Abstract
Factors that influence speciation rates among groups of organisms are integral to deciphering macroevolutionary processes; however, they remain poorly understood. Here, we use molecular phylogenetic data and divergence time estimates to reconstruct the pattern and tempo of speciation within a widespread and homogeneous bird family (white-eyes, Zosteropidae) that contains an archetypal “great speciator.” Our analyses show that the majority of this species-rich family constitutes a clade that arose within the last 2 million years, yielding a per-lineage diversification rate among the highest reported for vertebrates (1.95–2.63 species per million years). However, unlike most rapid radiations reported to date, this burst of diversification was not limited in geographic scope, but instead spanned the entire Old World tropics, parts of temperate Asia, and numerous Atlantic, Pacific, and Indian Ocean archipelagos. The tempo and geographic breadth of this rapid radiation defy any single diversification paradigm, but implicate a prominent role for lineage-specific life-history traits (such as rapid evolutionary shifts in dispersal ability) that enabled white-eyes to respond rapidly and persistently to the geographic drivers of diversification.
Keywords: diversification rate, speciation, Zosterops, white-eye, evolution
Disparity in diversification rates among groups of organisms is well documented and provides unique opportunities for studying evolutionary processes underlying the genesis of biological diversity (1–7). A few groups of organisms that diversified recently and rapidly have contributed disproportionately to speciation theory by providing comparatively accessible opportunities to evaluate factors that drive speciation (1–4). These groups are also characterized by a restricted geography (e.g., archipelagos, lakes, mountain tops, etc.) circumscribed by the more extensive distribution shared by the group and its close relatives. The reduced faunas, stark geographic boundaries, and limited set of earth history influences of these confined geographies make such systems attractive for studies of diversification.
Island settings are especially well known for variable speciation rates. For example, some island bird taxa spread across scattered insular landscapes with little or no differentiation, whereas others appear to have diversified rapidly across the same geographies (8). In the wake of island biogeographic theory and its antecedents (9–11), recognition of this pattern in birds resulted in characterization of a set of “great speciator” lineages (12), as well as a famous paradox: how can these lineages show such high degrees of differentiation across oceanic islands when their excellent dispersal ability should limit differentiation?
Discussions of this paradox have appealed to intermediate dispersal ability (8, 12) or to evolutionary shifts in dispersal ability (12) to explain the seeming conflict between large geographic range, implying good dispersal ability, and the high degree of morphological differentiation between nearby islands, implying poor dispersal ability. Evaluating the great speciator hypothesis, and deciphering its paradox, depends on using methods that can reconstruct phylogenetic relationships, identify natural groups of species, and estimate divergence times and associated speciation rates. Interpretations are vastly different if a lineage has diversified by a slow accumulation of species over time versus a rapid burst of speciation in the wake of a phase of active dispersal, or if a great speciator lineage is not a natural group and instead includes multiple independent radiations. The limited geographic scope and lack of available phylogenetic methods in the original studies of great speciator lineages have hindered interpretations to date.
The Zosterops [griseotinctus] species group in the white-eye family Zosteropidae is a classic example of a great speciator (12), inhabiting numerous islands in the southwest Pacific and exhibiting dramatic interisland differences in behavior and morphology across water gaps as narrow as 2 km (13, 14). However, the taxonomic limits of the Z. [griseotinctus] species group are uncertain (8), and understanding evolutionary patterns and processes in white-eyes more broadly requires accounting for 2 factors that have long confounded systematists: large distribution and similar morphology (15–17). The family Zosteropidae (≈100 species) is distributed across the entire Old World tropics, parts of temperate Eurasia, and numerous archipelagos in the Atlantic, Indian, and Pacific Oceans, and the largest genus (Zosterops) contains ≈80 similar-looking species found across the range of the family. As Ernst Mayr recognized more than 50 years ago, broad geographic range and homogeneous appearance complicate efforts to identify natural groups within white-eyes: “I know of no other group of birds in which close relatives, for example the subspecies of Zosterops atrifrons or the semispecies of the superspecies griseotincta, may differ more from each other than do distantly related species. Indeed some Oriental species are almost indistinguishable from African forms, from which they must have been isolated since remote times.” (15).
However, if the historic challenges of range and morphological homogeneity can be addressed, these very characteristics provide an ideal backdrop for examining the pattern and tempo of speciation across varied geographical scales and forms. Recent fieldwork across the Old World, combined with molecular phylogenetic methods, allows us to assess the pattern and tempo of white-eye speciation across its broad geography. Here, we reconstruct evolutionary relationships within the griseotinctus species group, and within Zosteropidae more broadly, enabling assessment of diversification rates both within this species group and across the family. Results from these phylogenetic analyses are then used to revisit theory surrounding diversification rate differences in birds, and to begin disentangling potential causes of the presumed high historical speciation rate within a great speciator.
Results
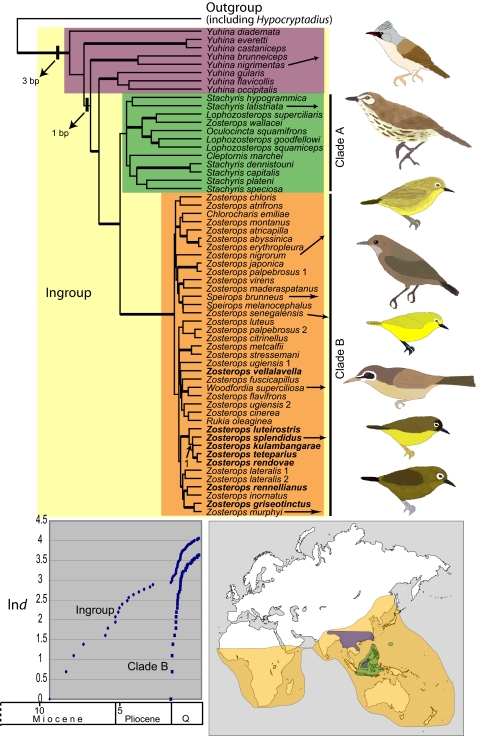
Using model-based phylogenetic methods, we analyzed nuclear and mitochondrial DNA sequence data to reconstruct evolutionary relationships among white-eyes, assess geographic and temporal patterns of diversification, and assess diversification rates within the Z. [griseotinctus] species complex. All analyses reconstructed white-eyes and Philippine members of the babbler (Timaliidae) genus Stachyris within the Asian babbler genus Yuhina [8/9 species sampled; see Fig. 1, supporting information (SI) Results, and Fig. S1]. White-eyes separated into 2 distinct groups in the phylogeny. Three small genera (Lophozosterops, Oculocincta, and Cleptornis), Zosterops wallacei, and the Philippine Stachyris formed a weakly supported clade (Clade A, see Fig. 1) sister to a well-supported clade that included the majority of white-eye species (Clade B, see Fig. 1). This latter clade contained all Zosterops species except Z. wallacei, and species from 4 other genera (Woodfordia, Rukia, Chlorocharis, and Speirops) scattered among the Zosterops. These 4 genera occur on oceanic islands or are restricted to high mountains, and were historically placed in separate genera based on morphological differences from typical white-eyes. One genus previously placed in Zosteropidae (Hypocryptadius) appears to have affinities outside the family, although denser outgroup sampling is necessary to determine its true relationships.
Fig. 1.
Phylogenetic and geographic relationships of white-eyes. Tree produced by rate-smoothing the ML phylogeny, calibrated with an age of 560,000 years before present at node 1 based on estimates of island age within the New Georgia Group of the Solomon Archipelago. Thickened branches indicate posterior probability ≥0.95 and bootstrap proportion ≥70; 2 inferred insertion/deletion events are labeled near the base of the tree. Bold species names indicate members of Z. [griseotinctus] species complex, all of which inhabit the New Georgia Island Group in the Solomon Archipelago except Z. rennellianus, which is endemic to Rennell Island, and the nominate Z. griseotinctus, which inhabits the Louisiade Islands off eastern New Guinea. Colors on map indicate distribution of Yuhina (purple), Clade A (green), and Clade B (orange). Lineage through time plots track the number of extant lineages (ln) for the entire ingroup and within Clade B alone. The time scale at the bottom applies to the LTT plot as well as the phylogeny. The Quaternary (Q) includes the Pleistocene and Holocene epochs. Bird images illustrate general patterns of diversity within the major clades.
Maximum uncorrected pairwise divergence for the ND2 gene within Clade B was only 7.1%. Based on recent taxonomic treatments of Zosterops and its allies (18, 19), this clade includes ≈80 biological species and >200 described subspecies. The combination of low molecular divergence and high species diversity within Clade B is unparalleled among birds. This level of molecular divergence is exceeded within single described avian species (20, 21); however, Zosterops ranks among the most species-rich bird genera.
Maximum divergence times estimated from oceanic island ages placed initial diversification of the genus Yuhina in the Miocene [95% confidence interval (CI), 6.31–8.06 million years); see Table S1) and diversification of the Zosteropidae around the Miocene/Pliocene boundary (CI, 4.46–5.57 million years). Clade A diversified first, but contains only ≈24 biological species, assuming taxonomic assignments of missing taxa are correct. Clade B, which includes most Zosterops species and 4 other genera (Fig. 1), began diversifying in the early Pleistocene. Notably, this radiation, which spans much of the Old World, is not much older than very local radiations within the group, such as the well-known white-eye radiation within the New Georgia Islands, Solomon Archipelago (Fig. 1). A conservative estimate of diversity in Clade B (80 species) and a Yule model of diversification yielded a diversification rate of 2.24 taxa per million years (CI, 1.95–2.63), which far exceeds most published estimates for speciose radiations (1, 3–7). For comparison, Weir and Schluter (22) estimated an average age of 3 million years for sister-species pairs of Neotropical birds, older than all of Clade B (Fig. 1), and a survey of avian diversification rates recovered a maximum estimate of 0.5 species per million years for Dendroica warblers (23). Published diversification rates exceeding those in white-eyes are rare and found in geographically restricted groups, such as Andean Lupinus (5), and cichlid fish in Lake Victoria and Lake Malawi (2). In addition to the geographic scope of the white-eye radiation, results here expand the taxonomic scope of hyperdiversifications beyond fish and plants to include birds.
Lineage-through-time (LTT) plots track the temporal accumulation of lineages in a clade and revealed striking differences between the entire ingroup (including Clade B) and Clade B alone (Fig. 1). For the entire ingroup (Yuhina, Stachyris, and Zosteropidae), the LTT plot fluctuates over time, but analysis of internode distances indicated no significant departure from a constant rates (CR) model (g = −0.627). In contrast, lineage accumulation within Clade B displays an initial steep slope and marked decrease in accumulation rate toward the present. The convexity of the LTT plot for Clade B is also reflected in the gamma statistic (g = −5.684). This high negative value could indicate a decreasing diversification rate over time, incomplete taxon sampling, or over-dispersed taxon sampling (24). The latter 2 conditions exist in this study: we sampled ≈50% of biological species and intentionally biased sampling toward potentially older lineages (e.g., aberrant genera, unique taxa, geographic extremes, etc.). A Monte Carlo CR test (24) indicated that incomplete taxon sampling alone could not account for the extreme gamma statistic (critical value = −2.75). Additional simulations revealed that our gamma statistic would be marginally significant (P = 0.05), even if our taxon sampling were maximally over-dispersed (i.e., if included, the taxa we omitted would only create nodes younger than all other nodes in our tree). Thus, our extreme gamma statistic represents a real slow down in diversification rates, rather than an artifact of incomplete or over-dispersed sampling. Although diversification rates will always be a balance between speciation and extinction (D = S − E), simulations have shown that the pattern seen in Clade B (“explosive early”) was likely produced by a decrease in speciation rates over time, rather than an increase in the rate of extinction (25). Thus, extant white-eye diversity resulted from extremely high speciation rates early in the Pleistocene that subsided toward the present, and our use of a constant-rate pure-birth model over the entire radiation may underestimate the diversification rate early in the radiation and overestimate the rate closer to the present.
Diversification rate assessment relies on accurate estimates of age and diversity. Dating nodes in molecular phylogenies is contentious and carries many assumptions (26, 27), but our estimate based on a Pacific island age is supported by 2 independent dating measures. A separate study of regional white-eye diversification used a different set of taxa and genes, and used an Indian Ocean island for calibration (28), but the basal node in their Zosterops radiation is shared with our phylogeny and was estimated at 1.84 million years, just inside our CI for that node (1.40–1.89 million years). Also, if we disregard the possibility that rates of molecular evolution might vary across bird lineages and apply to our data the minimum rate of ND2 divergence calculated for another bird group (29), the resulting age range for Clade B (2.0–2.3 million years) only slightly exceeds the estimates based on island ages. Classification of allopatric island taxa varies extensively depending on the species concept used, and can have a large effect on diversity estimates (30). We used species totals from sources that explicitly state adherence to the Biological Species Concept (18, 19), and are thus conservative estimates of species number. Polyphyletic species in our phylogeny (Fig. 1) and cryptic diversity in island white eyes (31) suggest that species diversity may in fact be higher than these estimates. Although changes in taxonomy or dating methods may shift the precise range of diversification rates, the diversification rate recovered for Zosterops is so much higher than in most other groups that these changes do not have a large effect. For example, the oldest divergence date estimated from the 3 methods (2.3 million years) yields a diversification rate of 1.6 species per million years. Also, even if the diversity in Clade B is halved and the oldest date is assumed, the rate drops to 1.3 species per million years, still more than twice that of any bird group examined so far (23).
Discussion
The classic great speciator, Z. [griseotinctus], was not reconstructed as monophyletic. Its spread and colonization within the restricted geography of the New Georgia Group and the Louisiades is considerably more complex than had been appreciated, owing to the broad geographic mixing of lineages. Nonetheless, our results serve to reinforce interest in diversification rates among white-eyes, albeit at a far greater geographic scale. Recent, rapid speciation was not restricted to northern Melanesia; it occurred across multiple continents and island groups, and, at finer scales, produced marked variation between sister taxa separated by the narrowest of water gaps as well as far-flung, morphologically aberrant montane and island endemics (e.g., Chlorocharis, Speirops, and Woodfordia). Also, this diversification occurred across a geographic theater greatly eclipsing that of its parent lineage, which had been evolving for >5 million years before the first speciation events within Zosterops (Fig. 1).
Diversification rates among white-eyes stand out as exceptional among vertebrates, and the broad geographic scope of this rapid radiation compels a reassessment of factors that influence diversification rates. White-eyes lack major ecomorphological differences among species, thus obviating purely adaptive explanations for the high diversification rate. Although the underlying drivers of most speciation in birds should be similar across lineages, namely isolation, variation, and fixation of novel traits (32), the rate at which these processes generate new species is likely determined by interplay between taxon-specific characteristics and the geography and earth history that a taxon experiences through time (33). Geography and earth history have clearly had a role in white-eye speciation, but the combination of extreme rate and varied geography suggests a prominent role for taxon-specific characteristics that enabled white-eyes to respond rapidly and persistently to the geographic drivers of diversification.
Factors capable of influencing high speciation rates in white-eyes include sociality, rapid morphological evolution, short generation time, generalist ecology, and dispersal ability. Social living birds, such as white-eyes, have increased potential for successful colonization by means of group dispersal, and can maintain viscous population genetic structure (34). Studies of recently founded white-eye populations underscore the potential for rapid differentiation; Zosterops lateralis populations founded within the last 200 years are morphologically distinguishable from source populations (35). Rapid change in Zosterops may in part be attributable to generation times that rank among the shortest in birds (SI Results). Also, some newly established white-eye populations have broader diets than parent populations; however, this generalist diet is not achieved through individuals becoming generalists, but rather different individuals specializing on different foods (36). Van Valen (37) demonstrated increased morphological variability in island bird populations that exploited broader niches compared with mainland populations. Examples of behavioral specialization independent of morphological variation are rare but include the Cocos Finch (Pinarolaxes inornata), part of the spectacular radiation of Galapagos Finches (38). Speculation about the potential role this phenomenon might have in the persistence and diversification of island radiations is enticing, but more data are needed.
Change in dispersal ability or propensity was an early explanation offered for the paradox of the great speciators (12), and, in light of our phylogenetic results, is now more persuasive. Consider that Z. griseotinctus colonized Long Island (≈300 km from source population) within the last 350 years after its volcanic defaunation (39), and Z. lateralis colonized New Zealand (≈1,500 km from source population) and Norfolk Island (≈700 km from source population) within the last 2 centuries (34). However, within this same clade, Zosterops rennellianus does not appear to have made the 20-km jump to Bellona, and Zosterops teteparius and Zosterops rendovae have not been recorded from their respective neighboring islands, a mere 2-km flight away (8). The requisite water-crossing ability needed by the ancestors of Z. rennellianus, Z. teteparius, and Z. rendovae to reach their islands, and that Z. lateralis and Z. griseotinctus still possess, must have been rapidly selected out of the modern populations of the former 3 species. A broad correlation between dispersal ability and clade diversity in birds (40) suggests that our results may be an extreme case of a more common trend. However, our results also imply that this correlation may emerge from an underlying driver of diversification, namely evolutionary shifts in dispersal ability.
Students of island biotas, going back to at least Sir Joseph Hooker and his discussions of Galapagos weeds (41), have often deduced evolutionary losses of dispersal ability in island populations of both animals and plants. The chapter entitled “They can't go home again” in Carlquist's book Island Life describes examples of insular endemic populations with reduced morphological adaptations for dispersal compared with mainland relatives presumed to resemble their ancestors [e.g., flightless insular birds and insects with reduced or no wings, closely related to mainland birds and insects with normal wings and flight, or insular dandelion relatives (family Asteraceae) with small heavy seed balls, related to mainland dandelions with light fluffy wind-born seed balls; see ref. 42]. Two recent studies illustrate how reduced dispersal ability can evolve very rapidly, within <1 decade and in only a few generations, when there is selection against dispersal: dandelion relatives evolving heavier and denser seed balls, in one case on islands off British Columbia (43, 44), in the other case on virtual islands of French urban trees planted in a sea of concrete sidewalks (45). More germane to the current discussion, birds (46) and butterflies (47) provide many examples of “behavioral flightlessness,” i.e., normal wings and power of flight, but behavioral reluctance to disperse, especially across water. The endemic insular Zosterops populations that we discuss in this article all exemplify behavioral flightlessness; none has reduced wings or reduced power of flight.
Of particular relevance to this article are evolutionary changes in dispersal ability during colonization cycles (10, 11, 48), as the balance shifts between evolutionary costs and benefits of dispersive behavior. Dispersal ability can be favored under conditions when habitat becomes available, and is opposed when that habitat is unavailable or becomes filled up. Such cycles tend to be autocatalytic in their initial stages, when suitable new habitat is first reached (49). Autocatalytic expansions and then retractions of human populations who leave written or oral testimony of their deeds and motives provide a compelling example. For example, Polynesians exploded over the Pacific Ocean in a burst of long-distance voyaging beginning approximately A.D. 1000, occupied every habitable Pacific island from Hawaii to New Zealand and Easter, and then abandoned long-distance voyaging when there were no more empty Pacific islands to colonize and, hence, no demographic payoff to warrant the hazards of voyaging (50). Similarly, Vikings exploded over the North Atlantic Ocean and Western Europe for a few centuries beginning in A.D. 793; then, they also abandoned long-distance voyaging when there were no more North Atlantic islands to colonize, and when mainland Europeans and Native Americans were able to resist Viking attacks (51). These Polynesian and Viking colonization cycles involved mainly cultural rather than genetic changes in dispersal, whereas the changes in our island Zosterops populations were presumably genetic, as known to be true for French urban dandelion relatives. However, the phenomenon of changes in the cost/benefit balance for dispersal during a colonization cycle is similar in both sets of cases.
Regardless of how white-eyes managed to disperse and speciate so rapidly across such a vast set of geographic settings, the rate of diversification and scale of dispersion recovered in our analyses identify Zosterops as an extreme case among birds, and more broadly across all vertebrates. The high rate of diversification raises the question of whether white-eyes, and other great speciators, are indeed categorically different from other taxa in their tempo of speciation, or lie at the extreme end of a continuous distribution of diversification rates. Future study of other widely distributed, diverse bird lineages such as Turdus, Pachycephala, and Myzomela may begin to enable comparative studies that can further assess the nature of great speciators.
More generally, in addition to expanding the geographic and taxonomic scope of hyperdiversifications and implicating a complex interaction between intrinsic and extrinsic drivers of rapid speciation, our data also provide broad support for an emerging synthesis of island biogeographic hypotheses (52). The pattern and tempo of diversification recovered for the white-eyes do not fit comfortably within any single diversification paradigm (e.g., dispersal, vicariance, equilibrium island biogeography, etc.) and underscore the importance of casting a broad net, in terms of taxonomy, geography, and theory, in modern diversification studies.
Materials and Methods
Taxon and Character Sampling.
Taxon sampling (Table S2) included individuals from 42 species of Zosteropidae and 23 species of Timaliidae. A forktail (Enicurus) was used as the outgroup taxon. For fresh tissue samples, genomic DNA was extracted, amplified, and sequenced by using standard protocols. We sequenced the entire second and third subunits of mitochondrial nicotinamide adenine dinucleotide dehydrogenase (ND2 and ND3) and the fifth intron of the nuclear gene transcription growth factor (TGFB2). DNA from 7 species was obtained from toe-pads clipped from museum skins. By using taxon-specific primers (Table S3) and amplifying short fragments of DNA, we sequenced portions of all 3 genes for these species. The computer program Sequencher 4.7 (Genecodes) was used to reconcile chromatograms of complementary fragments and align sequences across taxa.
Phylogenetic Analysis.
Maximum likelihood (ML) analyses were performed for each gene, as well as the combined data, by using PAUP*4.0b10 (53). Heuristic searches used TBR branch-swapping and 10 random taxon addition sequences. For each likelihood analysis, we used MrModeltest 2.2 (54) to determine the appropriate model of evolution and parameter estimates. We assessed support for nodes in the ML tree with nonparametric bootstrapping (500 replicates) in PhyML (55). MrBayes 3.1.2 (56) was used to estimate model parameters from the data and to evaluate support for specific relationships in the phylogeny. A mixed model approach was implemented to account for the potential difference in evolutionary model parameters between data partitions (codon positions and nuclear intron DNA). The program MrModeltest 2.2 (54) determined the appropriate evolutionary model for each partition. All parameters (except topology) were unlinked between partitions. We ran 2 sets of 4 Markov chains for 10 million generations. All samples before reaching stationarity were discarded. Markov chains were sampled every 1,000 generations. These subsamples, minus the burn-in generations, were used to create 50% majority-rule consensus trees.
Divergence Time Estimation.
The sequence data were tested for clocklike evolution with a χ2 test of likelihood ratios. The ML tree was converted to an ultrametric tree with penalized likelihood (57) in the program r8s (58), using the cross-validation procedure to determine the optimal smoothing parameter. An undated tree was initially produced for estimation of relative divergence times of various clades and for use in various diversification rate programs (see below). A bootstrapping procedure estimated SDs of node depths; 2 times the SD was added and subtracted from the mean to obtain a CI.
The fossil record provides no useful calibrations for white-eyes, and so geological calibrations were necessary. Instead, we used Solomon Island ages for calibration. We focused on the islands of the New Georgia Group, because they have endemic forms on each of the major islands. Within the New Georgia Group, the northern islands (Vellalavella, Kolombangara, and New Georgia) are volcanic in origin, but island ages can only be placed generally in the Plio-Pleistocene (59). The southern islands were formed from uplifted fore arc basement that was rapidly raised from abyssal depths due to subduction of the Australian plate. This process left deposits of limestone and marine sediment on each island, which can be used to estimate the time since the islands emerged from the ocean. Deposits on Ranongga exhibit an unconformity <300,000 years ago and then a rapid shallowing (60). The exact date of emergence is impossible to pinpoint, but the transition from abyssal depths after 300,000 years ago places bounds on the range. Holocene uplift rates of dated reef material can also be used to estimate island age. The rates from different localities over most of Ranongga range from 1 to 3 mm per year, with slightly higher values in the southern highlands (60). If a median, and constant, rate were assumed (2 mm per year), Ranongga (currently 854 m above sea level) would have emerged from the ocean ≈425,000 years ago.
To compensate for an unknown level of polymorphism in the ancestral population (26, 61), we estimated the genetic diversity in extant populations on the islands of Kolombangara, New Georgia, and Kohingo, which we then used as a proxy for ancestral polymorphism.
Diversification Rate.
The temporal distribution of speciation events was visualized with LTT plots by using the program Genie (62). Lineage accumulation was tested for departure from CR by calculating the gamma statistic (24). The CR test assumes full taxon sampling, a condition that is not met in this study. The effects of incomplete sampling were investigated with the Monte Carlo CR test (24), by simulating 10,000 trees in the program Phylogen (63). The trees were generated according to a CR, birth only model until 80 extant taxa existed (estimated number of species in Clade B). The trees were then pruned down to 39 extant taxa (the number of tips sampled in Clade B), gamma statistics calculated, and a critical value produced for a 1-tailed 95% CI. This value was then compared with the actual gamma statistic from Clade B. Then, we modified this simulation to examine the effects of overdispersed taxon sampling in Laser (64), by pruning out the youngest speciation events until 39 taxa remained, and using the distribution of resulting gamma statistics to test our empirical results (R code supplied by Dan Rabosky).
Measures of diversification rate that take into account internode distances (such as the Kendall-Moran estimator) also assume that the phylogeny is completely sampled. Because we had no objective criterion for placing missing taxa on the phylogeny, we assumed a Yule process and used a simple estimate of diversification rate [ln(N) − ln(N0)]/T that only uses initial diversity (N0 = 2), extant diversity (N), and time since origin (T). For extant diversity, we decided to use traditional classifications based on biological species (≈80 species in Clade B), which should give a conservative estimate of diversification rate.
Supplementary Material
Acknowledgments.
We thank A.T. Peterson, J. Cracraft, and G.F. Barrowclough for helpful comments on early drafts of the manuscript, D. Rabosky for assistance with diversification analyses, and numerous institutions and field workers that provided samples used in the study. This work was supported by National Geographic Society Grant CRE 7464-03 (to C.E.F.) and National Science Foundation Grant DEB-0743576 (to R.G.M.). This work was also supported by the University of Kansas General Research Fund (R.G.M.), the F. M. Chapman Memorial Fund, and the L. C. and L. J. Sanford Funds of the American Museum of Natural History (C.E.F.), and the University of Washington Burke Museum of Natural History (C.E.F. and C.E.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FJ460769–FJ460972).
This article contains supporting information online at www.pnas.org/cgi/content/full/0809861105/DCSupplemental.
References
- 1.Grant PR, Grant BR. Adaptive radiation of Darwin's finches. Am Sci. 2002;90:130–139. [Google Scholar]
- 2.Meyer A. Phylogenetic relationships and evolutionary processes in East African cichlid fishes. Trends Ecol Evol. 1993;8:279–284. doi: 10.1016/0169-5347(93)90255-N. [DOI] [PubMed] [Google Scholar]
- 3.Lovette IJ, Bermingham E, Rickleffs RE. Clade-specific morphological diversification and adaptive radiation in Hawaiian songbirds. Proc R Soc London B. 2002;269:37–42. doi: 10.1098/rspb.2001.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldwin BG, Sanderson MJ. Age and rate of diversification of the Hawaiian silversword alliance (Compositae) Proc Natl Acad Sci USA. 1998;95:9402–9406. doi: 10.1073/pnas.95.16.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes C, Eastwood R. Island radiation on a continental scale: Exceptional rates of plant diversification after uplift of the Andes. Proc Natl Acad Sci USA. 2006;103:10334–10339. doi: 10.1073/pnas.0601928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klak C, Reeves G, Hedderson T. Unmatched tempo of evolution in Southern African semi-desert ice plants. Nature. 2003;427:63–65. doi: 10.1038/nature02243. [DOI] [PubMed] [Google Scholar]
- 7.Wiens JJ, Engstrom TN, Chippendale PT. Rapid diversification, incomplete isolation, and the “speciation clock” in North American salamanders (genus Plethodon): Testing the hybrid swarm hypothesis of rapid radiation. Evolution. 2006;60:2585–2603. [PubMed] [Google Scholar]
- 8.Mayr E, Diamond J. The Birds of Northern Melanesia: Speciation, Ecology, and Biogeography. Oxford: Oxford Univ Press; 2001. [Google Scholar]
- 9.MacArthur RH, Wilson EO. An equilibrium theory of insular zoogeography. Evolution. 1963;17:373–387. [Google Scholar]
- 10.Wilson EO. Adaptive shift and dispersal in a tropical ant fauna. Evolution. 1959;13:122–144. [Google Scholar]
- 11.Wilson EO. The nature of the taxon cycle in the Melanesian ant fauna. Am Nat. 1961;95:169–193. [Google Scholar]
- 12.Diamond JM, Gilpin ME, Mayr E. Species-distance relation for birds of the Solomon Archipelago, and the paradox of the great speciators. Proc Natl Acad Sci USA. 1976;73:2160–2164. doi: 10.1073/pnas.73.6.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayr E. Systematics and the Origin of Species. New York: Columbia Univ Press; 1942. [Google Scholar]
- 14.Diamond J. Geographic variation in vocalisations of the white-eye superspecies Zosterops [griseotinctus] in the New Georgia Group. Emu. 1998;98:70–74. [Google Scholar]
- 15.Mayr E. Relationships among Indo-Australian Zosteropidae (Aves) Breviora. 1965;228:1–6. [Google Scholar]
- 16.Mees GF. An attempt at a natural classification of certain Zosteropidae of the Indo-Australian archipelago. Zool Meded. 1953;32:57–68. [Google Scholar]
- 17.Moreau RE. Variation in the western Zosteropidae (Aves) Bull Br Mus Nat Hist. 1957;4:309–433. [Google Scholar]
- 18.Dickinson EC, editor. The Howard and Moore Complete Checklist of the Birds of the World. Princeton: Princeton Univ Press; 2003. [Google Scholar]
- 19.Sibley CG, Monroe BL., Jr . Distribution and Taxonomy of Birds of the World. New Haven, CT: Yale Univ Press; 1990. [Google Scholar]
- 20.Marks BD, Hackett SJ, Capparella AP. Historical relationships among Neotropical lowland forest areas of endemism as determined by mitochondrial DNA sequence variation within the wedge-billed woodcreeper (Aves: Dendrocolaptidae: Glyphorynchus spirurus) Mol Phylogenet Evol. 2002;24:153–167. doi: 10.1016/s1055-7903(02)00233-6. [DOI] [PubMed] [Google Scholar]
- 21.Zou F, Lim HC, Marks BD, Moyle RG, Sheldon FH. Molecular phylogenetic analysis of the Grey-cheeked Fulvetta (Alcippe morrisonia) of China and Indochina: A case of remarkable genetic divergence in a “species.”. Mol Phylogenet Evol. 2007;44:165–174. doi: 10.1016/j.ympev.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Weir JT, Schluter D. The latitudinal gradient in recent speciation and extinction rates of birds and mammals. Science. 2007;315:1574–1576. doi: 10.1126/science.1135590. [DOI] [PubMed] [Google Scholar]
- 23.Price T. Speciation in Birds. Greenwood Village, CO: Roberts; 2008. [Google Scholar]
- 24.Pybus OG, Harvey PH. Testing macro-evolutionary models using incomplete molecular phylogenies. Proc R Soc London B. 2000;267:2267–2272. doi: 10.1098/rspb.2000.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabosky DL, Lovette IJ. Explosive evolutionary radiations: Decreasing speciation or increasing extinction through time? Evolution. 2008;62:1866–1875. doi: 10.1111/j.1558-5646.2008.00409.x. [DOI] [PubMed] [Google Scholar]
- 26.Arbogast BS, Edwards SV, Wakeley J, Beerli P, Slowinski JB. Estimating divergence times from molecular data on phylogenetic and population genetic timescales. Ann Rev Ecol Evol Syst. 2002;33:707–740. [Google Scholar]
- 27.Sanderson MJ, Doyle JA. Sources of error and confidence intervals in estimating the age of angiosperms from rbcL and 18S rDNA data. Am J Bot. 2001;88:1499–1516. [PubMed] [Google Scholar]
- 28.Warren BH, Bermingham E, Prys-Jones RP, Thébaud C. Immigration, species radiation and extinction in a highly diverse songbird lineage: White-eyes on Indian Ocean islands. Mol Ecol. 2006;15:3769–3786. doi: 10.1111/j.1365-294X.2006.03058.x. [DOI] [PubMed] [Google Scholar]
- 29.Arbogast BS, et al. The origin and diversification of Galapagos mockingbirds. Evolution. 2006;60:370–382. [PubMed] [Google Scholar]
- 30.Peterson AT, Navarro-Sigüenza AG. Alternate species concepts as bases for determining priority conservation areas. Cons Biol. 1999;13:427–431. [Google Scholar]
- 31.Phillimore AB, et al. Complex patterns of genetic and phenotypic divergence in an island bird and the consequences for delimiting conservation units. Mol Ecol. 2008;17:2839–2853. doi: 10.1111/j.1365-294X.2008.03794.x. [DOI] [PubMed] [Google Scholar]
- 32.Barraclough TG, Vogler AP. Detecting the geographical pattern of speciation from species-level phylogenies. Am Nat. 2000;155:419–434. doi: 10.1086/303332. [DOI] [PubMed] [Google Scholar]
- 33.Cracraft J. A nonequilibrium theory for the rate-control of speciation and extinction and the origin of macroevolutionary patterns. Syst Zool. 1982;31:348–365. [Google Scholar]
- 34.Clegg SM, et al. Genetic consequences of sequential founder events by an island-colonizing bird. Proc Natl Acad Sci USA. 2002;99:8127–8132. doi: 10.1073/pnas.102583399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clegg SM, et al. Microevolution in island forms: The roles of drift and directional selection in morphological divergence of a passerine bird. Evolution. 2002;56:2090–2099. doi: 10.1111/j.0014-3820.2002.tb00134.x. [DOI] [PubMed] [Google Scholar]
- 36.Scott SN, Clegg SM, Blomberg SP, Kikkawa J, Owens IPF. Morphological shifts in island-dwelling birds: The roles of generalist foraging and niche expansion. Evolution. 2003;57:2147–2156. doi: 10.1111/j.0014-3820.2003.tb00392.x. [DOI] [PubMed] [Google Scholar]
- 37.Van Valen L. Morphological variation and width of ecological niche. Am Nat. 1965;99:377–390. [Google Scholar]
- 38.Werner TK, Sherry TW. Behavioral specialization in Pinaroloxias inornata, the “Darwin's finch” of Cocos Island, Costa Rica. Proc Natl Acad Sci USA. 1987;84:5506–5510. doi: 10.1073/pnas.84.15.5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diamond J, Pimm SL, Gilpin ME, LeCroy M. Rapid evolution of character displacement in myzomelid honeyeaters. Am Nat. 1989;134:675–708. [Google Scholar]
- 40.Phillimore AB, Freckleton RP, Orme CDL Owens IPF. Ecology predicts large-scale patterns of phylogenetic diversification in birds. Am Nat. 2006;168:220–229. doi: 10.1086/505763. [DOI] [PubMed] [Google Scholar]
- 41.Hooker JD. On the vegetation of the Galapagos Archipelago. Trans Linn Soc London. 1849;20:235–262. [Google Scholar]
- 42.Carlquist S. Island Life. Garden City, NY: Natural History Press; 1965. [Google Scholar]
- 43.Cody ML, Overton JN. Short-term evolution of reduced dispersal in island plant populations. J Ecol. 1996;84:53–61. [Google Scholar]
- 44.Cody ML. Plants on Islands. Berkeley, CA: Univ of California Press; 2006. [Google Scholar]
- 45.Cheptou P-O, Carrue O, Rouifed S, Cantarel A. Rapid evolution of seed dispersal in an urban environment in the weed Crepis sancta. Proc Natl Acad Sci USA. 2008;105:3796–3799. doi: 10.1073/pnas.0708446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diamond J. Flightlessness and fear of flying in island species. Nature. 1981;293:507–508. [Google Scholar]
- 47.Holloway JD. The Lepidoptera of Norfolk Island. The Hague: Junk; 1977. [Google Scholar]
- 48.Ricklefs RE, Cox GW. Taxon cycles in the West Indian avifauna. Am Nat. 1972;106:195–219. [Google Scholar]
- 49.Keegan WF, Diamond J. Colonization of islands by humans: A biogeographical perspective. Adv Archaeol Meth Theor. 1987;10:49–92. [Google Scholar]
- 50.Kirch PB. On the Road of the Winds. Berkeley, CA: Univ of California; 2000. [Google Scholar]
- 51.Diamond J. Collapse: How Societies Choose to Fail or Succeed. New York: Viking/Penguin; 2005. [Google Scholar]
- 52.Heaney LR. Is a new paradigm emerging for oceanic island biogeography? J Biogeogr. 2007;34:753–757. [Google Scholar]
- 53.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony *and other methods. Sunderland, MA: Sinauer; 2002. [Google Scholar]
- 54.Nylander JAA. MrModeltest. Uppsala: Evolutionary Biology Centre; 2004. version 2. [Google Scholar]
- 55.Guindon S, Gascue O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 56.Ronquist F, Huelsenbeck JP. MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 57.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 58.Sanderson MJ. r8s: Inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 59.Cowley S, Mann P, Coffin MF, Shipley TH. Oligocene to Recent tectonic history of the Central Solomon intra-arc basin as determined from marine seismic reflection data and compilation of onland geology. Techtonophysics. 2004;389:267–307. [Google Scholar]
- 60.Mann P, Taylor FW, Lagoe M, Quarles A, Burr G. Accelerating late Quaternary uplift of the New Georgia Island Group (Solomon island arc) in response to subduction of the recently active Woodlark spreading center and Coleman seamount. Techtonophysics. 1998;295:259–306. [Google Scholar]
- 61.Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pybus OG, Rambaut A. GENIE: Estimating demographic history from molecular phylogenies. Bioinformatics. 2002;18:1404–1405. doi: 10.1093/bioinformatics/18.10.1404. [DOI] [PubMed] [Google Scholar]
- 63.Rambaut A. PhyloGen: Phylogenetic tree simulator package. Oxford: Oxford Univ Press; 2002. version 1.1. [Google Scholar]
- 64.Rabosky DL. LASER: A maximum likelihood toolkit for detecting temporal shifts in diversification rates. Evol Bioinform Online. 2006;2:257–260. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.