Abstract
The extensive alternative splicing in higher eukaryotes has initiated a debate whether alternative mRNA isoforms are generated by an inaccurate spliceosome or are the consequence of highly degenerate splice sites within the human genome. Here, we established a quantitative assay to evaluate the accuracy of splice-site pairing by determining the number of incorrect exon-skipping events made from constitutively spliced pre-mRNA transcripts. We demonstrate that the spliceosome pairs exons with an astonishingly high degree of accuracy that may be limited by the quality of pre-mRNAs generated by RNA pol II. The error rate of exon pairing is increased by the effects of the neurodegenerative disorder spinal muscular atrophy because of reduced levels of Survival of Motor Neuron, a master assembler of spliceosomal components. We conclude that all multi-intron-containing genes are alternatively spliced and that the reduction of SMN results in a general splicing defect that is mediated through alterations in the fidelity of splice-site pairing.
Keywords: pre-mRNA splicing, spinal muscular atrophy, splice-site pairing, splicing fidelity, survival of motor neuron
A critical step in pre-mRNA splicing is the recognition and correct pairing of 5′ and 3′ splice sites. Given the complexity of higher eukaryotic genes and the relatively low level of splice-site conservation (1), the precision of the splicing machinery in choosing and pairing splice sites is impressive. Introns ranging in size from less than 100 up to 105 bases are removed efficiently. At the same time, a large number of alternative splicing events accompany the processing of pre-mRNAs (2, 3). In addition, minor perturbations, such as single base mutations, can frequently lead to aberrant splicing (4), predominantly exon skipping and alternative splice-site activation (2, 3). Although the correct sequence context is imperative for splice-site selection, a number of splicing factors have also been implicated in maintaining splicing accuracy, including core components of the spliceosome, as well as Isy1, Prp8p, Slu7p, and Sky1p (5–8). More recent studies have shown that Prp16 and Prp22p act as ATP-dependent proofreading factors for the first and second step of splicing, respectively (8, 9). However, even with proofreading steps at the catalytic core, many alternative mRNA isoforms are generated through alternative splice-site pairing. The sheer number of alternative mRNA isoforms has triggered an ongoing debate as to whether the majority of these transcripts are generated by mistake or with a biological purpose (10). As of today, biological functions for mRNA isoforms have been demonstrated in a large number of the cases studied (11, 12). Yet, it is possible that a significant number of the mRNA isoforms that survive quality-control steps, such as nonsense-mediated decay (NMD) (13), nonstop decay (NSD) (14), or no-go decay (NGD) (15), are ultimately translated without an obvious biological function.
These considerations raise the question whether the apparent promiscuity of the splicing machinery has evolved to increase variation of a limited genome. Thus, increased proteomic output through alternative splicing may come at the cost of periodically generating isoforms that initially do not have biological functions in the cell (16). To generate these variant patterns of splicing, the strength of interactions between the splicing machinery and particular exons must be kept within an optimal energetic range so that the extent of inclusion and exclusion of exons may vary. For example, a slight shift in the binding equilibrium could increase or decrease inclusion of an exon. In principle, fluctuating levels of interaction can be reached by providing suboptimal binding sites for the spliceosome, by reducing the specificity of the spliceosome, or by a combination of both. However, reducing the specificity of the spliceosome to increase proteome diversification could be problematic, as there may be housekeeping processes that cannot afford reduced levels of some gene products. These considerations suggest that for certain genes, the spliceosome should be capable to carry out repeated intron-removal events with high specificity and fidelity. However, the degree to which the fidelity of splice-site pairing influences alternative splicing is not known.
Using quantitative real-time PCR we show that the ability of the spliceosome to ligate flanking exons is exceedingly high, rivaling the remarkable levels of accurate substrate selection displayed by RNA polymerases or ribosomes. The low error rates of splice-site pairing suggest that the accuracy of splicing may be limited by the fidelity of transcription. We conclude that the high levels of alternative splicing are consequences of splice sites that have evolved to offer a weak binding potential for components of the spliceosome. Interestingly, the effects of the neurodegenerative disorder spinal muscular atrophy (SMA) increase the error rate of splicing. Thus, perturbed pre-mRNA splicing in SMA is mediated in part through alterations of the intrinsic splice-site pairing fidelity of the spliceosome.
Results
Defining a Standard for Constitutive Splicing.
It has been demonstrated that RNA transcription and translation work with a high degree of fidelity (17–19). Defining errors during the process of RNA transcription is conceptually straightforward because the incorporation of new nucleotides is template-directed. Similarly, translation errors are easily defined because aminoacylated tRNAs are matched with their respective codons through anticodon interactions. Unlike transcription or translation, it is difficult to define what constitutes an error in splicing. As splice sites are very degenerate in higher eukaryotes, the location of “correct” exon/intron junctions is not clearly defined, making it tricky to differentiate between splice-site selection errors and designated alternative splicing events. However, stringent selection criteria can be applied to identify genes that, by nearly all measures, are only constitutively spliced. For such genes, any deviation from the constitutive splicing pattern can be used to approximate erroneous splice-site pairing.
The following criteria were required for selection of endogenous test genes: First, only one type of EST database entry should be recorded for such genes across all eukaryotic lineages. This would indicate that genes are constitutively spliced with no known alternative splicing pattern, an important feature when evaluating the base-line frequency of incorrect splicing or mis-splicing. Second, the test gene should not have splice junction-sequence similarity to other genes, thus reducing the potential to amplify a false-positive signal. Third, the test gene should be highly conserved at the protein level and ubiquitously expressed. High levels of conservation would further support the notion that a gene is constitutively spliced. In addition, the ubiquitous expression would ensure that the gene would be easily detectable. Fourth, to avoid activation of mRNA surveillance mechanisms, all resulting aberrant mRNA isoforms of the test gene should maintain an ORF to prevent NSD or NGD, and the generation of any premature termination codon should be located within 50 nucleotides of the last exon/exon junction to avoid activation of NMD.
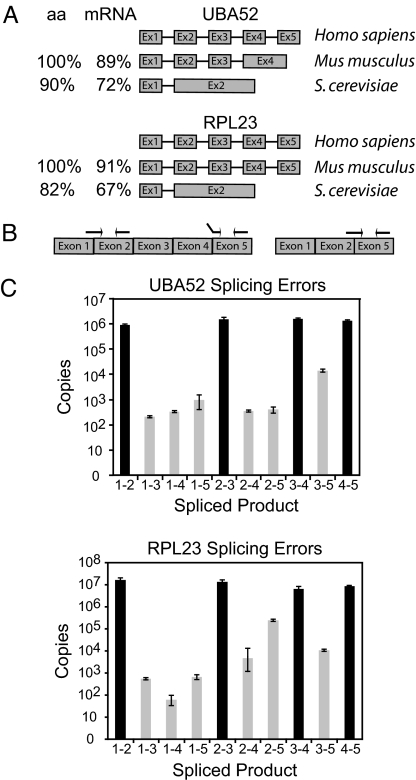
Applying these selection criteria, we identified ubiquitin A-52 ribosomal protein fusion product 1 (UBA52) and ribosomal protein-L 23 (RPL23) as excellent candidates for exclusive constitutive splicing in the human genome. The protein and mRNA sequences of both genes are highly conserved (Fig. 1A), and only one spliced mRNA product is found for either UBA52 or RPL23 from yeast to human. UBA52 is essential for targeting cellular proteins for degradation by the 26S proteasome (20) and RPL23 is an integral component of the ribosome, a highly conserved molecular machine (21). Both genes contain five exons, potentially generating eight different mRNA isoforms through alternative exon ligation.
Fig. 1.
Error rates of pre-mRNA splicing. (A) The exon/intron structures and phylogenetic conservation [amino acid (aa) and mRNA] of UBA52 and RPL23 are shown. (B) Experimental design using exon junction primers amplifying all mRNA isoforms generated through alternative exon ligation. Correct priming of the 1–2 splice variant is shown on the left. The middle set of primers shows incorrect priming of the 2–5 exon junction primer. The primers on the far right show correct priming of the 2–5 exon junction primer set. (C) Bar graph showing the total copy number detected for every exon skipping (gray) and constitutive splicing (black) event for all possible exon junctions in UBA52 and RPL23. cDNAs used to determine the error rate were generated with a mixture of random hexamers. Error rates are summarized in Table 1.
The Fidelity of Splice-site Pairing.
To determine the number of each constitutive and alternative exon ligation isoform, we performed real-time PCR using exon/exon junction primers that span the 3′ and 5′ end of ligated exons (Fig. 1B) (Tables S1 and S2). Crucial for the success of this assay is the specificity of the exon junction primers because half of the exon junction primer is always complementary to the constitutively spliced product (Fig. 1B). After optimizing location, annealing temperature, and primer concentrations, each primer set performed within a specificity range of 3 to 7 orders of magnitude (Fig. S1). cDNA generated from HeLa cell total RNA was then used to evaluate the abundance of each UBA52 or RPL23 mRNA isoform. All primer pairs assaying for the constitutively spliced UBA52 mRNAs were measured within 2-fold of the 106 input molecules, (Fig. 1C, primers 1–2, 2–3, 3–4, and 4–5). These results demonstrate that no significant PCR amplification and cDNA synthesis biases were introduced in our approach. Importantly, signals for all other mRNA isoforms were detected, however, at significantly reduced levels ranging between 50 and more than 104 copies. These copy number measurements reflect mRNA isoform quantities because all but the UBA52 1–3 and RPL23 1–5 splicing error levels are at least one order of magnitude greater than the potential number of molecules generated from mis-priming (Fig. S1). To adequately reflect the number of molecules generated from erroneous splice-site pairing, the expected number of mis-priming events was subtracted from each primer-pair measurement (Fig. 1C). We conclude that all possible exon-skipping mRNA isoforms are generated from UBA52 and RPL23 pre-mRNAs, albeit at drastically different rates (Table 1).
Table 1.
Error rate of splicing for skipping events
| 1–3 | 1–4 | 1–5 | 2—4 | 2–5 | 3–5 | |
|---|---|---|---|---|---|---|
| UBA52 | 1.6 × 10−4* | 2.5 × 10−4 | 7.4 × 10−4 | 2.7 × 10−4 | 2.9 × 10−4 | 1.9 × 10−3 |
| RPL23 | 4.7 × 10−5 | 5.7 × 10−6 | 5.4 × 10−5# | 4.5 × 1−4 | 2.3 × 10−2 | 9.4 × 10−4 |
The numbers represent the splicing error rate for each mis-splicing event. The standard error calculated from at least 6 repetitions is less than 10%.
*, limited by primer specificity (Fig. S1).
The most abundant alternative mRNA isoforms detected in both genes lack several internal exons (1–5 for UBA52 and 2–5 for RPL23, Fig. 1C), indicating that a significant fraction of pre-mRNAs was processed after the terminal exon had been transcribed. Although it is established that multiple factors contribute to the efficiency of exon recognition (22), we observe a weak correlation between the splice-site strength of erroneous exon ligations and the frequency of erroneous splice-site pairing (Fig. S2). However, even with such weak correlations, it is difficult to conclude that any single splicing element preferentially or exclusively induces splicing errors.
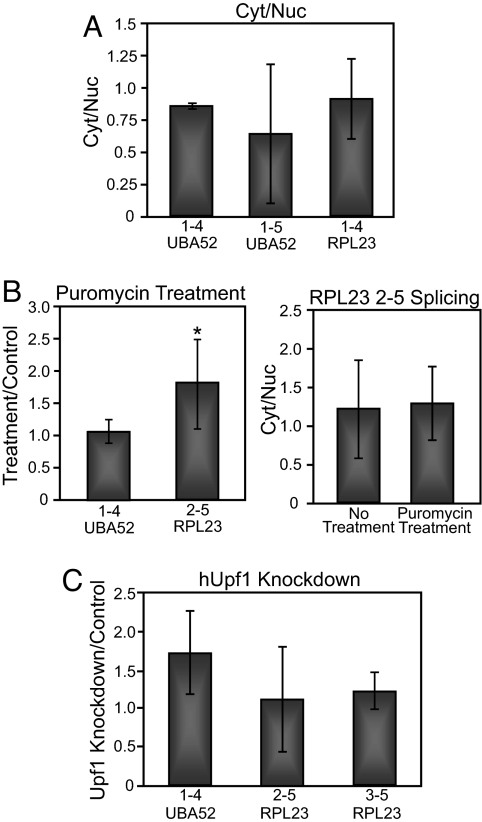
To confirm that the low exon-skipping error rates observed were not due to biased cDNA synthesis, we examined the fidelity of splice-site pairing in cDNA generated using either oligo(dT) or random hexamers. In addition to detecting the same number of constitutively spliced molecules regardless of primer-pair location along the gene (Fig. 1C), alternative splicing analyses using oligo(dT) primed cDNAs generated comparable error rates, thus excluding cDNA synthesis as a source of biasing transcript representation (Fig. S3 and Table S3). To test the possibility that erroneously spliced mRNAs are preferentially retained in the nucleus, we determined UBA52 and RPL23 mRNA isoform abundances from cytoplasmic and nuclear fractions. No significant differences in error frequencies were detected between nuclear and cytoplasmic mRNA populations (Fig. 2A), demonstrating that all UBA52 and RPL23 splicing errors are exported just as efficiently as the correctly spliced mRNAs.
Fig. 2.
Inhibition of translation and NMD does not affect the error rate of splicing. (A) Error analysis was performed and compared between RNA samples isolated from cytoplasmic and nuclear fractions. No statistically significant difference in the error rate is detected between nuclear and cytoplasmic samples, implying uniform export and half-life kinetics between mRNA isoforms tested. Amplification of nuclear U6 snRNA was used to confirm efficient nuclear and cytoplasmic fractionation (Fig. S4). (B) Comparison of splicing errors in the presence or absence of the translation inhibitor puromycin. Error analysis was performed without (left panel) or with (right panel) nuclear/cytoplasmic fractionation. (C) The effect of hUpf1 knockdown on the error rate of splicing. hUpf1 knockdown was confirmed by RT-PCR, Western Blot, and splicing analysis of NMD target genes that are differentially spliced upon hUpf1 knockdown (Fig. S5). *, P < 0.05
To determine whether UBA52 and RPL23 splicing errors are selectively degraded by the translational-dependent mRNA surveillance mechanisms NSD, NGD, or NMD, we inhibited translation by puromycin treatment before harvest. Inhibition of translation did not alter the measured error rate, even when analyzing cytoplasmic and nuclear fractions (Fig. 2B). Finally, we performed RNAi against hUpf1 to determine whether incorrectly spliced mRNAs are degraded by NMD. Consistent with our gene selection criteria, most splicing errors analyzed did not change with knockdown of hUpf1 (Fig. 2C). Only one of the errors tested displayed a statistically significant change in the error rate (Fig. 2C, 1–4 error), however, the measured 1.5-fold change is much lower than expected if this transcript were degraded by the NMD. These results support the interpretation that the steady-state levels measured for each UBA52 and RPL23 mRNA isoform reflect frequencies of alternative pre-mRNA splicing. In summary, the real-time PCR analysis suggests that the spliceosome recognizes and removes introns with a high degree of accuracy.
It is well-established that extensive alternative splicing is observed between different tissues and cell types (2). In most documented cases, these changes are explained through cell type-specific regulatory factors that are differentially expressed or activated in their cellular environment. However, it is unknown whether these different cellular environments also influence the accuracy of constitutive splice-site pairing. To address this question, we compared splice-site pairing error frequencies between HeLa cells and a fibroblast cell line. Similar error rate frequencies were observed between these cell lines, supporting the notion that cell type-specific alternative splicing is largely mediated through differential expression and activation of splicing factors (Fig. S6).
The Splice-site Pairing Error Rate Is Increased in SMA.
The error rate of splice-site pairing is low enough in which random mistakes in splicing are unlikely to cause disease. However, there is a potential for an increase in the error rate of splice-site pairing if mutations occur in genes responsible for maintaining the correct concentration or number of functional spliceosomes. One example that links pre-mRNA splicing to human disease is SMA, an autosomal recessive neuromuscular disorder caused by the deletion of one of the two copies of the Survival of Motor Neuron (SMN) gene (23). In SMA patients, the remaining SMN2 copy is unable to compensate for the loss of SMN1. The low amounts of SMN produced from SMN2 are adequate for a fetus to develop, but insufficient to maintain healthy motor neurons throughout life (24). SMN is the central component of the SMN complex, which is required for snRNP recycling, reassembly, and maintenance of high snRNP concentrations (25, 26). Previous work has shown that depletion of SMN recapitulated the SMA phenotype in zebrafish. Significantly, rescue of SMN-depleted animals was achieved by the injection of purified snRNPs, suggesting a critical role for snRNPs to the SMA phenotype (27). This proposal was recently supported by the demonstration that reduced SMN results in altered levels of snRNPs, which alter the splicing profile (26, 28).
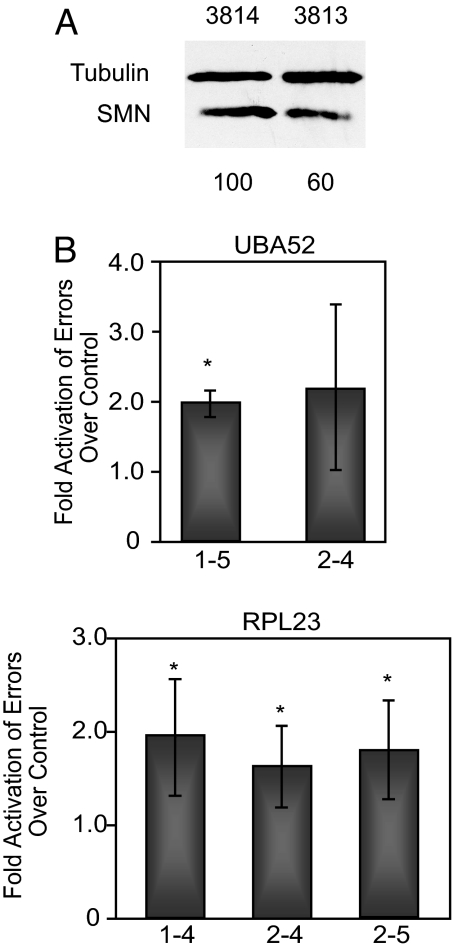
To test whether the accuracy of pre-mRNA splice-site pairing is modulated in SMA, we used our quantitative real-time PCR splicing assay. SMA patient fibroblast (3813) and control fibroblast (3814) cell lines were tested for alterations in the relative abundance of UBA52 and RPL23 mRNA isoforms. Patient fibroblast cell lines consistently exhibited an ≈2-fold higher error rate compared with control cell lines (Fig. 3B). This reduction in accuracy is highly significant (P <0.005) and consistent with the approximate 2-fold reduction in SMN protein levels observed for these patient/control cell lines (Fig. 3A). An increase in the splicing error rate can also be recapitulated by RNAi-mediated knockdown of SMN in HeLa cells (Fig. 4B). Together, these results demonstrate that the reduction in SMN and its associated reduction in snRNP levels (26, 28) perturb pre-mRNA splicing accuracy. The fact that increased errors were detectable for all constitutively spliced introns tested suggests that the reduction of SMN causes a general splicing defect. Thus, SMA likely influences pre-mRNA splicing of many genes, an interpretation consistent with a recent microarray analysis of SMN-deficient mouse tissues (28).
Fig. 3.
SMA patient fibroblast cells show an increase in the error rate of splicing. (A) Western blot highlighting the difference in SMN levels between SMA patient (3813) and control (3814) cells. The quantitation shown below was normalized to tubulin levels. (B) Bar graph illustrating the relative increase of the splicing error rate in SMA patient cells. *, P < 0.05
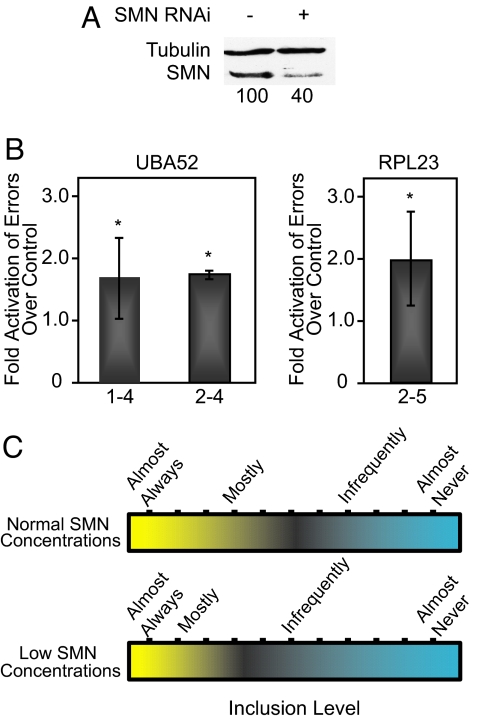
Fig. 4.
Decreased SMN concentrations increase the error rate of splicing. (A) Western blot confirming SMN knockdown in HeLa cells. The quantitation shown below was normalized to tubulin levels. SMN knockdown is also confirmed by RT-PCR (Fig. S7). (B) Bar graph showing the relative increase in splicing errors when cells are treated with RNAi against SMN. (C) Model showing the effect of normal versus reduced SMN concentrations on the recognition and inclusion of exons. At normal concentrations the scale of recognition is shown (Upper). When SMN concentrations are reduced, some alternatively spliced exons are less frequently included (Lower). *, P < 0.05.
Efficient exon recognition depends on multiple parameters, such as splice-site strength, splicing regulators, the exon/intron architecture, transcription, and the concentration of spliceosomal components (22). Given the variation within each of these parameters, the recognition potential of exons is expected to span a wide range (Fig. 4C). Alternative exons, represented within the center of this distribution, represent exons with the greatest chance of being altered by minor changes in splicing efficiency. In the SMA cell culture model analyzed, a reduction in SMN and its accompanying reduction in snRNP concentrations (26, 28) has the potential to trigger a change from preferential exon inclusion to exclusion, thus shifting the spectrum of exon recognition (Fig. 4C). SMA is characterized by the progressive loss of motor neurons (24). Interestingly, SMN expression levels fluctuate dramatically during motor neuron development (29), implying that pre-mRNA splicing perturbations may be exacerbated during this crucial phase of establishing contacts between neighboring cells.
Discussion
Our experiments show that the spliceosome removes introns with a high level of fidelity, rivaling the accuracy of substrate selection displayed during RNA transcription and mRNA translation (17–19). One important conclusion that can be drawn from these observations is that the extensive alternative splicing associated with the majority of higher eukaryotic genes is not based on an intrinsic inability of the splicing machinery to carry out faithful intron removal. Rather, alternative splicing as documented in EST databases is, for the most part, a consequence of suboptimal binding sites for components of the splicing machinery or for splicing regulators.
The splice-site pairing error analysis was carried out evaluating one of the most common and predominant forms of mis-splicing, alternative exon exclusion (2, 3). However, cryptic splice-site activation is also an abundant alternative splicing pathway. The focus on alternative exon exclusion was largely driven by the experimental design considerations and limitations. Given the defined splice sites flanking a set of exons, alternative exon exclusion events can be assayed for because the sequence of alternative exon/exon junctions can be derived. However, in the case of cryptic splice-site selection, alternative exon/exon junctions can only be predicted if such events have been recorded within EST databases. As the goal of our study was to determine the accuracy of the spliceosome in the context of the strongest splice signals, we only selected to evaluate conserved genes with no alternative isoforms reported in EST databases, thus excluding known cryptic splice-site activation events.
In our experimental design, we considered that the measured levels of mRNA isoforms could reflect the combined activities of pre-mRNA splicing and RNA surveillance. Upf1 depletion, cellular fractionation, and translation inhibition experiments served to exclude the possibility that the major RNA surveillance pathways NMD, NSD, and NGD do contribute to the low levels of mRNA isoforms measured. However, we cannot exclude the possibility that some of the UBA52 and RPL23 mRNA isoforms are selectively targeted by another mRNA surveillance mechanism. For example, the recently identified rapid mammalian deadenylation-dependent decay pathway (30) may modulate UBA52 and RPL23 mRNA isoform abundance, although the uniform 3′ UTR of the isoforms generated would argue against selected isoform degradation by that mechanism.
The Accuracy of Splice-site Pairing May Be Limited by the Fidelity of Transcription.
Remarkably, the lowest error rate detected was one error for every 105 splicing events, a frequency approximated by several other intron removal events (Fig. 1C, Table 1, and Fig. S2). In comparison, the transcription and translation machineries make a mistake once in every 103–105 nucleotide or amino acid insertion step (17–19). With such a fidelity, transcription of UBA52 or RPL23 results in nucleotide mis-incorporations once in every 3–300 pre-mRNA copies. Thus, it is possible that a significant fraction of the UBA52 or RPL23 isoforms detected in our experiments resulted from transcription errors. In principle, skipping of any internal exon could be the consequence of nucleotide mis-incorporation at the essential AG at the 3′ splice site, or the essential GU at the 5′ splice site. Using the low mis-incorporation frequency of one nucleotide for every 105 nucleotides, it is expected that 1 of every 25,000 exons transcribed contains a single splice-site mutation that induces alternative splicing. This calculation is a conservative estimate when considering that most exons require not only correctly transcribed splice sites, but also correctly transcribed splicing enhancers and silencers (22). Given their low abundance (Table 1 and Fig. 1C), it is highly likely that UBA52 mRNA isoforms 1–3-4–5, 1–2-4–5, and 1–4-5 and the RPL23 mRNA isoforms 1–3-4–5, 1–4-5, and 1–5 are a direct result of nucleotide mis-incorporations at the invariable nucleotides of the 3′ or 5′ splice sites. These considerations suggest that the accuracy of splice-site pairing may be limited by the fidelity of transcription.
Are All Multi-intron Genes Alternatively Spliced?.
Our splice-site pairing error analysis demonstrated that essentially all UBA52 and RPL23 mRNA isoforms generated from alternative exon inclusion are made in detectable quantities. Considering the constitutive nature of these genes, this is a surprising finding. However, we observed a great variation in the representation of these errors ranging from 1–1,000 errors in 105 splicing events. It is expected that these differences represent variations in the strength and efficiency of these particular exons as splicing substrates. Independent from whether our data uncovered the exact limits imposed by transcription fidelity, there will be transcription errors that result in nucleotide mis-incorporation at one of the splice sites. This realization argues, at least from a theoretical point of view, that there will always be alternative splicing, albeit with drastically different efficiencies. In combination with our observations, these considerations strongly suggest that all multi-intron–containing pre-mRNAs, including constitutively spliced genes, such as UBA52 and RPL23, generate multiple alternatively spliced mRNA isoforms. Although the frequency of mis-splicing events are relatively low for each event, the total number of mistakes for a given gene might actually be relatively high when considering the large number of introns that must be spliced out in some cases. In other words, the greater the number of introns in a gene, the greater the chance for a mistake in splice-site pairing. As an extreme example the human titin gene, which contains 363 exons, is considered (31). In theory, titin's exon/intron architecture permits the generation of approximately 65,000 different exon junctions. Using the conservative estimate of 1 error per 105 splicing events (Table 1) and ignoring documented alternative splicing and the influence of co-transcriptional splicing, it is expected that only 1 of 3 titin pre-mRNAs will produce a fully spliced copy of the titin gene. However, with an average number of 9 exons per human gene, the influence of erroneous splice-site pairing is expected to be minimal. In most cases, it is anticipated that the activities of NMD, NSD, and NGD mRNA quality-control steps limit the translation of potentially harmful mRNA isoforms (13). The small fractions of splicing errors that may evade RNA surveillance are likely tolerated in the cell because their infrequent occurrence renders them biologically irrelevant.
Here, we demonstrate that the spliceosome is capable of removing introns faithfully and with high accuracy. Quantitative analysis of all possible alternative exon exclusion patterns demonstrated that, in some cases, a splicing mistake is made only once in over 105 intron removal events. Several conclusions can be drawn from these results. First, the spliceosome recognizes and pairs splice sites with an astonishingly high degree of accuracy that may be limited by the quality of pre-mRNAs generated by RNA pol II. Second, the high levels of alternative splicing observed in the human genome are the consequence of suboptimal splicing signals. These observations suggest that the splicing machinery is not directly involved in the evolution of genes, but that its main function is to increase the coding potential of the genome. Third, all multi-intron pre-mRNAs are likely to undergo alternative splicing, albeit with various efficiencies that dictate the biological impact of the resulting mRNA isoform. Finally, a splicing error analysis of the human genetic disease SMA demonstrates that reduction of SMN results in a general splicing defect that is expected to be amplified in developing motor neurons.
Materials and Methods
RNA Isolation and cDNA Preparation.
RNA was isolated from cells using TRIzol (Invitrogen). This was followed by phenol chloroform extraction and IPA precipitation at room temperature. Total RNA was treated with DNA-free Kit (Ambion). The DNase-treated RNA was reverse transcribed using iScript (Bio-Rad), or MLV-RT (Promega) and oligo(dT) primer overnight. Cytoplasmic and nuclear RNA were fractionated using the procedure described by Sandri-Goldin (32).
Real-time PCR.
PCR was performed using iQ SYBR Green Supermix (Bio-Rad) at 100–300 nM of each primer. Plasmid DNA was used to seed the reaction for all standard curves made, at either 107, 106, 105, 104, 103, or 102 copies. To determine the number of copies of mRNA in cDNA, three concentrations of cDNA were used for every sample. All cDNA samples were run concurrently with standard curves for both the correctly spliced and the “erroneously spliced” mRNA to which it was compared. To determine the number of “erroneously spliced” copies, cDNA was run concurrently with a standard curve created by 10-fold dilutions of plasmid controls. The quantity of errors for each skipping event was calculated by interpolation from their respective standard curve. All samples were run with at least four dilutions to ensure a linear range of detection. The real-time PCR conditions were: 95 °C for 5 min followed by 40 cycles of 95 °C for 10 s, and an annealing/extension step for 10 s (temperature varied with primer set; Table S1 and Table S2). Following PCR, a melting curve was performed starting at 55 °C, and increasing 0.5 °C every 10 s for 80 cycles. Reactions were run on an agarose gel after PCR to check for contamination. All reactions were carried out in duplicate or triplicate and performed at least 3 separate times. All of the primer sets and annealing temperatures can be found in Tables S1 and S2. Methods to determine primer specificity are detailed in SI Materials and Methods.
Protein Harvest and Westerns.
Total protein was harvested using RIPA buffer. Samples were electrophoresed in an SDS-polyacrylamide gel and transferred to a nitrocellulose membrane (Bio-Rad). Westerns were developed with Supersignal West Pico Chemiluminescence Substrate (Pierce). The hUpf1 antibody was kindly provided by the Lykke-Andersen lab and the SMN antibody was from BD Biosciences.
Cell Lines and Tissue Culture.
We used the SMA patient model cell line (3813 cells) and a control cell line from the mother of the affected individual (3814 cells). The mother is a carrier for the SMA disease. HeLa cervical carcinoma cells were used for all RNAi treatments and to determine the baseline error rate. RNAi conditions and sequences are described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Elizabeth von Hasseln for helping to optimize RPL23 primer concentrations and annealing temperatures. We are grateful to Jens Lykke-Anderson for donating anti-hUpf1 antibodies and to Benjamin Blencowe for providing primer sequences to analyze splicing with hUpf1 knockdown. This work was supported by the National Institutes of Health grant GM62287 (K.J.H.), DOD grant BC052878 (K.J.H.), and Families of SMA grant HER0607 (K.J.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813128106/DCSupplemental.
References
- 1.Burge CB, Tuschl T, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2nd Ed. Cold Spring Harbor, New York: CSHL Press; 1999. pp. 525–560. [Google Scholar]
- 2.Holste D, Huo G, Tung V, Burge CB. HOLLYWOOD: A comparative relational database of alternative splicing. Nucleic Acids Res. 2006;34:D56–62. doi: 10.1093/nar/gkj048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim N, Alekseyenko AV, Roy M, Lee C. The ASAP II database: Analysis and comparative genomics of alternative splicing in 15 animal species. Nucleic Acids Res. 2007;35:D93–98. doi: 10.1093/nar/gkl884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 5.Chua K, Reed R. The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature. 1999;402:207–210. doi: 10.1038/46086. [DOI] [PubMed] [Google Scholar]
- 6.Dagher SF, Fu XD. Evidence for a role of Sky1p-mediated phosphorylation in 3′ splice site recognition involving both Prp8 and Prp17/Slu4. RNA. 2001;7:1284–1297. doi: 10.1017/s1355838201016077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- 8.Villa T, Guthrie C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev. 2005;19:1894–1904. doi: 10.1101/gad.1336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayas RM, Maita H, Staley JP. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorek R, Shamir R, Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 12.Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 14.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frischmeyer PA, et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 16.Boue S, Letunic I, Bork P. Alternative splicing and evolution. Bioessays. 2003;25:1031–1034. doi: 10.1002/bies.10371. [DOI] [PubMed] [Google Scholar]
- 17.Cochella L, Green R. Fidelity in protein synthesis. Curr Biol. 2005;15:R536–540. doi: 10.1016/j.cub.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 18.Jeon C, Agarwal K. Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc Natl Acad Sci USA. 1996;93:13677–13682. doi: 10.1073/pnas.93.24.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nesser NK, Peterson DO, Hawley DK. RNA polymerase II subunit Rpb9 is important for transcriptional fidelity in vivo. Proc Natl Acad Sci USA. 2006;103:3268–3273. doi: 10.1073/pnas.0511330103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berchtold MW, Berger MC. Isolation and analysis of a human cDNA highly homologous to the yeast gene encoding L17A ribosomal protein. Gene. 1991;102:283–288. doi: 10.1016/0378-1119(91)90091-o. [DOI] [PubMed] [Google Scholar]
- 21.Starkey CR, Menon RP, Prabhu S, Levy LS. Primary sequence and evolutionary conservation of ribosomal protein genes from the domestic cat. Biochem Biophys Res Commun. 1996;220:648–652. doi: 10.1006/bbrc.1996.0458. [DOI] [PubMed] [Google Scholar]
- 22.Hertel KJ. Combinatorial control of exon recognition. J Biol Chem. 2008;283:1211–1215. doi: 10.1074/jbc.R700035200. [DOI] [PubMed] [Google Scholar]
- 23.Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy- determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 24.Lefebvre S, et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 25.Baccon J, Pellizzoni L, Rappsilber J, Mann M, Dreyfuss G. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J Biol Chem. 2002;277:31957–31962. doi: 10.1074/jbc.M203478200. [DOI] [PubMed] [Google Scholar]
- 26.Gabanella F, et al. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS ONE. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winkler C, et al. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev. 2005;19:2320–2330. doi: 10.1101/gad.342005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tizzano EF, Cabot C, Baiget M. Cell-specific survival motor neuron gene expression during human development of the central nervous system: Implications for the pathogenesis of spinal muscular atrophy. Am J Pathol. 1998;153:355–361. doi: 10.1016/S0002-9440(10)65578-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol Cell. 2006;24:943–953. doi: 10.1016/j.molcel.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 31.Bang ML, et al. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- 32.Sandri-Goldin RM. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.