Abstract
Roundworms of the genus Ascaris are common parasites of the human gastrointestinal tract. A battery of selective inhibitors protects them from host enzymes and the immune system. Here, a metallocarboxypeptidase (MCP) inhibitor, ACI, was identified in protein extracts from Ascaris by intensity-fading MALDI-TOF mass spectrometry. The 67-residue amino acid sequence of ACI showed no significant homology with any known protein. Heterologous overexpression and purification of ACI rendered a functional molecule with nanomolar equilibrium dissociation constants against MCPs, which denoted a preference for digestive and mast cell A/B-type MCPs. Western blotting and immunohistochemistry located ACI in the body wall, intestine, female reproductive tract, and fertilized eggs of Ascaris, in accordance with its target specificity. The crystal structure of the complex of ACI with human carboxypeptidase A1, one of its potential targets in vivo, revealed a protein with a fold consisting of two tandem homologous domains, each containing a β-ribbon and two disulfide bonds. These domains are connected by an α-helical segment and a fifth disulfide bond. Binding and inhibition are exerted by the C-terminal tail, which enters the funnel-like active-site cavity of the enzyme and approaches the catalytic zinc ion. The findings reported provide a basis for the biological function of ACI, which may be essential for parasitic survival during infection.
Keywords: ascariasis, crystal structure, host resistance, immunolocalization, metallocarboxypeptidase inhibitor
More than a quarter of the human population is affected by soil-transmitted helminthes, which impair nutrition and the immune response toward widespread pandemics such as AIDS and tuberculosis (1, 2). The roundworm Ascaris lumbricoides is the most common human parasite of the gastrointestinal tract. It causes ascariasis (3), which has a worldwide distribution with highest prevalence in tropical and subtropical regions and in areas with inadequate sanitation. Ascariasis is triggered by the ingestion of parasite eggs. These evolve to larvae that migrate through different tissues and return to the small intestine, where they mature to adult male and female worms. At this stage, females deposit thousands of eggs daily, which are secreted with the feces, thus contributing to soil contamination and spreading of the infection [for details, see supporting information (SI) Fig. S1]. During its life cycle, Ascaris threatens human health with nonspecific abdominal symptoms, intestinal obstruction and perforation, biliary colic, gallstone formation, liver abscesses, pancreatitis, and pulmonary eosinophilia (4, 5). A nearly identical nematode species, Ascaris suum, is found in the pig. It has a tremendous impact on livestock farming and can also infect primates and humans, giving rise to a similar disease pattern to A. lumbricoides (6–8).
As part of the parasite defense strategy, Ascaris roundworms secrete a series of inhibitors to target digestive and immune-related host proteases, among others pepsin, trypsin, chymotrypsin/elastase, cathepsins, and metallocarboxypeptidases (MCPs) (9–16). MCPs are zinc-containing exoproteases that catalyze the hydrolysis of C-terminal amino acids from proteins and peptides. They perform a large variety of physiologically relevant functions in organisms of different phyla (17). These enzymes have been grouped into the funnelin tribe of proteases and are subdivided into A/B- and N/E-type MCPs (18). Human A/B-type funnelins include the digestive enzymes CPA1, CPA2, and CPB1, and mast cell CPA3, which is related to inflammatory processes (19, 20). The biological action of MCPs is specifically modulated through protein inhibitors. To date, seven such MCP inhibitors have been described from potato and tomato (PCI and MCPI; 38 and 39 residues, respectively) (21, 22), medical leech Hirudo medicinalis (LCI; 66 residues) (23), the ticks Rhipicephalus bursa and Haemaphysalis longicornis (TCI and HlTCI; 75 and 77 residues, respectively) (24, 25), rat and human latexin (alias ECI; 222 and 223 residues, respectively) (26, 27), and the intestinal parasites A. lumbricoides and A. suum (ACI) (12, 13). Although the former inhibitors have been studied extensively in terms of activity and structure, ACI has hitherto only been studied for its amino acid sequence. We present here its cloning, heterologous expression, purification, and three-dimensional structure in complex with a MCP, unveiling its mechanism of inhibition. We also report its target specificity and in vivo localization in Ascaris worms, which lead to a deeper understanding of the life-threatening disease ascariasis and may pave the way for drug and vaccine development.
Results and Discussion
Identification, Sequencing, and Cloning of ACI from Ascaris.
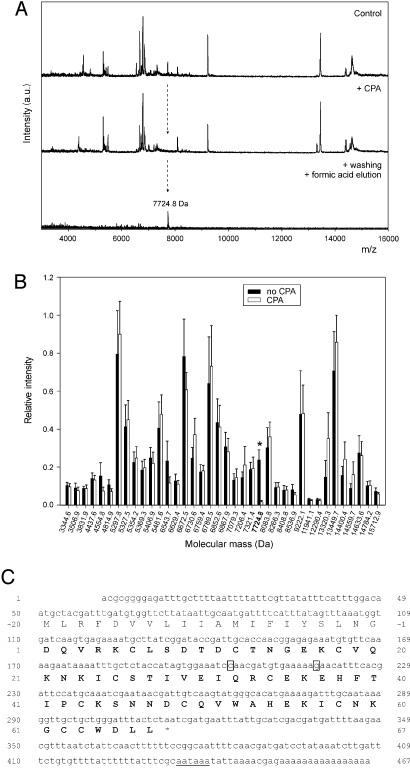
Initial recombinant overexpression trials of ACI on the basis of the reported amino acid sequence (12) produced only minute yields and two distinct forms (termed A and B; see Fig. S2) of identical molecular mass (7,502.5 Da) and 8 cysteine residues forming four disulfide bonds. Of these two, only form B inhibited bovine carboxypeptidase A1 (bCPA1), albeit weakly (Ki = 5.9 ± 0.2 μM) compared with other reported MCP inhibitors (17). We thus attempted to analyze ACI directly from a crude homogenate of Ascaris worms. After assessing the presence of inhibitory activity against bCPA1 (see SI Materials and Methods), the extract was subjected to intensity fading MALDI-TOF mass spectrometry (28). This procedure rendered a molecular mass of 7,724.8 Da for the Ascaris MCPI (Fig. 1 A and B), which did not match that of the reported sequence (12). The inhibitor was purified by using a combination of affinity chromatography and reversed-phase HPLC. Its 30 N-terminal residues matched the sequence reported (12). However, the protein had 10 rather than 8 cysteine residues, and subsequent analysis of peptides obtained by digestion with endoproteinase Lys-C revealed that the C terminus (-GCCWDLL67) had been erroneously determined (-LPWGL65) (12). Therefore, the cDNA of ACI was cloned (Fig. 1C), and it was found to consist of 467 nucleotides, with a deduced protein sequence of 87 residues. The first 20 residues correspond to a signal peptide that precedes the aspartate residue found at the N terminus of the purified inhibitor. The calculated molecular mass of the deduced mature protein was consistent with that of natural ACI determined by MALDI-TOF mass spectrometry, with five disulfide bonds. Sequence similarity searches revealed no homology with any other reported sequence with the exception of the C terminus, which showed certain resemblance to the C termini of other reported MCPIs (see below and Fig. 3D).
Fig. 1.
Identification of natural ACI by intensity-fading MALDI-TOF mass spectrometry and cDNA cloning of ACI. (A) Mass spectra of an Ascaris extract before (Control) and after the addition of CPA–Sepharose resin (+ CPA). (Bottom) Mass spectrum corresponding to the inhibitor recovered after washing and acidification of the sample. (B) Plot of the relative intensities of the molecular ions present in the mass spectrum of the Ascaris extract before and after interaction with CPA. Ten independent experiments were conducted to draw the plot. The molecular mass of the inhibitor identified by intensity fading MALDI-TOF mass spectrometry is labeled with an asterisk. (C) Nucleotide and deduced amino acid sequence of ACI. The full-length cDNA sequence was assembled from two overlapping clones obtained by 5′- and 3′-RACE techniques (nucleotides 1–315 and 179–467, respectively). It contains a 49-nucleotide 5′-untranslated sequence, an ORF of 264 nucleotides, and a 154-nucleotide 3′-untranslated region. A canonical polyadenylation signal, AATAAA (underlined), is detected 12 nucleotides upstream from the poly(A)+ tail. The amino acid sequence of mature ACI is printed in bold. Two isoforms (boxed) are found at the triplets codifying for residues at positions 32 and 37, caa/gaa (glutamine/glutamate) and gaa/caa (glutamate/glutamine), respectively.
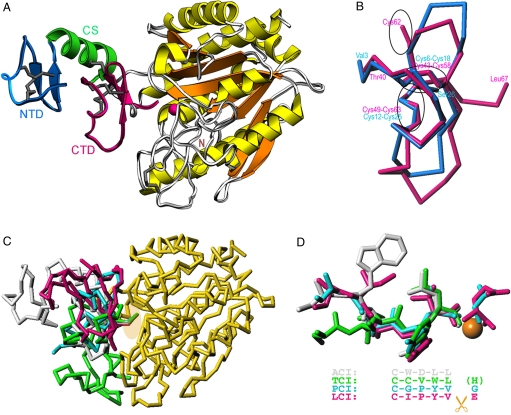
Fig. 3.
Structure of ACI in its complex with hCPA1. (A) Richardson plot of the complex of ACI (NTD in blue, CS in green, and CTD in magenta) and hCPA1 (α-helices in yellow, β-strands in orange, coils in white) in standard orientation for funnelins (18). Regular secondary structure elements (ribbons for the α-helix; arrows for β-strands) and disulfide bonds are shown for ACI. The catalytic zinc ion of the metalloenzyme is shown as a magenta sphere. (B) Superimposition of the NTD (blue Cα trace) and the CTD (magenta Cα trace) of ACI. The position of the disulfide bonds is pinpointed by ellipsoids. (C) Overlay of the structures of the exogenous inhibitors analyzed to date in complex with funnelins, PCI [cyan; Protein Data Bank (PDB) code 4cpa], TCI (green; PDB code 1zlh), LCI (magenta; PDB code 1dtd), and ACI (white; PDB ID code 3fju), after superimposition of the respective protease moieties. For clarity, only hCPA1 (yellow) in its complex with ACI is further depicted. An ellipsoid pinpoints the C termini of the inhibitors. (D) Close-up view of C in the same orientation showing only the C termini of the inhibitors in the same color as in C. The structural equivalence of the positions of the tails is provided by the alignment. While the terminal residue is severed upon complex formation and occupies the S1′ pocket in PCI and LCI, in TCI it is cleaved but not present in the active site. In ACI it is not cut at all as it occupies subsite S1.
Heterologous Expression and Purification of ACI.
Recombinant ACI was overexpressed in Escherichia coli as a fusion protein (Fig. S3), whose cleavage left a glycine residue at the N terminus of the inhibitor protein (molecular mass of 7,781.8 Da). A final reversed-phase HPLC step rendered a unique peak with a retention time equivalent to that of natural ACI. The typical yield was ≈10 mg of pure recombinant ACI per L of cell culture.
Conformational Stability and Activity of ACI.
Circular dichroism and NMR spectroscopy experiments showed that the conformations of natural and recombinant ACI were indistinguishable. Both molecules maintained a well-folded conformation in a wide range of chaotropic reagents and temperature and only became denatured by the simultaneous presence of denaturing and reducing agents (Figs. S4 and S5). This high stability may be attributed to the five disulfide bonds, which strongly constrain the ACI structure, as reported for PCI, LCI, and TCI (29–31). Equilibrium dissociation constants for the complexes of natural ACI and recombinant ACI with a selection of MCPs were indistinguishable (data not shown). This agreement revealed that ACI is a tight binding, competitive inhibitor of A/B-type but not N/E-type funnelins, with Ki values in the nanomolar range (Table 1). Within this potency, the inhibitor showed a distinct specificity for digestive enzymes (CPA1, CPA2, and CPB1) and mast cell CPA3 over CPA4 and plasma CPB2 (also called TAFIa), which displayed significantly higher (10–20 times) Ki values.
Table 1.
Inhibition constants (Ki) of ACI against MCPs of the A/B and N/E type of funnelins
| Carboxypeptidase | Ki, nM |
|---|---|
| Bovine CPA1 | 2.4 ± 0.3 |
| Human CPA1 | 1.6 ± 0.2 |
| Human CPA2 | 2.5 ± 0.2 |
| Human CPA3 | 2.1 ± 0.3 |
| Human CPA4 | 23.9 ± 3.1 |
| Human CPB1 | 3.4 ± 0.3 |
| Human CPB2/TAFIa | 42.0 ± 1.7 |
| Human CPN | NI |
| Drosophila CPD-I | NI |
Data are shown as mean ± SD. NI, no inhibition at 100 μM inhibitor concentration.
Immunolocalization of ACI in Ascaris.
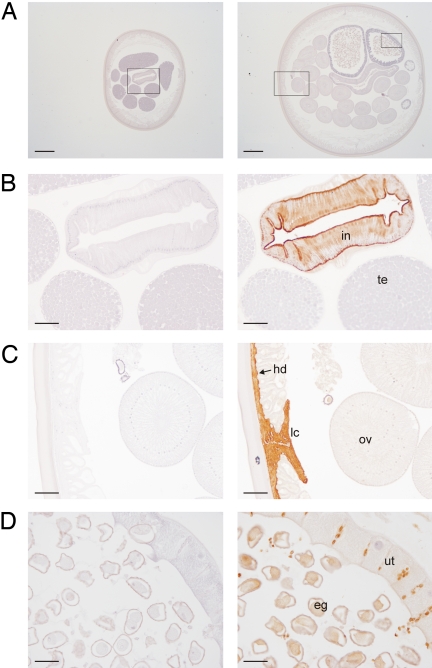
Specific antibodies raised against recombinant ACI were used to detect the protein in extracts from adult male and female Ascaris tissues by Western blot analysis. The inhibitor was found in the intestine and body wall of both male and female worms and in the ovary and uterus of female worms (Fig. S6). Immunohistochemistry assays confirmed these results. Antibodies strongly recognized the inhibitor in the intestine and hypodermis (body wall) of male and female worms (Fig. 2). In the body wall the signal was especially strong around the lateral chords, which contain the excretory canals. In females, the immunostaining was weaker in the uterus, ovary, and oviduct, whereas it was strong in the fertilized eggs. No labeling was detected in the male reproductive system (i.e., testis, vas deferens, and seminal vesicle).
Fig. 2.
Immunohistochemical localization of ACI in Ascaris. (A) Sections of adult male (Left) and female (Right) worms incubated with preimmune serum. Regions that are shown amplified in B, C, and D are inside squares. (Scale bars, 500 μm.) (B) Close-up view of a section of an adult male worm incubated with preimmune serum (Left) and anti-ACI antibody (Right). (Scale bars, 100 μm.) (C and D) Close-up views of a section of an adult female worm incubated with preimmune serum (Left) and anti-ACI antibody (Right). [Scale bars, 100 μm (C) and 50 μm (D).] Abbreviations: in, intestine; te, testis; lc, lateral chord; hd, hypodermis; ov, ovary; ut, uterus; and eg, eggs.
Structure of the ACI in Complex with Human CPA1.
The crystal structure analysis of human CPA1 (hCPA1) in its complex with recombinant ACI (Fig. 3A) reveals that the protease conforms to the classic α/β-hydrolase fold of A/B- and N/E-type zinc-dependent MCPs of the funnelin tribe of proteases (for a detailed description of the hCPA1 structure, see SI Materials and Methods) (17, 18, 32). ACI consists of structurally similar tandem modules, an N-terminal domain (NTD; Val-3I–Ser-26I; inhibitor residues carry an I suffix) and a C-terminal domain (CTD; Thr-40I–Leu-67I), which are linked by a connecting segment (CS; Thr-27I–Phe-39I) (Fig. 3 A and B). Each domain comprises a short N-terminal segment in extended conformation followed by a 90° turn that leads to a double, intertwined 1,4-turn. Thereafter, the chain folds back to enter a C-terminal β-ribbon (Fig. 3A and Fig. S7A). Intradomain stabilization is provided by two internal disulfide bonds per domain. The CS includes an α-helix (Ile-28I–His-38I) that is linked to the second β-ribbon strand of the CTD via a fifth disulfide bond (Cys-34I–Cys-62I). The two domains interact with each other through the N terminus and the tip plus the second strand of the β-ribbon of the NTD, the CS helix, and the N-terminal segment in extended conformation and the double 1,4-turn of the CTD (Fig. 3A and Fig. S7A). The domain architecture of ACI resembles that of latexin, in which two tandem cystatin-like modules are linked by a helical segment (33). However, there is no structural similarity with latexin beyond this coincidence in the modular arrangement, and no significant structural homology was found with any structure reported. We conclude that ACI conforms to a new fold and that this inhibitor may have arisen by gene duplication.
Structural Determinants of hCPA1 Inhibition Through ACI.
ACI inhibits hCPA1 by blocking access to the active-site cleft through an interface of 690 Å2 by forming 35 close contacts, including 12 hydrogen bonds and 6 hydrophobic interactions (see Table S1). Complex formation involves the C-terminal tail (Trp-64I–Leu-67I) and the tip (Asn-59I–Lys-60I) and the center (Ile-57I) of the N-terminal β-ribbon strand of the CTD and the end of the CS (Glu-37I–Phe-39I) of ACI, i.e., the NTD is not involved. Intervening segments of hCPA1 include the funnel-rim loop connecting strand β3 with helix α2, Lβ3α2, two regions of Lα4α5, as well as Lβ5β6, Lα7α8, and Lβ8α9. The main determinant of inhibition is the C-terminal stretch of ACI, which extends and penetrates the active-site cleft (Fig. 3A and Fig. S7B). This extended conformation is held together by the second disulfide bond of the CTD and a π-stacking interaction of Trp-64I with Asn-59I Nδ2 (Fig. S7B). ACI occupies the active-site cleft subsites S3, S2, and S1, and the C-terminal carboxylate oxygen atoms coordinate the catalytic zinc ion of the mature protease moiety in a slightly asymmetric bidentate manner (2.06 Å and 2.41 Å away, respectively). On the primed side of the cleft, an acetate ion lies next to the zinc, partially occupying the S1′ specificity pocket and mimicking the C terminus of a substrate. The acetate establishes a double salt bridge with Arg-145 Nη1 (2.77 Å) and Nη2 (2.94 Å) of the protease. One of its carboxylate oxygens binds Tyr-248 (2.62 Å) and the other hydrogen bonds Asn-144 Nδ2 (2.86 Å).
The way in which ACI inhibits hCPA1 resembles the action of PCI, LCI, and TCI but not latexin (33–37). Although the latter completely shields the active site of its target MCP in a nonspecific manner by contacting funnel rim elements, the other three inhibitors interact with these elements, but their C-terminal tails protrude into the active-site cleft in a substrate-like manner, as found for ACI (Fig. 3 C and D). Superposition of the C-terminal residues of the four inhibitors, which come from evolutionary distant species, reveals moderate sequence similarity. However, while the C-terminal residue is cleaved off in PCI, LCI, and TCI upon complex formation, it remains unaltered in ACI. This happens because the last residue of ACI matches the penultimate residues of PCI, LCI, and TCI (Fig. 3D).
Conclusions
Here, we have comprehensively analyzed the inhibitor ACI in the intestinal parasite Ascaris, the first to target metallopeptidases. Its preferential inhibition of MCPs found in the intestine and mast cells of the intestinal mucosa of the host would explain that Ascaris can survive in a hostile environment for many years by evading or dampening host responses that might otherwise kill it or trigger its expulsion. These data are consistent with the specific localization of ACI in the intestine and body wall of male and female Ascaris worms and in fertilized eggs. Such localization is compatible with the protection of adult worms and eggs in the host intestine and of the larvae during migration.
Structural analysis revealed that ACI consists of two linked homologous tandem domains. Inhibition of hCPA1 involves only the CTD and the preceding CS but not the NTD. This domain may target other digestive enzymes present in the host gut, as reported, for example, for the bifunctional α-amylase/trypsin inhibitor from the Indian finger millet Ragi (38).
Materials and Methods
A detailed description of procedures is provided in SI Materials and Methods and Table S2. Briefly, the ACI inhibitor was identified in an extract of Ascaris by intensity fading MALDI-TOF mass spectrometry as described in ref. 28. The inhibitor was subsequently isolated by CPA–Sepharose affinity chromatography and reversed-phase HPLC and sequenced by automated Edman degradation. The full-length ACI cDNA was cloned by RT-PCR and 5′/3′-RACE techniques as described in ref. 24. Recombinant ACI was overexpressed in E. coli Origami cells by using the pET-32a-derived vector, pET-32a-ACI, which contains a thioredoxin-hexahistidine fusion followed by a tobacco-etch virus (TEV) protease cleavage site. The fusion protein was purified by Ni2+ affinity chromatography and subsequently digested with TEV protease. The inhibitor was purified to homogeneity by RP-HPLC. Natural and recombinant ACI were analyzed by CD and NMR spectrometry under native and denaturing conditions. Their inhibitory activity was tested by spectophotometric assays with selected chromogenic substrates as described in ref. 24. Natural ACI was detected in adult male and female Ascaris dissected in different tissues or cut in slides by Western blotting or immunohistochemistry by using standard protocols. The ACI–hCPA1 complex was prepared by incubating equimolar quantities of enzyme and inhibitor. Crystals were obtained by using the sitting-drop vapor diffusion method. The structure was solved by Patterson search by using the coordinates of unbound hCPA1 as a searching model.
Supplementary Material
Acknowledgments.
We acknowledge Sílvia Bronsoms and Tibisay Guevara for excellent technical assistance, Joaquim Castellà for advice on the dissection of the Ascaris worms, and Robin Rycroft for helpful contributions to the manuscript. We acknowledge the help provided by European Molecular Biology Laboratory and European Synchrotron Radiation Facility synchrotron local contacts. This work was supported by the following grants: EU FP6 Strep Project LSHG-2006-018830 “CAMP”; EU FP7 Collaborative Project 223101 “AntiPathoGN”; BIO2007-68046, BIO2006-02668, and the CONSOLIDER-INGENIO 2010 Project “La Factoría de Cristalización” (CSD2006-00015) from Spanish public agencies. Funding was provided by European Synchrotron Radiation Facility for data collection. L.S. is a recipient of a predoctoral fellowship from the Ministry for Science and Innovation, Spain. J.L.A. is beneficiary of a “Juan de la Cierva” research contract awarded by the same ministry.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2FJU). The nucleotide sequence reported in this paper has been deposited in the GenBank database (accession number EU057973).
This article contains supporting information online at www.pnas.org/cgi/content/full/0812623106/DCSupplemental.
References
- 1.Bentwich Z, et al. Can eradication of helminthic infections change the face of AIDS and tuberculosis? Immunol Today. 1999;20:485–487. doi: 10.1016/s0167-5699(99)01499-1. [DOI] [PubMed] [Google Scholar]
- 2.Keiser J, Utzinger J. Efficacy of current drugs against soil-transmitted helminth infections: Systematic review and meta-analysis. J Am Med Assoc. 2008;299:1937–1948. doi: 10.1001/jama.299.16.1937. [DOI] [PubMed] [Google Scholar]
- 3.Crompton DW. Ascaris and ascariasis. Adv Parasitol. 2001;48:285–375. doi: 10.1016/s0065-308x(01)48008-0. [DOI] [PubMed] [Google Scholar]
- 4.Das CJ, Kumar J, Debnath J, Chaudhry A. Imaging of ascariasis. Australas Radiol. 2007;51:500–506. doi: 10.1111/j.1440-1673.2007.01887.x. [DOI] [PubMed] [Google Scholar]
- 5.Stephenson LS, Latham MC, Ottesen EA. Malnutrition and parasitic helminth infections. Parasitology. 2000;121:S23–S38. doi: 10.1017/s0031182000006491. [DOI] [PubMed] [Google Scholar]
- 6.Nadler SA, Hudspeth DS. Phylogeny of the Ascaridoidea (Nematoda: Ascaridida) based on three genes and morphology: Hypotheses of structural and sequence evolution. J Parasitol. 2000;86:380–393. doi: 10.1645/0022-3395(2000)086[0380:POTANA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 7.Phills JA, Harrold AJ, Whiteman GV, Perelmutter L. Pulmonary infiltrates, asthma and eosinophilia due to Ascaris suum infestation in man. N Engl J Med. 1972;286:965–970. doi: 10.1056/NEJM197205042861802. [DOI] [PubMed] [Google Scholar]
- 8.Pritchard DI, et al. Laboratory infection of primates with Ascaris suum to provide a model of allergic bronchoconstriction. Clin Exp Immunol. 1983;54:469–476. [PMC free article] [PubMed] [Google Scholar]
- 9.Babin DR, Peanasky RJ, Goos SM. The isoinhibitors of chymotrypsin/elastase from Ascaris lumbricoides: The primary structure. Arch Biochem Biophys. 1984;232:143–161. doi: 10.1016/0003-9861(84)90530-7. [DOI] [PubMed] [Google Scholar]
- 10.Grasberger BL, Clore GM, Gronenborn AM. High-resolution structure of Ascaris trypsin inhibitor in solution: Direct evidence for a pH-induced conformational transition in the reactive site. Structure. 1994;2:669–678. doi: 10.1016/s0969-2126(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 11.Gronenborn AM, Nilges M, Peanasky RJ, Clore GM. Sequential resonance assignment and secondary structure determination of the Ascaris trypsin inhibitor, a member of a novel class of proteinase inhibitors. Biochemistry. 1990;29:183–189. doi: 10.1021/bi00453a025. [DOI] [PubMed] [Google Scholar]
- 12.Homandberg GA, Litwiller RD, Peanasky RJ. Carboxypeptidase inhibitors from Ascaris suum: The primary structure. Arch Biochem Biophys. 1989;270:153–161. doi: 10.1016/0003-9861(89)90017-9. [DOI] [PubMed] [Google Scholar]
- 13.Homandberg GA, Peanasky RJ. Characterization of proteins from Ascaris lumbricoides which bind specifically to carboxypeptidase. J Biol Chem. 1976;251:2226–2233. [PubMed] [Google Scholar]
- 14.Huang K, Strynadka NC, Bernard VD, Peanasky RJ, James MN. The molecular structure of the complex of Ascaris chymotrypsin/elastase inhibitor with porcine elastase. Structure. 1994;2:679–689. doi: 10.1016/s0969-2126(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 15.Kageyama T. Molecular cloning, expression and characterization of an Ascaris inhibitor for pepsin and cathepsin E. Eur J Biochem. 1998;253:804–809. doi: 10.1046/j.1432-1327.1998.2530804.x. [DOI] [PubMed] [Google Scholar]
- 16.Ng KK, et al. Structural basis for the inhibition of porcine pepsin by Ascaris pepsin inhibitor-3. Nat Struct Biol. 2000;7:653–657. doi: 10.1038/77950. [DOI] [PubMed] [Google Scholar]
- 17.Arolas JL, Vendrell J, Aviles FX, Fricker LD. Metallocarboxypeptidases: Emerging drug targets in biomedicine. Curr Pharm Des. 2007;13:347–364. doi: 10.2174/138161207780162980. [DOI] [PubMed] [Google Scholar]
- 18.Gomis-Rüth FX. Structure and mechanism of metallocarboxypeptidases. Crit Rev Biochem Mol Biol. 2008;43:319–345. doi: 10.1080/10409230802376375. [DOI] [PubMed] [Google Scholar]
- 19.Lyons PJ, Callaway MB, Fricker LD. Characterization of carboxypeptidase A6, an extracellular matrix peptidase. J Biol Chem. 2008;283:7054–7063. doi: 10.1074/jbc.M707680200. [DOI] [PubMed] [Google Scholar]
- 20.Wei S, et al. Identification and characterization of three members of the human metallocarboxypeptidase gene family. J Biol Chem. 2002;277:14954–14964. doi: 10.1074/jbc.M112254200. [DOI] [PubMed] [Google Scholar]
- 21.Hass GM, Hermodson MA. Amino acid sequence of a carboxypeptidase inhibitor from tomato fruit. Biochemistry. 1981;20:2256–2260. doi: 10.1021/bi00511a029. [DOI] [PubMed] [Google Scholar]
- 22.Hass GM, et al. The amino acid sequence of a carboxypeptidase inhibitor from potatoes. Biochemistry. 1975;14:1334–1342. doi: 10.1021/bi00677a036. [DOI] [PubMed] [Google Scholar]
- 23.Reverter D, et al. A carboxypeptidase inhibitor from the medical leech Hirudo medicinalis: Isolation, sequence analysis, cDNA cloning, recombinant expression, and characterization. J Biol Chem. 1998;273:32927–32933. doi: 10.1074/jbc.273.49.32927. [DOI] [PubMed] [Google Scholar]
- 24.Arolas JL, et al. A carboxypeptidase inhibitor from the tick Rhipicephalus bursa: Isolation, cDNA cloning, recombinant expression, and characterization. J Biol Chem. 2005;280:3441–3448. doi: 10.1074/jbc.M411086200. [DOI] [PubMed] [Google Scholar]
- 25.Gong H, et al. Characterization of a carboxypeptidase inhibitor from the tick Haemaphysalis longicornis. J Insect Physiol. 2007;53:1079–1087. doi: 10.1016/j.jinsphys.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Liu Q, et al. Cloning, tissue expression pattern and genomic organization of latexin, a human homologue of rat carboxypeptidase A inhibitor. Mol Biol Rep. 2000;27:241–246. doi: 10.1023/a:1010971219806. [DOI] [PubMed] [Google Scholar]
- 27.Normant E, Martres MP, Schwartz JC, Gros C. Purification, cDNA cloning, functional expression, and characterization of a 26-kDa endogenous mammalian carboxypeptidase inhibitor. Proc Natl Acad Sci USA. 1995;92:12225–12229. doi: 10.1073/pnas.92.26.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yanes O, Villanueva J, Querol E, Aviles FX. Detection of noncovalent protein interactions by “intensity fading” MALDI-TOF mass spectrometry: Applications to proteases and protease inhibitors. Nat Protoc. 2007;2:119–130. doi: 10.1038/nprot.2006.487. [DOI] [PubMed] [Google Scholar]
- 29.Arolas JL, Bronsoms S, Ventura S, Aviles FX, Calvete JJ. Characterizing the tick carboxypeptidase inhibitor: Molecular basis for its two-domain nature. J Biol Chem. 2006;281:22906–22916. doi: 10.1074/jbc.M602301200. [DOI] [PubMed] [Google Scholar]
- 30.Chang JY, Li L, Canals F, Aviles FX. The unfolding pathway and conformational stability of potato carboxypeptidase inhibitor. J Biol Chem. 2000;275:14205–14211. doi: 10.1074/jbc.275.19.14205. [DOI] [PubMed] [Google Scholar]
- 31.Salamanca S, et al. The unfolding pathway of leech carboxypeptidase inhibitor. J Biol Chem. 2002;277:17538–17543. doi: 10.1074/jbc.M200040200. [DOI] [PubMed] [Google Scholar]
- 32.Vendrell J, Querol E, Aviles FX. Metallocarboxypeptidases and their protein inhibitors: Structure, function and biomedical properties. Biochim Biophys Acta. 2000;1477:284–298. doi: 10.1016/s0167-4838(99)00280-0. [DOI] [PubMed] [Google Scholar]
- 33.Pallares I, et al. Structure of human carboxypeptidase A4 with its endogenous protein inhibitor, latexin. Proc Natl Acad Sci USA. 2005;102:3978–3983. doi: 10.1073/pnas.0500678102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arolas JL, et al. The three-dimensional structures of tick carboxypeptidase inhibitor in complex with A/B carboxypeptidases reveal a novel double-headed binding mode. J Mol Biol. 2005;350:489–498. doi: 10.1016/j.jmb.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 35.Rees DC, Lipscomb WN. Refined crystal structure of the potato inhibitor complex of carboxypeptidase A at 2.5 Å resolution. J Mol Biol. 1982;160:475–498. doi: 10.1016/0022-2836(82)90309-6. [DOI] [PubMed] [Google Scholar]
- 36.Reverter D, et al. Structure of a novel leech carboxypeptidase inhibitor determined free in solution and in complex with human carboxypeptidase A2. Nat Struct Biol. 2000;7:322–328. doi: 10.1038/74092. [DOI] [PubMed] [Google Scholar]
- 37.Sanglas S, et al. Structure of activated thrombin-activatable fibrinolysis inhibitor, a molecular link between coagulation and fibrinolysis. Mol Cell. 2008;31:598–606. doi: 10.1016/j.molcel.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 38.Strobl S, et al. A novel strategy for inhibition of α-amylases: Yellow meal worm α-amylase in complex with the Ragi bifunctional inhibitor at 2.5-Å resolution. Structure. 1998;6:911–921. doi: 10.1016/s0969-2126(98)00092-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.