Abstract
Brown-rot fungi such as Postia placenta are common inhabitants of forest ecosystems and are also largely responsible for the destructive decay of wooden structures. Rapid depolymerization of cellulose is a distinguishing feature of brown-rot, but the biochemical mechanisms and underlying genetics are poorly understood. Systematic examination of the P. placenta genome, transcriptome, and secretome revealed unique extracellular enzyme systems, including an unusual repertoire of extracellular glycoside hydrolases. Genes encoding exocellobiohydrolases and cellulose-binding domains, typical of cellulolytic microbes, are absent in this efficient cellulose-degrading fungus. When P. placenta was grown in medium containing cellulose as sole carbon source, transcripts corresponding to many hemicellulases and to a single putative β-1–4 endoglucanase were expressed at high levels relative to glucose-grown cultures. These transcript profiles were confirmed by direct identification of peptides by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Also up-regulated during growth on cellulose medium were putative iron reductases, quinone reductase, and structurally divergent oxidases potentially involved in extracellular generation of Fe(II) and H2O2. These observations are consistent with a biodegradative role for Fenton chemistry in which Fe(II) and H2O2 react to form hydroxyl radicals, highly reactive oxidants capable of depolymerizing cellulose. The P. placenta genome resources provide unparalleled opportunities for investigating such unusual mechanisms of cellulose conversion. More broadly, the genome offers insight into the diversification of lignocellulose degrading mechanisms in fungi. Comparisons with the closely related white-rot fungus Phanerochaete chrysosporium support an evolutionary shift from white-rot to brown-rot during which the capacity for efficient depolymerization of lignin was lost.
Keywords: cellulose, fenton, lignin, cellulase, brown-rot
Lignocellulose in vascular plant cell walls is one of the largest sinks for fixed global carbon and is increasingly eyed as a potential feedstock in biofuels and new biomaterials portfolios (1). Relatively few organisms can efficiently convert the recalcitrant polymer blend in lignocellulose to monomeric components (2). The principal exceptions are basidiomycetes, which attack wood through 2 main decay types called white-rot and brown-rot. Wood-decaying basidiomycetes are essential contributors to carbon cycling in forest soils, and brown-rot fungi are additionally important because they are a major cause of failure in wooden structures. White-rot fungi degrade all components of plant cell walls, including cellulose, hemicellulose, and lignin. Although they cannot grow on lignin alone, they have the unique ability to degrade a large proportion of it completely to CO2 and H2O. This biodegradative strategy exposes the structural polysaccharides of plant cell walls, thus making them susceptible to hydrolysis by cellulases and hemicellulases. Brown-rot fungi employ a different approach; although they modify lignin extensively, the products remain in situ as a polymeric residue (3, 4). Given the incomplete ligninolysis that occurs during brown-rot, it remains unclear how these fungi gain access to plant cell wall polysaccharides. However, it seems probable that the 2 decay types share at least some mechanisms, because molecular phylogeny, morphological considerations, and substrate preference suggest that brown-rot fungi have repeatedly evolved from white-rot fungi (5). Indeed, the 2 major experimental organisms for studies of brown-rot, Postia placenta and Gloeophyllum trabeum, are distantly related species that represent independent origins of brown-rot (5). Any similarities in their decay mechanisms must represent either general mechanisms of wood decay common to white-rot and brown-rot species, or convergently evolved brown-rot mechanisms. Moreover, P. placenta is closely related to the model white-rot fungus, Phanerochaete chrysosporium, so comparisons between these species may provide insight into the mechanistic basis of transitions from white-rot to brown-rot.
White-rot fungi produce complex ligninolytic systems that are thought to depend in part on extracellular oxidative enzymes, especially peroxidases, laccases, and other oxidases. It remains an open question whether brown-rot fungi possess any of these ligninolytic components. White-rot fungi also secrete complete, synergistically acting cellulase systems that include both endo- and exo-acting enzymes. These exocellobiohydrolases and endoglucanases often share architectures that include separate catalytic and cellulose-binding domains. In contrast, relatively few cellulases have been described in brown-rot fungi (6). It has been long recognized that cellulose depolymerization appears to occur before the substrate porosity has increased enough to admit cellulases (7), and more recent studies (8) have shown that the amorphous regions within cellulose microfibrils are cleaved by P. placenta, resulting in rapid depolymerization but little weight loss. One possibility consistent with these observations is that brown-rot fungi attack cellulose with low molecular weight oxidants that act in conjunction with a limited set of relatively small cellulases.
The hydroxyl free radical, generated via Fenton chemistry (H2O2 + Fe2+ + H+ → H2O + Fe3+ + ·OH), has long been implicated as 1 of the small oxidants that contributes to polysaccharide depolymerization during brown-rot. Current models for hydroxyl radical participation have been reviewed (6) and typically involve generation of this highly reactive oxidant at or near the substrate. Key requirements for Fenton systems include mechanisms for extracellular H2O2 generation and for reduction of Fe3+ to Fe2+, which might be accomplished by extracellular fungal metabolites such as hydroquinones or by extracellular enzymes such as cellobiose dehydrogenase.
We report here analyses of the P. placenta draft genome together with transcript profiles and mass spectrometric identification of extracellular proteins. Consistent with a unique strategy for cellulose degradation, we observed a dramatic absence of conventional cellulase genes and most class II fungal peroxidases and a rich diversity of genes potentially supporting generation of extracellular reactive oxygen species.
Results
Carbohydrate Active Enzymes.
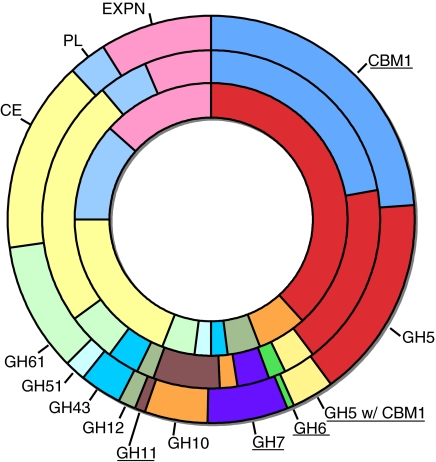
Given the well-known efficiency with which brown-rot fungi rapidly depolymerize and degrade cellulose, the P. placenta genome revealed remarkably few, if any, conventional cellulases. Of 17,173 proteins predicted in the dikaryotic genome, 242 unique genes encode potential carbohydrate-active enzymes [(9); http://www.cazy.org], of which 228 (94%) have at least 1 potential ortholog (BLASTP bit score ≥100) in P. chrysosporium. These putative CAZY genes include 144 glycoside hydrolases (GH), 10 carbohydrate esterases (CE), 75 glycosyltransferases (GT), 7 expansin-like proteins (EXPN), and 6 polysaccharide lyases (PL) [complete CAZY list within the National Center for Biotechnology Information Gene Expression Omnibus (GEO accession no. GSE12540 si_table_1.xls)]. In distinct contrast to all cellulolytic fungal aerobes, exocellobiohydrolases CBH2 (GH6) and CBH1 (GH7), and cellulose-binding endoglucanases are missing in the P. placenta genome (Fig. 1). Also absent are family 1 carbohydrate binding modules (CBM1). These highly conserved cellulose-binding domains are fused to functionally diverse CAZYs in a wide range of cellulolytic microbes. Surprisingly then, the repertoire of recognizable cellulolytic enzymes in P. placenta appears limited to just 2 potential endoglucanases (1,4-β-glucanases) and several β-glucosidases. In contrast to cellulolytic saprophytes (e.g., Trichoderma reesei, Aspergillus spp. or Neurospora crassa) and aggressive plant pathogens (e.g., Fusarium graminearum or Magnaporthe grisea), the overall number and distribution of GHs in P. placenta are similar to those in the ectomycorrhizal symbiont Laccaria bicolor, the human pathogen Cryptococcus neoformans, and the biotrophic plant pathogen Ustilago maydis (supporting information (SI) Table S1). Phylogenetic analyses of P. placenta and P. chrysosporium genomes indicate that the transition from white-rot to brown-rot has been associated with multiple independent reductions including the GH families 6, 7, 10, 11 and 61 (Figs. 1 and 2; Table S1). Thus, the transition from white-rot to brown-rot has been associated with multiple independent reductions in the GH families.
Fig. 1.
Distribution of various CAZymes in P. placenta (inner ring), T. reesei (middle ring), and P. chrysosporium (outer ring). Proteins not found in P. placenta are underlined. Comparisons with additional species are listed in Table S1. CBM1, family 1 carbohydrate binding modules; GH#, modules within individual glycoside hydrolase families; GH5 (CBM1), glycoside hydrolase family 5 modules associated with family 1 carbohydrate binding modules; GT, glycosyl transferases; CE, carbohydrate esterases; PL, polysaccharide lyases; EXPN, expansin-related proteins.
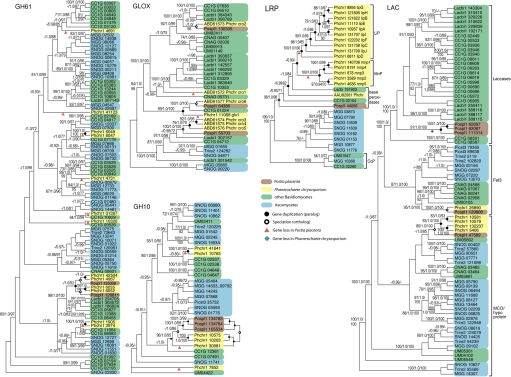
Fig. 2.
Phylogenies of glycoside hydrolase (GH 61, GH10), glyoxal oxidase/copper radical oxidase (GLOX), laccase (LAC) and related multicopper oxidase, and low redox peroxidase (LRP) and related class II fungal peroxidases from complete genomes of P. placenta (Pospl1), P. chrysosporium (Phchr1), C. cinerea (CC1G), L. bicolor (Lacbi1), C. neoformans (CNAG), U. maydis (UM), M. grisea (MGG), Stagonospora nodorum (SNOG), T. reesei (Trire2), and Pichia stipitis (Picst3). Datasets were assembled by using BLAST (with qUniProtKB query sequences Q5XXE5, O60206, P36218, Q00023, O14405, Q01772 and Q12718), with a cut-off value of E-06. Parsimony (PAUP* 4.0; 10,000 heuristic searches, 1000 bootstrap replicates), maximum likelihood (RAxML; 1000 bootstrap replicates, with models suggested by ProtTest), and Bayesian (MrBayes v3.1.2; 2 runs of 4 chains, 10 million generations each, with mixed protein models) support values are indicated in the order MP/PP/ML. Topologies shown are from Bayesian phylogenetic analyses. An alternative topology from parsimony analysis is shown for part of the GH10 phylogeny. Inferred gene losses, duplication events (paralogy), and speciation events (orthology) are indicated within Postia and Phanerochaete only.
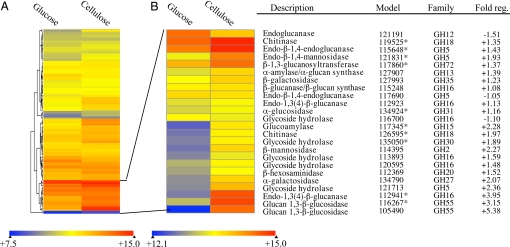
Microarrays representing 12,438 unique alleles were used to examine P. placenta transcript levels in basal salts medium containing either glucose or wood-derived microcrystalline cellulose as the sole carbon sources. In total, 290 gene models showed >2-fold transcript accumulation, and of these, 255 increased in cellulose medium and 35 increased in glucose medium (GEO accession no. GSE12540 si_table_3.xls). Transcripts of 99 GH-encoding genes significantly increased (P < 0.01) in the cellulose medium, and of these, 18 increased >2-fold (Fig. 3). Twenty-one GH transcripts significantly increased in the glucose-containing medium, but none exceeded a 2-fold change. In addition, shotgun liquid chromatography coupled tandem mass spectrometry (LC-MS/MS) identified 26 specific CAZYs in the extracellular fluid of P. placenta grown in basal salts supplemented with ball-milled aspen wood, microcrystalline cellulose, or cotton (Table S2). The CAZY genes expressed in cellulose included laminarinases, chitinases, and various hemicellulases (endoxylanases, β-xylosidases, L-α-arabinofuranosidases, endo-β-mannanases, and β-mannosidases). It is unclear whether any of these enzymes could directly attack crystalline cellulose.
Fig. 3.
Regulation of CAZY-encoding genes. (A) Expression profile of 144 glycoside hydrolase-encoding genes in media containing glucose versus microcrystalline cellulose as sole carbon sources. In B, a cluster of 24 of highly expressed genes is expanded and the color scale recalibrated to illustrate differences in transcript accumulation. Expression ratios were derived from comparisons of glucose-grown versus cellulose-grown mycelia. Analysis is based on 3 biological replicates per culture medium. Quantile normalization and robust multiarray averaging (RMA) were applied to the entire dataset. ANOVA showed 120 GH-encoding genes, including all 24 above, were significantly regulated (P < 0.01). Reciprocals of ratios <1.0 are multiplied by −1. Asterisks indicate proteins identified by LC-MS/MS. A detailed listing of CAZYs with statistical analyses of expression data are presented in SI Appendix and GEO accession GSE12540 si_table_1.xls.
Extracellular H2O2 Generation.
Gene models potentially supporting Fenton chemistry through the generation of extracellular H2O2 include copper radical oxidases and GMC oxidoreductases (Table S3). Results summarized here focus on those genes with expression patterns that are consistent with a role in cellulose depolymerization, and detailed information for all genes is available within GEO accession no. GSE12540 si_table_6.xls.
On the basis of overall sequence similarity to P. chrysosporium glyoxal oxidase (GLOX) and conservation of catalytic residues (10), 3 P. placenta models were identified as copper radical oxidases (CROs). GLOX is 1 of 7 CROs in P. chrysosporium and physiologically coupled to lignin peroxidase (LiP) via H2O2 generation. Of particular relevance to potential Fenton systems, CRO genes encoding proteins Ppl56703 and Ppl130305 are up-regulated in microcrystalline cellulose, and Ppl56703 peptides were detected in aspen-grown cultures. The P. chrysosporium cro1 and glx1 genes are not closely related, and they do not have orthologs in P. placenta, which suggests that there have been 2 independent losses of these CRO lineages in Postia (Fig. 2). Ppl56703 is orthologous to the cro3-4-5 lineage in P. chrysosporium, which therefore represents a Phanerochaete-specific expansion of the gene family. As in the case of the GHs, evolution of brown-rot is associated with a reduced diversity of CROs.
Catalytically distinct from CROs, GMC oxidoreductases (InterPro IPR000172) include various alcohol and sugar oxidases. Among the former, P. placenta protein model Ppl118723 is similar to G. trabeum methanol oxidase (GenBank DQ835989) (> 85% amino acid identity over full length). Recent immunolocalization studies strongly implicate the G. trabeum alcohol oxidase as a source of H2O2 (11) to support Fenton chemistry. Suggesting a similar role in P. placenta, microarray analysis revealed high transcript levels and a sharp increase in transcription of the gene encoding Ppl118723 in cellulose-grown cultures relative to noncellulolytic cultures. Comparatively high transcript levels in cellulose- and glucose-grown cultures were also observed for genes encoding Ppl128830 and Ppl108489, models tentatively identified as glucose-1-oxidases based on conserved key residues (12). Peptides corresponding to these putative gox genes were detected in extracellular filtrates (GEO accession no. GSE12540 si_table_6.xls and si_table_11.xls). Aryl-alcohol oxidase, an extracellular GMC oxidoreductase cooperating with aryl-alcohol dehydrogenases for continuous peroxide supply in some white-rot fungi (12) does not seem to be involved in cellulose attack by P. placenta because the corresponding models were not or only slightly up-regulated. Another extracellular GMC oxidoreductase, pyranose-2-oxidase, has been implicated in lignocellulose degradation in P. chrysosporium (13), but no orthologs were detected in P. placenta.
Iron Reduction and Homeostasis.
Protein model Ppl124517 was identified as a putative quinone reductase (QRD). In the brown-rot fungus G. trabeum, a QRD may drive extracellular Fenton systems via redox cycling of secreted fungal quinones (6). Transcription of the P. placenta QRD gene was significantly up-regulated in cellulose medium (GEO accession no. GSE12540 si_table_3.xls), which is consistent with a role for cellulolytic Fenton chemistry involving quinone redox-cycling. In this connection, up-regulation of the genes encoding phenylalanine ammonia lyase (Ppl112824) and a putative quinate transporter (Ppl44553) may also be relevant by virtue of their respective roles in the biosynthesis and transport of essential quinones.
In addition to hydroquinone-based iron reduction systems, low molecular weight glycoproteins (GLPs) that can act as iron reductases have been hypothesized as components of extracellular Fenton systems in G. trabeum and P. chrysosporium (14). Four P. placenta models show significant similarity (>48% amino acid identity) to P. chrysosporium glp1 and glp2, and the gene encoding Ppl128974 is significantly up-regulated on microcrystalline cellulose medium (GEO accession no. GSE12540 si_table_7.xls). The sequence corresponding to another fungal protein implicated in Fe3+ reduction, CDH (6), appears to be absent in P. placenta.
In addition to its pivotal role in a wide range of cellular processes, iron homeostasis must play a central role in modulating a functioning Fenton system. The P. placenta genome features numerous genes potentially involved in iron transport and redox state (GEO accession no. GSE12540 si_table_7.xls). In addition to 7 ferric reductases, 2 iron permease genes were identified one of which lies immediately downstream from a canonical yeast ferroxidase ortholog (Fet3). Transcripts of these adjacent genes were among the most highly up-regulated in cellulose medium (GEO accession no. GSE12540 si_table_3.xls).
Modification of Lignin and Other Aromatic Compounds.
Genes encoding the class II secretory peroxidases lignin peroxidase (LiP), manganese peroxidase (MnP) and versatile peroxidase (VP) were not detected in the P. placenta genome (Table S3). Peroxidase model Ppl44056 lacks residues involved in Mn2+ binding and oxidation of aromatic compounds (15), and superimposition of protein models strongly suggests that Ppl44056 is a low redox potential peroxidase (Fig. S1). Consistent with this structural evidence, phylogenetic analyses of class II peroxidase genes from Postia, Phanerochaete, and other fungal genomes suggest that Ppl44056 is not closely related to LiP and MnP, but is part of an assemblage of “basal peroxidases” that includes the novel peroxidase (NoP) of P. chrysosporium and peroxidases from Coprinopsis cinerea and L. bicolor (Fig. 2) (16). The backbone of the class II peroxidase phylogeny is not strongly supported, but analyses of broadly sampled datasets (16) suggest that the LiP and MnP gene lineages of P. chrysosporium were independently derived from the basal peroxidases before the divergence of Postia and Phanerochaete. If so, then the absence of LiP and MnP in P. placenta may reflect additional instances of gene loss.
Laccases have been suggested to play a role in lignin modification by white-rot fungi but have not previously been demonstrated in brown-rot fungi. The precise role of these enzymes remains uncertain, but numerous studies have demonstrated laccase-catalyzed oxidation of phenolic and nonphenolic lignin model substrates, particularly in the presence of low molecular weight mediators. The results from P. placenta belie the usual picture of brown-rot in that models Ppl62097 and Ppl111314 are likely laccases sensu stricto (17) (Fig. 2). Transcript levels of the genes encoding Ppl89382 and Ppl111314 appear differentially regulated by decreasing slightly (−1.08-fold) and increasing (+2.29-fold), respectively, on cellulose medium relative to glucose medium (GEO accession no.GSE12540 si_table_7.xls). These enzymes could contribute to hydroxyl radical generation by oxidizing hydroquinones as described in ref. 18. Interestingly, laccase genes are absent from the genome of P. chrysosporium (19), suggesting that laccase (sensu stricto) is not a core component of fungal wood decay mechanisms and is certainly not essential for white-rot.
Other up-regulated genes potentially involved in quinone redox-cycling, and oxidation of lignin derived products include those encoding “polyphenol oxidase” (Ppl114245), i.e., tyrosinase or catechol oxidase related to typical laccases and various oxidoreductases of uncertain function (Ppl107061, Ppl28683, Ppl34850, Ppl61437, Ppl24981) (GEO accesssion no. GSE12540 si_table_3.xls).
Oxalate Metabolism.
In addition to pH effects on a wide range of enzymes, extracellular accumulation of oxalate by P. placenta may affect ferric iron availability and thereby impact hydroxyl radical formation (20; reviewed in ref. 6). A metabolic shunt between the citric acid and glyoxylate cycles is central to oxalic-acid accumulation by the brown-rot fungus Fomitopsis palustris (21). Analysis of the P. placenta genome demonstrates a functional glyoxylate shunt and substantially extends our understanding of the number, structure, and transcription of key genes (Fig. S2; SI Appendix; GEO accession no. GSE12540 si_table_8.xls).
Cytochrome P450 Monooxygenases.
P450s have various roles in secondary metabolism and are thought to be involved in biodegradation of lignin and various xenobiotic compounds. The P. placenta genome features an impressive set of 236 P450 genes (SI Appendix, GEO acccesion no. GSE12540 si_figure_3.jpg), compared with 149 in P. chrysosporium, and expansions of certain families (CYP64, CYP503, CYP5031 and CYP617) were observed. Genes encoding Ppl110015 (CYP53) and Ppl128850 (CYP503) were significantly up-regulated in cellulose medium (GEO accession no. GSE12540 si_table_3.xls). The former is highly conserved in fungi and thought to catalyze benzoate hydroxylation.
Other.
The genome was systematically examined for genes involved in oxidative phosphorylation, stress-related genes, signal transduction, and regulatory genes, particularly those potentially controlling glycoside hydrolase expression and mating type (complete listings and analysis in SI Appendix and Figs. S3–S5).
Discussion
Analysis of the P. placenta genome elucidated a repertoire of genes and expression patterns distinct from those of other known cellulose-degrading microbes. The overall number of CAZY-encoding genes in P. placenta, 242, is not particularly low, and among these, the number of glycosyl transferases, 75, is fairly typical. However, the genome completely lacks cellulose-binding domains and the number of GHs is relatively low owing in part to the paucity of cellulases. No exocellobiohydrolases and only 2 potential β-1,4 endoglucanase genes were identified. One putative EG (Ppl115648) is expressed at relatively high levels.
Comparisons with genomes of other cellulolytic microbes reveal a strikingly distinct set of glycoside hydrolase genes in P. placenta. Among aerobes, only the cellulolytic gliding bacterium Cytophaga hutchinsonii lacks exocellobiohydrolases and endoglucanases fused to cellulose-binding domains (22). The precise mechanism used by C. hutchinsonii is somewhat mysterious, but it has been suggested that cellulose chains are peeled away from the polymer and transported into the periplasm (23). There, nonprocessive endoglucanases might readily degrade the cellulose. Such a mechanism seems unlikely in P. placenta because all evidence suggests that cellulose depolymerization by brown-rot fungi occurs at a distance from the advancing hyphae. In contrast, C. hutchinsonii is in direct contact with cellulose.
Possibly, the CBM-less β-1–4-endoglucanase Ppl115648, which is clearly expressed in cellulose-containing media (Fig. 3), may possess processive activity that enables it to liberate the cellobiose that β-glucosidases then hydrolyze to assimilable glucose. Indeed, the accumulation of putative β-glucosidase transcripts and the corresponding proteins that we observed are consistent with the availability of cellobiose in our cultures. Precedents for crystalline cellulose hydrolysis by β-1,4-endoglucanases within GH family 5 have been reported (24, 25), but it seems unlikely that the Ppl115648 endoglucanase alone can account for the efficient cellulose depolymerization by P. placenta. Other GHs and/or hypothetical proteins, perhaps some of those expressed in microcrystalline cellulose cultures (Fig. 3; GEO accession no. GSE12540 si_table_1.xls), may be necessary for the complete breakdown of cellulose. Heterologous expression of P. placenta GH-encoding genes followed by biochemical characterization of the purified proteins may resolve this question.
Many investigations of white-rot and brown-rot mechanisms have implicated the participation of low molecular weight oxidants, particularly Fenton-generated hydroxyl radicals. As recently reviewed (6), 3 somewhat overlapping mechanisms of oxidative degradation have been advanced. One view emphasizes the importance of CDH. In the case of P. placenta, CDH is absent. Another view invokes the role of low molecular weight glycopeptides that catalyze extracellular iron reduction. Initially identified in P. chrysosporium (14), potential orthologs of these glycopeptide-encoding genes were identified in P. placenta, and in 1 case, increased transcript levels were observed in cellulose medium. Accordingly, a role for these glycoproteins in a P. placenta Fenton system is possible. The third mechanism involves extracellular quinone redox cycling (26). Evidence supporting this system includes cellulose induction of genes encoding QRD, quinate transporter, phenylalanine ammonia lyase, and laccase. However, the importance of hydroquinone-driven Fenton chemistry in P. placenta remains unclear because this fungus secretes high levels of oxalate (27), and Fe3+-oxalate chelates are poorly reducible by hydroquinones (28).
The elevated expression in cellulose medium of Fet3 and Ftr1, components of the high-affinity iron-uptake system, may be at least partially explained by such chelates. Whereas cellulose itself may sequester Fe3+ (29), the generation of Fe3+-oxalate and potentially other redox active iron-chelates might also contribute to lower the effective concentration of bioavailable iron that is accessible to the organism. Thus, cellulolytic conditions might turn on the high-affinity iron-uptake system to ensure proper levels of intracellular iron.
Also compatible with Fenton mechanisms is the observed cellulose-induced expression of structurally divergent oxidases (e.g., copper radical oxidases, glucose-1-oxidases, and methanol oxidases) and putative iron reductases. Given the significant number of secreted hemicellulases observed, wood decay by P. placenta likely involves attack by oxidative and hydrolytic mechanisms. Elevated hemicellulase expression may reflect increased substrate exposure and availability, relative to cellulose and lignin, especially early in the decay process. Products of the hydrolytic attack could in turn serve as candidate substrates for copper radical oxidases and GMC oxidoreductases, thereby generating extracellular H2O2. Similarly, methanol released via demethoxylation of lignin (3, 4) may play an important role in H2O2 generation as a substrate for methanol oxidase. Such a role is consistent with our observed expression patterns and with previous investigations with G. trabeum (11). Of course hydroxyl radical may also play an important role early in decay, and it has been demonstrated to preferentially attack hemicellulose in wood (30). Interestingly, ·OH attack on cellulose oxidizes chain ends (31) and the depolymerized material becomes less amenable to cellulase action (30), providing a plausible explanation for the lack of exocellobiohydrolase genes in this fungus.
Comparison of the P. placenta and P. chrysosporium genomes indicates that the derivation of brown-rot is characterized largely by the contraction or loss of multiple gene families that are thought to be important in typical white-rot, such as cellulases, LiPs, MnPs, CROs, CDH, and pyranose-2-oxidase. This general pattern of simplification is consistent with the view (32) that brown-rot fungi, having evolved novel mechanisms for initiating cellulose depolymerization, have cast off much of the energetically costly lignocellulose-degrading apparatus that is retained in white-rot fungi, such as P. chrysosporium.
Materials and Methods
Genome Sequencing, Assembly, and Annotation.
A pure whole-genome shotgun approach was used to sequence P. placenta strain MAD-698-R (USDA, Forest Mycology Center, Madison, WI). The 7.2X coverage assembly was produced from sequenced paired reads by using JAZZ assembler. By using an array of gene predictors in the JGI annotation pipeline, a total of the 17,173 gene models were predicted and annotated for this dikaryotic fungus. Predicted genes, supporting evidence, annotations, and analyses are available through interactive visualization and analysis tools from the JGI genome portal www.jgi.doe.gov/Postia. Detail regarding the assembly, repetitive elements, ESTs and annotation, are provided separately (SI Appendix).
Mass Spectrometry.
Soluble extracellular protein was concentrated from shake cultures containing ball-milled aspen, microcrystalline cellulose (Avicel), or de-waxed cotton as previously reported (33). Sample preparation and LC-MS/MS analysis were performed as described (www.biotech.wisc.edu/ServicesResearch/MassSpec/ingel.htm). Peptides were identified by using a Mascot search engine (Matrix Science) against protein sequences of 17,173 predicted gene models described above. Complete listings of CAZYs and oxidative enzymes, including peptide sequences and scores, are provided in SI Appendix and GEO accession no. GSE12540 si_table_11.xls.
Expression Microarrays.
Roche NimbleGen arrays were designed to assess expression of 12,438 genes during growth on microcrystalline cellulose or on glucose as sole carbon sources (Fig. S6). The corresponding set of coding regions was manually annotated to include only the ‘best allelic model’ among CAZY-encoding genes (GEO accession no. GSE12540 si_table_1.xls). Methods are detailed in SI Appendix and all data deposited under GEO accession no. GSE12540.
Supplementary Material
Acknowledgments.
We thank Sally Ralph (FPL) for preparation of ball-milled aspen. This work was supported by the U.S. Department of Energy's Office of Science, Biological and Environmental Research Program, and University of California, Lawrence Berkeley National Laboratory Contract DE-AC02–05CH11231; Lawrence Livermore National Laboratory Contract DE-AC52–07NA27344; Los Alamos National Laboratory Contract DE-AC02–06NA25396; University of Wisconsin Grant DE-FG02–87ER13712; Forest Products Laboratory, U.S. Department of Agriculture, Cooperative State Research, Education, and Extension Services Grant 2007–35504-18257; National Institutes of Health Grant GM060201 (to University of New Mexico); Centro de Investigaciones Biológicas (Madrid) EU-project NMP2–2006-026456; Ministry of Education Czech Republic Grant LC06066.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The annotated genome is available on an interactive web portal at http://www.jgi.doe.gov/Postia. The sequences reported in this paper have been deposited in the GenBank database (accession nos. ABWF00000000 and FL595400-FL633513). The model and microarray results reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE12540).
This article contains supporting information online at www.pnas.org/cgi/content/full/0809575106/DCSupplemental.
References
- 1.U.S. DOE. Breaking the Biological Barriers to Cellulosic Ethanol: A Joint Research Agenda. Washington, DC: U.S. Department of Energy; 2006. DOE/SC-0095 (DOE) [Google Scholar]
- 2.Eriksson K-EL, Blanchette RA, Ander P. Microbial and Enzymatic Degradation of Wood and Wood Components. Berlin: Springer-Verlag; 1990. [Google Scholar]
- 3.Niemenmaa O, Uusi-Rauva A, Hatakka A. Demethoxylation of [O(14)CH (3)]-labelled lignin model compounds by the brown-rot fungi Gloeophyllum trabeum and Poria (Postia) placenta. Biodegradation. 2007;19:555–565. doi: 10.1007/s10532-007-9161-3. [DOI] [PubMed] [Google Scholar]
- 4.Yelle DJ, Ralph J, Lu F, Hammel KE. Evidence for cleavage of lignin by a brown rot basidiomycete. Environ Microbiol. 2008;10:1844–1849. doi: 10.1111/j.1462-2920.2008.01605.x. [DOI] [PubMed] [Google Scholar]
- 5.Hibbett DS, Donoghue MJ. Analysis of character correlations among wood decay mechanisms, mating systems, and substrate ranges in homobasidiomycetes. Syst Biol. 2001;50(2):215–242. [PubMed] [Google Scholar]
- 6.Baldrian P, Valaskova V. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol Rev. 2008;32(3):501–521. doi: 10.1111/j.1574-6976.2008.00106.x. [DOI] [PubMed] [Google Scholar]
- 7.Cowling EB, Brown W. Structural features of cellulosic materials in relation to enzymatic hydrolysis. In: Hajny GJ, Reese ET, editors. Cellulases and Their Applications. Washington, DC: American Chemical Society; 1969. pp. 152–187. American Chemical Society Advances in Chemistry Series 95. [Google Scholar]
- 8.Kleman-Leyer K, Kirk TK. Changes in the molecular size distribution of cellulose during attack by white-rot and brown-rot fungi. Appl Environ Microbiol. 1992;58:1266–1270. doi: 10.1128/aem.58.4.1266-1270.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280(Pt 2):309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whittaker MM, Kersten PJ, Cullen D, Whittaker JW. Identification of catalytic residues in glyoxal oxidase by targeted mutagenesis. J Biol Chem. 1999;274(51):36226–36232. doi: 10.1074/jbc.274.51.36226. [DOI] [PubMed] [Google Scholar]
- 11.Daniel G, et al. Characteristics of Gloeophyllum trabeum alcohol oxidase, an extracellular source of H2O2 in brown rot decay of wood. Appl Environ Microbiol. 2007;73(19):6241–6253. doi: 10.1128/AEM.00977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varela E, Martinez JM, Martinez AT. Aryl-alcohol oxidase protein sequence: A comparison with glucose oxidase and other FAD oxidoreductases. Biochim Biophys Acta. 2000;1481(1):202–208. doi: 10.1016/s0167-4838(00)00127-8. [DOI] [PubMed] [Google Scholar]
- 13.de Koker TH, Mozuch MD, Cullen D, Gaskell J, Kersten PJ. Pyranose 2-oxidase from Phanerochaete chrysosporium: Isolation from solid substrate, protein purification, and characterization of gene structure and regulation. Appl Environ Microbiol. 2004;70:5794–5800. doi: 10.1128/AEM.70.10.5794-5800.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka H, et al. Characterization of a hydroxyl-radical-producing glycoprotein and its presumptive genes from the white-rot basidiomycete Phanerochaete chrysosporium. J Biotechnol. 2007;128(3):500–511. doi: 10.1016/j.jbiotec.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 15.Martinez AT. Molecular biology and structure-function of lignin-degrading heme peroxidases. Enzyme Microb Technol. 2002;30:425–444. [Google Scholar]
- 16.Morgenstern I, Klopman S, Hibbett DS. Molecular evolution and diversity of lignin degrading heme peroxidases in the Agaricomycetes. J Mol Evol. 2008;66(3):243–257. doi: 10.1007/s00239-008-9079-3. [DOI] [PubMed] [Google Scholar]
- 17.Hoegger PJ, Kilaru S, James TY, Thacker JR, Kües U. Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 2006;273(10):2308–2326. doi: 10.1111/j.1742-4658.2006.05247.x. [DOI] [PubMed] [Google Scholar]
- 18.Guillen F, Gomez-Toribio V, Martinez MJ, Martinez AT. Production of hydroxyl radical by the synergistic action of fungal laccase and aryl alcohol oxidase. Arch Biochem Biophys. 2000;383(1):142–147. doi: 10.1006/abbi.2000.2053. [DOI] [PubMed] [Google Scholar]
- 19.Martinez D, et al. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol. 2004;22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- 20.Varela E, Tien M. Effect of pH and oxalate on hydroquinone-derived hydroxyl radical formation during brown rot wood degradation. Appl Environ Microbiol. 2003;69(10):6025–6031. doi: 10.1128/AEM.69.10.6025-6031.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munir E, Yoon JJ, Tokimatsu T, Hattori T, Shimada M. A physiological role for oxalic acid biosynthesis in the wood-rotting basidiomycete Fomitopsis palustris. Proc Natl Acad Sci USA. 2001;98(20):11126–11130. doi: 10.1073/pnas.191389598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie G, et al. Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii. Appl Environ Microbiol. 2007;73(11):3536–3546. doi: 10.1128/AEM.00225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson DB. Three microbial strategies for plant cell wall degradation. Ann N Y Acad Sci. 2008;1125:289–297. doi: 10.1196/annals.1419.026. [DOI] [PubMed] [Google Scholar]
- 24.McCarter SL, et al. Exploration of cellulose surface-binding properties of Acidothermus cellulolyticus Cel5A by site-specific mutagenesis. Appl Biochem Biotechnol. 2002;98–100:273–287. doi: 10.1385/abab:98-100:1-9:273. [DOI] [PubMed] [Google Scholar]
- 25.Tsai CF, Qiu X, Liu JH. A comparative analysis of two cDNA clones of the cellulase gene family from anaerobic fungus Piromyces rhizinflata. Anaerobe. 2003;9(3):131–140. doi: 10.1016/S1075-9964(03)00087-8. [DOI] [PubMed] [Google Scholar]
- 26.Cohen R, Suzuki MR, Hammel KE. Differential stress-induced regulation of two quinone reductases in the brown rot basidiomycete Gloeophyllum trabeum. Appl Environ Microbiol. 2004;70(1):324–331. doi: 10.1128/AEM.70.1.324-331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaneko S, Yoshitake K, Itakura S, Tanaka H, Enoki A. Relationship between production of hydroxyl radicals and degradation of wood, crystalline cellulose, and lignin-related compound or accumulation of oxalic acid in cultures of brown-rot fungi. J Wood Sci. 2005;51:262–269. [Google Scholar]
- 28.Jensen KA, Jr, Houtman CJ, Ryan ZC, Hammel KE. Pathways for extracellular Fenton chemistry in the brown rot basidiomycete Gloeophyllum trabeum. Appl Environ Microbiol. 2001;67(6):2705–2711. doi: 10.1128/AEM.67.6.2705-2711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu G, Goodell B. Mechanisms of wood degradation by brown-rot fungi: Chelator-mediated cellulose degradation and binding of iron by cellulose. J Biotechnol. 2001;87(1):43–57. doi: 10.1016/s0168-1656(00)00430-2. [DOI] [PubMed] [Google Scholar]
- 30.Ratto M, Ritschkoff A, Viikari L. The effect of oxidative pretreatment on cellulose degradation by Poria placenta and Trichoderma reesei. Appl Microbiol Biotechnol. 1997;48:53–57. [Google Scholar]
- 31.Kirk TK, Ibach R, Mozuch MD, Conner AH, Highley TL. Characteristics of cotton cellulose depolymerized by a brown-rot fungus, by acid, or by chemical oxidants. Holzforschung. 1991;45:239–244. [Google Scholar]
- 32.Worrall JJ, Anagnost SE, Zabel RA. Comparison of wood decay among diverse lignicolous fungi. Mycologia. 1997;89:199–219. [Google Scholar]
- 33.Vanden Wymelenberg A, et al. Computational analysis of the Phanerochaete chrysosporium v2.0 genome database and mass spectrometry identification of peptides in ligninolytic cultures reveals complex mixtures of secreted proteins. Fungal Genet Biol. 2006;43:343–356. doi: 10.1016/j.fgb.2006.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.