Abstract
Migration on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) that does not correlate with formula molecular weights, termed “gel shifting,” appears to be common for membrane proteins but has yet to be conclusively explained. In the present work, we investigate the anomalous gel mobility of helical membrane proteins using a library of wild-type and mutant helix-loop-helix (“hairpin”) sequences derived from transmembrane segments 3 and 4 of the human cystic fibrosis transmembrane conductance regulator (CFTR), including disease-phenotypic residue substitutions. We find that these hairpins migrate at rates of −10% to +30% vs. their actual formula weights on SDS-PAGE and load detergent at ratios ranging from 3.4–10 g SDS/g protein. We additionally demonstrate that mutant gel shifts strongly correlate with changes in hairpin SDS loading capacity (R2 = 0.8), and with hairpin helicity (R2 = 0.9), indicating that gel shift behavior originates in altered detergent binding. In some cases, this differential solvation by SDS may result from replacing protein-detergent contacts with protein-protein contacts, implying that detergent binding and folding are intimately linked. The CF-phenotypic V232D mutant included in our library may thus disrupt CFTR function via altered protein-lipid interactions. The observed interdependence between hairpin migration, SDS aggregation number, and conformation additionally suggests that detergent binding may provide a rapid and economical screen for identifying membrane proteins with robust tertiary and/or quaternary structures.
Keywords: detergent binding, gel shifting, membrane proteins, SDS-PAGE, sodium dodecyl sulfate
Determination of protein molecular weight (MW) via polyacrylamide gel electrophoresis (PAGE) in the presence of sodium dodecyl sulfate (SDS) is a universally used method in biomedical research. The technique requires that the proteins under investigation on the gel and the proteins used for MW calibration adopt equivalent shapes after SDS treatment. This condition is achieved not by the complete unfolding of the proteins but rather by the aggregation of SDS molecules at hydrophobic protein sites to induce “reconstructive denaturation,” where proteins adopt a conformational mixture of α-helix and random coil [reviewed in (1)]. The resultant protein-detergent complexes consist of helical SDS-coated polypeptide regions spatially separated by flexible and uncoated linkers, termed “necklace and bead” structures (2). Individual sizes of the micellar “beads” can vary and seem to be determined by amino acid sequence (3). A further proviso for MW estimation by SDS-PAGE is a consistent amount of detergent binding among proteins (4). Maximum SDS-binding levels are generally quoted at 1.4 g SDS/g protein, following the work of Reynolds and Tanford that identified this weight ratio as nearly invariant for a variety of globular and erythrocyte ghost proteins (4). Subsequent investigations of SDS loading have refined this saturation value for globular proteins to as much as 1.5–2 g detergent/g protein (5).
Peptide segments that contain an abundance of hydrophobic residues—such as the transmembrane (TM) sequences of membrane proteins—have helical regions expected to be embedded within the SDS micelle interior, while amphipathic segments common to globular polypeptides orient their hydrophobic faces toward the detergent tails and hydrophilic portions toward the micelle surface [reviewed in (1)]. Membrane proteins in some cases load 2-fold greater amounts of SDS than globular polypeptides; examples include the 3.4 g SDS/g stoichiometry of human erythrocyte membrane glycoprotein glycophorin (6) and 4.5 g SDS/g bound to CP-B2, a membrane protein from R. rubrum chromatophores (7). Large SDS/protein aggregate stoichiometry is nevertheless not universal among membrane-soluble polypeptides: intact cytochrome b5 binds 1.2 g SDS/g protein (8); the KcsA potassium channel tetramer 0.7 g SDS/g (9); and the human red cell glucose transporter Glut1 1.7 g SDS/g (10).
Protein tertiary structure may affect both detergent-loading levels and polypeptide-SDS-PAGE migration rates. Disulfide bonds, for example, reduce SDS binding to globular proteins by up to 2-fold (11), and have been linked to the anomalously fast migration of unreduced vs. reduced lysozyme, presumably because the intact disulfide bonds impose a more compact shape on the enzyme (12). Similarly, in a previous study with helix-loop-helix or “hairpin” model membrane proteins based on portions of the TM domain of the human cystic fibrosis transmembrane conductance regulator (CFTR), we noted that increased SDS-PAGE migration rates were traceable to a compact S-S bridged conformation (13). The E. coli β-barrel membrane protein OmpA also binds less SDS as a folded than fully denatured species (14, 15), and moves through gels according to structural compactness, with the folded form migrating faster than the completely denatured protein (16).
While these overall observations suggest that there may be a specific relationship between protein structure and SDS loading, a unifying source for atypical membrane protein SDS-PAGE migration has not emerged, and questions as to whether this detergent “denatures” membrane proteins and/or their oligomeric forms are often posed (17). In the present work, we establish a quantitative relationship between gel migration behavior and SDS aggregate stoichiometry using a library of helical hairpins derived from CFTR. We find that gel shifts strongly correlate (R2 = 0.8) with changes in the SDS-loading capacity of these miniature membrane proteins, indicating that altered detergent binding explains anomalous SDS-PAGE behavior. Our results reveal a distinction between two CF-phenotypic mutants studied: V232D binds significantly less SDS than the WT protein while P205S SDS binding is indistinguishable from WT, indicating that CFTR dysfunction may arise variously as a consequence of altered protein-lipid interactions or via altered intra-protein contacts.
Results
SDS-PAGE Mobility of Hairpins vs. Full-length Helical Membrane Proteins.
To gain an understanding of the commonality and magnitude of anomalous SDS-PAGE migration behavior observed in full-length polypeptides, we surveyed electrophoretic migration data obtained from the literature for a non-redundant dataset of helical membrane proteins. The apparent MWs among this group deviate widely from formula MW (Table 1) with gel shifts (i.e., migration on PAGE that does not correspond to formula MW) ranging from −46% (migration faster than formula MW) observed for the 11-mer of the F-type ATPase c subunit to +48% (slower than formula MW) displayed by the phospholamban monomer. In the present work, we undertook to determine whether such gel shifts could originate in changes in the stoichiometry of protein-detergent complexes. Hairpin sequences derived from TM segments 3 and 4 of CFTR were examined because this system offers several advantages: the helix-loop-helix construct represents the minimum model of membrane protein tertiary structure, and its modest size permits the detection of gel mobility differences that may be masked in larger membrane proteins. Further, we can routinely express and purify milligram quantities of WT and mutant hairpins (13, 18–20). For the present study, we examined WT and 11 hairpin mutants with replacements at various sites in the predicted structure of CFTR TM3/4 (Fig. 1A). This set of substitutions was designed to alter residue size, charge, and hydropathy, and care was taken not to bias the data set toward replacements of a specific type.
Table 1.
Gel shifts of various helical membrane proteins on SDS-PAGE
| Protein* | MW (kDa) |
Gel shift dMW, %† | Ref. | |
|---|---|---|---|---|
| App. | Formula | |||
| I. tartaricus F-type ATPase c subunit | 41 | |||
| Undecamer | 53 | 97 | −46 | |
| Monomer | 6.5 | 8.8 | −26 | |
| Archaeal ammonium transporter Amt-1 | 42 | |||
| Trimer | 90 | 134 | −33 | |
| Monomer | 33 | 45 | −27 | |
| E. coli lactose permease | 33 | 47 | −30 | 43 |
| Vitamin B12 transporter BtuC | 26 | 35 | −26 | 44 |
| H+/Cl− exchange transporter ClC | 38 | 51 | −25 | 45 |
| Potassium channel KcsA tetramer | 60 | 76 | −21 | 46 |
| Phospholamban | 47 | |||
| Pentamer | 29 | 30 | −17 | |
| Monomer | 9 | 6.1 | +48 | |
| Particulate methane monooxygenase | 48 | |||
| pmoA subunit | 24 | 28 | −14 | |
| pmoB subunit | 47 | 46 | 2 | |
| pmoC subunit | 22 | 30 | −27 | |
| Glycerol facilitator channel GlpF | 29 | 34 | −15 | 49 |
| E. coli MscS channel | 50 | |||
| Dimer | 52 | 62 | −16 | |
| Monomer | 27 | 31 | −12 | |
| Multidrug transporter AcrB | 100 | 115 | −13 | 51 |
| Glycerol-3-phosphate transporter GlpT | 45 | 50 | −10 | 52 |
| Spinach aquaporin | 31 | 32 | −3 | 53 |
| Rhomboid family protease GlpG | 31 | 31 | 0 | 54 |
| M. tuberculosis MscL channel monomer | 20 | 16 | +26 | 55 |
| β2-adrenergic receptor | 62 | 47 | +30 | 56 |
| M. thermoautotrophicum MthK tetramer | 200 | 149 | +34 | 57 |
*Proteins were selected from a list of non-redundant helical membrane proteins based on the availability of published SDS-PAGE data and are arranged in approximate order of faster-to-slower than expected migration rates. The helical nature of each protein has been confirmed in a high-resolution structure. Oligomeric data were obtained only from gels where extrinsic cross-linking agents were absent.
†The migration distances of proteins on SDS-PAGE gels reported in the papers examined, compared to standards on these gels. dMW = (apparent MW − formula MW)/formula MW × 100%. Negative dMWs indicate movement faster (at a lower apparent MW) than expected and vice-versa.
Fig. 1.
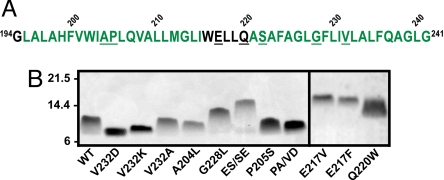
Hairpin sequences and SDS-PAGE analysis. (A) Amino acid sequence of the WT TM3/4 hairpin. Residues predicted to be in helical (green text) or loop (black text) regions of CFTR are shown (34). Residues substituted in this work are underlined. (B) Representative SDS-PAGE of helical hairpin mutants. Positions of MW standards (in kDa) are indicated. This panel is a composite of two gels, as indicated by the solid line between lanes. PA/VD and ES/SE denote the P205A/V232D and E217S/S222E hairpins, respectively.
Gel mobilities of WT and mutant TM3/4 hairpins were assessed in a commercially available neutral-pH SDS-PAGE system (Fig. 1B). We noted that certain hairpins ran as more diffuse bands than others (e.g., P205S and Q220W vs. V232K and E217V, see Fig. 1B). Spreading of bands on SDS-PAGE can reflect heterogeneity in protein species or in protein-detergent complexes; given that each sample loaded is fully purified, it is possible that the latter explanation may lead to the comet-like appearance of certain bands. Hairpin apparent MWs were determined with reference to globular protein standards and normalized by formula MWs to obtain gel shift values (dMWs, Table 2). Five mutants (A204L, P205A/V232D, V232A, P205S, and Q220W) migrated as WT within statistical significance; 2 were faster (V232D and V232K); and 4 were slower (G228L, E217V, E217F, and E217S/S222E). The hairpin gel shifts approximate those of several full-length membrane proteins (Table 1) but do not completely recapitulate the entire range of the intact polypeptides.
Table 2.
Gel shifts, SDS binding, helicity, and column MW of TM3/4 hairpins
| Hairpin* | Gel shift (dMW, %) | Bound SDS (g/g) | Helicity (MRE X 103)† | Column MW (mut-wt, %)‡ |
|---|---|---|---|---|
| V232Dcf | -11 ± 2.6 | 3.4 ± 0.9 | −17 ± 1.2 | +19 ± 1.5 |
| V232K | -10 ± 3.0 | 3.8 ± 0.6 | −16 ± 1.1 | +5.6 ± 1.6 |
| A204L | −2.2 ± 2.3 | 6.0 ± 0.7 | −18 ± 1.3 | +3.4 ± 1.6 |
| P205A/V232Dcf | +0.12 ± 5.2 | 4.7 ± 0.4 | −19 ± 2.6 | +21 ± 0.6 |
| WT | +0.42 ± 4.5 | 5.4 ± 1.4 | −18 ± 2.2 | 0.0 ± 0.79 |
| V232A | +3.6 ± 3.7 | 5.2 ± 0.4 | −18 ± 0.9 | +6.1 ± 1.9 |
| P205Scf | +4.7 ± 6.0 | 4.7 ± 1.0 | −18 ± 0.7 | +4.4 ± 1.6 |
| Q220W | +6.3 ± 2.4 | 5.0 ± 0.7 | -21 ± 2.7 | -4.9 ± 1.3 |
| G228L | +14 ± 5.1 | 6.9 ± 1.4 | -23 ± 1.7 | +14 ± 2.6 |
| E217V | +28 ± 1.3 | 6.7 ± 1.0 | -25 ± 1.7 | +32 ± 4.0 |
| E217F | +29 ± 2.8 | 9.4 ± 1.9 | -28 ± 2.6 | +17 ± 1.1 |
| E217S/S222E | +29 ± 7.6 | 10 ± 2.3 | -25 ± 2.8 | +35 ± 0.9 |
| Glycophorin§ | - | 3.4 ± 0.6 | −9.5 ± 1.2 | +83 ± 5.1 |
*Mutant hairpins are listed in order of increasing gel shift. The mean ± standard deviation of 3–6 independent experiments is shown. Values in each column that significantly differ from WT (P ≤ 0.05 in unpaired 2-tailed t tests) are underlined. The cf superscript denotes substitutions linked to cystic fibrosis in the CF mutation database (http://www.genet.sickkids.on.ca/cftr).
†Helicity approximated as mean residue ellipticity (MRE) at 222 nm, in units of deg cm2 dmol−1.
‡Change in apparent vs. formula MW estimated from SEC-HPLC retention times (column dMW), normalized to the WT value. Positive values indicate increased hydrodynamic radii vs. WT and vice-versa. The column dMW of the WT TM3/4 hairpin was measured as −29 ± 0.56 %.
§Detergent loading, helicity, and column dMW of human glycophorin.
TM3/4 Hairpins Load Varying Amounts of SDS.
Free and bound detergent was separated using size exclusion-chromatography (SEC) on silica gel columns (21, 22). This technique provides a means to separate free and bound SDS, and also estimates hairpin hydrodynamic volume (vide infra). Hairpins eluted from 8.93 ± 0.01 mL to 9.45 ± 0.02 mL (each within the 9.00–9.75-ml fraction, termed the peak fraction) as determined by absorbance at 280 nm (Fig. S1), and protein determination indicated that the majority of protein eluted in the peak fraction. We found that WT TM3/4 bound 5.4 ± 1.4 g SDS/g protein (Table 2), comparable to the 4.5 g/g loading value reported for the unglycosylated membrane protein CP-B2 under similar buffer conditions (7). This level of detergent binding exceeds by approximately 2-fold the 1.5–2 g SDS/g range expected of the globular proteins used for gel and column MW calibration. The mutant hairpins ranged in loading levels from 3.4–10 g SDS/g, with V232D binding significantly fewer SDS molecules than WT, and the E217F and E217S/S222E mutants binding significantly more. All hairpins load more detergent than do intact cytochrome b5, KcsA, and Glut1 [range 0.7–1.7 g SDS/g protein, (8–10)], and the E217F and E217S/S222E mutants are apparently the highest reported binders among the admittedly limited numbers of membrane proteins evaluated to date. This difference does not appear to arise from the assay methodology used here, as the SDS loading of our glycophorin control, 3.4 g ± 0.6 g SDS/g is essentially identical to the previously reported value (6).
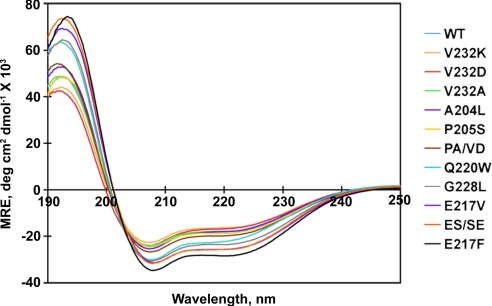
The Helical Content of TM3/4 Hairpins Is Increased in Slow-Migrating Mutants.
The circular dichroism (CD) spectrum of WT and each mutant TM3/4 hairpin in mobile phase buffer was obtained to ensure that hairpin secondary structure was consistent with “reconstructive denaturation” by SDS to a helix-coil mixture. All hairpin spectra were characteristic of α-helix, with dual minima at 208 nm and 222 nm (Fig. 2). However, the magnitudes of these minima, approximating the percentage of helical structure in the hairpin, varied in a mutant-dependent manner (Table 2). Comparison with gel shift data indicates that mutants migrating slower than WT on SDS-PAGE have increased helicity while those moving faster than WT on gels appear slightly—but not significantly—reduced; comparable trends have been noted previously in the TM3/4 hairpin system (13, 18, 20). CD spectra determined for the glycophorin control were found to closely approximate the spectrum previously reported for the pure protein in SDS (23).
Fig. 2.
CD profiles for all hairpins in 0.3% SDS and 50 mM sodium phosphate, pH 7. Mutants are arranged (right of panel) in order of increasing helicity at 222 nm. PA/VD and ES/SE denote the P205A/V232D and E217S/S222E hairpins, respectively. Spectra shown are the average of 3–6 independent experiments. See Table 2 for a list of helicity values.
Interrelationship of Detergent Binding, PAGE Migration, and Helicity in TM3/4 Hairpins.
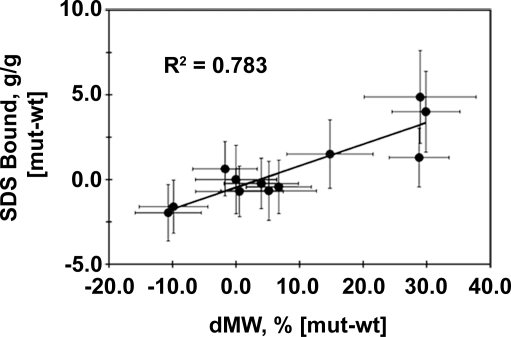
Regression analysis was used to evaluate the ability of changes in SDS-binding levels to explain hairpin gel shift behavior. Comparisons were carried out using detergent loading (in g SDS/g) and gel shift (in dMW, %) values normalized to WT to eliminate reference to the globular standards used for MW estimation. Linear fitting of SDS loading vs. gel shift returned a correlation coefficient (R2) of 0.783 (Fig. 3), indicating that approximately 80% of hairpin SDS-PAGE behavior is attributable to altered SDS/protein aggregate stoichiometry. This result demonstrates that anomalous SDS-PAGE behavior in membrane proteins can arise from altered detergent binding.
Fig. 3.
Correlation of SDS binding and PAGE mobility. The relationship between bound SDS and gel shift is shown with the correlation coefficient (R2) of the best fit line. Trendline P value is 0.001. The variations in mutant-WT values were propagated from the standard deviations of each mean using standard formulae.
SDS loading also directly correlates at a level of approximately 75% with hairpin helicity, and helicity directly correlates with gel shift at a level of approximately 90% (Fig. S2). The former result is consistent with the typical view of SDS binding to polypeptide segments as associated with induction of helical conformation [reviewed in (1)]. As well, adoption of helical structure concomitant with loading of SDS onto previously uncoated polypeptide regions would be expected to render the hairpin more rod-like and reduce gel mobility (20).
Role of Hairpin Hydropathy in SDS Binding.
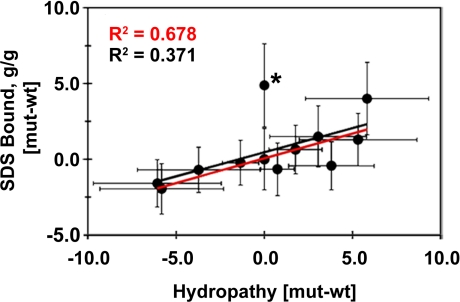
Since SDS generally loads at hydrophobic sites (1), and because changes in residue hydropathy were suggested to influence SDS binding in the globular protein αΑ-crystallin (24), we examined the relationship of hairpin hydropathy and detergent loading, helicity, and gel migration (Fig. 4). More hydrophobic mutants generally do bind more SDS in the TM3/4 system, and thereby exhibit reduced gel mobility and increased helicity. In the presence of the iso-hydropathic mutant E217S/S222E, correlations between each parameter and hydropathy range from R2 values of 0.371 (for bound SDS vs. hydropathy, Fig. 4) to 0.582 (for gel shift vs. hydropathy, Fig. S3). This mutant, however, is an outlier in all cases, and correlation coefficients rise to values of 0.678 to 0.755 when it is eliminated from curve fitting. Hydropathy is therefore a strongly contributing factor—but not always the only descriptor—of anomalous detergent loading by TM3/4 hairpins.
Fig. 4.
Correlation of SDS binding and hydropathy. Mutation-dependent changes in bound SDS are plotted as a function of hydropathy changes. Mutant-WT hydropathy values were calculated individually with 6 common scales (35–40) and the results averaged, such that positive hydropathy indicates increased apolarity and negative hydropathy increased hydrophilicity. The E217S/S222E mutant is marked with an asterisk. Correlation coefficients (R2) of the best fit line in the absence of E217S/S222E (red) or with all mutants (black) are shown. Trendline P value in the absence of E217S/S222E is 0.002.
Hydrodynamic Volume of Hairpin-SDS Aggregates.
Elution on SEC-HPLC also allowed the relationship between hairpin compactness and SDS binding to be assessed independent of the mass/charge parameter exploited in SDS-PAGE sieving. On SEC, elution position depends on hydrodynamic volume (a combination of size and shape); less compact species are retained for shorter times than more compact species due to their exclusion from pores in the column matrix. Although it binds approximately 2× more detergent than expected of the globular MW standards, the TM3/4 WT hairpin exhibits no gel shift under the SDS-PAGE conditions used, and correspondingly displays a compact size on SEC [≈30% smaller than its formula MW (Table 2)]. Conversely, even though it migrates faster than WT on SDS-PAGE, the V232D hairpin reports a radius approximately 20% larger than WT (Table 2). Of the remaining hairpins, nine decreased in apparent compactness vs. WT (range from +3% to +35%, see Table 2), while Q220W appeared somewhat more compact than WT. The glycophorin control eluted with a column dMW of +83%, consistent with dimerization in SDS (25).
Discussion
SDS Binding and Hairpin Conformation.
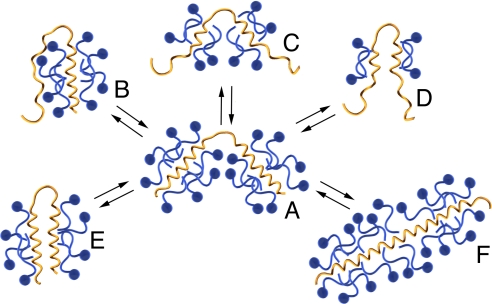
The nature of membrane protein gel shifts on SDS-PAGE is of interest both because of the universality of the technique and because these shifts appear to be a relatively common occurrence. What has remained mysterious is the underlying cause of these differences in migration. The work described here clearly establishes that altered SDS loading is the major determinant of anomalous membrane protein PAGE migration behavior. Our results further suggest an intimate link between altered SDS binding and hairpin conformation that has two interrelated components, analogous to the two stages of membrane protein folding (26): (i) coating of TM regions by detergent acyl chains and adoption of helical conformation; and (ii) competition between protein-protein and protein-detergent contacts (27). Operation of these two effects in a concerted manner with SDS loading allows us to rationalize the trends in our hairpin data, as illustrated in Fig. 5.
Fig. 5.
Interrelationship between hairpin conformation and detergent binding. Hairpins (yellow) loaded with SDS molecules (blue) are represented. The number of SDS molecules on each hairpin, and the number of turns of helical structure, are intended to illustrate relationships between relative levels of detergent binding and/or helicity in a quantitative manner. The necklace and bead structure typical of an unfolded membrane protein with its TM segments fully coated with detergent acyl chains (A) is shown at the center. Potential alterations in SDS loading accompanying hydropathy reductions (B–D) are shown at the top; in some instances, regions with reduced hydropathy may no longer intercalate with lipid acyl chains but instead may partition closer to the micelle surface (B) or remain uncoated (C and D). In cases where detergent-TM domain interactions remain constant, conformational changes may also alter SDS loading stoichiometry (E and F). Interconversions among all types of hairpin-detergent complexes are possible. See text for further discussion.
SDS loading onto the TM regions of a hairpin unfolded to “necklace and bead” structure (Fig. 5A) may be altered by hydropathy reductions, resulting in TM regions partitioned toward the micelle surface, and/or in incompletely formed or frayed helices (Fig. 5 B–D). If an initially compact hairpin (Fig. 5E) undergoes a change in partitioning concomitant with loss of detergent aggregation (Fig. 5D), faster than WT gel shifts would be expected even though hydrodynamic radius may increase. However, even if SDS/protein stoichiometry (and by extension, gel shift) remains unchanged, increases in the conformational flexibility of non-coated regions may alter the hairpin's hydrodynamic radius (compare Fig. 5 B–E)–a potential explanation for the as-WT gel shift but increased hydrodynamic radius relative to WT of the P205A/V232D mutant. In other instances, decreased helicity compared to the necklace and bead situation (Fig. 5, compare A to B–D) may be diagnostic of incomplete TM segment solvation by SDS acyl chains.
Conversely, when hydropathy is unchanged, and/or replacements do not affect TM helix burial in the detergent micelle, protein-detergent complex stoichiometry may be dependent on tertiary contacts (Fig. 5 compare A to E); here, altered SDS binding may arise by replacing protein-protein contacts with protein-detergent contacts, where an alteration in hairpin sequence facilitates permeation of the detergent between them. Induction of additional helical structure in hairpin regions previously uncoated by the detergent, such as binding in loop regions, may also accompany tertiary contact changes in some instances (Fig. 5F). It is possible that the 9–10 g SDS/g stoichiometry of E217F and the iso-hydropathic E217S/S222E mutant are representative of detergent loading by a fully “denatured” helical membrane protein (such as the “caterpillar” structure in Fig. 5F).
The model in Fig. 5 A–F is broadly consistent with our SEC-HPLC studies on hairpin hydrodynamic radii, which imply that gel shift values are not always correlated with hairpin compactness (Table 2). For example, V232D and P205A/V232D display larger than WT hydrodynamic radii on SEC-HPLC (+19% and +21%, respectively)—even though each Asp-containing mutant migrates faster or as-WT on PAGE.
Oligomeric States of Membrane Proteins on SDS-PAGE.
Competition between protein-protein and protein-detergent interactions in SDS does provide an explanation as to why none of the hairpins tested migrate as fast through PAGE as do large multimeric complexes of full-length membrane proteins. Because of hairpin size (two TM segments) and lack of oligomer formation in SDS, each is limited in the number of protein-protein contacts that may preclude SDS binding. Larger apparent MW decreases may therefore require SDS-persistent contacts between more than 2 TM helices. Support for this contention can be seen in the gel migration patterns of phospholamban (Table 1). As the oligomeric state of phospholamban increases, its gel shift changes from positive to negative (i.e., its apparent MW becomes smaller compared with its formula MW), implying that fewer detergent molecules are bound per monomer as the complex assembles. Whether protein-protein interactions indeed displace protein-detergent interactions during oligomerization is an intriguing question that merits further investigation.
The observed dependences of hairpin gel shift on hydropathy and helicity additionally caution that gel shifting should not be considered as an absolute indicator of oligomeric state, compactness, or degree of foldedness independent of additional experiments. For example, on SDS-PAGE calibrated with globular protein standards, peptides and/or full-length proteins with large SDS aggregate stoichiometry may appear to run as dimers when they are in reality monomeric; migration of the phospholamban monomer at approximately 1.5× its formula MW (dMW of +48%, Table 1) illustrates this possibility.
SDS Aggregation as a Mimic of Protein-Lipid Interactions in CFTR.
The utility of detergents in characterizing membrane proteins rests on their ability to act as a reasonable stand-in for the lipid bilayer, with protein-detergent contacts mimicking protein-lipid interactions. Our observation that the V232D mutant binds less SDS than WT TM3/4 may therefore indicate that the dysfunction in the full-length CFTR molecule caused by this mutation arises from altered protein-lipid associations. The increase in polarity of the Val-to-Asp substitution may necessitate sequestration of the side chain from lipid, potentially followed by changes in helix-helix contacts that accommodate the polar residue (13, 28). On the other hand, the as-WT stoichiometry of the P205S protein-SDS complex suggests that this lesion could disrupt CFTR folding and/or dynamics by directly altering protein-protein interactions while maintaining native protein-lipid contacts. In stating these potential consequences of disease mutations in CFTR, however, we note that these possibilities are based on our experiments in SDS; this detergent does not necessarily recapitulate bilayer conditions (29–31).
Conclusion
Since the SDS weight ratios bound by hairpins exceed by up to 10-fold those reported for other membrane proteins, our results imply that incomplete denaturation by SDS may be widespread; preliminary work in our laboratory suggests that the trends described herein hold true for WT and mutant analogues of other helical hairpins (TM1/2 and TM9/10 of CFTR). These results further suggest that MW determination for membrane proteins on SDS-PAGE is ultimately likely to require calibration with membrane-soluble standards; such a set has been used to estimate MWs for TM-mimic peptides (32, 33). However, an important caveat for such standards (and the proteins compared with them) is that each must represent a fully denatured conformation that binds SDS at a consistent level. Nevertheless, the links established here among hairpin PAGE migration, SDS aggregation number, hydrodynamic radius, and conformation offer a practical advantage, that is, SDS-PAGE analysis can provide a rapid and economical initial screen to identify membrane proteins with SDS-resistant tertiary and/or quaternary structures.
Materials and Methods
Expression and Purification of CFTR Helical Hairpins.
TM3/4 hairpins were expressed and purified as previously described (13). Mutations were introduced using the QuikChange site-directed mutagenesis kit (Stratagene). The TM3/4 hairpins produced have a Cys-to-Ala replacement at residue 225 and each carries an identical N-terminal and C-terminal extension (GSGMKETAAAKFERQHMDSPDLGTDDDDKAM and LEHHHHHH) containing the S-tag epitope used for Western blot detection and the His6 tag used for purification.
PAGE Analysis.
Gel boxes and 12% NuPAGE Bis-Tris gels were purchased from Invitrogen. Gels were run at 140 V for 100 min under reducing conditions in LDS-loading buffer and MES running buffer, pH 7.3, following the manufacturer's protocols, but samples were not boiled before gel loading. Proteins were visualized with Coomassie Blue staining. Hairpin apparent MW values were estimated from the migration rates of Mark-12 markers (Invitrogen), and gel shift values (dMW) were calculated by normalizing apparent MW by theoretical MW (see SI Materials and Methods for details). Mutants with negative values of dMW have faster than expected electrophoretic mobility and vice versa.
Size-Exclusion Chromatography (SEC).
SEC-HPLC was performed on a 7.8 mm × 300 mm BioSep SEC-S2000 column (Phenomenex) equilibrated and run at a rate of 0.5 mL/min with 0.3% high-purity SDS (BioUltra, Fluka), 50 mM Na phosphate, pH 7 (mobile-phase buffer), using a Waters 600 pump. The column was pre-equilibrated with at least 10 volumes of mobile phase buffer at before sample injection. Purified hairpins (0.25–0.4 mg) were dissolved in 300 μL mobile-phase buffer and allowed to equilibrate at room temperature overnight before injection. Human glycophorin (predominantly glycophorin A, Sigma), was dissolved in mobile-phase buffer to a concentration of 0.8 mg/mL and equilibrated at room temperature overnight before injection of 0.24 mg aliquots. Each hairpin or glycophorin sample was microcentrifuged at maximum speed for 15 min at room temperature before injection. Fractions were collected for all proteins for 45 min at 1.5 min/0.75 mL intervals and elution detected by absorbance at 280 nm.
Hairpin apparent MW values were estimated from SEC-HPLC elution volumes by comparison to the elution volumes of globular protein standards (22). Protein standards from the LMW Gel Filtration Calibration Kit (GE Healthcare) and separately purchased insulin B chain (Sigma) were used for calibration (see SI Materials and Methods for details). Each hairpin eluted at a significantly different volume than WT (P ≤ 0.01). Normalization of column apparent MW to formula MW was performed as above for gel shifts. The formula MW of glycophorin was estimated at 31 kDa (25).
SDS and Protein Determination.
Fractions collected before, during, and after protein elution were diluted 1/5 with ultrapure water before SDS and protein determination. Determination of SDS was performed using quantitative methylene blue extraction essentially as described [(4), details in SI Materials and Methods]. The Micro BCA assay (Pierce) was used for protein determination according to the manufacturer's directions. Standard curves were prepared using manufacturer-supplied BSA in mobile-phase buffer diluted 1/5 with ultrapure water. The SDS concentrations of each protein-free fraction collected during an individual HPLC run were averaged to provide the free SDS concentration, and bound SDS determined for the peak fraction: SDSbound = SDStotal, peak fraction – SDSfree. Peak fraction bound SDS values were divided by total protein to obtain the weight ratio of hairpin detergent loading.
Circular Dichroism (CD) Spectroscopy.
Far-UV CD measurements were performed on undiluted peak fractions (typical protein concentrations 8–10 μM) at room temperature on a Jasco 720 CD spectrometer using a 0.1-mm cuvette. Three scans were accumulated for each spectrum with a response time of 4 s, a bandwidth of 2 nm, and a scan speed of 50 nm/min from 250 to 190 nm. Background spectra without protein were subtracted.
Supplementary Material
Acknowledgments.
The authors wish to thank Fiona Cunningham for assistance in preparation of Table 1 and David Tulumello for helpful discussions. This work was supported, in part, by grants to C.M.D. from the Canadian Institutes of Health Research (CIHR FRN5810), and the Canadian Cystic Fibrosis Foundation. A.R. is the recipient of a Research Training Center (RESTRACOMP) award from the Hospital for Sick Children, and held a postdoctoral award from the CIHR Strategic Training Program in Protein Folding: Principles and Diseases. V.G.N. holds a Masters' Training Award from the Fonds de la Recherche en Santé du Quebec (FRSQ). G.C. was the recipient of a University of Toronto Scholarship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813167106/DCSupplemental.
References
- 1.Imamura T. In: Encyclopedia of Surface and Colloid Science. Somasundaran P, editor. New York: Taylor & Francis; 2006. pp. 5251–5263. [Google Scholar]
- 2.Shirahama K, Tsujii K, Takagi T. Free-boundary electrophoresis of sodium dodecyl sulfate-protein polypeptide complexes with special reference to SDS-polyacrylamide gel electrophoresis. J Biochem (Tokyo) 1974;75:309–319. doi: 10.1093/oxfordjournals.jbchem.a130398. [DOI] [PubMed] [Google Scholar]
- 3.Ibel K, et al. Protein-decorated micelle structure of sodium-dodecyl-sulfate–protein complexes as determined by neutron scattering. Eur J Biochem. 1990;190:311–318. doi: 10.1111/j.1432-1033.1990.tb15578.x. [DOI] [PubMed] [Google Scholar]
- 4.Reynolds JA, Tanford C. Binding of dodecyl sulfate to proteins at high binding ratios: Possible implications for the state of proteins in biological membranes. Proc Natl Acad Sci USA. 1970;66:1002–1007. doi: 10.1073/pnas.66.3.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanford C. The hydrophobic effect: Formation of micelle and biological membranes. 2nd Ed. New York: Wiley; 1980. [Google Scholar]
- 6.Grefrath SP, Reynolds JA. The molecular weight of the major glycoprotein from the human erythrocyte membrane. Proc Natl Acad Sci USA. 1974;71:3913–3916. doi: 10.1073/pnas.71.10.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyake J, Ochiai-Yanagi S, Kasumi T, Takagi T. Isolation of a membrane protein from R rubrum chromatophores and its abnormal behavior in SDS-polyacrylamide gel electrophoresis due to a high binding capacity for SDS. J Biochem. 1978;83:1679–1686. doi: 10.1093/oxfordjournals.jbchem.a132080. [DOI] [PubMed] [Google Scholar]
- 8.Robinson NC, Tanford C. The binding of deoxycholate, Triton X-100, sodium dodecyl sulfate, and phosphatidylcholine vesicles to cytochrome b5. Biochemistry. 1975;14:369–378. doi: 10.1021/bi00673a025. [DOI] [PubMed] [Google Scholar]
- 9.Chill JH, Louis JM, Miller C, Bax A. NMR study of the tetrameric KcsA potassium channel in detergent micelles. Protein Sci. 2006;15:684–698. doi: 10.1110/ps.051954706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallsten M, Lundahl P. Binding of sodium dodecyl sulphate to an integral membrane protein and to a water-soluble enzyme: Determination by molecular-sieve chromatography with flow scintillation detection. J Chrom. 1990;512:3–12. doi: 10.1016/s0021-9673(01)89469-3. [DOI] [PubMed] [Google Scholar]
- 11.Pitt-Rivers R, Impiombato FS. The binding of sodium dodecyl sulphate to various proteins. Biochem J. 1968;109:825–830. doi: 10.1042/bj1090825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunker AK, Kenyon AJ. Mobility of sodium dodecyl sulphate - protein complexes. Biochem J. 1976;153:191–197. doi: 10.1042/bj1530191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Therien AG, Grant FE, Deber CM. Interhelical hydrogen bonds in the CFTR membrane domain. Nat Struct Biol. 2001;8:597–601. doi: 10.1038/89631. [DOI] [PubMed] [Google Scholar]
- 14.Dornmair K, Kiefer H, Jahnig F. Refolding of an integral membrane protein. OmpA of Escherichia coli. J Biol Chem. 1990;265:18907–18911. [PubMed] [Google Scholar]
- 15.Ohnishi S, Kameyama K, Takagi T. Characterization of a heat modifiable protein, Escherichia coli outer membrane protein OmpA in binary surfactant system of sodium dodecyl sulfate and octylglucoside. Biochim Biophys Acta. 1998;1375:101–109. doi: 10.1016/s0005-2736(98)00145-x. [DOI] [PubMed] [Google Scholar]
- 16.Kleinschmidt JH, Wiener MC, Tamm LK. Outer membrane protein A of E. coli folds into detergent micelles, but not in the presence of monomeric detergent. Protein Sci. 1999;8:2065–2071. doi: 10.1110/ps.8.10.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renthal R. An unfolding story of helical transmembrane proteins. Biochemistry. 2006;45:14559–14566. doi: 10.1021/bi0620454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi MY, Cardarelli L, Therien AG, Deber CM. Non-native interhelical hydrogen bonds in the cystic fibrosis transmembrane conductance regulator domain modulated by polar mutations. Biochemistry. 2004;43:8077–8083. doi: 10.1021/bi0494525. [DOI] [PubMed] [Google Scholar]
- 19.Therien AG, Glibowicka M, Deber CM. Expression and purification of two hydrophobic double-spanning membrane proteins derived from the cystic fibrosis transmembrane conductance regulator. Protein Expr Purif. 2002;25:81–86. doi: 10.1006/prep.2001.1612. [DOI] [PubMed] [Google Scholar]
- 20.Wehbi H, Rath A, Glibowicka M, Deber CM. Role of the extracellular loop in the folding of a CFTR transmembrane helical hairpin. Biochemistry. 2007;46:7099–7106. doi: 10.1021/bi602570u. [DOI] [PubMed] [Google Scholar]
- 21.Moller JV, le Maire M. Detergent binding as a measure of hydrophobic surface area of integral membrane proteins. J Biol Chem. 1993;268:18659–18672. [PubMed] [Google Scholar]
- 22.le Maire M, et al. Gel chromatography and analytical ultracentrifugation to determine the extent of detergent binding and aggregation, and Stokes radius of membrane proteins using sarcoplasmic reticulum Ca2+-ATPase as an example. Nat Protoc. 2008;3:1782–1795. doi: 10.1038/nprot.2008.177. [DOI] [PubMed] [Google Scholar]
- 23.Schulte TH, Marchesi VT. Conformation of human erythrocyte glycophorin A and its constituent peptides. Biochemistry. 1979;18:275–280. doi: 10.1021/bi00569a006. [DOI] [PubMed] [Google Scholar]
- 24.de Jong WW, Zweers A, Cohen LH. Influence of single amino acid substitutions on electrophoretic mobility of sodium dodecyl sulfate-protein complexes. Biochem Biophys Res Commun. 1978;82:532–539. doi: 10.1016/0006-291x(78)90907-5. [DOI] [PubMed] [Google Scholar]
- 25.Furthmayr H, Marchesi VT. Subunit structure of human erythrocyte glycophorin A. Biochemistry. 1976;15:1137–1144. doi: 10.1021/bi00650a028. [DOI] [PubMed] [Google Scholar]
- 26.Engelman DM, et al. Membrane protein folding: Beyond the two stage model. FEBS Lett. 2003;555:122–125. doi: 10.1016/s0014-5793(03)01106-2. [DOI] [PubMed] [Google Scholar]
- 27.Joh NH, et al. Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature. 2008;453:1266–1270. doi: 10.1038/nature06977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bond PJ, Holyoake J, Ivetac A, Khalid S, Sansom MS. Coarse-grained molecular dynamics simulations of membrane proteins and peptides. J Struct Biol. 2007;157:593–605. doi: 10.1016/j.jsb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Russ WP, Engelman DM. TOXCAT: A measure of transmembrane helix association in a biological membrane. Proc Natl Acad Sci USA. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duong MT, Jaszewski TM, Fleming KG, MacKenzie KR. Changes in apparent free energy of helix-helix dimerization in a biological membrane due to point mutations. J Mol Biol. 2007;371:422–434. doi: 10.1016/j.jmb.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sulistijo ES, MacKenzie KR. Sequence dependence of BNIP3 transmembrane domain dimerization implicates side-chain hydrogen bonding and a tandem GxxxG motif in specific helix-helix interactions. J Mol Biol. 2006;364:974–990. doi: 10.1016/j.jmb.2006.09.065. [DOI] [PubMed] [Google Scholar]
- 32.Gratkowski H, Lear JD, DeGrado WF. Polar side chains drive the association of model transmembrane peptides. Proc Natl Acad Sci USA. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choma C, Gratkowski H, Lear JD, DeGrado WF. Asparagine-mediated self-association of a model transmembrane helix. Nat Struct Biol. 2000;7:161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- 34.Riordan JR, et al. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 35.Liu LP, Deber CM. Guidelines for membrane protein engineering derived from de novo designed model peptides. Biopolymers. 1998;47:41–62. doi: 10.1002/(SICI)1097-0282(1998)47:1<41::AID-BIP6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 36.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 37.Eisenberg D, Weiss RM, Terwilliger TC, Wilcox W. Hydrophobic moments and protein structure. Faraday Discuss Chem Soc. 1982;17:109–120. [Google Scholar]
- 38.Engelman DM, Steitz TA, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 39.Hessa T, et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 40.White SH, Wimley WC. Membrane protein folding and stability: Physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 41.Neumann S, Matthey U, Kaim G, Dimroth P. Purification and properties of the F1F0 ATPase of Ilyobacter tartaricus, a sodium ion pump. J Bacteriol. 1998;180:3312–3316. doi: 10.1128/jb.180.13.3312-3316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blakey D, et al. Purification of the Escherichia coli ammonium transporter AmtB reveals a trimeric stoichiometry. Biochem J. 2002;364:527–535. doi: 10.1042/BJ20011761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newman MJ, Foster DL, Wilson TH, Kaback HR. Purification and reconstitution of functional lactose carrier from Escherichia coli. J Biol Chem. 1981;256:11804–11808. [PubMed] [Google Scholar]
- 44.de Veaux LC, Clevenson DS, Bradbeer C, Kadner RJ. Identification of the btuCED polypeptides and evidence for their role in vitamin B12 transport in Escherichia coli. J Bacteriol. 1986;167:920–927. doi: 10.1128/jb.167.3.920-927.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maduke M, Pheasant DJ, Miller C. High-level expression, functional reconstitution, and quaternary structure of a prokaryotic ClC-type chloride channel. J Gen Physiol. 1999;114:713–722. doi: 10.1085/jgp.114.5.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heginbotham L, Odessey E, Miller C. Tetrameric stoichiometry of a prokaryotic K+ channel. Biochemistry. 1997;36:10335–10342. doi: 10.1021/bi970988i. [DOI] [PubMed] [Google Scholar]
- 47.Wegener AD, Jones LR. Phosphorylation-induced mobility shift in phospholamban in sodium dodecyl sulfate-polyacrylamide gels: Evidence for a protein structure consisting of multiple identical phosphorylatable subunits. J Biol Chem. 1984;259:1834–1841. [PubMed] [Google Scholar]
- 48.Andrade SL, Dickmanns A, Ficner R, Einsle O. Expression, purification, and crystallization of the ammonium transporter Amt-1 from Archaeoglobus fulgidus. Acta Crystallogr. 2005;61:861–863. doi: 10.1107/S1744309105027004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manley DM, et al. Secondary structure and oligomerization of the E. coli glycerol facilitator. Biochemistry. 2000;39:12303–12311. doi: 10.1021/bi000703t. [DOI] [PubMed] [Google Scholar]
- 50.Sukharev S. Purification of the small mechanosensitive channel of Escherichia coli (MscS): The subunit structure, conduction, and gating characteristics in liposomes. Biophys J. 2002;83:290–298. doi: 10.1016/S0006-3495(02)75169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pos KM, Diederichs K. Purification, crystallization and preliminary diffraction studies of AcrB, an inner-membrane multi-drug efflux protein. Acta Crystallogr D Biol Crystallogr. 2002;58:1865–1867. doi: 10.1107/s0907444902013963. [DOI] [PubMed] [Google Scholar]
- 52.Auer M, et al. High-yield expression and functional analysis of Escherichia coli glycerol-3-phosphate transporter. Biochemistry. 2001;40:6628–6635. doi: 10.1021/bi010138+. [DOI] [PubMed] [Google Scholar]
- 53.Karlsson M, et al. Reconstitution of water channel function of an aquaporin overexpressed and purified from Pichia pastoris. FEBS Lett. 2003;537:68–72. doi: 10.1016/s0014-5793(03)00082-6. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–180. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 55.Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Structure of the MscL homolog from Mycobacterium tuberculosis: A gated mechanosensitive ion channel. Science. 1998;282:2220–2226. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- 56.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 57.Jiang Y, et al. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417:515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.