SLA1 snoRNP complex did not reveal stable interaction with either the SL RNA or the Sm protein complex. In yeast, physical interaction between Gar1p and the SMN protein, involved in Sm complex assembly, aids in snoRNP biogenesis (47, 73). Although SMN homologs have not been identified with T. brucei, similar interactions may link the subsequent steps in SL RNP assembly and Sm protein-dependent modifications. The loss of cap 1 methylation results in delays in SL RNA processing, suggesting that the nucleotide modification is a signal for further processing. By linking cap 1 formation to pseudouridylation, the SL RNA processing complex may facilitate completion of Sm complex-independent modifications early in SL RNP formation.
Maturation of the cap 4 structure downstream of cap 1 is dependent on Sm complex formation (see Fig. 6). TbMTr2 and TbMTr3 cap ribose MTases are nucleoplasmic (1, 2, 78). The TbMTr1 complex supports a minimal two-step process for cap 4 formation in SL RNA biogenesis separated by Sm complex formation.
For TbMTr1−/− cells, a dramatic shift from 50% cap 0 substrate to 100% mRNA cap 4 was interpreted as the result of a delay in substrate maturation, with subsequent MTases acting efficiently on cap 1-less substrate (77). The presence of cap 1 and cap 2 in TbMTAP−/− mRNA indicates that some underprocessed substrates bypassed TbMTr2 and TbMTr3. A discriminatory mechanism may exist for incorporating mature SL cap substrates on mRNAs; however, the viability of SL cap ribose MTase knockout lines attests to the permissiveness of the trans splicing and translation systems. The role of TbMTAP may be direct, perhaps through interaction with substrate SL RNA, or indirect, by exacerbating SL processing delays through the disruption of TbMTr1 complex formation.
The linkage of the two modifying functions may herald similar specializations in other organisms. The proposed scaffold function of TbMTAP linking TbMTr1 to SLA1 snoRNP fits with WD40 domain proteins facilitating protein-protein interactions but does not exclude additional functional roles. TbMTAP may be instrumental in targeting the SL RNA to the correct subnuclear region, and it may regulate cap 1 methylation and/or ψ28 formation. The decrease in ψ28 formation in the absence of TbMTr1 or TbMTAP is of particular interest. If ψ28 reduction is indicative of the delay in substrate processing or a deficiency that is carried onto mRNA remains to be determined.
The clustering of RNA processing enzymes is detected in several systems, including complexes for RNA editing (59) and RNA turnover (19, 69). The assembly of processing centers for cellular RNA biogenesis in the nucleolus or Cajal bodies aids in the processing of cellular RNAs by spatially linking activities for multiple modifications. With multiple H/ACA snoRNAs guiding pseudouridine formation, the specialization of modifying snoRNPs by direct linkage of additional enzymatic activities streamlines the processing of specific RNAs. The TbMTr1 complex may be specific to the highly transcribed and actively consumed SL RNA or to the first of many such associations to be described. As cap ribose methylation and pseudouridylation are conserved in higher eukaryotic snRNAs, such linkages would facilitate the biogenesis of these highly modified transcripts.
Supplementary Material
Acknowledgments
This work was supported by NIH grant AI056034. J.R.Z. was supported by USPHS National Research Service Award GM07104.
We thank Arthur Günzl for the PTP vector and advice on complex purification, Jay Bangs for pKO vectors, Michael Oberholzer and Thomas Seebeck for pMOT vectors, Kent Hill for YTAT cells and for use of the Zeiss fluorescence microscope, and Robert Hitchcock and Isadora Ruvalcaba-Trejo for stimulating discussions.
Footnotes
Published ahead of print on 22 December 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arhin, G. K., H. Li, E. Ullu, and C. Tschudi. 2006. A protein related to the vaccinia virus cap-specific methyltransferase VP39 is involved in cap 4 modification in Trypanosoma brucei. RNA 1253-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arhin, G. K., E. Ullu, and C. Tschudi. 2006. 2′-O-Methylation of position 2 of the trypanosome spliced leader cap 4 is mediated by a 48 kDa protein related to vaccinia virus VP39. Mol. Biochem. Parasitol. 147137-139. [DOI] [PubMed] [Google Scholar]
- 3.Bakin, A., and J. Ofengand. 1993. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyl transferase center: analysis by the application of a new sequencing technique. Biochemistry 329754-9762. [DOI] [PubMed] [Google Scholar]
- 4.Ballarino, M., M. Morlando, F. Pagano, A. Fatica, and I. Bozzoni. 2005. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 255396-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bangs, J. D., P. F. Crain, T. Hashizume, J. A. McCloskey, and J. C. Boothroyd. 1992. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 2679805-9815. [PubMed] [Google Scholar]
- 6.Barth, S., A. Hury, X. H. Liang, and S. Michaeli. 2005. Elucidating the role of H/ACA-like RNAs in trans-splicing and rRNA processing via RNA interference silencing of the Trypanosoma brucei CBF5 pseudouridine synthase. J. Biol. Chem. 28034558-34568. [DOI] [PubMed] [Google Scholar]
- 7.Berriman, M., E. Ghedin, C. Hertz-Fowler, G. Blandin, H. Renauld, D. C. Bartholomeu, N. J. Lennard, E. Caler, N. E. Hamlin, B. Haas, U. Bohme, L. Hannick, M. A. Aslett, J. Shallom, L. Marcello, L. Hou, B. Wickstead, U. C. Alsmark, C. Arrowsmith, R. J. Atkin, A. J. Barron, F. Bringaud, K. Brooks, M. Carrington, I. Cherevach, T. J. Chillingworth, C. Churcher, L. N. Clark, C. H. Corton, A. Cronin, R. M. Davies, J. Doggett, A. Djikeng, T. Feldblyum, M. C. Field, A. Fraser, I. Goodhead, Z. Hance, D. Harper, B. R. Harris, H. Hauser, J. Hostetler, A. Ivens, K. Jagels, D. Johnson, J. Johnson, K. Jones, A. X. Kerhornou, H. Koo, N. Larke, S. Landfear, C. Larkin, V. Leech, A. Line, A. Lord, A. Macleod, P. J. Mooney, S. Moule, D. M. Martin, G. W. Morgan, K. Mungall, H. Norbertczak, D. Ormond, G. Pai, C. S. Peacock, J. Peterson, M. A. Quail, E. Rabbinowitsch, M. A. Rajandream, C. Reitter, S. L. Salzberg, M. Sanders, S. Schobel, S. Sharp, M. Simmonds, A. J. Simpson, L. Tallon, C. M. Turner, A. Tait, A. R. Tivey, S. Van Aken, D. Walker, D. Wanless, S. Wang, B. White, O. White, S. Whitehead, J. Woodward, J. Wortman, M. D. Adams, T. M. Embley, K. Gull, E. Ullu, J. D. Barry, A. H. Fairlamb, F. Opperdoes, B. G. Barrell, J. E. Donelson, N. Hall, C. M. Fraser, et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309416-422. [DOI] [PubMed] [Google Scholar]
- 8.Boothroyd, J., D. Campbell, and R. Sutton. 1985. Expression of surface antigen genes in Trypanosoma brucei involves a novel system of discontinuous transcription, p. 61-66. In R. Lerner, R. Channock, and F. Brown (ed.), Vaccines 85: modern approaches to vaccines. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 9.Collins, K. 2000. Mammalian telomeres and telomerase. Curr. Opin. Cell Biol. 12378-383. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham, I. 1977. New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J. Protozool. 24325-329. [DOI] [PubMed] [Google Scholar]
- 11.Darzacq, X., B. E. Jády, C. Verheggen, A. M. Kiss, E. Bertrand, and T. Kiss. 2002. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridinylation guide RNAs. EMBO J. 212746-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darzacq, X., N. Kittur, S. Roy, Y. Shav-Tal, R. H. Singer, and U. T. Meier. 2006. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 173207-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decatur, W. A., and M. J. Fournier. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278695-698. [DOI] [PubMed] [Google Scholar]
- 14.Decatur, W. A., and M. J. Fournier. 2002. rRNA modifications and ribosome function. Trends Biochem. Sci. 27344-351. [DOI] [PubMed] [Google Scholar]
- 15.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 111475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dossin, F. D. M., and S. Schenkman. 2005. Actively transcribing RNA polymerase II concentrates on spliced leader genes in the nucleus of Trypanosoma cruzi. Eukaryot. Cell 4960-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dragon, F., J. E. G. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eng, J., A. McCormack, and J. I. Yates. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5976-989. [DOI] [PubMed] [Google Scholar]
- 19.Estévez, A. M., T. Kempf, and C. Clayton. 2001. The exosome of Trypanosoma brucei. EMBO J. 203831-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Florens, L., and M. P. Washburn. 2006. Proteomic analysis by multidimensional protein identification technology. Methods Mol. Biol. 328159-175. [DOI] [PubMed] [Google Scholar]
- 21.Freistadt, M. S., G. A. M. Cross, and H. D. Robertson. 1988. Discontinuously synthesized mRNA from Trypanosoma brucei contains the highly methylated 5′ cap structure, m7GpppA*A*C(2′-O)mU*A. J. Biol. Chem. 26315071-15075. [PubMed] [Google Scholar]
- 22.Furuichi, Y., and A. J. Shatkin. 2000. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 55135-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haanstra, J. R., M. Stewart, V. D. Luu, A. van Tuijl, H. V. Westerhoff, C. Clayton, and B. M. Bakker. 2008. Control and regulation of gene expression: quantitative analysis of the expression of phosphoglycerate kinase in bloodstream form Trypanosoma brucei. J. Biol. Chem. 2832495-2507. [DOI] [PubMed] [Google Scholar]
- 24.Hall, M. P., and C. K. Ho. 2006. Functional characterization of a 48 kDa Trypanosoma brucei cap 2 RNA methyltransferase. Nucleic Acids Res. 345594-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hertz-Fowler, C., C. S. Peacock, V. Wood, M. Aslett, A. Kerhornou, P. Mooney, A. Tivey, M. Berriman, N. Hall, K. Rutherford, J. Parkhill, A. C. Ivens, M. A. Rajandream, and B. Barrell. 2004. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 32D339-D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hitchcock, R. A., S. Thomas, D. A. Campbell, and N. R. Sturm. 2007. The promoter and transcribed regions of the Leishmania tarentolae spliced leader RNA gene array are devoid of nucleosomes. BMC Microbiol. 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jády, B. E., and T. Kiss. 2001. A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 20541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, P. J., J. M. Kooter, and P. Borst. 1987. Inactivation of transcription by UV irradiation of T. brucei provides evidence for a multicistronic transcription unit including a VSG gene. Cell 51273-281. [DOI] [PubMed] [Google Scholar]
- 28a.Kuge, H., G. G. Brownlee, P. D. Gershon, and J. D. Richter. 1998. Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 263208-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lafontaine, D. L. J., C. Bousquet-Antonelli, Y. Henry, M. Caizergues-Ferrer, and D. Tollervey. 1998. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 12527-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamb, J. R., V. Fu, E. Wirtz, and J. D. Bangs. 2001. Functional analysis of the trypanosomal AAA protein TbVCP with trans-dominant ATP hydrolysis mutants. J. Biol. Chem. 27621512-21520. [DOI] [PubMed] [Google Scholar]
- 31.Lapeyre, B., and S. K. Purushothaman. 2004. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol. Cell 16663-669. [DOI] [PubMed] [Google Scholar]
- 32.Liang, X.-H., L. Liu, and S. Michaeli. 2001. Identification of the first trypanosome H/ACA RNA that guides pseudouridine formation on rRNA. J. Biol. Chem. 27640313-40318. [DOI] [PubMed] [Google Scholar]
- 33.Liang, X.-H., Y.-X. Xu, and S. Michaeli. 2002. The spliced leader-associated RNA is a trypanosome-specific sn(o) RNA that has the potential to guide pseudouridine formation on the SL RNA. RNA 8237-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang, X. H., S. Uliel, A. Hury, S. Barth, T. Doniger, R. Unger, and S. Michaeli. 2005. A genome-wide analysis of C/D and H/ACA-like small nucleolar RNAs in Trypanosoma brucei reveals a trypanosome-specific pattern of rRNA modification. RNA 11619-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lubben, B., P. Fabrizio, B. Kastner, and R. Luhrmann. 1995. Isolation and characterization of the small nucleolar ribonucleoprotein particle snR30 from Saccharomyces cerevisiae. J. Biol. Chem. 27011549-11554. [DOI] [PubMed] [Google Scholar]
- 36.MacCoss, M. J., W. H. McDonald, A. Saraf, R. Sadygov, J. M. Clark, J. J. Tasto, K. L. Gould, D. Wolters, M. Washburn, A. Weiss, J. I. Clark, and J. R. Yates III. 2002. Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. USA 997900-7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mair, G., E. Ullu, and C. Tschudi. 2000. Cotranscriptional cap 4 formation on the Trypanosoma brucei spliced leader RNA. J. Biol. Chem. 27528994-28999. [DOI] [PubMed] [Google Scholar]
- 38.Mandelboim, M., S. Barth, M. Biton, X.-H. Liang, and S. Michaeli. 2003. Silencing of Sm proteins in Trypanosoma brucei by RNAi captured a novel cytoplasmic intermediate in SL RNA biogenesis. J. Biol. Chem. 27851469-51478. [DOI] [PubMed] [Google Scholar]
- 39.Mandelboim, M., C. L. Estraño, C. Tschudi, E. Ullu, and S. Michaeli. 2002. On the role of exon and intron sequences in trans-splicing utilization and cap 4 modification of the trypanosomatid Leptomonas collosoma SL RNA. J. Biol. Chem. 27735210-35218. [DOI] [PubMed] [Google Scholar]
- 40.Martínez-Calvillo, S., S. Yan, D. Nguyen, M. Fox, K. Stuart, and P. J. Myler. 2003. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell 111291-1299. [DOI] [PubMed] [Google Scholar]
- 41.Matera, A. G., R. M. Terns, and M. P. Terns. 2007. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8209-220. [DOI] [PubMed] [Google Scholar]
- 42.McDonald, W. H., R. Ohi, D. T. Miyamoto, T. J. Mitchison, and J. R. Yates III. 2002. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectrom. 219245-251. [Google Scholar]
- 43.Meier, U. T. 2006. How a single protein complex accommodates many different H/ACA RNAs. Trends Biochem. Sci. 31311-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mittra, B., J. R. Zamudio, J. M. Bujnicki, J. Stepinski, E. Darzynkiewicz, D. A. Campbell, and N. R. Sturm. 2008. The TbMTr1 spliced leader RNA cap 1 2′-O-ribose methyltransferase from Trypanosoma brucei acts with substrate specificity. J. Biol. Chem. 2833161-3172. [DOI] [PubMed] [Google Scholar]
- 45.Oberholzer, M., S. Morand, S. Kunz, and T. Seebeck. 2006. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol. 145117-120. [DOI] [PubMed] [Google Scholar]
- 46.Palfi, Z., G. L. Xu, and A. Bindereif. 1994. Spliced leader-associated RNA of trypanosomes. Sequence conservation and association with protein components common to trans-spliceosomal ribonucleoproteins. J. Biol. Chem. 26930620-30625. [PubMed] [Google Scholar]
- 47.Pellizzoni, L., J. Baccon, B. Charroux, and G. Dreyfuss. 2001. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 111079-1088. [DOI] [PubMed] [Google Scholar]
- 48.Peng, J., J. E. Elias, C. C. Thoreen, L. J. Licklider, and S. P. Gygi. 2002. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 243-50. [DOI] [PubMed] [Google Scholar]
- 49.Perry, K. L., K. P. Watkins, and N. Agabian. 1987. Trypanosome mRNAs have unusual “cap 4” structures acquired by addition of a spliced leader. Proc. Natl. Acad. Sci. USA 848190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pintard, L., F. Lecointe, J. M. Bujnicki, C. Bonnerot, H. Grosjean, and B. Lapeyre. 2002. Trm7p catalyses the formation of two 2′-O-methyltransferases in yeast tRNA anticodon loop. EMBO J. 211811-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rashid, R., B. Liang, D. L. Baker, O. A. Youssef, Y. He, K. Phipps, R. M. Terns, M. P. Terns, and H. Li. 2006. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol. Cell 21249-260. [DOI] [PubMed] [Google Scholar]
- 52.Reference deleted.
- 53.Roberts, T. G., N. R. Sturm, B. K. Yee, M. C. Yu, T. Hartshorne, N. Agabian, and D. A. Campbell. 1998. Three small nucleolar RNAs identified from the spliced leader-associated RNA locus in kinetoplastid protozoans. Mol. Cell. Biol. 184409-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruan, J.-P., S. Shen, E. Ullu, and C. Tschudi. 2007. Evidence for a capping enzyme with specificity for the trypanosome spliced leader RNA. Mol. Biochem. Parasitol. 156246-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruan, J.-P., E. Ullu, and C. Tschudi. 2007. Characterization of the Trypanosoma brucei cap hypermethylase Tgs1. Mol. Biochem. Parasitol. 15566-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruggero, D., S. Grisendi, F. Piazza, E. Rego, F. Mari, P. H. Rao, C. Cordon-Cardo, and P. P. Pandolfi. 2003. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 299259-262. [DOI] [PubMed] [Google Scholar]
- 57.Salinas, K., S. Wierzbicki, L. Zhou, and M. E. Schmitt. 2005. Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component. J. Biol. Chem. 28011352-11360. [DOI] [PubMed] [Google Scholar]
- 58.Schimanski, B., T. N. Nguyen, and A. Günzl. 2005. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot. Cell 41942-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simpson, L., S. Sbicego, and R. Aphasizhev. 2003. Uridine insertion/deletion RNA editing in trypanosome mitochondria: a complex business. RNA 9265-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stagg, S. M., P. LaPointe, and W. E. Balch. 2007. Structural design of cage and coat scaffolds that direct membrane traffic. Curr. Opin. Struct. Biol. 17221-228. [DOI] [PubMed] [Google Scholar]
- 61.Sturm, N. R., and D. A. Campbell. 1999. The role of intron structures in trans-splicing and cap 4 formation for the Leishmania spliced leader RNA. J. Biol. Chem. 27419361-19367. [DOI] [PubMed] [Google Scholar]
- 62.Sturm, N. R., J. Fleischmann, and D. A. Campbell. 1998. Efficient trans-splicing of mutated spliced leader exons in Leishmania tarentolae. J. Biol. Chem. 27318689-18692. [DOI] [PubMed] [Google Scholar]
- 63.Tabb, D. L., W. H. McDonald, and J. R. Yates III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 121-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Terns, M. P., and R. M. Terns. 2002. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 1017-39. [PMC free article] [PubMed] [Google Scholar]
- 65.Tkacz, I. D., S. Cohen, M. Salmon-Divon, and S. Michaeli. 2008. Identification of the heptameric Lsm complex that binds U6 snRNA in Trypanosoma brucei. Mol. Biochem. Parasitol. 16022-31. [DOI] [PubMed] [Google Scholar]
- 66.Ullu, E., and C. Tschudi. 1995. Accurate modification of the trypanosome spliced leader cap structure in a homologous cell-free system. J. Biol. Chem. 27020365-20369. [DOI] [PubMed] [Google Scholar]
- 67.Ullu, E., and C. Tschudi. 1991. Trans splicing in trypanosomes requires methylation of the 5′ end of the spliced leader RNA. Proc. Natl. Acad. Sci. USA 8810074-10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uzureau, P., J. P. Daniels, D. Walgraffe, B. Wickstead, E. Pays, K. Gull, and L. Vanhamme. 2008. Identification and characterization of two trypanosome TFIIS proteins exhibiting particular domain architectures and differential nuclear localizations. Mol. Microbiol. 691121-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vanacova, S., and R. Stefl. 2007. The exosome and RNA quality control in the nucleus. EMBO Rep. 8651-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vulliamy, T., R. Beswick, M. Kirwan, A. Marrone, M. Digweed, A. Walne, and I. Dokal. 2008. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc. Natl. Acad. Sci. USA 1058073-8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 27540174-40179. [DOI] [PubMed] [Google Scholar]
- 72.Watkins, N. J., A. Gottschalk, G. Neubauer, B. Kastner, P. Fabrizio, M. Mann, and R. Luhrmann. 1998. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 41549-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whitehead, S. E., K. W. Jones, X. Zhang, X. Cheng, R. M. Terns, and M. P. Terns. 2002. Determinants of the interaction of the spinal muscular atrophy disease protein SMN with the dimethylarginine-modified box H/ACA small nucleolar ribonucleoprotein GAR1. J. Biol. Chem. 27748087-48093. [DOI] [PubMed] [Google Scholar]
- 74.Yang, P. K., C. Hoareau, C. Froment, B. Monsarrat, Y. Henry, and G. Chanfreau. 2005. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol. Cell. Biol. 253295-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang, P. K., G. Rotondo, T. Porras, P. Legrain, and G. Chanfreau. 2002. The Shq1p.Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J. Biol. Chem. 27745235-45242. [DOI] [PubMed] [Google Scholar]
- 76.Yoon, A., G. Peng, Y. Brandenburger, O. Zollo, W. Xu, E. Rego, and D. Ruggero. 2006. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science 312902-906. [DOI] [PubMed] [Google Scholar]
- 77.Zamudio, J. R., B. Mittra, S. Foldynová-Trantírková, G. M. Zeiner, J. Lukeš, J. M. Bujnicki, N. R. Sturm, and D. A. Campbell. 2007. The 2′-O-ribose methyltransferase for cap 1 of spliced leader RNA and U1 small nuclear RNA in Trypanosoma brucei. Mol. Cell. Biol. 276084-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zamudio, J. R., B. Mittra, G. M. Zeiner, M. Feder, J. M. Bujnicki, N. R. Sturm, and D. A. Campbell. 2006. Complete cap 4 formation is not required for viability in Trypanosoma brucei. Eukaryot. Cell 5905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zeiner, G. M., S. Foldynová, N. R. Sturm, J. Lukeš, and D. A. Campbell. 2004. SmD1 is required for spliced leader RNA biogenesis. Eukaryot. Cell 3241-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zeiner, G. M., R. A. Hitchcock, N. R. Sturm, and D. A. Campbell. 2004. 3′-end polishing of the kinetoplastid spliced leader RNA is performed by SNIP, a 3′→5′ exonuclease with a motley assortment of small RNA substrates. Mol. Cell. Biol. 2410390-10396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zeiner, G. M., N. R. Sturm, and D. A. Campbell. 2003. Exportin 1 mediates nuclear export of the kinetoplastid spliced leader RNA. Eukaryot. Cell 2222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zeiner, G. M., N. R. Sturm, and D. A. Campbell. 2003. The Leishmania tarentolae spliced leader contains determinants for association with polysomes. J. Biol. Chem. 27838269-38275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.