Abstract
The lipid A portion of lipopolysaccharide, the major component of the outer leaflet of the outer membrane of Gram-negative bacteria, is toxic to humans. Modification of lipid A by enzymes often reduces its toxicity. The outer-membrane protein LpxR from Salmonella typhimurium is a lipid A-modifying enzyme. It removes the 3′-acyloxyacyl moiety of the lipid A portion of lipopolysaccharide in a Ca2+-dependent manner. Here, we present the crystal structure of S. typhimurium LpxR, crystallized in the presence of zinc ions. The structure, a 12-stranded β-barrel, reveals that the active site is located between the barrel wall and an α-helix formed by an extracellular loop. Based on site-directed mutagenesis and modeling of a substrate on the active site, we propose a catalytic mechanism similar to that of phospholipase A2, in which a Ca2+ forms the oxyanion hole and a histidine activates a water molecule (or a cascade of two water molecules) that subsequently attacks the carbonyl oxygen of the scissile bond.
Keywords: crystal structure, endotoxin, lipopolysaccharide, outer-membrane protein, phospholipase
Gram-negative bacteria possess an outer membrane that functions as a permeability barrier and protects the bacteria against harmful components from the environment. It is an asymmetric bilayer with phospholipids and lipopolysaccharide (LPS) in the inner and outer leaflet, respectively. LPS usually consists of three covalently linked domains: lipid A, the core, and the O-antigen (1). Lipid A forms the hydrophobic membrane anchor and is responsible for the endotoxic activity of LPS. Lipid A of Salmonella typhimurium and Escherichia coli is a β-1′,6-linked disaccharide of glucosamine, phosphorylated at the 1 and 4′ positions and acylated at the 2, 3, 2′, and 3′ positions with R-3-hydroxymyristic acid (3-OH C14:0) (Fig. 1A). The hydroxyl groups of the 2′- and 3′-linked fatty acyl chains are further esterified with laurate (C12:0) and myristate (C14:0), respectively. Lipid A bearing two phosphate groups and six acyl chains that are 12–14 carbons in length are the most efficient in activating the innate immune system. However, a number of pathogenic bacteria have been shown to synthesize LPS with modified lipid A anchors that show reduced endotoxicity (2).
Fig. 1.
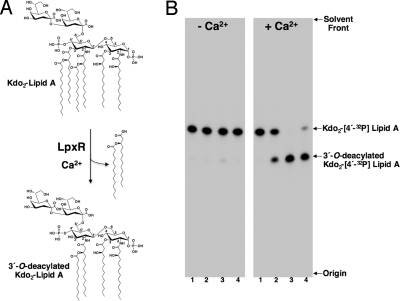
LpxR-catalzyed deacylation of Kdo2–lipid A. (A) In the presence of Ca2+, the outer-membrane enzyme LpxR catalyzes the hydrolysis of the ester linkage at the 3′ position of Kdo2–lipid A, releasing the intact 3′-acyloxyacyl group. (B) Enzymatic activity of purified LpxR. Purified and refolded LpxR was assayed for 3′-O-deacylase activity by using Kdo2–[4′-32P]lipid A as the substrate. Membranes from E. coli K-12 strain HMS174(DE3)/pLpxR1 (0.001 mg/mL) served as the positive control (lane 2). Purified LpxR at a concentration of 0.001 and 0.0001 mg/mL served as the protein source in assays spotted in lanes 3 and 4, respectively. The no enzyme control is shown in lane 1. Assays were carried out for 30 min at 30 °C with 2.5 μM Kdo2–[4′-32P]lipid A either in the presence or absence of 5 mM CaCl2. The reaction products were separated by TLC and detected with PhosphorImager analysis.
Several Gram-negative bacteria, including S. typhimurium, covalently modify their LPS in response to environmental stimuli in a regulated fashion [supporting information (SI) Fig. S1]. Three of these LPS-modifying enzymes, PagP, PagL, and LpxR, are embedded in the outer membrane. PagP catalyzes the addition of a phospholipid-derived palmitate chain to the hydroxyl of the R-3-hydroxymyristate chain at the 2 position of lipid A (3, 4). PagL catalyzes the removal of a single R-3-hydroxymyristate chain at position 3 of lipid A (5). LpxR, which has been identified recently, is a 3′-O-deacylase and removes the complete 3′-acyloxyacyl moiety of lipid A (6) (Fig. 1A). Unlike PagP and PagL, expression of LpxR is not under the control of the two-component regulatory system PhoP/PhoQ. LpxR activity appears to be Ca2+-dependent. Orthologs of S. typhimurium LpxR are found in the genomes of several other Gram-negative pathogens, including Vibrio cholerae, Yersinia enterocolitica, E. coli O157:H7 and Helicobacter pylori. Interestingly, Salmonella LpxR remains dormant within the outer membrane under diverse growth conditions. However, introduction of an lpxR-encoding plasmid into E. coli K-12 or S. typhimurium does lead to limited lipid A deacylation in whole cells (6). How LpxR activity is regulated within the outer membrane remains unknown but is of great interest because removal of the 3′-O-linked fatty acyl chains would substantially alter recognition of the LPS by the innate immune system. In H. pylori, which produces a tetraacylated lipid A of low immunological activity, LpxR remains in an activated state within the outer membrane resulting in the production of an LPS that is completely deacylated at the 3′ position. H. pylori lpxR mutants synthesize a hexaacylated lipid A bearing an acyloxyacyl moiety at the 3′ position (7). Perhaps Helicobacter LpxR promotes survival by producing a lipid A structure with lower endotoxicity, helping to evade the innate immune response. In the case of H. pylori, persistence is critical because the human stomach is the only ecological niche of this unusual bacterium. Alternatively, in organisms that have a more diverse lifestyle, such as Salmonella, LpxR activity is more tightly controlled in the outer membrane but may be activated in microenvironments within the human host, contributing to their survival.
Both the PagP and PagL crystal structures have been elucidated, and for PagP an NMR structure has also been determined. PagP and PagL are eight-stranded β-barrel proteins, both with a serine hydrolase active site (8–10). In PagP, the catalytic site is present in extracellular loops, and its residues are pointing toward the β-barrel axis. Furthermore, in the PagP structures solved to date, the triad is not in an active conformation because the histidine is too far away from the serine and aspartic acid residues. Probably, upon substrate binding, the proper conformation of the active site will be induced. In PagL, the catalytic site seems to be in the active constellation.
To date, the catalytic site of LpxR is unknown. Here, we present the crystal structure of the 32-kDa S. typhimurium LpxR. We demonstrate that the crystallized protein is active, and we present mutagenesis data on the residues in the active site. The structure, the mutagenesis data, and a modeled Kdo2–lipid A onto the active site of LpxR provide insight into the catalytic mechanism.
Results and Discussion
Production, Folding, and Activity of Recombinant LpxR.
LpxR from S. typhimurium was produced in E. coli and refolded from inclusion bodies. Folding was monitored by SDS/PAGE analysis. Like many other outer-membrane proteins, LpxR displayed heat modifiability, i.e., the heat-denatured form had a lower electrophoretic mobility than the correctly folded form. Folded LpxR was purified by using ion-exchange and gel-filtration chromatography. The refolded, purified LpxR showed a specific activity of 9.15 × 102 nmol/min per mg (corresponding with a turnover of 0.5 s−1) resulting in a 100-fold increase of deacylase activity relative to membrane fractions isolated from E. coli overexpressing the protein (9.1 nmol/min per mg). As shown in Fig. 1B, refolded LpxR also retained its requirement for Ca2+ for removing the 3′-acyloxyacyl group from the 32P-labeled Kdo2–lipid A substrate. Therefore, we concluded that the purified LpxR is correctly folded in vitro into its active conformation and suitable for structure determination.
Overall Structure of LpxR.
The asymmetric unit of the crystallized LpxR in the P21212 space group contains two LpxR molecules, 415 water molecules, nine zinc ions, four glycerol molecules and eight detergent (C10E5) molecules. The two molecules in the asymmetric unit are very similar (rmsd for all protein atoms is only 0.15 Å) and seem to form a symmetric dimer (Fig. S2). However, based on the program PISA of the EMBL-EBI an LpxR dimer is not likely to be present in solution (for details, see SI Text). The presence of LpxR as monomers in solution is also supported by size exclusion chromatography (for details, see SI Text).
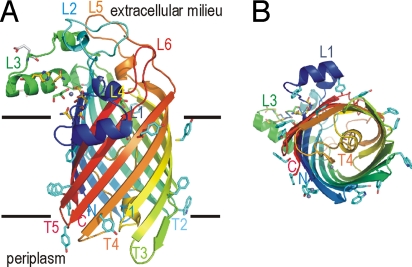
The overall structure of an LpxR molecule consists of a 12-stranded antiparallel β-barrel, with a shear number of 14 (Fig. 2). Generally, in outer-membrane protein structures the N and C termini of the β-barrel are present in the periplasm, and the periplasmic turns are short whereas the extracellular loops are longer. LpxR also has these general features. Remarkably, however, the periplasmic turn 4 (T4) is unusually long. T4 consists of residues Asp190–Glu213 that form a loop inside the β-barrel, closing it like a plug at the periplasmic side. The extracellular loops L1–L3 consist of α-helical regions. A topology based on the crystal structure is shown in Fig. S3.
Fig. 2.
Overall structure of LpxR. (A) Ribbon representation of LpxR. The ribbon is colored with a gradient from the N terminus in blue to the C terminus in red. Aromatic residues located at the membrane boundaries are shown as cyan sticks. Membrane boundaries are indicated by black lines. The extracellular side is located at the top of the figure, and the periplasmic side is at the bottom. Active-site residues are shown as yellow sticks. Zn2+ atoms are shown as gray spheres, and glycerol molecules are shown as sticks in green. Loops, turns, the N terminus, and the C terminus are labeled. (B) LpxR viewed from the periplasmic side of the protein. The representation of the structure is similar to that in A. Figs. 2, 3, 4, S2, and S5 were prepared with PyMOL (23).
Active Site of LpxR.
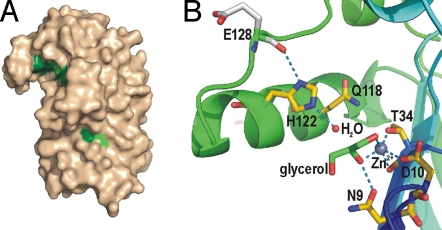
From earlier studies it is not known to which class of hydrolases LpxR belongs. Sequence alignment of LpxR homologues reveals a number of conserved residues (Fig. S4). Most of these residues, i.e., N9, D10, Y/F33, T/S34, G36, Q/H57, P62, F/Y79, Q118, H122, and W133, are located in a cleft between loop L3 and the barrel wall (Fig. 3 and Fig. S5). Interestingly, the cleft is present at the boundary of the hydrophobic and the hydrophilic regions of the membrane at the extracellular side of the molecule, where the scissile bond of the substrate LPS is located. Outside of this cleft, several other conserved residues are present, i.e., G106, G109, Q136, N/D139, G188, F/Y220, R/K226, and G258. To identify catalytically important residues for deacylase activity of LpxR several mutant LpxR proteins were generated with substitution of highly conserved residues, i.e., N9A, D10A, T34A, T34S, Q118A, H122A, and H122Q. Additionally, we substituted two nonconserved serines in the active site cleft, i.e., S61A and S63A (both adjacent to P62). The N9A, D10A, T34A, H122A, and H122Q mutant proteins were catalytically inactive, whereas the Q118A mutant protein displayed only 6% residual activity (Table S1). The T34S, S61A and S63A mutant proteins displayed >50% residual activity. In conclusion, S61 and S63 are not important for activity, whereas N9, D10, T34, and H122 are essential. Furthermore, the mutagenesis revealed that the hydroxyl group of T34 is important because the residue could be replaced by Ser without loss of activity. The observation that LpxR was fully inhibited by diethylpyrocarbonate proved that a histidine, presumably H122, is essential for catalysis (11). Furthermore, we found that dissolved imidazole (100 mM) restored the catalytically inactive H122A mutant protein to ≈30% of the wild-type activity by complementing the absent imidazole ring.
Fig. 3.
Active site of LpxR. (A) Surface representation of LpxR in wheat color. Positions of fully conserved residues are shown in dark green. Residues that are largely conserved are shown in light green. (B) Close up of the active site of LpxR. The orientation of LpxR is identical to that in Fig. 2A. Zn2+ ions are shown as gray spheres, a water molecule as a red sphere, and a glycerol molecule is represented as sticks with green carbons. Oxygen atoms are colored red and nitrogen blue. Short distances, i.e., H bonds or shorter distances between noncovalently bonded atoms, are shown as blue dashed lines.
As described, Ca2+ is essential for LpxR activity. Other divalent cations, such as Sr2+ and Cd2+ but not Zn2+, can replace Ca2+ without complete loss of catalytic activity (6). We expected that in the crystals, obtained in the presence of Zn2+, a Zn2+ would bind to the active site of LpxR, in a fashion similar to the way Ca2+ would bind. Only after calculation of an anomalous difference map, a low-anomalous peak was visible between T34 and D10, which may be caused by partial occupation by Zn2+ and other divalent cations. Another ligand of the Zn2+ is a glycerol molecule (Fig. 3B). This glycerol also makes a hydrogen bond to N9. In total, the Zn2+ is coordinated by five ligands. The OG1 of T34 is 1.8 Å away from Zn2+, O2 of glycerol 2.1 Å, O of T34 2.4 Å, OD2 of D10 2.5 Å, and O1 of glycerol 2.5 Å in molecule A. The Nε2 of H122, to which a water molecule is bound, is separated from the Zn2+ by 6.50 Å in chain A and by 6.66 Å in chain B. In phospholipase A2 (PLA2) Ca2+ is also essential for activity (12). In several PLA2 structures (2bg6, 1kp4, 2aa2, 1tgm, and 1kqu) the Ca2+, which forms the oxyanion hole, is separated on average by 6.3 Å from the Nε2 of the active site histidine, which acts as a base activating a water molecule. This average distance is close to the distance between the presumed catalytic Ca2+ (Zn2+ in our current structure) and H122 in LpxR. Therefore, it is tempting to speculate that LpxR displays a catalytic mechanism similar to that proposed for PLA2.
Modeling of Kdo2–Lipid A onto the Active Site of LpxR.
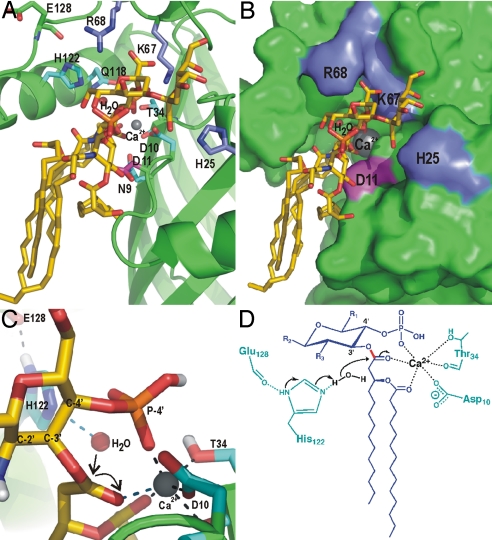
S. typhimurium LpxR can cleave both tetraacylated (Kdo2–lipid IVA) and hexaacylated (Kdo2–lipid A) lipid A substrates; thus, the presence of an acyl chain, rather than an acyloxyacyl group, at the 3′ position is sufficient for enzymatic activity. However, the Kdo sugars are required for cleavage because lipid IVA does not serve as a substrate (6). To gain further insight into the catalytic mechanism of LpxR and into the substrate specificity, we modeled Kdo2–lipid A onto the active site of Salmonella LpxR by using the program HADDOCK2.0 (13, 14). Several restraints were used to keep a Ca2+ at the approximate position of the Zn2+ in our structure. The only restraint that was used between Kdo2–lipid A and the active site of LpxR was between the scissile carbonyl oxygen and the Ca2+. We analyzed the 10 best models of the lowest score cluster, of which a representative model is shown in Fig.4. Clustering statistics are provided in Table S2. The docking results show that Kdo2–lipid A fits well into the active site of LpxR, and no major conformational changes in LpxR are required to bind Kdo2–lipid A. The largest intermolecular energy contributions come from Ca2+ (with an energy of −337 kcal/mol), K67 (−108), D11 (−48), H25 (−46), and R68 (−38). This energy value represents the intermolecular potential energy [i.e., sum of van der Waals and electrostatics (Coulomb) energies] between the protein (including Ca2+) and the substrate. K67 and R68, and H25 make salt bridges with the carboxyl groups of the Kdo sugars 1 and 2, respectively. D11 makes a hydrogen bond to the amide of the 2′ N-linked acyloxyacyl chain. These interactions are the only direct hydrophilic interactions and are probably key to the specificity of LpxR from S. typhimurium toward LPS. Because K67, D11, H25, and R68 are not conserved among LpxR homologs, other LpxR homologs must have evolved other ways to bind LPS. In addition to the H bonds and salt bridges, many interactions between hydrophobic residues in the barrel wall of LpxR and the acyl chains of the substrate are present. These interactions have relatively low intermolecular energy contributions (−15 kcal/mol in total by 17 residues) and are far less specific. The flexibility of the acyl chains of the substrate allows for binding to different hydrophobic residues, which explains why the hydrophobic residues in the barrel wall near the acyl chains of the substrate need not be conserved, as is seen for LpxR.
Fig. 4.
Modeling of Kdo2–lipid A into the active site of LpxR and the proposed catalytic mechanism. (A) View of Kdo2–lipid A, which is shown in sticks with yellow carbons, modeled onto LpxR. Fully conserved residues are shown as sticks in cyan. Residues that are probably involved in binding of the Kdo sugars, i.e., K67, R68, and H25 are shown as blue sticks. D11 is shown in magenta. The calcium ion is shown as a gray sphere. (B) LpxR is shown as a surface representation in green with K67, R68, H25, D11, Kdo2–lipid A, and the calcium colored as in A. (C) Closeup of the catalytic site of the modeling result. The representation is the same as in A, with the exception that the view angle is different and that polar hydrogens are shown in white. (D) Proposed catalytic mechanism for LpxR. The substrate is shown in blue, protein residues are shown in cyan, the water and calcium are shown in black. The scissile bond is shown in red. Black arrows indicate the movement of the electron pairs.
The high interaction energy contribution of Ca2+ can be explained by its interaction with three ligands from the substrate Kdo2–lipid A, in each of the 10 models analyzed. These are (i) the carbonyl oxygen of the scissile bond, (ii) the carbonyl oxygen of the secondary acyl chain, and (iii) the negatively charged oxygen of the nearby phosphate. Additionally, in most models, the Ca2+ interacts with three ligands from LpxR, as has been observed for Zn2+ in the crystal structure. This coordination is much more favorable for Ca2+ than the coordination that has been found in the crystal structure, in which Zn2+ was coordinated by three ligands of LpxR and only two of a glycerol molecule. The average number of ligands bound to Ca2+ found in the Protein Data Bank is 6.6, whereas this number is 4.3 for Zn2+. In this way Kdo2–lipid A may regulate the activity of LpxR because it is involved in the formation of the oxyanion hole by coordination of the essential Ca2+. Binding of the phosphate of the substrate to the Ca2+ of the oxyanion hole has been observed in PLA2. Evidently, the conserved residues D10, T34, and H122, which were found to be essential for activity, do not contribute to the intermolecular energy at all because these residues are involved in binding of either Ca2+ or a water molecule. The conserved and essential N9, however, does not contribute to binding of the substrate, water, or Ca2+ in our modeling studies. Possibly N9 is important for stabilizing the transient negatively charged carbonyl oxygen during the transition state, which we did not account for in the modeling. Unfortunately, our modeling studies also did not yield any conclusive insight about the roles of other conserved residues Y/F33, G36, Q/H57, P62, F/Y79, Q118, and W133. Possibly they are important for the integrity of the active-site conformation.
In summary, based on the crystal structure, the analogy to PLA2, and the supporting modeling studies, we propose that H122 is an essential catalytic-site residue acting as a base. Its position is stabilized by the carbonyl oxygen of E128. Ca2+, which is coordinated by D10 and T34, is required for making the oxyanion hole possibly together with N9. The proposed catalytic mechanism of LpxR is shown in Fig. 4 C and D. Although we have gained clear insights into the structure and catalytic mechanism of Salmonella LpxR, it remains unclear how the enzyme is activated within the outer membrane. Given that changes in the lipid A domain of LPS directly influence its endotoxic potential, the role that LpxR plays in remodeling of the bacterial surface merits further investigation.
Materials and Methods
Cloning, Expression, in Vitro Folding, and Purification of LpxR.
Wild-type LpxR was overproduced from the constructed plasmid pLpxR1 (6) in E. coli strain HMS147(DE3) (Novagen). For structure determination, mature LpxR was produced as inclusion bodies in E. coli BL21Star(DE3) (Novagen) by using the autoinduction method (see SI Text for more details). Inclusion bodies were purified as described in ref. 15. Inclusion bodies were dissolved in 8 M urea, 10 mM glycine (pH 8.3) to a concentration of 10 mg/mL protein. LpxR was then refolded by a quick, 10-fold dilution in 4.0% n-dodecyl-β-d-maltoside (DDM) (wt/vol) and 20 mM glycine (pH 8.3), followed by incubation at 37 °C for 18 h.
For subsequent purification of LpxR, the refolding solution was diluted at least 3-fold with binding buffer [20 mM ethanolamine (pH 10), 0.08% C10E5 (vol/vol)]. The sample was applied to a 1-mL ResourceQ column (GE Healthcare), which was eluted with a linear gradient of 0–250 mM sodium chloride in 30 mL. Collected fractions were analyzed by SDS/PAGE. Fractions containing LpxR were pooled and concentrated to 10 mg/mL by using Centricon YM-3 modules (Amicon, Millipore). Concentrated protein was then loaded onto a Tricorn Superdex 75 10/300 gel filtration column (GE Healthcare) that was equilibrated with a buffer containing 20 mM Tris·HCl (pH 9.0), 150 mM NaCl, and 0.08% C10E5 (vol/vol). Then, the fractions containing LpxR were pooled and concentrated to 12 mg/mL. The concentrated protein was dialyzed three times for 24 h by using a 3.5-kDa cutoff dialysis membrane against a 1,000-fold volume of buffer containing 2 mM Tris·HCl (pH 9.0) and 0.08% C10E5 (vol/vol).
A size-exclusion chromatography experiment was performed with a Tricorn Superdex 200 10/300 column (GE Healthcare) with 20 mM Tris (pH 7.5), 100 mM NaCl, 1 mM DTT, 1 mM EDTA, and 0.05% DDM (wt/vol) as the eluent.
Crystallization.
For crystallization of LpxR we used the hanging-drop vapor-diffusion method. Crystals belonging to the P2 space group grew with 14% PEG 6000 (wt/vol), 10% glycerol (vol/vol), 50 mM Tris·HCl (pH 8.5), and 5 mM ZnCl2 at 18 °C. Crystals that belong to the P21212 space group were obtained with 10% PEG 6000 (wt/vol), 20% glycerol (vol/vol), 50 mM Tris·HCl (pH 8.5), and 5 mM ZnCl2 at 37 °C. Both crystals had crystal dimensions of ≈200 μm × 100 μm × 20 μm. Crystals were harvested from the drops with a cryoloop and directly cooled into liquid nitrogen.
Data Collection and Structure Determination.
Datasets were obtained at 100 K on charge-coupled device detectors at the beamlines ID29 for the P2 crystal form and ID14-EH3 for the P21212 space group, both at the European Synchrotron Radiation Facility (ESRF). Data of the crystals belonging to the P2 space group were collected at λ = 1.28203, at which the anomalous signal of liganded zinc is at a maximum. For the crystals belonging to the P21212 space group, data were collected at λ = 0.931. The diffraction of P2 crystals showed concentric elongated spots. This was probably caused by movement of the crystal in the cryostream. For data processing, we used the program XDS (16). Table S3 summarizes data collection information. For both crystal forms, by using the single-wavelength anomalous dispersion (SAD) data and the programs ShelxC/D and E (17) in the CCP4 suite, we located seven Zn2+ sites in the asymmetric unit, calculated phases, and performed density modification, yielding poorly interpretable electron density maps. DMMULTI in the CCP4 suite was used to perform multicrystal and NCS averaging (see SI Text for more details). This yielded a much higher quality electron density map, which allowed us to build the LpxR structure by using the program COOT (18). The structure was refined against the highest-resolution data P21212 of 1.9 Å resolution by using REFMAC 5.0 (19). Refinement statistics are summarized in Table S3, and the electron density is shown in Fig. S6.
Site-Directed Mutagenesis.
Single amino acid substitutions were made by using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions with plasmid pLpxR2 as the template. pLpxR2 is a derivative of the low-copy expression vector pWSK29 containing a wild-type copy of lpxR (6). Oligonucleotides used for mutagenesis are listed in Table S4. Washed membrane preparations containing mutant LpxR proteins were prepared as described in ref. 6. Expression of recombinant LpxR proteins was monitored by SDS/PAGE analysis, where each mutant protein showed a similar level of expression and heat modifiability, which is indicative of correct folding.
Assay of LpxR 3′-O-Deacylase Activity.
LpxR activity was measured at 30 °C as described in ref. 6 with 2.5 μM Kdo2–[4′-32P]lipid A as the substrate (20). To determine the specific activity of LpxR, either purified LpxR (5 × 10−5 mg/mL) or washed membranes (0.001 mg/mL) were assayed in the linear range of assay, and the enzymatic activity (nmol/min) was calculated by determining the percentage of the substrate converted to the deacylated reaction product.
Modeling of Kdo2–Lipid A onto the Active Site of LpxR.
Models of Kdo2–lipid A docked onto LpxR were created with HADDOCK2.0 (13, 14). The coordinates of LpxR were taken from chain A of the crystal structure, and the Kdo2–lipid A coordinates were based on those of the LPS molecule that was cocrystallized with FhuA (21). Ambiguous interaction restraints were defined, based on the proposed catalytic mechanism. We used the following restraints: T34(OG1)–Ca2+, T34(O)–Ca2+, D10(OD2)–Ca2+, H122–H2O, and Kdo2–lipid A(carbonyl oxygen of scissile bond)–Ca2+. One thousand rigid-body docking solutions were generated from an ensemble of 20 Kdo2–lipid A conformations presampled by simulated annealing refinement in DMSO. The best 200 solutions based on the HADDOCK score were then refined with Kdo2–lipid A treated as fully flexible, whereas only the interface region of LpxR was flexible. All 200 models were finally refined in explicit solvent (DMSO was used to mimic the hydrophobic environment imposed by the membrane) and clustered based on rmsd criteria. The intermolecular potential energy was calculated as the sum of Van der Waals and electrostatic (Coulomb) energies of the interactions between the protein and Kdo2–lipid A. Nonbonded interactions were calculated with the OPLS force field (22) by using a cutoff of 8.5 Å. The electrostatic potential was calculated by using a shift function, whereas a switching function (between 6.5 and 8.5 Å) was used to define the Van der Waals potential. Partial charges on Kdo2–lipid A were defined by homology to functional groups already defined in the OPLS force field.
Supplementary Material
Acknowledgments.
We thank the European Synchrotron Radiation Facility in Grenoble for providing synchrotron radiation facilities and the beam-line scientists at the beam lines ID14-EH3 and ID29 for their help. We thank Joost Ballering for help with growing crystals and Janneke Heeres and Arne Visscher for assistance in building the structure. This work was supported by the Council for Chemical Sciences of the Netherlands Organization for Scientific Research (NWO-CW), by a NWO-CW Veni grant (to L.R.) and a NWO-CW Vici grant (to A.M.J.J.B.), and by National Institutes of Health Grants AI064184 and AI76322 (to M.S.T.) and GM-51310 (to C.R.H.R.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3FID).
This article contains supporting information online at www.pnas.org/cgi/content/full/0813064106/DCSupplemental.
References
- 1.Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trent MS, Stead CM, Tran AX, Hankins JV. Diversity of endotoxin and its impact on pathogenesis. J Endotoxin Res. 2006;12:205–223. doi: 10.1179/096805106X118825. [DOI] [PubMed] [Google Scholar]
- 3.Guo L, et al. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 4.Bishop RE, et al. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trent MS, Pabich W, Raetz CR, Miller SI. A PhoP/PhoQ-induced Lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J Biol Chem. 2001;276:9083–9092. doi: 10.1074/jbc.M010730200. [DOI] [PubMed] [Google Scholar]
- 6.Reynolds CM, et al. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3′-acyloxyacyl moiety of lipid A. J Biol Chem. 2006;281:21974–21987. doi: 10.1074/jbc.M603527200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stead CM, Beasley A, Cotter RJ, Trent MS. Deciphering the unusual acylation pattern of Helicobacter pylori lipid A. J Bacteriol. 2008;190:7012–7021. doi: 10.1128/JB.00667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahn VE, et al. A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. EMBO J. 2004;23:2931–2941. doi: 10.1038/sj.emboj.7600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang PM, et al. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc Natl Acad Sci USA. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rutten L, et al. Crystal structure and catalytic mechanism of the LPS 3-O-deacylase PagL from Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2006;103:7071–7076. doi: 10.1073/pnas.0509392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hnizda A, Santrucek J, Sanda M, Strohalm M, Kodicek M. Reactivity of histidine and lysine side chains with diethylpyrocarbonate: A method to identify surface-exposed residues in proteins. J Biochem Biophys Methods. 2008;70:1091–1097. doi: 10.1016/j.jbbm.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Bahnson BJ. Structure, function, and interfacial allosterism in phospholipase A2: Insight from the anion-assisted dimer. Arch Biochem Biophys. 2005;433:96–106. doi: 10.1016/j.abb.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 13.Dominguez C, Boelens R, Bonvin AM. HADDOCK: A protein–protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 14.de Vries SJ, et al. HADDOCK versus HADDOCK: New features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 15.Dekker N, Merck K, Tommassen J, Verheij HM. In vitro folding of Escherichia coli outer-membrane phospholipase A. Eur J Biochem. 1995;232:214–219. doi: 10.1111/j.1432-1033.1995.tb20801.x. [DOI] [PubMed] [Google Scholar]
- 16.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 17.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 18.Emsley P, Cowtan K. COOT: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 19.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 20.Tran AX, et al. Periplasmic cleavage and modification of the 1-phosphate group of Helicobacter pylori lipid A. J Biol Chem. 2004;279:55780–55791. doi: 10.1074/jbc.M406480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Siderophore-mediated iron transport: Crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 22.Jorgensen WL, Tirado-Rives J. The OPLS (optimized potentials for liquid simulations) potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 23.DeLano WL. Unraveling hot spots in binding interfaces: Progress and challenges. Curr Opin Struct Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.