Abstract
Objective. To identify whether a shared genetic influence accounts for the occurrence of OA at different skeletal sites.
Methods. Multivariate modelling of data on prevalent radiographic OA at the hand (DIP, PIP and CMC joints), hip and knee joints assessed in 992 monozygotic and dizygotic female twin participants from the TwinsUK Registry.
Results. OA at all the five joint sites was heritable. Genetic influences were strongly correlated among joints in the hand; however, there was little evidence of common genetic pathways to account for the co-occurrence of OA at the hand, hip and knee.
Conclusions. While genetic influences are important in explaining the variation in occurrence of OA at the hand, hip and knee, there is no evidence that common or shared genetic factors determine the occurrence of disease across all these skeletal sites. The findings suggest that there are important aetiological differences in the disease that are site-specific in women. These results have implications for the design of studies examining the genetic basis of OA as well as for strategies aimed at preventing and treating the disease.
Keywords: Osteoarthritis, Site, Twin, Hand, Hip, Knee, Structural equation modelling, Genetic linkage, Genetic association
Introduction
OA describes a set of age-related, pathological and radiological changes in joints that share common features across different skeletal sites in the body. Twin and family studies have demonstrated a significant contribution of genetic factors (39–70%) [1]. With the advent of genetic linkage and association studies the contribution made by a number of specific genetic loci to the disease is being increasingly well defined [2].
However, there is uncertainty as to whether OA represents a single common phenotype or whether the disease is more heterogeneous, with different genetic and environmental risk factors in operation at different skeletal sites. The common pathological and radiological patterns of disease, together with the frequent occurrence of disease at multiple joint sites in some individuals (so-called ‘generalized OA’), argue in favour of the existence of a single disease phenotype. However, it is well recognized that traditional epidemiological risk factors for the disease differ between body sites. For example, BMI has been shown to be a risk factor for OA of the hand and knee [3], but not the hip [4, 5]. Individual radiographic features of OA (e.g. osteophytes and joint space narrowing) differ in their heritability both within a body site and across body sites [1]. This evidence favours a more heterogeneous disease in which the contribution of individual genes in OA differs between joints sites. Whether or not OA is best explained by a common genetic process has important implications for the design of studies examining the precise nature of the genetic contribution to the disease.
Twin studies provide a well-defined set of methods for teasing apart the relative genetic and environmental contributions to individual traits as well as assessing, through multivariate analysis, the extent to which genetic influences are shared between different traits. Here we used a structural equation modelling approach in monozygotic (MZ) and dizygotic (DZ) twins selected from the TwinsUK Registry to determine whether the pattern of occurrence of hand, hip and knee OA can be explained by shared genetic influences.
Methods
Healthy female twin volunteers were selected from the TwinsUK Registry and standard radiographs of the hand, hip and knee were taken as previously reported [6, 7]. Written consent was obtained from the twins. The project was approved by the St Thomas’ Hospital ethics committee. The radiographs were taken between 1995 and 2000 and were classified using the Kellgren and Lawrence scoring system at the knee and hip, and separately at the PIP, DIP and CMC joints on the left and right sides. The score combines measures of joint space narrowing, osteophytes, sclerosis and cyst formation in each joint area. At each site, measures were scored by a single observer after their reliability had been validated. The score for each joint site was taken as the sum of the Kellgren and Lawrence scores on each side. Details of scoring and rater validation are given elsewhere [6, 7].
The analysis used structural equation modelling, a standard approach that is used widely in twin analysis as implemented in Mx software [8]. The method assigns variation among traits in a twin population to additive genetic factors (A) and factors in the environment that may be common to members of a twin pair (C) or unique to individual twins (E). Models containing combinations of the variance components can be constructed based on the known underlying correlation structure in twin populations (namely that A correlates 1 in MZ twins and 0.5 in DZ twins; C correlates 1 in both MZ and DZ twins; and E is uncorrelated in both types of twin). The fit of each model can be assessed by comparing the observed phenotypic correlations among twins with correlations expected under the model through the chi-squared statistic. The significance of each variance component within the model can be assessed by measuring the deterioration in chi-square when components are removed. E (which represents an error term) is retained in all models that are considered and in this analysis genetic dominance effects were not considered. This method has been widely used [9, 10] and has been performed successfully in TwinsUK for the study of a variety of traits [11, 12].
The analysis first examined the intraclass correlation for each of the five body sites separately in the MZ and DZ twins. Structural equation modelling was carried out to assess the heritability of OA at each body site (univariate analysis).
Modelling was then extended to take into account the pattern of correlation among all five site variables within the twins (multivariate analysis) and to examine (i) the extent to which observed correlation among traits might be accounted for by shared genetic and environmental factors and (ii) whether there was evidence to suggest a common underlying genetic phenotype for OA across all body sites.
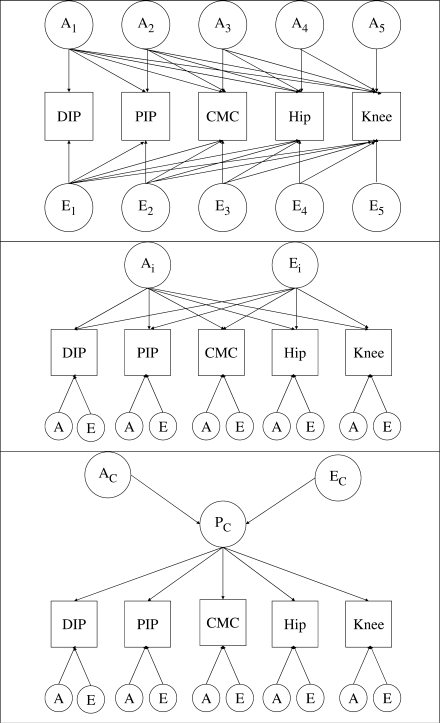
Three multivariate models were considered and are illustrated in Fig. 1:
The Cholesky decomposition model: this model includes five independent latent genetic and environmental variables that factor on each phenotype.
The independent pathway model: this model considers the data to be explained by a single shared genetic and a single shared environmental factor.
The common pathway model: this model considers a single shared latent phenotype, determined in turn by latent genetic and environment factors.
The Cholesky decomposition provides the fullest potential explanation of the data under the twin model. Parameter estimates from this model were used to calculate the genetic and environmental correlation among the traits. The independent pathway and common pathway models can be regarded as providing potentially simpler explanations of the data. Both are submodels of the Cholesky model and their suitability was compared using the significance of the chi-squared deterioration in fit when compared with Cholesky. The independent and common pathway models are not nested within each other. Their suitability can be indirectly compared by examining the Akaike information criterion [AIC, defined as model χ2 minus 2 x degrees of freedom (df)]. Lower values of AIC indicate the most suitable model.
Fig. 1.
The multivariate models of the five OA sites. Models containing additive (A) genetic and unique environmental (E) components are depicted. The top panel illustrates the Cholesky model, the middle panel the independent pathway model and the lower panel the common pathway model. The Cholesky model contains 5 genetic factors (A1–A5) and five environmental factors (E1–E5) loading sequentially on the five OA phenotypes. The independent pathway model contains one common genetic factor (Ac) and one common environmental factor (Ec) shared by all five OA phenotypes, together with site-specific genetic and environmental components (labelled A and E). The common pathway model contains one shared underlying phenotype (Pc) itself determined by a genetic and environmental components (Ac and Ec), together with site-specific genetic and environmental components (labelled A and E).
For the analysis, continuous variables representing each of five joints (PIP, DIP, CMC, hip and knee) were constructed from the residuals of regression analyses after the inclusion of age. Data were transformed to approximate a normal distribution.
Results
Complete data on hand, hip and knee radiographic OA were available in 992 individual twins comprising 153 MZ and 343 DZ twin pairs. Their characteristics, together with the distribution of OA at the five joint sites, are shown in Table 1.
Table 1.
Characteristics of the female twins
| MZ | DZ | |
|---|---|---|
| Number of individual twins | 306 | 686 |
| Age, yrs, median (range) | 55.9 (36.0, 69.7) | 52.1 (24.2, 70.3) |
| Height, cm, mean (S.D.) | 160.9 (5.8) | 162.7 (5.7) |
| Weight, kg, mean (S.D.) | 64.4 (10.7) | 65.5 (11.6) |
| DIP OA (Grade 2 or more at any joint),% | 29 | 21 |
| PIP OA (Grade 2 or more at any joint),% | 11 | 8 |
| CMC OA (Grade 2 or more at any joint),% | 24 | 17 |
| Hip OA (Grade 2 or more, either left or right),% | 9 | 9 |
| Knee OA (Grade 2 or more, either left or right),% | 23 | 20 |
Grade 2 or more—Kellgren and Lawrence score.
The intraclass correlations were higher among MZ than DZ twins consistent with a genetic effect. Univariate analysis confirmed the presence of a genetic influence at each of the five anatomical sites (DIP, PIP, CMC, hip and knee) considered separately. The most appropriate model for the data contained only additive genetic and unique environmental components (the AE model). The intraclass correlations and heritabilities derived from the AE models are shown in Table 2.
Table 2.
Intraclass correlation and heritability of OA at the five skeletal sites
| RMZ (95% CI) | RDZ (95% CI) | Heritability, % (95% CI) | |
|---|---|---|---|
| DIP | 0.65 (0.59, 0.72) | 0.32 (0.25, 0.39) | 65 (57, 73) |
| PIP | 0.58 (0.51, 0.66) | 0.25 (0.17, 0.32) | 53 (44, 62) |
| CMC | 0.74 (0.70, 0.80) | 0.21 (0.14, 0.29) | 68 (60, 75) |
| Hip | 0.39 (0.25, 0.51) | 0.09 (0.00, 0.18) | 28 (15, 40) |
| Knee | 0.39 (0.27, 0.51) | 0.20 (0.11, 0.30) | 37 (28, 48) |
Intraclass correlation (R) for MZ and DZ twins, the heritability estimates and 95% CIs are given for each joint site; calculated from the univariate AE model.
Genetic and environmental correlations derived from the AE Cholesky model are shown in Table 3, along with the phenotypic correlations for OA at each site. The results of multivariate modelling are shown in Table 4. Of the Cholesky models, the AE model provided the most suitable explanation of the data. The results indicate that a degree of genetic correlation exists between DIP and PIP joint sites (r = 0.23). However, there is little genetic correlation between OA at the hip and the knee.
Table 3.
Phenotypic, genetic and environmental correlations for OA among the five skeletal sites
| DIP | PIP | CMC | Hip | Knee | |
|---|---|---|---|---|---|
| Phenotypic correlation | |||||
| DIP | 1.000 | ||||
| PIP | 0.479 | 1.000 | |||
| CMC | 0.315 | 0.201 | 1.000 | ||
| Hip | 0.069 | 0.075 | 0.034 | 1.000 | |
| Knee | 0.074 | 0.080 | 0.079 | 0.043 | 1.000 |
| Genetic correlation | |||||
| DIP | 1.000 | ||||
| PIP | 0.230 | 1.000 | |||
| CMC | 0.093 | 0.015 | 1.000 | – | |
| Hip | 0.036 | 0.081 | 0.180 | 1.000 | |
| Knee | −0.008 | −0.110 | −0.006 | −0.039 | 1.000 |
| Environmental correlation | |||||
| DIP | 1.000 | ||||
| PIP | 0.201 | 1.000 | |||
| CMC | 0.130 | 0.091 | 1.000 | ||
| Hip | 0.018 | 0.006 | 0.078 | 1.000 | |
| Knee | 0.042 | 0.092 | 0.066 | 0.033 | 1.000 |
The genetic and environmental correlations represent the correlation in additive genetic factors and unique environmental factors, respectively, taken from the AE model.
Table 4.
Results of multivariate modelling for the five skeletal sites simultaneously
| χ2 | df | AIC | |
|---|---|---|---|
| Cholesky | |||
| ACE | 108.3 | 50 | 8.3 |
| AE | 111.6 | 65 | −18.4 |
| CE | 211.5 | 65 | 81.5 |
| E | 533.3 | 80 | 373.3 |
| Independent pathway | |||
| ACE | 116.9 | 65 | −13.1 |
| AE | 118.3 | 75 | −31.7 |
| CE | 221.7 | 75 | 71.7 |
| E | 536.2 | 85 | 366.2 |
| Common pathway | |||
| ACE | 121.7 | 73 | −24.3 |
| AE | 122.2 | 79 | −35.8 |
| CE | 226.3 | 79 | 68.3 |
| E | 536.2 | 85 | 366.2 |
‘A’ represents additive genetic factors; ‘C’ common environmental factors; ‘E’ unique environmental factors. AIC represents the Akaike information criterion that describes the balance between goodness of fit and parsimony of each model: the lowest value is in bold typeface.
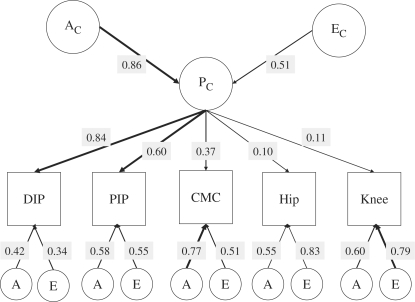
When the independent and common pathway models were compared with the Cholesky models, the AE common pathway model was found to offer the most suitable explanation of the data with the lowest value of AIC at −35.8 (Table 4). This AE common pathway model explains the variance at each site in terms of unique A and E contributions as well as a contribution from the ‘common OA phenotype’ (Pc).
The parameter estimates derived from the AE common pathway model are shown in Fig. 2. To obtain the contribution that Pc and unique A and E make to the variance in a trait, squares of the path coefficient are taken. Thus, the model postulates the existence of an underlying OA phenotype (Pc) with a heritability 74% (0.862) that chiefly explains the co-occurrence of hand OA. At the DIP joint, the common phenotype Pc accounts for 71% (0.842) of the variation, and at the PIP joint it accounts for 37% (0.602) of the variation. At the CMC joint, the common phenotype accounts for 14% of the variation. The heritability of the variation in the phenotype that is not accounted for by Pc is 18% (0.422) at the DIP joint, 34% (0.582) at the PIP joint and 60% (0.772) at the CMC joint.
Fig. 2.
Path coefficients of the common pathway model for all five OA variables. The figure shows parameter estimates for the path coefficients of the common pathway AE model, selected as the most appropriate depiction of the data. Thickness of the arrows represents the strength of the association. The squares of the path coefficients provide an estimate of the variance explained by common and specific genetic and environmental components.
These results suggest that the ‘common OA phenotype’ does not account for significant variation in the propensity to OA at the hip and the knee in this model. Indeed, Pc accounts for only 1% of the variance in hip and knee OA. Effectively, this model can be interpreted as indicating that the ‘common OA phenotype’ is not an important component of OA at these large joints, but it does seem to play a role in hand OA.
Discussion
The results presented here provide evidence of a genetic overlap between OA at the small joints of the hand but not the large joints of the lower limb. The DIP and PIP joints have considerable sharing of genetic factors, with a lesser degree of genetic overlap at the CMC joints. Using structural equation modelling, we could find little evidence of the phenotype ‘generalized OA’ or indication of a systemic disease. If it does exist, this study suggests that it must be rare.
There are a number of possible mechanisms accounting for these observations. One suggestion is that joint shape is an important determinant of disease risk and the widely different shapes of individual joints (for example, at the CMC joint compared with the PIP and DIP joints) may be determined by different sets of genes. Instability at the trapeziometacarpal joint, for example, has been shown to predict development of CMC joint OA [13]. An alternative explanation is that the dominating genetic influences on the disease are manifest through gene–environment interaction. Thus the influence of repeated micro-trauma at the knee, for example, may have an influence on development of disease at that site [14]. Equally, the development of hand OA in women may reflect a hormonal influence. Variation in collagen composition at different sites provides an alternative explanation. Animal work has provided evidence for different cartilage response to injury at different areas in the same joint [15]. Similarly, it is possible that the properties of chondrocytes, and the local anabolic and catabolic growth factors that determine their resistance to apoptosis, differ by skeletal site.
Limitations of this analytical approach need to be considered. The sample comprised only females and those twins for whom a complete set of radiographs was available. There is nothing to suggest that these twins differed in their characteristics from the remainder of those registered in the TwinsUK sample. Results cannot be extrapolated to males. The representativeness of the twin sample as a whole has been questioned in the past: however, we have shown twins to be similar to singletons for a wide range of health and lifestyle factors including OA [16]. As this is a representative sample of the healthy population the prevalence of radiographic OA—particularly at the hip—is relatively low, making conclusions drawn regarding hip OA less robust. It would be interesting to see whether a sample of symptomatic subjects with hip OA demonstrated similarly low genetic correlations at the large joints. This was a cross-sectional survey of an age-related condition and the traits were subject to right censorship. A longitudinal approach in which time is included as a variable might allow identification of the temporal, as well as the spatial, clustering in the evolution of this age-related condition.
The models examined here provide only a descriptive representation of this particular data set and results would need to be validated independently if these findings are to be truly generalizable. This is particularly true with regard to men: results from women may not be extrapolated to the opposite sex. Nevertheless, from the perspective of planning future genetic studies of OA, our analysis presents evidence that the genetic influence on OA differs by anatomical site and that only PIP and DIP joints can be usefully considered as reflecting the same underlying genetic tendency.
These results may shed light on why the currently published studies of OA genetics show inconsistent findings [17]. They may also provide part of the explanation for site-specific differences in the influence of individual risk factors for the disease and in responses to treatment. The findings have implications for future epidemiological research. Design of large-scale collaborative studies of the future that are needed to unravel the precise genetic loci involved in OA should take into account the diversity of gene action at individual anatomical sites.

Acknowledgements
The authors are grateful for the contribution of David Hunter, Leto Antoniades, Mathew Matson, Toby Andrew and Deborah Hart. They would also like to thank the twin participants from the TwinsUK Registry.
Funding: This work was supported by grants from the Wellcome Trust, Medical Research Council, Chronic Diseases Research Foundation and the Arthritis Research Campaign. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Disclosure statement: The authors have declared no conflicts of interest.
References
- 1.Spector TD, MacGregor AJ. Risk factors for osteoarthritis: genetics. Osteoarthritis Cartilage. 2004;12(Suppl. A):S39–44. doi: 10.1016/j.joca.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Loughlin J. The genetic epidemiology of human primary osteoarthritis: current status. Expert Rev Mol Med. 2005;7:1–12. doi: 10.1017/S1462399405009257. [DOI] [PubMed] [Google Scholar]
- 3.Cicuttini FM, Baker JR, Spector TD. The association of obesity with osteoarthritis of the hand and knee in women: a twin study. J Rheumatol. 1996;23:1221–6. [PubMed] [Google Scholar]
- 4.Sturmer T, Gunther KP, Brenner H. Obesity, overweight and patterns of osteoarthritis: the Ulm Osteoarthritis Study. J Clin Epidemiol. 2000;53:307–13. doi: 10.1016/s0895-4356(99)00162-6. [DOI] [PubMed] [Google Scholar]
- 5.Gelber AC, Hochberg MC, Mead LA, Wang NY, Wigley FM, Klag MJ. Body mass index in young men and the risk of subsequent knee and hip osteoarthritis. Am J Med. 1999;107:542–8. doi: 10.1016/s0002-9343(99)00292-2. [DOI] [PubMed] [Google Scholar]
- 6.MacGregor AJ, Antoniades L, Matson M, Andrew T, Spector TD. The genetic contribution to radiographic hip osteoarthritis in women: results of a classic twin study. Arthritis Rheum. 2000;43:2410–6. doi: 10.1002/1529-0131(200011)43:11<2410::AID-ANR6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 7.Spector TD, Cicuttini F, Baker J, Loughlin J, Hart D. Genetic influences on osteoarthritis in women: a twin study. Br Med J. 1996;312:940–3. doi: 10.1136/bmj.312.7036.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neale MC, Boker SM, Xie G, Maes HH. 2006. Mx: Statistical modeling, 7th edn. VCU Box 900126, Richmond VA23298: Department of Psychiatry. [Google Scholar]
- 9.Martin NG, Eaves LJ. The genetical analysis of covariance structure. Heredity. 1977;38:79–95. doi: 10.1038/hdy.1977.9. [DOI] [PubMed] [Google Scholar]
- 10.Neale MC, Cardon L. Dordrecht: Kluwer Academic Publishers; 1992. Methodology for genetic studies in twins and families. [Google Scholar]
- 11.de Lange M, Snieder H, Ariens RA, Spector TD, Grant PJ. The genetics of haemostasis: a twin study. Lancet. 2001;357:101–5. doi: 10.1016/S0140-6736(00)03541-8. [DOI] [PubMed] [Google Scholar]
- 12.Williams FM, Cherkas LF, Spector TD, MacGregor AJ. A common genetic factor underlies hypertension and other cardiovascular disorders. BMC Cardiovasc Disord. 2004;4:20. doi: 10.1186/1471-2261-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter DJ, Zhang Y, Sokolove J, Niu J, Aliabadi P, Felson DT. Trapeziometacarpal subluxation predisposes to incident trapeziometacarpal osteoarthritis (OA): the Framingham Study. Osteoarthritis Cartilage. 2005;13:953–7. doi: 10.1016/j.joca.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Brandt KD, Radin EL, Dieppe PA, van de PL. Yet more evidence that osteoarthritis is not a cartilage disease. Ann Rheum Dis. 2006;65:1261–4. doi: 10.1136/ard.2006.058347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoop R, Buma P, van der Kraan PM, et al. Differences in type II collagen degradation between peripheral and central cartilage of rat stifle joints after cranial cruciate ligament transection. Arthritis Rheum. 2000;43:2121–31. doi: 10.1002/1529-0131(200009)43:9<2121::AID-ANR24>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 16.Andrew T, Hart DJ, Snieder H, de Lange M, Spector TD, MacGregor AJ. Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin Res. 2001;4:464–77. doi: 10.1375/1369052012803. [DOI] [PubMed] [Google Scholar]
- 17.Ryder JJ, Garrison K, Song F, et al. Genetic associations in peripheral joint osteoarthritis and spinal degenerative disease: a systematic review. Ann Rheum Dis. 2007;67:584–91. doi: 10.1136/ard.2007.073874. [DOI] [PubMed] [Google Scholar]