Abstract
Hepatic ischemia-reperfusion (I/R) injury is an important complication of liver surgery and transplantation. Mitochondrial function is central to this injury. To examine alterations in mitochondrial function during I/R, we assessed the mitochondrial proteome in C57Bl/6 mice. Proteomic analysis of liver mitochondria revealed 234 proteins with significantly altered expression after I/R. From these, 13 proteins with the greatest expression differences were identified. One of these proteins, peroxiredoxin-6 (Prdx6), has never before been described in mitochondria. In hepatocytes from sham-operated mice, Prdx6 expression was found exclusively in the cytoplasm. After ischemia or I/R, Prdx6 expression disappeared from the cytoplasm and appeared in the mitochondria, suggesting mitochondrial trafficking. To explore the functional role of Prdx6 in hepatic I/R injury, wild-type and Prdx6-knockout mice were subjected to I/R injury. Prdx6-knockout mice had significantly more hepatocellular injury compared with wild-type mice. Interestingly, the increased injury in Prdx6-knockout mice occurred despite reduced inflammation and was associated with increased mitochondrial generation of H2O2 and dysfunction. The mitochondrial dysfunction appeared to be related to complex I of the electron transport chain. These data suggest that hepatocyte Prdx6 traffics to the mitochondria during I/R to limit mitochondrial dysfunction as a protective mechanism against hepatocellular injury.
Keywords: mitochondrial respiration, hepatocytes
ischemia-reperfusion (i/r) injury is a major complication of transplantation, liver resection, and hemorrhagic shock (1, 5, 8, 20, 23). The injury resulting from warm hepatic I/R has a biphasic pattern. The early phase involves the production of reactive oxygen species (ROS) by Kupffer cells and hepatocytes and leads to an early oxidant-dependent injury (6, 13, 35). The late phase is characterized by a robust inflammatory cascade, culminating in the recruitment of neutrophils to the liver (11, 32).
The extent to which the initial cellular injury contributes to propagation of the inflammatory response and further tissue damage is poorly understood. There is considerable evidence suggesting that ROS generation is central to the injury response. ROS generated during early reperfusion often overwhelm endogenous antioxidant defenses, leading to induction of proinflammatory mediators (12), as well as direct damage to intracellular structures (25). Mitochondria are known to be a major source as well as a target for ROS. It was demonstrated that treatment of hepatocytes with tert-butyl hydroperoxide, an analog of short-chain lipid hydroperoxides formed during oxidative stress, increased mitochondrial ROS production (29, 30). The mitochondrial origin of the ROS burst during oxidant injury in hepatocytes supports the idea that mitochondria play a key role in hepatic I/R injury, since mitochondrial dysfunction is the main cause of hepatocellular death due to necrosis or apoptosis (19). Complexes I and III of the mitochondrial electron transport chain are major sources of ROS (3, 4, 39, 42). The activities of these enzyme complexes are also considered rate-limiting steps for the mitochondrial respiratory chain.
There are a number of endogenous antioxidants that protect the cell from ROS-induced damage. Superoxide dismutase, catalase, and glutathione peroxidase have been well studied. A relatively new group of antioxidants, the peroxiredoxins (Prdxs), belong to the rapidly growing family of the thiol-specific antioxidant proteins that are highly conserved from bacteria to mammals (7, 34). The Prdxs are divided into two categories, the 1-Cys and 2-Cys Prdxs, based on the number of cysteine (Cys) residues directly involved in the reduction of peroxides. Mammalian cells express six Prdx isoforms (Prdxs1-6), which are divided into three categories. Four of the six mammalian Prdxs (Prdxs1-4) belong to the “typical” 2-Cys subgroup and have additional conserved Cys residues in the COOH-terminal region (34). Prdx5 is the single representative of the “atypical” 2-Cys Prdx class in mammalians (34). The Prdx6 isoform encloses only the NH2-terminal-conserved Cys and is, therefore, classified as a 1-Cys Prdx (16). Prdx6 uses glutathione and ascorbate as electron donors and is the only member of the Prdx family that has the ability to remove H2O2 and phospholipid hydroperoxide and is, therefore, able to reduce the accumulation of phospholipid hydroperoxides in plasma membranes (24, 28). Prdx6 expression has not previously been documented in mitochondria. However, Prdx6 has been described in other cell compartments, including the cytoplasm, secretory organelles, and lyosomes (10, 44).
In the present study, we examined changes to the hepatic mitochondrial proteasome during I/R injury. Our data show that the antioxidant Prdx6 shuttles from the cytoplasm to the mitochondria in hepatocytes during I/R injury. Furthermore, our data suggest that this translocation of Prdx6 helps to stabilize mitochondrial function during I/R injury.
MATERIALS AND METHODS
Animals and hepatic I/R injury model.
This project was approved by the University of Cincinnati Animal Care and Use Committee and was in compliance with the National Institutes of Health guidelines. Male C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME) and Prdx6 null mice (University of Pennsylvania, Philadelphia, PA), weighing 21–25 g, were used in all experiments. The generation of Prdx6(−/−) mice is described elsewhere (27). The animals underwent either sham surgery or partial hepatic ischemia, as previously described (22). Briefly, mice were anesthetized with pentobarbital sodium (60–90 mg/kg ip). A midline laparotomy was performed, and an atraumatic clip was used to interrupt blood supply to the left lateral and median lobe of the liver (∼70% ischemia of the liver). The caudal lobes retained intact portal and arterial inflow and venous outflow, preventing intestinal venous congestion. After 90 min of partial hepatic ischemia, the clip was removed, initiating hepatic reperfusion. Mice were killed after 0, 1, or 8 h of reperfusion. Sham control mice underwent the same protocol without vascular occlusion. Body temperature was maintained between 34 and 36°C during the whole procedure.
Subcellular fractionation.
Livers were perfused with 37°C Krebs-Henseleit buffer (freshly added 1.3 mM CaCl2) at a flow of 5 ml/min for 15 min, equilibrated with O2/CO2 (19:1) with a Silastic tubing oxygenator. Afterwards, livers were excised and washed in cold (4°C) phosphate-buffered saline. Mitochondria from whole liver were isolated according to a commercial protocol (G-Bioscience, St. Louis, MO), with slight modifications. Briefly, 500 mg of liver tissue were washed with 10 ml of ice-cold phosphate-buffered saline, before the tissue was minced with a razor blade. The minced tissue was homogenized on ice in a glass Dounce homogenizer (15 strokes, pestle A; 15 strokes, pestle B) in homogenization buffer (5 ml/g tissue). The nuclear fraction and cell debris were removed by centrifugation (1,550 g, 10 min, 4°C), and the supernatant was transferred to a new centrifuge tube. By sequential centrifugation (24,700 g, 30 min and 10 min, 4°C) of this supernatant, the mitochondria were sedimented and resuspended in suspension buffer (5 ml/g tissue), whereas the remaining supernatant (cytosolic fraction) was collected. All fractions were stored at −80°C. For the two-dimensional (2D) gel analysis, the enriched mitochondrial pellet was resuspended in a 2D gel extraction/urea lysis buffer [extraction buffer: 10 mM Tris, pH 7.6, 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), freshly added PMSF, leupeptin, PIC, DTT, and phosphatase inhibitor cocktail II (Sigma); urea lysis buffer: 9 M urea, 4% CHAPS, 2% DTT, 0.1% SDS] by mixing end over end (1 ml, 30 min, 4°C). The mitochondrial suspension was centrifuged (24,700 g, 5 min, 4°C), and the crude mitochondrial enriched supernatant was transferred to a new centrifugation tube and stored at −20°C.
Hepatocyte isolation.
Hepatocytes were isolated by non-recirculating collagenase perfusion through the portal vein. Livers were perfused in situ for 5 min with Ca2+- and Mg2+-free Hank's balanced salt solution (pH 7.4) with 50 U/ml collagenase at a flow rate initiated at 8 ml/min, which was then immediately increased to 55 ml/min. The liver was excised, minced, and strained through a steel mesh and brought to a final volume of 25 ml in Williams' medium and collected by centrifugation at 50 g for 2 min at 4°C. Cells were washed three times in 25 ml of Williams' medium (50 g for 2 min at 4°C). The cell pellet was resuspended in 11 ml L15C medium (Leibovitz tissue culture medium, pH 7.4; 18 mM HEPES; 1.5 mg/ml glucose; 0.5 μg/ml insulin; 2.0 mg/ml BSA; 100 μg/ml penicillin/streptomycin) and followed by adding 10 ml Percoll solution (9 parts Percoll/1 part 10× Hanks balanced salt solution). After spinning at 50 g for 15 min at 4°C, the supernatant was removed, and the isolated hepatocytes were washed two times in 15 ml Williams' medium (50 g for 2 min at 4°C). Viability was checked by trypan blue exclusion.
2D gel electrophoresis.
Proteins from whole liver mitochondria were separated by a 2D electrophoresis, using precast immobilized pH gradient strips (IPG strips, Amersham, Piscataway, NJ) with pH ranges 4–7 (narrow range). Mitochondrial protein concentration was determined using the GenoTech Non-Interfering Protein Assay (G-Biosciences, St. Louis, MO). For isoelectric focusing, the samples were loaded on the IPG strip by in-gel sample rehydration, using a urea-thiourea mixture as solubilizing agent overnight. One-hundred fifty micrograms of protein were loaded per sample on the first-dimension gel. Isoelectric focusing on an IPG-Phor isoelectrofocusing unit (Amersham/GE healthcare, Piscataway, NJ) was conducted at 20°C, focused for 22 h [0–300 V in 1 min (gradient), 300 V for 2 h; 300–3,500 V in 2 h (gradient), 3,500 V for 20 h] for a total of 77.9 kVh. IPG strips were equilibrated three times for 15 min in freshly prepared equilibration buffer (respectively, 40 ml per equilibration; equilibration buffer: 6 M urea, 112 mM Tris-acetate, 30% vol/vol glycerol, 5% wt/vol SDS, 0.01% wt/vol bromphenol blue, adjust pH to 8.8 with acetic acid, freshly add DTT or IAA) before the second-dimension SDS-PAGE was carried out. Strips were embedded on top of 10T, 2.2C duracryl double gels (220 × 220 × 1.5 mm; Genomic Solutions, Ann Arbor, MI) after filling the chambers of the vertical SDS-PAGE unit (Investigator 2D gel running system, Genomic Solution) with appropriated buffers (anode buffer: 210 mM Tris-acetate, pH 8.9 with acetic acid; cathode buffer: 100 mM Tris, 100 mM tricine, 0.1% wt/vol SDS). The second-dimensional electrophoresis was carried out at 8°C with constant power (15 W) for 6 h. The protein spots were visualized by silver staining, using a silver stain kit (Genomic Solutions), scanned (Image Scanner II, Amersham/GE Healthcare), and digital quantified with the differential display image analysis software Z3 (Compugen, Tel-Aviv, Israel). All 2D gels were run a minimum of three independent times under each condition to ensure reproducibility.
Protein digestion and peptide mass spectrometry.
Each spot was assigned a value in parts per million corresponding to the single-spot volume as a proportion of the total spot volume of all spots in the gel, following background subtraction and removal of other artifacts. Differential expression between the groups was determined as a fold change, and proteins with the most appropriated change in expression between the groups were selected for analysis by peptide mass spectrometry (MS). Protein spots were excised, chopped, destained, and dehydrated from the gels and subjected to a tryptic digestion, as originally described by Shevchenko et al. (40) with modification as presented by Jarrold et al. (15). The extracted peptides were concentrated in a speed vac centrifuge (Speed-Vac plus, Savant, Ramsey, MN) to a final volume of 10–15 μl. Peptides were desalted and purified utilizing the C18 ZipTips (Millipore, Bedford, MA). Purified peptides were eluted by pipetting up 2.5 μl of 0.3% trifluoroacetic acid in 60% vol/vol acetonitrile, followed by peptide concentration to a final volume of 0.5 μl by speed vac centrifugation. After incorporation of the concentrated peptides in 1 μl matrix solution (0.1% trifluoroacetic acid in 50% vol/vol acetonitrile + 5 mM ammonium phosphate monobasic + 5 μg/μl α-cyano-4-hydroxy-cinnamic acid), they were directly loaded onto a matrix-assisted laser desorption/ionization (MALDI) target plate (Opti-TOF 384 well plates, Applied Biosystems, Foster City, CA). MALDI-MS analysis of the respective peptides was performed on a 4800 MALDI-time-of-flight (TOF)-TOF instrument from Applied Biosystems (Foster City, CA), operated in reflector-positive mode. Proteins were subsequently identified using the MASCOT search algorithm (Matrix Science, Boston, MA) (33) from a combination of peptide mass fingerprint profiles and MS/MS sequencing data of the 10 most abundant from each digest. Criteria for protein identification included the total ion scores, a minimum of two MS/MS spectra, and the percent sequence coverage.
Western blot analysis.
Protein concentrations of subcellular fractions, either from whole liver tissue or from isolated hepatocytes, were estimated by the BCA protein assay kit (Pierce, Rockford, IL). Samples were separated in precast, denaturing 8–12% SDS-polyacrylamide gel (Pierce, Rockford, IL) and transferred to a 0.1-μm-pore nitrocellulose membrane. Nonspecific binding sites were blocked with Tris-buffered saline with 0.1% Tween 20 [TBST; 40 mM Tris (ph 7.5), 300 mM NaCl] containing either 5% nonfat dry milk or 5% BSA. Incubation of the primary antibodies [cyclooxygenase (COX) IV, cytochrome c (Cell Signaling Technology, Danvers, MA); histone H1.0, Prdx6 (Abcam, Cambridge, MA); IκB-α (Santa Cruz Biotechnology, Santa Cruz, CA)] was performed in TBST containing either 5% nonfat dry milk or 5% BSA overnight at 4°C. The membranes were washed in TBST three times for 15 min and incubated with secondary antibodies conjugated to horseradish peroxidase. Immunoreactive proteins were visualized by autoradiography, using the enhanced chemiluminescence Western Blotting Detection Reagent (Amersham, GE Healthcare Bio-Sciences).
Liver neutrophil accumulation.
Liver myeloperoxidase content was assessed by methods described elsewhere (36). Briefly, 100 mg liver tissue were homogenized in 2 ml buffer A (3.4 mmol/l KH2HPO4 and 16 mmol/l Na2HPO4, pH 7.4) for 30 s on ice. After centrifugation at 10,000 g for 20 min at 4°C, the supernatant was discarded, and the pellet was resuspended in 10 volumes of buffer B (43.2 mmol/l KH2HPO4, 6.5 mmol/l Na2HPO4, 10 mmol/l EDTA, and 0.5% hexadecyl-trimethylammonium, pH 6.0). The tissue suspension was sonicated for 30 s on ice and incubated for 2 h at 60°C. After centrifugation at 10,000 g for 5 min at 4°C, the supernatant was collected, and an aliquot was mixed with buffer C [43.3% wt/vol 3,3′,5,5′-tetramethylbenzidine, 3.2 ml N,N-dimethylformamide, 12.0 ml dH2O, 2.6 ml buffer D (77,4 mmol/l KH2HPO4, 2.6 mmol/l Na2HPO4, 10% hexadecyltrimethyl-ammonium, add dH2O to a final volume of 130 ml, pH 5.4), add freshly 1 μl H2O2] and incubated at 37°C for 15 min, and the optical density was measured at 655 nm.
Blood and tissue analysis.
Blood was obtained by cardiac puncture for analysis of serum alanine aminotransferase (ALT) as an index of hepatocellular damage. Measurements of serum ALT were made using a colorimetric diagnostic kit (Wiener Laboratorios, Rosario, Argentina), according to the manufacturer's instructions. For histological examination, liver tissues were fixed in 10% neutral buffered formalin, embedded in paraffin before cut, and stained with hematoxylin and eosin.
Fluorescence-based assay of mitochondrial respiration.
Hepatocyte mitochondria were isolated as previously outlined with minor modifications (9, 18, 43). All steps were carried out at 4°C. Briefly, 500 mg of liver were minced and washed in isolation buffer I (210 mM mannitol, 70 mM sucrose, 5 mM HEPES, 1 mM EGTA, and 0.5% fatty acid-free BSA, pH 7.4) until the homogenate was blood free. Five volumes of isolation buffer I were added, and the tissue homogenized using a smooth glass grinder with Teflon pestle driven by a power drill. The homogenate was then adjusted to eight volumes of isolation buffer I and centrifuged at 700 g for 10 min at 4°C. The supernatant was filtered through two layers of cheesecloth and recentrifuged for 10 min at 14,000 g to precipitate the mitochondrial fraction. The supernatant was discarded; mitochondria were washed by resuspension in 4 ml of isolation buffer I and centrifuged at 10,000 g for 10 min at 4°C. This washing step was repeated in isolation buffer II (210 mM mannitol, 70 mM sucrose, 10 mM MgCl2, 5 mM K2HPO4, 10 mM 3-(N-morpholino)propanesulfonic acid, and 1 mM EGTA, pH 7.4). Finally, mitochondria were resuspended in 450 μl isolation buffer II. Protein measurements were preformed to adjust equal amounts of mitochondrial proteins. One-nanomole A65N-1 oxygen probe (Luxcel Bioscience, Cork, Ireland), supplied as dry reagent, was reconstituted in 1 ml of respiration buffer (250 mM sucrose, 15 mM KCl, 1 mM EGTA, 5 mM MgCl2, and 30 mM K2HPO4, pH 7.4) and diluted to a concentration of 100 nM. One-hundred microliters of this solution were transferred into a 96-well plate (100 pmol of probe per well). Fifty microliters of the mitochondrial proteins were added to each well, followed by 50 μl of substrate (12.5:12.5 mM glutamate-malate final concentration) without or with ADP (1.65 mM final concentration). As positive control, respiratory chain uncoupling was tested by using carbonyl cyanide 4-(trifluoromethoxy) phenyldydrazone (final concentration 100 nmol/mg protein), dissolved in dimethyl sulfoxide (final dimethyl sulfoxide content was not more than 0.5% vol/vol). Finally, 100 μl of heavy mineral oil were added to each well to seal the samples from ambient oxygen. Measurement was done in a Victor3V (PerkinElmer precisely, 1420 Multilabel counter) at 30°C for 90 min in a time-resolved fluorescence mode. Instrument settings were as follows, 340/642 nm excitation/emission filters, a delay of 30 μs, and a measurement window of 100 μs. After completion of fluorescence measurements, time profiles of fluorescence intensity were analyzed using WorkOut2 (PerkinElmer) software. Rates of oxygen consumption based on the known relationship between probe fluorescence quenching and oxygen concentration were determined (17). Briefly, fluorescence traces were converted into oxygen concentration profiles using the following transformation:
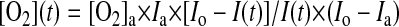 |
where [O2]a is oxygen concentration in air-saturated buffer (235 μM at 30°C); I(t), Io, and Ia are fluorescence signal of the probe at time t, signal in air-saturated buffer (baseline signal without enzyme), and signal in deoxygenated buffer (maximal signal), respectively. Rates of change of dissolved oxygen were subsequently determined from the initial slopes of these concentration profiles, similar to the electrode system.
Mitochondrial respiration and energy coupling.
Mitochondrial oxygen consumption was measured polarographically with a computer-controlled Clark-type oxygen electrode (Hansatech Instruments, Norfolk, UK). The respiratory mixture, consisting of 0.5 ml of a KCl respiratory buffer [140 mM KCl, 0.1 mM EDTA, 2.5 mM KH2PO4, 2.5 mM MgCl2, and 0.05% bovine serum albumin, in 5 mM HEPES, pH 7.4 (KOH)] and 150 μg of mitochondrial protein were equilibrated at 37°C under constant stirring. Substrate-stimulated state 4 respiration (ADP limited) was determined after adding 3 mM glutamate/3 mM malate or 6 mM sodium succinate. The rate of state 3 respiration was then determined following the addition of 0.2 mM ADP. The respiratory control ratio (RCR) was calculated as the ratio of state 3 to state 4 respiration.
Measurement of mitochondrial reactive oxygen using luminol.
H2O2-induced luminol chemiluminescence was measured as described elsewhere (37), using a Berthold Autolumat Plus luminometer. Luminol will react with H2O2 and peroxynitirite, but not with superoxide (38). H2O2 specificity is determined by inhibiting chemiluminescence with catalase. The reaction mixture is composed of 100 μg of mitochondrial protein, 5 μM luminol, and 10 μg/ml horseradish peroxidase in a final volume of 1.0 ml KCl respiratory buffer. Chemiluminescence was initiated by the addition of either 3 mM glutamate/3 mM malate or 6 mM sodium succinate, and monitored at 37°C. The H2O2 luminescence signal was completely quenched by 500 units catalase/ml; values shown are catalase-inhibited chemiluminescence values.
Statistical analysis.
All data are expressed as means ± SE. Data were analyzed with a one-way analysis of variance with subsequent Student's t-test in a two-tailed analysis. Differences were considered significant when P < 0.05.
RESULTS
Hepatic mitochondrial proteome analysis during I/R.
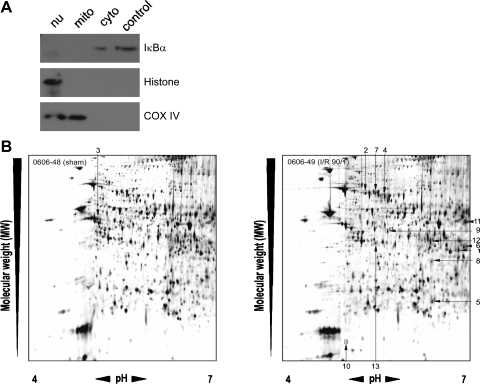
Because the functional state of mitochondria are critical to the fate of the liver after I/R (19), we examined changes to the hepatic mitochondrial proteasome during I/R. Liver mitochondria were isolated from sham-operated mice and mice undergoing 90 min of ischemia and 1 h of reperfusion. Mitochondrial fractions were pure, as there was no evidence of contamination with cytoplasmic (IκBα) or nuclear (histone H1.0) proteins (Fig. 1A). Mitochondrial protein fractions were then separated using 2D-PAGE, and gels were silver stained (Fig. 1B). Image analysis was performed to determine relative protein expression between sham and I/R groups. Two-hundred thirty-four protein spots were increased or decreased in expression more than twofold compared with mitochondria from sham mice. From these, 13 proteins were selected for identification based on the greatest differences between sham and I/R groups calculated by the Z3-Gel analysis software (Fig. 1B). Protein spots were digested with trypsin, analyzed by MS, and identified using the Mascot database. Table 1 shows the sequence information of the identified proteins, the number of matched peptides, and the total ion score for the peptides, as an indicator for accurate protein identifications with scores above 100, based on the results from the Mascot search algorithm. After I/R, there was an increased mitochondrial expression of hydroxyacyl-coenzyme A dehydrogenase, two heat shock protein 70-related proteins, transmembrane protein 4, glutamate dehydrogenase 1, sorbitol dehydrogenase precursor, fumarylacetoacetase, methionine adenosyltransferase Iα, and Prdx6. One protein, ATP5b, was found to have decreased expression after I/R. Some of these identified proteins are related to the energy metabolism, whereas others are linked to the antioxidant metabolism, like methionine adenosyltransferase Iα, sorbitol dehydrogenase, and Prdx6. Because Prdx6 has never been previously described in mitochondria, we focused on the functional significance of this protein for our subsequent studies.
Fig. 1.
Analysis of the hepatic mitochondrial proteome during ischemia-reperfusion (I/R). A: the purity of mitochondrial fractions was determined by staining for a cytoplasmic (cyto) protein (IκBα), a nuclear (nu) protein (histone), and a mitochondrial (mito) protein [cyclooxygenase (COX) IV]. Data are representative of at least three independent experiments. B: two-dimensional gel electrophoresis of hepatic mitochondrial proteins from mice undergoing sham surgery (left) or 90 min of ischemia followed by 1 h of reperfusion (90/1; right). Numbers refer to the proteins selected for identification based on fold expression change from sham. Identification of these proteins is provided in Table 1. Data are representative of three independent experiments.
Table 1.
Liver mitochondrial protein identification
| Spot No. | Protein Name | Accession No. | MW | pI | Peptides | Total Ion Score |
|---|---|---|---|---|---|---|
| 1 | Sorbitol dehydrogenase precursor | gi 1009706 | 40,066 | 6.60 | 8 | 154 |
| 2 | Heat shock 70-kDa protein 8 | gi 27805925 | 71,195 | 5.49 | 14 | 279 |
| 3 | Atp5b protein | gi 23272966 | 56,632 | 5.24 | 8 | |
| 4 | Albumin 1 | gi 33859506 | 68,678 | 5.75 | 12 | 236 |
| 5 | Peroxiredoxin 6 | gi 6671549 | 24,811 | 5.98 | 13 | 308 |
| 6 | Fumarylacetoacetase | gi 50973 | 46,216 | 6.70 | 8 | 223 |
| 7 | Heat shock 70-kDa protein 9B (mortalin-2) | gi 21040386 | 73,808 | 6.04 | 14 | 180 |
| 8 | Hadha protein; hydroxyacyl-coenzyme A dehydrogenase/3-ketoacyl-coenzyme A thiolase | gi 20073317 | 43,885 | 8.40 | 6 | 253 |
| 9 | Methionine adenosyltransferase I, α | gi 19526790 | 43,481 | 5.51 | 9 | 355 |
| 10 | Transmembrane protein 4 | gi 9903607 | 20,754 | 4.95 | 9 | 161 |
| 11 | Glutamate dehydrogenase 1 | gi 6680027 | 61,298 | 8.05 | 13 | 424 |
| 12 | Sterol carrier protein 2 | gi 50356003 | 58,789 | 6.62 | 9 | 93 |
| 13 | Chitinase-related protein MCRP | gi 1336166 | 28,829 | 5.14 | 2 | |
| Positive control | Albumin (Bos Taurus) | gi 30794280 | 69,278 | 5.82 | 15 | 369 |
Protein spots from Figure 1 were cut, trypsin-digested, analyzed by mass spectroscopy, and identified using the Mascot database. MW, molecular weight; pI, isoelectric point.
Subcellular expression of Prdx6 in whole liver tissue and hepatocytes.
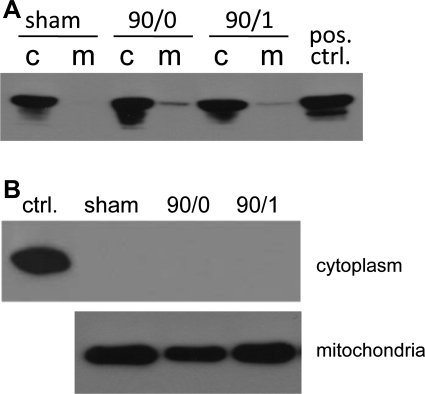
To determine the subcellular localization of Prdx6 and to determine whether this expression pattern is altered by I/R, we analyzed whole liver cytoplasmic and mitochondrial expression of Prdx6 by Western blot. As shown in Fig. 2A, Prdx6 expression was found uniformly in the cytoplasm. Interestingly, we found that, after ischemia or I/R, Prdx6 could be detected in the mitochondrial fraction, indicating trafficking of Prdx6 from the cytoplasm to the mitochondria. To ensure the physical integrity of the hepatic mitochondria isolated from postischemic livers, Western blot for cytochrome c was performed. These studies demonstrated that isolated mitochondria were intact, as no leakage of cytochrome c to the cytoplasm was detected (Fig. 2B).
Fig. 2.
Hepatic peroxiredoxin (Prdx) 6 expression in cytoplasmic and mitochondrial compartments during I/R. A: cytoplasmic (c) and mitochondrial (m) protein fractions were isolated from livers of mice undergoing sham surgery, 90 min of ischemia (90/0), and 90/1, and immunoblotted for Prdx6. Data are representative of three independent experiments. B: the integrity of isolated mitochondria was determined by immunobloting cytoplasmic and mitochondrial fractions for cytochrome c. Data are representative of three independent experiments.
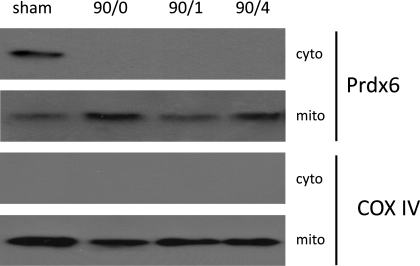
Because derangement of mitochondrial function in hepatocytes is the trigger for cell death, we next examined Prdx6 expression in subcellular fractions of hepatocytes isolated during I/R. In hepatocytes from sham-operated control mice, Prdx6 was found primarily in the cytoplasmic compartment with minimal Prdx6 detected in mitochondria (Fig. 3). However, after ischemia or I/R, Prdx6 expression in the cytoplasm was lost, and Prdx6 expression in mitochondria was markedly increased (Fig. 3). COX IV staining indicated equal loading of mitochondrial protein. The purity of the isolated fractions was done in the same way as described for whole liver tissue, and no contamination of the mitochondrial or cytoplasmic fractions was detectable (data not shown). Collectively, these data suggest that ischemic stress to hepatocytes induces shuttling of Prdx6 from the cytoplasm to the mitochondria.
Fig. 3.
Hepatocyte Prdx6 expression in cytoplasmic and mitochondrial compartments during I/R. Cytoplasmic and mitochondrial protein fractions were isolated from livers of mice undergoing sham surgery, 90/0, 90/1, and 90 min of ischemia and 4 h of reperfusion (90/4), and immunoblotted for Prdx6. The same subcellular fractions were immunoblotted for COX IV to serve as a loading control, as well as an index of mitochondrial integrity. Data are representative of three independent experiments.
Prdx6 protects against liver injury induced by I/R.
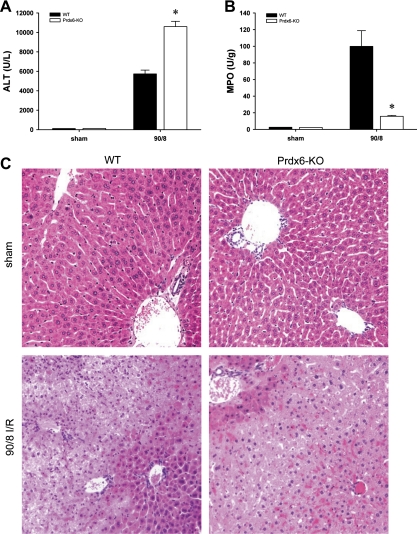
Our data demonstrate that, in hepatocytes, Prdx6 traffics from the cytoplasm to mitochondria during I/R. It is well established that mitochondrial function dictates hepatocyte survival. To determine whether the mitochondrial trafficking of Prdx6 in hepatocytes during I/R has an effect on hepatic injury, we examined the injury response of wild-type (WT) and Prdx6-knockout (KO) mice to I/R in vivo. Mice were subjected to 90 min of ischemia followed by 8 h of reperfusion, which is known to cause severe hepatocellular injury. Prdx6-KO mice had significantly greater liver injury, as determined by serum levels of ALT (Fig. 4A). Despite this increase in liver injury, Prdx6-KO mice had significantly less neutrophil accumulation, as measured by liver content of myeloperoxidase (Fig. 4B). These biochemical data were confirmed by histopathological analysis. Sham-operated WT and Prdx6-KO mice displayed normal liver architecture (Fig. 4C). After I/R in WT mice, there was marked neutrophilic inflammation and hepatocyte necrosis (Fig. 4C). However, in the Prdx6-KO mice, there was less evidence of neutrophilic infiltrates, yet hepatocellular necrosis was more severe than in WT mice (Fig. 4C). To determine if the increased liver injury observed might be a result of increased apoptosis, we stained liver sections for active caspase-3. There was no difference in the amount of staining for active caspase-3 in livers from WT and Pdx6-KO mice (data not shown). These data demonstrate that Prdx6 is an important regulator of the hepatic injury response to I/R.
Fig. 4.
Prdx6 protects against hepatic I/R injury. Wild-type (WT) and Prdx6-knockout (Prdx6-KO) mice were subjected to 90 min of ischemia followed by 8 h of reperfusion. A: hepatocellular injury was determined by measuring serum levels of alanine aminotransferase (ALT). B: neutrophil accumulation was determined by liver content of myeloperoxidase (MPO). Values are means ± SE with n = 3 per group. *P < 0.05 compared with sham. C: Liver histology. Normal hepatic architecture was observed in sham-operated WT mice and Prdx6-KO mice. After I/R, livers from WT mice had large areas of necrosis with neutrophilic infiltration, whereas livers from Prdx6-KO mice showed more widespread necrosis with little evidence of neutrophil accumulation. Original magnification was ×100.
Mitochondrial Prdx6 stabilizes mitochondrial respiratory chain function during liver I/R.
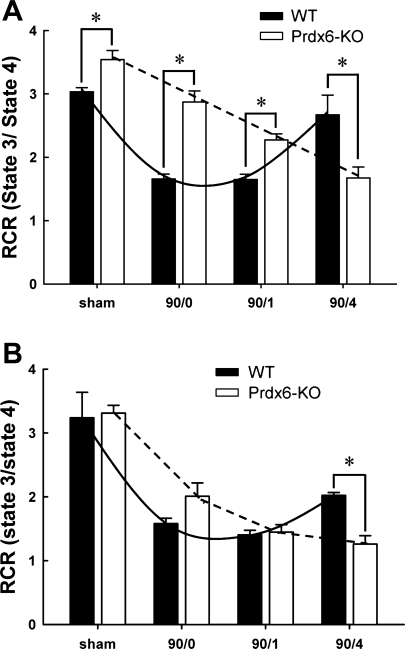
To determine if the increased liver injury observed in Prdx6-KO mice was attributable to increased mitochondrial dysfunction as a result of the loss of mitochondrial Prdx6, respiratory activities of liver mitochondria from WT and Prdx6-KO mice were evaluated in the presence of glutamate/malate (substrates for complex I) or succinate (substrate for complex II) and ADP. The RCR is a ratio of maximal respiration (state 3) to basal respiration (state 4) and represents an index of mitochondrial coupling and, therefore, function. After stimulation with glutamate/malate, mitochondria from Prdx6-KO mice showed a higher RCR baseline (Fig. 5A). Despite this, mitochondria from Prdx6-KO mice showed a precipitous decrease in RCR over the time course of I/R. In contrast, mitochondria from WT mice showed an initial decrease in RCR after ischemia or ischemia and 1 h of reperfusion, but then recovered by 4 h of reperfusion (Fig. 5A). A similar phenomenon was observed in mitochondria stimulated with succinate (Fig. 5B).
Fig. 5.
Effect of I/R on respiratory control ratio (RCR) in liver mitochondria from WT and Prdx6-KO mice. A: stimulation of complex I with glutamate/malate. B: stimulation of complex II with succinate. Values are means ± SE with n = 3 per group. *P < 0.05.
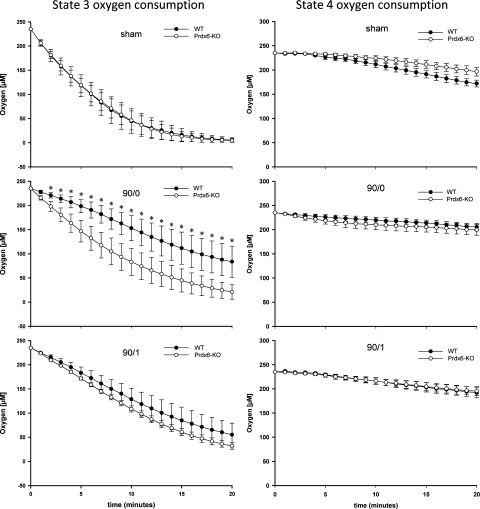
To further explore the importance of mitochondrial Prdx6 in stabilizing mitochondrial function, we examined mitochondrial oxygen consumption using a kinetic assay employing a long-decay, phosphorescent, oxygen probe (9). Because we found that complex I was more affected by the loss of Prdx6, we only used glutamate/malate as a substrate in this assay. As shown in Fig. 6, mitochondria from WT and Prdx6-KO mice had similar basal (state 4) oxygen consumption after sham surgery or I/R (Fig. 6). ADP-stimulated (state 3) oxygen consumption was similar between mitochondria from sham-operated WT and Prdx6-KO mice (Fig. 6). After ischemia, oxygen consumption by mitochondria from Prdx6-KO mice greatly exceeded that from WT mice. After ischemia and 1 h of reperfusion, oxygen consumption was again similar between WT and Prdx6-KO mitochondria (Fig. 6).
Fig. 6.
Effect of I/R on oxygen consumption in liver mitochondria from WT and Prdx6-KO mice. Kinetic analysis of oxygen consumption was conducted by fluorescence analysis of glutamate/malate-driven respiration of liver mitochondria from mice undergoing sham surgery, 90/0, and 90/1. Values are means ± SE with n = 3 per group. *P < 0.05 compared with Prdx6-KO.
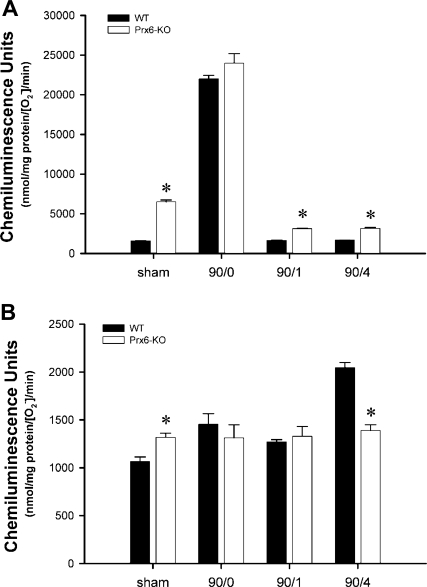
Mitochondrial H2O2 generation is increased during liver I/R in Prdx6-KO mice.
To determine whether the trafficking of Prdx6 to the mitochondria scavenges ROS generated by mitochondria during I/R, we measured mitochondrial H2O2 generation during I/R. Since we found that mitochondrial function during I/R was disturbed in the absence of Prdx6 (Figs. 5 and 6), we next conducted experiments to determine whether substrate-induced mitochondrial respiration was associated with increased generation of ROS and whether the presence of Prdx6 regulated this process. Mitochondria isolated from WT and Prdx6-KO mice after sham surgery, ischemia, or ischemia and 1 or 4 h of reperfusion were stimulated with glutamate/malate or succinate. We found that H2O2 generation by mitochondria from sham-operated mice increased significantly after stimulation with either substrate and that, in both cases, production was significantly higher in mitochondria from Prdx6-KO mice (Fig. 7). Mitochondria isolated after ischemia generated the highest levels of H2O2, and, under these conditions, there was no significant difference between mitochondria from WT or Prdx6-KO mice for either substrate. However, mitochondria from Prdx6-KO mice undergoing I/R generated significantly greater amounts of H2O2 than mitochondria from WT mice, only when stimulated with glutamate/malate, suggesting a preferential localization of Prdx6 to complex I (Fig. 7A). Interestingly, mitochondria from WT mice undergoing ischemia and 4 h of reperfusion and stimulated with succinate generated more H2O2 than Prdx6-KO mice (Fig. 7B). Collectively, these data suggest that trafficking of Prdx6 to the mitochondria during I/R helps to regulate mitochondrial respiratory-derived ROS formation, thereby reducing intracellular oxidative stress.
Fig. 7.
Role of Prdx6 in mitochondrial production of H2O2 during I/R. Liver mitochondria from WT and Prdx6-KO mice undergoing sham surgery, 90/0, 90/1, and 90/4 were analyzed in response to glutamate/malate (A) and succinate (B) stimulation. Values are means ± SE with n = 5 per group. *P < 0.05 compared with WT.
DISCUSSION
These studies are the first to demonstrate the expression of Prdx6 in mitochondria. We have shown using whole liver and isolated hepatocytes that Prdx6 is found predominantly in the cytoplasm under normal (sham) conditions. However, after ischemia or I/R, we found that Prdx6 becomes associated with mitochondria, presumably through recruitment from the cytoplasmic pool. These findings led us to evaluate the functional significance of this change in subcellular compartmentalization. Prdx6-KO mice were found to have significantly more liver injury than WT mice, suggesting that Prdx6 expression is required for regulating the hepatic injury response to I/R. One interesting finding of our in vivo studies was the observation that, despite nearly twice the level of hepatocellular injury, Prdx6-KO mice had greatly diminished neutrophil recruitment. This is a significant finding in that it provides evidence that severe hepatic I/R injury can occur independently of the inflammatory response. This is contrary to the current belief that the majority of liver injury occurring after several hours of reperfusion is mediated by recruited neutrophils (11, 14, 21). We observed a similar phenomenon in the I/R injury response in aged mice (31). While not addressed in the present studies, Prdx6 may directly affect neutrophil function as lack of Prdx6 affects neutrophil respiratory burst activity (A. B. Fisher, unpublished observations).
A limitation to our in vivo studies was the global deletion of Prdx6 in Prdx6-KO mice. Therefore, the increased injury observed cannot be directly ascribed to a lack of mitochondrial Prdx6. This led us to critically determine the role of Prdx6 in mitochondrial function in vitro using mitochondria isolated after ischemia or I/R. Our studies clearly demonstrate that lack of mitochondrial Prdx6 leads to mitochondrial dysfunction via disruption of mitochondrial respiration. We demonstrated increased oxygen consumption in mitochondria from Prdx6-KO mice using both static and kinetic assays. The ratio of ADP-stimulated to basal respiration (state 3/state 4) yields a RCR, which is a measurement of the functionality (coupling) of the mitochondria, and reflects the phosphorylation efficiency (2, 43). A greater RCR value is indicative of a greater degree of coupling between respiration and oxidative phospholylation and suggests an increased efficiency of electron transfer. We found that the RCR of mitochondria from Prdx6-KO mice steadily declined after ischemia and I/R, whereas the RCR of mitochondria from WT mice temporarily decreased during ischemia and early reperfusion, but then recovered to baseline values. Furthermore, using selective substrates, we found that Prdx6 appears to interact preferentially with complex I vs. complex III, as there were much more significant changes in respiration observed after stimulation of mitochondrial with glutamate/malate, a substrate for complex I.
A similar selectivity for complex I was observed in our studies of H2O2 generation. Complex I is considered a major source of ROS in liver mitochondria (3). Under physiological conditions, electrons carried by NADH or FADH2 (generated by the TCA cycle) generally enter the transport chain either through complex I or complex II, respectively. A greater amount of H2O2 production, which would be indicative of electron leak, might be expected in mitochondria from Prdx6-KO mice based on the higher oxygen consumption over time, as well as the observed decay of RCR values. We found that H2O2 production by mitochondria from Prdx6-KO mice undergoing I/R was much greater than their WT counterparts, only when stimulated with glutamate/malate. When succinate was provided as a substrate, there was little difference between WT and Prdx6-KO animals.
Although increased mitochondrial ROS production is thought to have an important role in oxidative tissue damage, the mechanisms leading to increased mitochondrial ROS are unclear. Recently, it was demonstrated that, within intact mitochondria, oxidation of glutathione to glutathione disulfide leads to glutathionylation of complex I (41). This correlates with increased superoxide formation, which is converted to H2O2 and can easily diffuse into the cytoplasm (26). The increase in mitochondrial ROS production by complex I after liver I/R injury could be due to a raise in glutathione oxidation. The Prdx6 shuttling to the mitochondria could, therefore, be a response to stop or delay the damage, since glutathione is the physiological electron donor for Prdx6, which is able to reduce hydroperoxides in the presence of GSH (24).
In summary, the present study describes novel new activities of Prdx6. We show, for the first time, that Prdx6 is expressed in hepatocyte mitochondria and that this expression appears to occur via recruitment subsequent to ischemic insult. Our subsequent studies demonstrate that lack of Prdx6 results in fulminant hepatocellular injury after I/R. This injury was independent of inflammation. Prdx6 was found to stabilize mitochondrial function during I/R by normalizing mitochondrial respiration and reducing mitochondrial generation of ROS. These effects appear to be related to a preferential impact of Prdx6 on complex I of the respiratory chain. Our data extend our understanding of the mechanisms of hepatic I/R injury and detail the important role of Prdx6 in this response. These findings are highly relevant to liver surgery and transplantation and may help identify new therapeutic targets.
GRANTS
This work was supported by National Institutes of Health Grants AG025881 and DK56029 to A. B. Lentsch.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Delva E, Camus Y, Nordlinger B, Hannoun L, Parc R, Deriaz H, Lienhart A, Huguet C. Vascular occlusions for liver resections. Operative management and tolerance to hepatic ischemia: 142 cases. Ann Surg 209: 211–218, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Estabrook RW Mitochondrial respiratory control and the polarographic measurement of ADP: O ratios. In: Methods in Enzymology, edited by Estabrook R. London: Academic, 1967, p. 41–47.
- 3.Genova ML, Ventura B, Giuliano G, Bovina C, Formiggini G, Parenti Castelli G, Lenaz G. The site of production of superoxide radical in mitochondrial Complex I is not a bound ubisemiquinone but presumably iron-sulfur cluster N2. FEBS Lett 505: 364–368, 2001. [DOI] [PubMed] [Google Scholar]
- 4.Han D, Williams E, Cadenas E. Mitochondrial respiratory chain-dependent generation of superoxide anion and its release into the intermembrane space. Biochem J 353: 411–416, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hawker F Liver dysfunction in critical illness. Anaesth Intensive Care 19: 165–181, 1991. [DOI] [PubMed] [Google Scholar]
- 6.He SQ, Zhang YH, Venugopal SK, Dicus CW, Perez RV, Ramsamooj R, Nantz MH, Zern MA, Wu J. Delivery of antioxidative enzyme genes protects against ischemia/reperfusion-induced liver injury in mice. Liver Transpl 12: 1869–1879, 2006. [DOI] [PubMed] [Google Scholar]
- 7.Hofmann B, Hecht HJ, Flohe L. Peroxiredoxins. Biol Chem 383: 347–364, 2002. [DOI] [PubMed] [Google Scholar]
- 8.Huguet C, Gavelli A, Bona S. Hepatic resection with ischemia of the liver exceeding one hour. J Am Coll Surg 178: 454–458, 1994. [PubMed] [Google Scholar]
- 9.Hynes J, Marroquin LD, Ogurtsov VI, Christiansen KN, Stevens GJ, Papkovsky DB, Will Y. Investigation of drug-induced mitochondrial toxicity using fluorescence-based oxygen-sensitive probes. Toxicol Sci 92: 186–200, 2006. [DOI] [PubMed] [Google Scholar]
- 10.Immenschuh S, Baumgart-Vogt E. Peroxiredoxins, oxidative stress, and cell proliferation. Antioxid Redox Signal 7: 768–777, 2005. [DOI] [PubMed] [Google Scholar]
- 11.Jaeschke H Mechanisms of Liver Injury. II. Mechanisms of neutrophil-induced liver cell injury during hepatic ischemia-reperfusion and other acute inflammatory conditions. Am J Physiol Gastrointest Liver Physiol 290: G1083–G1088, 2006. [DOI] [PubMed] [Google Scholar]
- 12.Jaeschke H Reactive oxygen and ischemia/reperfusion injury of the liver. Chem Biol Interact 79: 115–136, 1991. [DOI] [PubMed] [Google Scholar]
- 13.Jaeschke H, Bautista AP, Spolarics Z, Spitzer JJ. Superoxide generation by Kupffer cells and priming of neutrophils during reperfusion after hepatic ischemia. Free Radic Res Commun 15: 277–284, 1991. [DOI] [PubMed] [Google Scholar]
- 14.Jaeschke H, Farhood A, Smith CW. Neutrophils contribute to ischemia/reperfusion injury in rat liver in vivo. FASEB J 4: 3355–3359, 1990. [PubMed] [Google Scholar]
- 15.Jarrold B, DeMuth J, Greis K, Burt T, Wang F. An effective skeletal muscle prefractionation method to remove abundant structural proteins for optimized two-dimensional gel electrophoresis. Electrophoresis 26: 2269–2278, 2005. [DOI] [PubMed] [Google Scholar]
- 16.Kang SW, Baines IC, Rhee SG. Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J Biol Chem 273: 6303–6311, 1998. [DOI] [PubMed] [Google Scholar]
- 17.Lakowicz JR Principles of Fluorescence Spectroscopy. New York: Plenum, 1999.
- 18.Lapidus RG, Sokolove PM. Spermine inhibition of the permeability transition of isolated rat liver mitochondria: an investigation of mechanism. Arch Biochem Biophys 306: 246–253, 1993. [DOI] [PubMed] [Google Scholar]
- 19.Lemasters JJV Necrapoptosis and the mitochondrial permeability transition: shared pathways to necrosis and apoptosis. Am J Physiol Gastrointest Liver Physiol 276: G1–G6, 1999. [DOI] [PubMed] [Google Scholar]
- 20.Lemasters JJ, Thurman RG. Reperfusion injury after liver preservation for transplantation. Annu Rev Pharmacol Toxicol 37: 327–338, 1997. [DOI] [PubMed] [Google Scholar]
- 21.Lentsch AB, Kato A, Yoshidome H, McMasters KM, Edwards MJ. Inflammatory mechanisms and therapeutic strategies for warm hepatic ischemia/reperfusion injury. Hepatology 32: 169–173, 2000. [DOI] [PubMed] [Google Scholar]
- 22.Lentsch AB, Yoshidome H, Cheadle WG, Miller FN, Edwards MJ. Chemokine involvement in hepatic ischemia/reperfusion injury in mice: roles for macrophage inflammatory protein-2 and KC. Hepatology 27: 1172–1177, 1998. [DOI] [PubMed] [Google Scholar]
- 23.Liu DL, Jeppsson B, Hakansson CH, Odselius R. Multiple-system organ damage resulting from prolonged hepatic inflow interruption. Arch Surg 131: 442–447, 1996. [DOI] [PubMed] [Google Scholar]
- 24.Manevich Y, Sweitzer T, Pak JH, Feinstein SI, Muzykantov V, Fisher AB. 1-Cys peroxiredoxin overexpression protects cells against phospholipid peroxidation-mediated membrane damage. Proc Natl Acad Sci USA 99: 11599–11604, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martindale JL, Holbrook NJ. Cellular response to oxidative stress: signaling for suicide and survival. J Cell Physiol 192: 1–15, 2002. [DOI] [PubMed] [Google Scholar]
- 26.McCord JM Oxygen-derived free radicals in postischemic tissue injury. N Engl J Med 312: 159–163, 1985. [DOI] [PubMed] [Google Scholar]
- 27.Mo Y, Feinstein SI, Manevich Y, Zhang Q, Lu L, Ho YS, Fisher AB. 1-Cys peroxiredoxin knock-out mice express mRNA but not protein for a highly related intronless gene. FEBS Lett 555: 192–198, 2003. [DOI] [PubMed] [Google Scholar]
- 28.Monteiro G, Horta BB, Pimenta DC, Augusto O, Netto LE. Reduction of 1-Cys peroxiredoxins by ascorbate changes the thiol-specific antioxidant paradigm, revealing another function of vitamin C. Proc Natl Acad Sci USA 104: 4886–4891, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nieminen AL, Byrne AM, Herman B, Lemasters JJ. Mitochondrial permeability transition in hepatocytes induced by t-BuOOH: NAD(P)H and reactive oxygen species. Am J Physiol Cell Physiol 272: C1286–C1294, 1997. [DOI] [PubMed] [Google Scholar]
- 30.Nieminen AL, Saylor AK, Tesfai SA, Herman B, Lemasters JJ. Contribution of the mitochondrial permeability transition to lethal injury after exposure of hepatocytes to t-butylhydroperoxide. Biochem J 307: 99–106, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okaya T, Blanchard J, Schuster R, Kuboki S, Husted T, Caldwell CC, Zingarelli B, Wong H, Solomkin JS, Lentsch AB. Age-dependent responses to hepatic ischemia/reperfusion injury. Shock 24: 421–427, 2005. [DOI] [PubMed] [Google Scholar]
- 32.Okaya T, Lentsch AB. Cytokine cascades and the hepatic inflammatory response to ischemia and reperfusion. J Invest Surg 16: 141–147, 2003. [PubMed] [Google Scholar]
- 33.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567, 1999. [DOI] [PubMed] [Google Scholar]
- 34.Rhee SG, Chae HZ, Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med 38: 1543–1552, 2005. [DOI] [PubMed] [Google Scholar]
- 35.Rymsa B, Wang JF, de Groot H. O2•− release by activated Kupffer cells upon hypoxia-reoxygenation. Am J Physiol Gastrointest Liver Physiol 261: G602–G607, 1991. [DOI] [PubMed] [Google Scholar]
- 36.Schierwagen C, Bylund-Fellenius AC, Lundberg C. Improved method for quantification of tissue PMN accumulation measured by myeloperoxidase activity. J Pharmacol Methods 23: 179–186, 1990. [DOI] [PubMed] [Google Scholar]
- 37.Senft AP, Dalton TP, Nebert DW, Genter MB, Hutchinson RJ, Shertzer HG. Dioxin increases reactive oxygen production in mouse liver mitochondria. Toxicol Appl Pharmacol 178: 15–21, 2002. [DOI] [PubMed] [Google Scholar]
- 38.Shertzer HG, Clay CD, Genter MB, Chames MC, Schneider SN, Oakley GG, Nebert DW, Dalton TP. Uncoupling-mediated generation of reactive oxygen by halogenated aromatic hydrocarbons in mouse liver microsomes. Free Radic Biol Med 36: 618–631, 2004. [DOI] [PubMed] [Google Scholar]
- 39.Shertzer HG, Genter MB, Shen D, Nebert DW, Chen Y, Dalton TP. TCDD decreases ATP levels and increases reactive oxygen production through changes in mitochondrial F(0)F(1)-ATP synthase and ubiquinone. Toxicol Appl Pharmacol 217: 363–374, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858, 1996. [DOI] [PubMed] [Google Scholar]
- 41.Taylor ER, Hurrell F, Shannon RJ, Lin TK, Hirst J, Murphy MP. Reversible glutathionylation of complex I increases mitochondrial superoxide formation. J Biol Chem 278: 19603–19610, 2003. [DOI] [PubMed] [Google Scholar]
- 42.Turrens JF, Alexandre A, Lehninger AL. Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria. Arch Biochem Biophys 237: 408–414, 1985. [DOI] [PubMed] [Google Scholar]
- 43.Will Y, Hynes J, Ogurtsov VI, Papkovsky DB. Analysis of mitochondrial function using phosphorescent oxygen-sensitive probes. Nat Protoc 1: 2563–2572, 2006. [DOI] [PubMed] [Google Scholar]
- 44.Wu YZ, Manevich Y, Baldwin JL, Dodia C, Yu K, Feinstein SI, Fisher AB. Interaction of surfactant protein A with peroxiredoxin 6 regulates phospholipase A2 activity. J Biol Chem 281: 7515–7525, 2006. [DOI] [PubMed] [Google Scholar]