Abstract
Myxococcus xanthus is a bacterium that undergoes multicellular development requiring coordinate regulation of multiple signaling pathways. One pathway governs aggregation and sporulation of some cells in a starving population and requires C-signaling, whereas another pathway causes programmed cell death and requires the MazF toxin. In response to starvation, the levels of the bifunctional transcription factor/antitoxin MrpC and its related proteolytic fragment MrpC2 are increased, inhibiting the cell death pathway via direct interaction of MrpC with MazF. Herein, we demonstrate that MrpC2 plays a direct role in the transcriptional response to C-signaling. We show that MrpC2 binds to sequences upstream of the C-signal-dependent fmgA promoter. These sequences are present in other C-signal-dependent promoter regions, indicating a general role for MrpC2 in developmental gene regulation. Association of MrpC and/or MrpC2 with the fmgA promoter region in vivo requires FruA, a protein that is similar to response regulators of 2-component signal transduction systems, but may not be phosphorylated. DNA binding studies showed that this association likely involves an unusual mechanism for a response regulator in which FruA and MrpC2 bind cooperatively to adjacent sites upstream of the fmgA promoter. We propose that this unusual mechanism of combinatorial control allows coordination of morphogenetic C-signaling with starvation signaling and cell death, determining spatiotemporal gene expression and cell fate.
Keywords: bacterial development, C-signaling, cell fate, signal transduction, sporulation
Understanding how cells integrate many different signals to regulate genes and determine cell fates during multicellular development is a fundamental question. Myxococcus xanthus provides an attractive model to address this question because starvation initiates a relatively simple developmental process (1). Thousands of rod-shaped cells coordinate their movements to build fruiting bodies in which cells differentiate into dormant, spherical spores [supporting information (SI) Fig. S1]. However, not all cells form spores. Alternative fates are programmed cell death (PCD) (2) or persistence outside of fruiting bodies as peripheral rods (3).
Signals act at different times during the M. xanthus developmental process to control gene expression, coordinate cell movements, and determine cell fates. Starvation triggers the stringent response, which involves production of the second messenger (p)ppGpp (Fig. S1), and leads to activation of early developmental genes and secretion of protease activity that generates a mixture of peptides and amino acids known as A-signal (4). Extracellular A-signaling governs expression of many additional genes, including the mrp operon (Fig. S1) (5). Later, when cells begin to aggregate, C-signaling takes over. The C-signal appears to be a proteolytic cleavage product of CsgA that is associated with the cell surface (6–8). Because C-signaling requires cell alignment (9) and possibly end-to-end contact, it is paracrine or short-range signaling, which is common in eukaryotes but rare among bacteria (10). The short-range nature of C-signaling and its effects on cell movement and gene expression can explain its critical role in coordinating aggregation with sporulation (4, 11, 12). Cell alignment within a nascent fruiting body has been proposed to allow a high level of C-signaling and activation of genes required for sporulation.
FruA plays an important role in regulating genes important for aggregation and sporulation (Fig. S1). FruA is similar to response regulators of 2-component signal transduction systems (13). The N-terminal domain of FruA was proposed to be phosphorylated in response to C-signal (14), but the domain lacks 2 aspartate residues that are normally important for phosphorylation of a third aspartate residue, and the putative histidine protein kinase (HPK) has not been identified despite considerable effort. The C-terminal domain of FruA has been shown to bind to sites in the promoter regions of developmentally regulated genes (15–17). FruA positively regulates expression of these genes, but in the case of fmgA (formerly referred to as the Ω4400 locus), mutational analysis of the promoter region implied that an additional transcriptional activator is required (18).
Here, we report identification of the activator as MrpC2 and we name the gene at the Ω4400 locus fmgA (FruA- and MrpC2-regulated gene A). MrpC2 lacks the N-terminal 25 residues of MrpC and might be generated by proteolytic activity of a developmentally regulated protease (LonD) (Fig. S1) (19). MrpC is similar to transcription factors in the cAMP receptor protein (CRP) family (20), but a nucleotide effector has not been identified. Recently, MrpC was shown to interact with the toxin MazF, which mediates PCD during development (2). In addition to identifying MrpC2 as an activator of fmgA transcription, we show that FruA is required for association of MrpC and/or MrpC2 with the fmgA promoter region in vivo and that FruA and MrpC2 bind cooperatively to fmgA promoter region DNA in vitro. Cooperative binding of a response regulator and an independent transcription factor is an unusual mechanism of gene regulation. Preliminary results indicate that this mechanism is shared by other C-signal-dependent genes (see Discussion). We propose that cooperative binding of FruA and MrpC2 facilitates integration of positional information (via short-range C-signaling) with nutritional status and PCD, governing gene expression and cell fate, analogous to combinatorial control during development of multicellular eukaryotes.
Results
MrpC2 Binds to a cis-Regulatory Sequence in the fmgA Promoter Region.
Transcription from the fmgA promoter is important for development because aggregation of M. xanthus DK4292 containing Tn5 lac Ω4400 was delayed by ≈6 h compared to wild-type DK1622 (SI Text). Mutational analysis identified cis-regulatory sequences at −86 to −77 and −63 to −46 upstream of the fmgA promoter (Fig. 1A) (18), and subsequent analysis showed that the FruA DNA-binding domain (FruA-DBD) binds to the sequence between −86 and −77 (17). The sequence between −63 and −46 contains 2 elements found in other C-signal-dependent promoter regions; a 5-bp element (consensus GAACA) and a C box (consensus CAYYCCY; Y means C or T) (21–23). Mutations in this region nearly abolish fmgA promoter activity (Fig. 1A) (18), suggesting that a transcriptional activator binds to this region and perhaps to similar sequences in other C-signal-dependent promoter regions. To identify the putative activator, DNA-binding proteins were partially purified as described previously (24) from M. xanthus that had undergone 12 h of development, a time when fmgA is expressed (18). Proteins in the AS fraction were incubated with a 32P-labeled DNA fragment (−101 to +25) spanning the fmgA promoter region, and electrophoretic mobility shift assays (EMSAs) revealed a single shifted complex (Fig. 1A). EMSAs with DNA probes having a mutation between −63 and −46 eliminated or greatly reduced formation of the shifted complex, with the exception of the single base-pair change at −53, which also had a smaller effect on promoter activity in vivo (Fig. 1A) (18). The shifted complex appeared to be formed by a protein in the AS fraction that binds to sequences between −63 and −46 upstream of the fmgA promoter.
Fig. 1.
Binding of MrpC2 to the fmgA promoter region. (A) Effects of mutations on fmgA promoter activity in vivo and on DNA binding in vitro. Top summarizes mutational effects on developmental fmgA-lacZ expression (18). The number beneath each mutant sequence indicates the percentage of wild-type promoter activity. Bottom shows EMSAs performed with 32P-labeled fmgA DNA (6 nM) spanning from −101 to +25 and proteins in the AS fraction (0.7 μg/μl). The arrow indicates the shifted complex produced by incubating the WT DNA fragment with the AS fraction, and other lanes show the effects of mutations. (B) SDS/PAGE of protein purified from the AS fraction by using fmgA DNA (−101 to +25). The arrow indicates the major species in the affinity-purified protein (APP) after staining with silver. Numbers indicate the migration positions of molecular weight (kDa) standards. (C) EMSAs with 32P-labeled fmgA DNA (6 nM) spanning from −101 to +25 and proteins in the AS fraction or the APP. The arrow indicates the shifted complex produced with the WT DNA fragment. APP failed to form the shifted complex with a DNA fragment bearing the GAAC to TCCA mutation at −63 to −60 (mutant). (D) EMSAs with 32P-labeled fmgA DNA (1.2 nM) spanning from −101 to +25, WT or mutant as indicated (see A for mutations), and His10-MrpC2 (1 μM) or the AS fraction (0.7 μg/μl). The arrowhead and arrow indicate the shifted complexes produced by His10-MrpC2 and the AS fraction, respectively. Image is a composite from several experiments, but in each experiment the WT fmgA DNA served as a control, and the signal intensity of the shifted complexes was comparable to that shown.
To purify the putative activator protein from the AS fraction, DNA-affinity chromatography was performed with the fmgA promoter region (−101 to +25). The major protein species after purification had an apparent molecular weight of ≈30 kDa (Fig. 1B). The affinity-purified protein (APP) generated a shifted complex indistinguishable from that observed with the AS fraction (Fig. 1C). Also, like the AS fraction (Fig. 1A), APP failed to generate a shifted complex with mutant (−63 to −60) fmgA promoter region DNA (Fig. 1C). Therefore, APP was subjected to mass spectrometry analysis after protease digestion. Peptide sequences matching MrpC were the only significant matches to M. xanthus proteins predicted from the genome sequence. MrpC is similar to CRP-family transcription factors and was shown previously to be essential for development (20). A form of MrpC lacking the N-terminal 25 residues, called MrpC2, was identified previously in an AS fraction based on binding to the fruA promoter region (24). Our results suggested that MrpC2 in the AS fraction binds to the fmgA promoter region at a site (−63 to −46) important for promoter activity.
To test the idea that MrpC2 in the AS fraction was responsible for the shifted complex (Fig. 1A), antibodies against MrpC were added after the complex had been allowed to form. EMSAs revealed the formation of super-shifted complexes and loss of the original-shifted complex (Fig. S2), supporting the idea that MrpC2 in the AS fraction binds to fmgA promoter region DNA.
To confirm that MrpC2 binds to the fmgA promoter region, N-terminally His-tagged MrpC2 (His10-MrpC2) was expressed in E. coli and purified (Fig. S3A). His10-MrpC2 exhibited a similar pattern of binding to wild-type and mutant fmgA promoter-region DNA as seen with the AS fraction (Fig. 1D). The complex produced by His10-MrpC2 migrates more slowly than the complex produced by the AS fraction, presumably because of the 10 His residues plus 8 additional residues present in the His10-MrpC2 fusion protein. Mutations between −63 and −46 eliminated or reduced MrpC2 binding, with the exception of a single base-pair change at −53. These results, taken together with the effects of mutations in this region on fmgA promoter activity (Fig. 1A) (18), imply that MrpC2 binding to this region activates fmgA transcription. Because the region includes a 5-bp element and a C box, which are found in a similar arrangement upstream of other C-signal-dependent promoters (21–23), MrpC2 might directly activate other C-signal-dependent genes (see Discussion). Interestingly, mutations upstream of −63 appeared to enhance (−74 to −70) or reduce (−76 to −75) MrpC2 binding, whereas 2 mutations between −86 and −77, that impair binding of FruA-DBD (17), did not affect MrpC2 binding (Fig. 1D). We conclude that FruA and MrpC2 bind to adjacent, important, cis-regulatory sequences upstream of the fmgA promoter.
We tested nucleotides for an effect on DNA binding by MrpC2 because it is similar to CRP-family transcription factors (20); however, we found no evidence for a nucleotide effector (SI Text).
FruA Is Required for Association of MrpC and/or MrpC2 with the fmgA Promoter Region in Vivo.
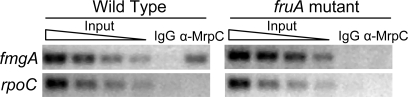
The proximity of the FruA and MrpC2 binding sites in the fmgA promoter region suggested that one protein might recruit the other or that the two proteins might bind cooperatively. Expression of fruA depends on MrpC2 (Fig. S1) (24), so neither transcription factor is expected to accumulate in a mrpC mutant. However, MrpC and MrpC2 accumulate normally in a fruA mutant (19) (data not shown), yet fmgA fails to be expressed (17). Why are MrpC and MrpC2 insufficient to activate fmgA transcription? We hypothesized that MrpC and MrpC2 fail to bind to the fmgA promoter region in the absence of FruA. To test this hypothesis, ChIP with polyclonal antibodies against MrpC (which also recognize MrpC2) (19) was used to measure the association of MrpC and/or MrpC2 with the fmgA promoter region (−101 to +155) integrated ectopically into the chromosome of wild-type or fruA mutant cells that had undergone development. DNA recovered from ChIP was subjected to PCR with primers designed to amplify the ectopic copy of the fmgA promoter region. The PCR analysis revealed that the fmgA promoter region was enriched by ChIP with antibodies against MrpC relative to control antibodies for wild type, but not for the fruA mutant (Fig. 2). Neither strain showed enrichment of rpoC coding region DNA (as a negative control). We conclude that FruA is required for association of MrpC and/or MrpC2 with the fmgA promoter region in vivo.
Fig. 2.
ChIP analysis of M. xanthus with the fmgA promoter region (−101 to +155) integrated ectopically in otherwise wild-type or fruA mutant backgrounds. After 18 h of development, cells were treated with formaldehyde and lysed, and cross-linked chromatin was immunoprecipitated with anti-MrpC antibodies or IgG as a control. DNA was amplified with appropriate primers for the fmgA promoter region at the ectopic chromosomal site or for the rpoC coding region as a negative control. A 2-fold dilution series of input DNA purified from 0.25%, 0.125%, 0.0625%, or 0.03125% of the total cellular extract before immunoprecipitation was used as a template in parallel PCRs to show that the PCR conditions were in the linear range of amplification for each primer set.
Cooperative Binding of MrpC2 and FruA to the fmgA Promoter Region.
The requirement for FruA for association of MrpC and/or MrpC2 with the fmgA promoter region in vivo is consistent with recruitment or cooperative binding. To distinguish between these models and to test the notion that FruA directly affects binding of MrpC2 to the fmgA promoter region, recombinant His-tagged FruA (FruA-His6) was purified (Fig. S3B) for analysis of DNA binding by EMSAs. When EMSAs were performed on 5% polyacrylamide gels, FruA-His6 appeared to bind to the fmgA promoter region weakly compared with His10-MrpC2, but there was a strong enhancement of shifted complex formation when both proteins were incubated with fmgA DNA (Fig. 3A). In the presence of both proteins, 2 complexes were observed. The abundant lower complex (LC) comigrated with the complexes formed by either protein alone, suggesting that the LC is a mixture of complexes composed of DNA bound by His10-MrpC2 or FruA-His6. The upper complex (UC) was suggestive of a complex of 2 protein molecules bound to DNA. Serendipitously, we discovered that the proportion of UC to LC increased if EMSAs were performed on 8% polyacrylamide gels (Fig. S4A). Under these conditions, FruA-His6 alone appeared to bind to the fmgA promoter region more strongly than His10-MrpC2 alone.
Fig. 3.
Enhancement of shifted complex formation. (A) The combination of FruA-His6 and His10-MrpC2 enhances complex formation. EMSAs with 32P-labeled fmgA DNA (1.2 nM) spanning from −101 to +25 with no protein, His10-MrpC2 (1 μM), FruA-His6 (3 μM), or both His10-MrpC2 (1 μM) and FruA-His6 (3 μM). Arrowheads indicate UC and LC. (B) The combination of FruA-DBD-His8 and His10-MrpC2 does not enhance complex formation. EMSAs are the same as in A except with FruA-DBD-His8 (14 μM). The arrowhead and arrow indicate the complexes produced by His10-MrpC2 and FruA-DBD-His8, respectively.
Recombinant FruA-His6 purified from E. coli is presumably not phosphorylated. Consistent with this notion, treatment of FruA-His6 with phosphatase from bacteriophage λ did not diminish its ability to bind DNA alone or to enhance formation of shifted complexes in combination with His10-MrpC2 (data not shown). Substitution of a glutamate residue for the putative phosphorylated-aspartate residue in the N-terminal domain of FruA (D59E), which in some response regulators mimics the phosphorylated-active form of the protein (25), did not change the behavior of FruA-His6 in EMSAs, and neither did treatment with small molecule phosphodonors (Fig. S4 B–G). These results, together with the striking enhancement of shifted complex formation by recombinant FruA-His6 in combination with His10-MrpC2 (Fig. 3A and Fig. S4A), suggest that FruA might be active without phosphorylation.
Interestingly, the C-terminal DNA-binding domain of FruA was insufficient to enhance shifted complex formation in combination with MrpC2. The complexes formed by the combination of proteins were indistinguishable from those produced when FruA-DBD-His8 or His10-MrpC2 alone was incubated with fmgA promoter region DNA (Fig. 3B). We conclude that the N-terminal domain of FruA contains an important determinant for enhancement of shifted complex formation in combination with MrpC2 and the fmgA promoter region.
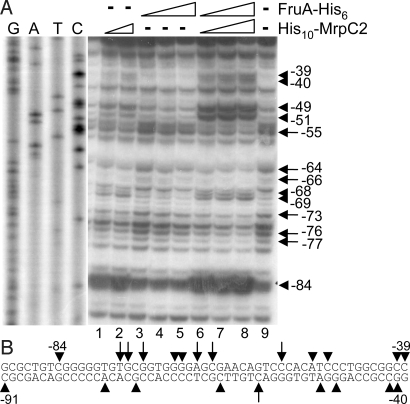
To characterize the enhanced DNA binding observed in the presence of His10-MrpC2 and FruA-His6, DNase I footprinting of complexes in solution was performed. Protected and hypersensitive sites were observed with His10-MrpC2 alone in the region spanning from −84 to −39 (Fig. 4A), a slightly larger region than mapped by EMSAs (Fig. 1). The hypersensitive sites suggest that His10-MrpC2 bends the DNA on binding. The protection and hypersensitivity in this region increased when FruA-His6 was present in combination with His10-MrpC2, but was not observed with FruA-His6 alone, suggesting that His10-MrpC2 binding was increased in the presence of FruA-His6. DNase I footprinting of the other strand with FruA-His6 alone revealed a hypersensitive site near −91 (Fig. S5), slightly upstream of a region that was previously shown to bind FruA-DBD-His8 (17). The intensity of this hypersensitive site increased when His10-MrpC2 was present in combination with FruA-His6, but hypersensitivity was not observed with His10-MrpC2 alone, suggesting that FruA-His6 binding was increased in the presence of His10-MrpC2. As observed with the other strand (Fig. 4A), there were protected and hypersensitive sites downstream of −91 with His10-MrpC2 alone, and these signals increased in the presence of both proteins (Fig. S5). In addition, hypersensitive sites were observed near −74 and −63 when both proteins were present, but not with either protein alone (Fig. S5), suggesting simultaneous binding of MrpC2 and FruA to the same DNA molecule. The DNase I footprinting results are summarized in Fig. 4B. These results demonstrate cooperative binding of FruA and MrpC2 to the fmgA promoter region, providing plausible explanations for the observed dependence of MrpC and/or MrpC2 on FruA for association with the fmgA promoter region in vivo (Fig. 2) and for the observed enhancement of shifted complex formation in vitro (Fig. 3A and Fig. S4A).
Fig. 4.
DNase I footprinting. (A) fmgA promoter region DNA (−139 to +25) was 5′-labeled at −139, incubated with 1 or 1.5 μM His10-MrpC2 (lanes 1–2), or with 1.5, 3, or 4.5 μM FruA-His6 (lanes 3–5), or with 0.5, 1, or 1.5 μM His10-MrpC2 in combination with 1.5, 3, and 4.5 μM FruA-His6 (lanes 6–8), or with no protein (lane 9), and subjected to DNase I footprinting. Lanes G, A, T, and C show sequence ladders generated by the same labeled primer used to generate the probe for DNase I footprinting. Arrows indicate sites protected from DNase I digestion, and arrowheads indicate hypersensitive sites. (B) Summary of protected and hypersensitive sites from A and Fig. S5.
To determine whether the binding sites for both His10-MrpC2 and FruA-His6 in the fmgA promoter region are important for enhanced formation of shifted complexes, EMSAs were performed with mutant DNA fragments expected to impair binding of one or the other protein. Mutations in the region from −86 to −77 greatly reduced binding of FruA-DBD-His8 (17) and greatly reduced the enhancement of shifted complex formation by the combination of FruA-His6 and His10-MrpC2 (Fig. 5). UC was undetectable, and LC was greatly diminished. We attempted to eliminate the FruA-His6 binding site without impairing His10-MrpC2 binding. A DNA fragment from −76 to −41 was insufficient for His10-MrpC2 binding (data not shown), indicating that the site required for His10-MrpC2 binding may partially overlap the site required for FruA-His6 binding. However, adding 5 bp of non-fmgA sequence (CACAA) to the upstream end allowed His10-MrpC2 binding (Fig. 5). No FruA-His6 binding was detected with this modified (+5 bp) −76 to −41 DNA fragment. In the presence of His10-MrpC2 and FruA-His6, UC was undetectable and very little enhancement of LC formation was observed. These results demonstrate the importance of the FruA-His6 binding site for enhanced formation of shifted complexes. Likewise, the His10-MrpC2 binding site is important, because a DNA fragment containing a mutation at −63 to −60, which eliminates detectable His10-MrpC2 binding, also abolished detectable enhancement of shifted complex formation (Fig. 5). Furthermore, both binding sites must be on the same DNA fragment. No enhancement of shifted complex formation was observed when His10-MrpC2 and FruA-His6 were added to a mixture of 2 DNA fragments (only one of which was 32P-labeled in each of two separate experiments) capable of binding only His10-MrpC2 [the modified (+5 bp) −76 to −41 fragment] or only FruA-His6 (a fragment spanning from −101 to −64) (Fig. S6). Supershift assays provided further evidence that both His10-MrpC2 and FruA-His6 are responsible for enhanced formation of shifted complexes with fmgA promoter region DNA (Fig. S7). These results support the interpretation that enhancement of shifted complex formation involves binding of both His10-MrpC2 and FruA-His6 to adjacent (possibly overlapping) sites upstream of the fmgA promoter, and, together with our footprinting and ChIP results, support a model in which FruA and MrpC2 bind cooperatively to regulate fmgA transcription during M. xanthus development.
Fig. 5.
Enhancement of shifted complex formation depends on binding sites for both FruA and MrpC2. EMSAs with 32P-labeled fmgA DNA (1.2 nM) spanning from −101 to +25, WT or mutant as indicated (see Fig. 1A for mutations), and His10-MrpC2 (1 μM) and/or FruA-His6 (3 μM) as indicated. The modified (+5 bp) −76 to −41 DNA fragment has non-fmgA sequence (CACAA) at its upstream end.
Discussion
We have discovered that a crucial cis-regulatory element in the fmgA promoter region is bound by MrpC2 and that association of MrpC and/or MrpC2 with the fmgA promoter region in vivo requires FruA. Our DNA binding studies revealed cooperative binding of FruA and MrpC2 to adjacent (possibly overlapping) sites upstream of the fmgA promoter. Our preliminary results, described below, indicate that several other C-signal-dependent promoter regions are cooperatively bound by FruA and MrpC2. Cooperative binding of a response regulator and an independent transcription factor is an unusual mechanism of gene regulation that has not been reported previously. Although FruA is similar to response regulators, it may be active without phosphorylation, and MrpC2 is a proteolytic fragment of MrpC, which functions not only as a transcription factor but also as an antitoxin in the regulation of PCD (2). Therefore, our evidence supports a model in which cooperative binding of 2 unusual transcription factors facilitates the coordination of multiple signaling pathways to ensure proper control of gene expression and cell fate during M. xanthus development.
Preliminary studies indicate that cooperative binding of MrpC2 and FruA is a conserved mechanism of gene regulation in response to C-signaling during M. xanthus development. The cis-regulatory element to which MrpC2 binds in the fmgA promoter region includes a 5-bp element and a C box. These 2 sequences are similarly arranged upstream of other C-signal-dependent promoters and are important for promoter activity (21–23), suggesting that MrpC2 may bind to these sites. Indeed, in the promoter region of the operon identified by Tn5 lac Ω4499 (22), MrpC2 binds near a 5-bp element, and in combination with FruA, formation of shifted complexes in EMSAs is greatly enhanced (unpublished data). In the promoter region of the dev operon (23), whose products are important for sporulation, MrpC2 binds to a region that includes a 5-bp element and 2 C-box-like sequences, and addition of FruA greatly enhances complex formation in EMSAs (S.M., P. Viswanathan, and L.K., unpublished data). In the promoter region of the gene identified by Tn5 lac Ω4403 (21), MrpC2 binds to a region that includes 2 5-bp elements in inverted orientation, and enhancement of shifted complex formation in combination with FruA is likewise observed (J. Lee, S.M., and L.K., unpublished data). Our preliminary studies, taken together with the evidence presented here for fmgA, indicate that cooperative binding of MrpC2 and FruA is a conserved mechanism of C-signal-dependent gene regulation.
Cooperative binding of MrpC2 and FruA to promoter regions of C-signal-dependent genes is an unusual mechanism of gene regulation. Typically, DNA-binding response regulators are phosphorylated by an HPK, and this enhances DNA binding (25). The bound response regulator recruits RNA polymerase to the promoter or facilitates another step during transcription initiation. To our knowledge, cooperative binding of a response regulator and an independent transcription factor has not been observed previously. Other mechanisms have been shown to allow response regulators to act in combination with proteins that are not independent transcription factors. For example, the response regulator RcsB interacts with RcsA and other auxiliary regulatory proteins, subjecting capsular polysaccharide synthesis to complex control in many bacteria (26). RcsA forms a heterodimer with RcsB and appears to stabilize the protein–DNA complex (27). In one report, RcsB has been shown to act in combination with another response regulator, PhoP, which is an independent transcription factor, but the mechanism is unknown (28). Our results establish that the response regulator FruA and the (sometimes) independent transcription factor MrpC2 can act in combination by binding cooperatively to promoter regions of C-signal-dependent genes. FruA and/or MrpC2 probably interact with RNA polymerase at the fmgA promoter. The 2 proteins occupy a location typical for Class I activators, which function by contacting the C-terminal domain of the α-subunits of RNA polymerase (29).
The detailed mechanism of cooperative binding of MrpC2 and FruA to the fmgA promoter region remains to be explored. The binding sites of the 2 proteins may partially overlap, because a 7-bp mutation at −83 to −77 impairs FruA-DBD-His8 binding (17) and DNA upstream of −76 is required for His10-MrpC2 binding (data not shown). The 2 proteins may interact with opposite faces of the DNA in a region of overlap, analogous to certain homeodomain proteins, which bind DNA cooperatively (30). As for some homeoprotein–DNA complexes, cooperativity might depend not only on protein–protein interactions, but on bending of the DNA by one or both proteins. Binding of either MrpC2 or FruA alone to the fmgA promoter region produced DNase I hypersensitivity indicative of DNA bending, and the combination of proteins increased the intensity and number of hypersensitive sites (Fig. 4 and Fig. S5), demonstrating cooperative binding. Likewise, EMSAs showed that the combination of proteins enhances formation of shifted complexes (Fig. 3A and Fig. S4A) and that this depends on sequences important for binding of each protein (Fig. 5) consistent with cooperative binding. The shifted complexes that were observed also depended on the percentage of polyacrylamide in gels used in the EMSAs, with 8% gels facilitating detection of FruA binding and detection of UC that presumably represents binding of FruA and MrpC2. The gel matrix influences stability of protein–DNA complexes during EMSAs (31). The enhancement of shifted complex formation by MrpC2 and FruA requires the N-terminal domain of FruA (Fig. 3B), suggesting that this domain interacts directly with MrpC2, but further studies will be needed to elucidate the detailed mechanism of cooperativity.
Several lines of evidence suggest that the N-terminal domain of FruA, which is similar to the receiver domain of response regulators that is typically phosphorylated by an HPK, might function without phosphorylation. First, the N-terminal domain of FruA lacks 2 aspartate residues that are highly conserved in receiver domains and normally play an important role in phosphorylation of a third aspartate residue (14, 25). Second, extensive efforts have failed to identify a cognate HPK. Third, FruA is not phosphorylated in vitro by heterologous HPKs, EnvZ or HepK from E. coli and Anabaena, respectively (R. Zhou and L.K., unpublished data). Fourth, a phosphomimetic D59E substitution in FruA did not increase DNA binding or enhancement of shifted complex formation in combination with MrpC2 (Fig. S4D). Fifth, treatment with small molecule phosphodonors, which activates many response regulators (25), did not increase DNA binding of FruA (Fig. S4 E–G). Although these results do not rule out the possibility that FruA is phosphorylated, our discovery of potent cooperative binding by recombinant (presumably unphosphorylated) FruA and MrpC2, which depends on the N-terminal domain of FruA, reveals an unusual function of a receiver domain that may not be phosphorylated. Receiver domains that cannot or need not be phosphorylated in order for the pseudoresponse regulator protein to function have been described in bacterial DNA-binding proteins (32–34) and in proteins that regulate circadian rhythms in bacteria (35, 36) and plants (37).
If C-signaling does not lead to phosphorylation of FruA, then what is the mechanism of signal transduction? Cooperative binding of FruA and MrpC2 to the fmgA promoter region and other promoter regions, together with activation of fruA transcription by MrpC2 (24), represents a coherent feed-forward regulatory loop design found commonly in regulatory networks because of its beneficial characteristics (38). C-signaling could affect production of MrpC2 and/or activity of FruA (Fig. S1). If there is an effect of C-signaling on FruA, it is likely posttranslational because a mutant defective in C-signaling accumulates FruA normally during development (14).
The complex regulation of MrpC2 production and the recent finding that MrpC plays a role in PCD make this unusual transcription factor/antitoxin an attractive target for regulation by C-signaling, which has not been examined. Mutants defective in C-signaling are defective in PCD (39). MrpC is phosphorylated by a cascade of Ser/Thr protein kinases (STPKs), presumably in response to an unknown signal during growth, inhibiting accumulation of MrpC and MrpC2 (19). Starvation conditions may remove the signal (Fig. S1), allowing MrpC and MrpC2 to accumulate. Recently, it was shown that the EspA signal transduction pathway influences the MrpC and MrpC2 concentrations (40), presumably providing another link to starvation (Fig. S1). Also, MrpC was shown to interact with the toxin MazF, inhibiting PCD (2). On the other hand, MrpC appears to directly activate mazF transcription. It is important to test whether MrpC2 differs from MrpC in either of these activities. The concentrations of MrpC, its phosphorylated or cleaved forms, and their interactions with MazF and at different promoters, may determine the fate of cells in a developing population of M. xanthus (2).
Commitment to form a spore has been hypothesized to involve induction of genes at the Ω7536 locus, which in turn depends on induction of the dev operon (41). Because dev appears to be regulated by cooperative binding of MrpC2 and FruA (S.M., P. Viswanathan, and L.K., unpublished data), we propose that commitment to sporulation is governed by these key transcription factors. MrpC is a major hub in the regulatory network, linked extensively to starvation (Fig. S1). Its direct involvement in commitment to sporulation might couple persistent starvation to the decision to form a spore. FruA is likewise a major hub in the regulatory network. Transcription of fruA is highly regulated (41), and it is unclear how much of this regulation feeds through MrpC. Short-range C-signaling contributes positional information (i.e., cell alignment in the nascent fruiting body) to the decision to sporulate (4, 11, 12), and it may do so through MrpC and/or a posttranslational effect on FruA, as discussed above. Commitment to sporulation may also be governed by a third activator of dev transcription, LadA, which likely responds to a signal and acts positively from a site downstream of the promoter (42). Combinatorial regulation of dev by ≥3 transcription factors that bind upstream and downstream of the promoter resembles regulation of developmental genes in multicellular eukaryotes.
Materials and Methods
Bacterial Strains and Plasmids.
Strains and plasmids used in this study are listed in Table S1.
Growth and Development.
E. coli containing plasmids were grown at 37 °C in Luria–Bertani medium (43) containing 200-μg/ml ampicillin. Growth and development of M. xanthus was as described (21).
Preparation of fmgA DNA Fragments.
The preparation of 32P-labeled DNA fragments for EMSAs and DNase I footprinting is described in SI Text.
EMSAs and DNase I Footprinting.
EMSAs were performed as described (17), except the binding reaction mixtures were incubated at 25 °C for 15 min. For footprinting, 0.2 units of DNase I (Promega) was added to the binding reaction mixture (20 μl) for 2 min at 25 °C. The mixture was the same as for EMSAs, except it contained 5 mM MgCl2, 0.5 mM CaCl2, 0.025 μg/μl double-stranded poly(dI-dC), and no glycerol. Reactions were stopped by adding 100 μl of solution containing 300 mM sodium acetate, 20 mM EDTA, 0.2% SDS, 0.02 μg/μl proteinase K, and 100 μg/ml yeast tRNA, and incubating at 52 °C for 15 min. After extraction with 100 μl of phenol (twice), DNA was precipitated with ethanol. The DNA was resuspended in formamide loading buffer (43), boiled for 3 min, subjected to electrophoresis on an 8% polyacrylamide gel containing 8 M urea, and visualized by autoradiography. Sequencing ladders were generated by using the SequiTherm EXCEL II DNA Sequencing Kit protocol (Epicentre Biotechnologies).
DNA-Affinity Chromatography.
A fmgA DNA fragment (−101 to +25) was synthesized by PCR with a 5′-biotin label at −101, bound to streptavidin beads, and DNA-affinity chromatography was performed with the AS fraction as described (42).
Preparation of MrpC2 and FruA.
His10-MrpC2 (19) and FruA-DBD-His8 (17) were purified as described previously from E. coli strains SMhisMrpC2 and EDYFruA, respectively. FruA-His6 was purified from E. coli SMFruAhis as described in SI Text.
ChIP.
M. xanthus strains MDY4400.DZF1 and MDY4400.FA were used for ChIP as described (17) with the following modifications: Anti-MrpC antibodies (500 ng) (19) or control IgG (500 ng) (Santa Cruz Biotechnology) were used for immunoprecipitation, 2-fold serial dilutions were made of the input DNA samples, and the primers used for PCR of the fmgA promoter region were the one for +25, described in SI Text, and one upstream (yielding a product of ≈180 bp) in the vector used for ectopic integration (5′-CTGCCAGGAATTGGGGATC-3′).
Supplementary Material
Acknowledgments.
We thank Sumiko Inouye (Robert Wood Johnson Medical School, Piscataway, NJ) for sending plasmids, protocols, and antibodies; Ruanbao Zhou (Michigan State University, East Lansing, MI) for providing OmpR; Yu Liu for help with purification of FruA; and Bill Henry, Dale Kaiser, Ann Stock, Lotte Sogaard-Andersen, David Arnosti, Rob Britton, and Mark Robinson for helpful comments. This research was supported by National Science Foundation Grant MCB-0744343 and by the Michigan Agricultural Experiment Station.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808516106/DCSupplemental.
References
- 1.Whitworth DE, editor. Myxobacteria: Multicellularity and Differentiation. Washington, DC: Am Soc Microbiol; 2008. [Google Scholar]
- 2.Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- 3.O'Connor KA, Zusman DR. Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. J Bacteriol. 1991;173:3318–3333. doi: 10.1128/jb.173.11.3318-3333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaiser D. Signaling in Myxobacteria. Annu Rev Microbiol. 2004;58:75–98. doi: 10.1146/annurev.micro.58.030603.123620. [DOI] [PubMed] [Google Scholar]
- 5.Sun H, Shi W. Analyses of mrp genes during Myxococcus xanthus development. J Bacteriol. 2001;183:6733–6739. doi: 10.1128/JB.183.23.6733-6739.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SK, Kaiser D. C-factor: A cell–cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell. 1990;61:19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- 7.Shimkets LJ, Rafiee H. CsgA, an extracellular protein essential for Myxococcus xanthus development. J Bacteriol. 1990;172:5299–5306. doi: 10.1128/jb.172.9.5299-5306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lobedanz S, Sogaard-Andersen L. Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus. Gene Dev. 2003;17:2151–2161. doi: 10.1101/gad.274203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SK, Kaiser D. Cell alignment required in differentiation of Myxococcus xanthus. Science. 1990;249:926–928. doi: 10.1126/science.2118274. [DOI] [PubMed] [Google Scholar]
- 10.Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Shimkets LJ. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu Rev Microbiol. 1999;53:525–549. doi: 10.1146/annurev.micro.53.1.525. [DOI] [PubMed] [Google Scholar]
- 12.Sogaard-Andersen L, et al. Coupling gene expression and multicellular morphogenesis during fruiting body formation in Myxococcus xanthus. Mol Microbiol. 2003;48:1–8. doi: 10.1046/j.1365-2958.2003.03399.x. [DOI] [PubMed] [Google Scholar]
- 13.Ogawa M, Fujitani S, Mao X, Inouye S, Komano T. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol Microbiol. 1996;22:757–767. doi: 10.1046/j.1365-2958.1996.d01-1725.x. [DOI] [PubMed] [Google Scholar]
- 14.Ellehauge E, Norregaard-Madsen M, Sogaard-Andersen L. The FruA signal transduction protein provides a checkpoint for the temporal coordination of intercellular signals in Myxococcus xanthus development. Mol Microbiol. 1998;30:807–817. doi: 10.1046/j.1365-2958.1998.01113.x. [DOI] [PubMed] [Google Scholar]
- 15.Ueki T, Inouye S. Activation of a development-specific gene, dofA, by FruA, an essential transcription factor for development of Myxococcus xanthus. J Bacteriol. 2005;187:8504–8506. doi: 10.1128/JB.187.24.8504-8506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueki T, Inouye S. Identification of a gene involved in polysaccharide export as a transcription target of FruA, an essential factor for Myxococcus xanthus development. J Biol Chem. 2005;280:32279–32284. doi: 10.1074/jbc.M507191200. [DOI] [PubMed] [Google Scholar]
- 17.Yoder-Himes D, Kroos L. Regulation of the Myxococcus xanthus C-signal-dependent Ω4400 promoter by the essential developmental protein FruA. J Bacteriol. 2006;188:5167–5176. doi: 10.1128/JB.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoder D, Kroos L. Mutational analysis of the Myxococcus xanthus Ω4400 promoter region provides insight into developmental gene regulation by C signaling. J Bacteriol. 2004;186:661–671. doi: 10.1128/JB.186.3.661-671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nariya H, Inouye S. A protein Ser/Thr kinase cascade negatively regulates the DNA-binding activity of MrpC, a smaller form of which may be necessary for the Myxococcus xanthus development. Mol Microbiol. 2006;60:1205–1217. doi: 10.1111/j.1365-2958.2006.05178.x. [DOI] [PubMed] [Google Scholar]
- 20.Sun H, Shi W. Genetic studies of mrp, a locus essential for cellular aggregation and sporulation of Myxococcus xanthus. J Bacteriol. 2001;183:4786–4795. doi: 10.1128/JB.183.16.4786-4795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viswanathan P, Kroos L. cis elements necessary for developmental expression of a Myxococcus xanthus gene that depends on C signaling. J Bacteriol. 2003;185:1405–1414. doi: 10.1128/JB.185.4.1405-1414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoder D, Kroos L. Mutational analysis of the Myxococcus xanthus Ω4499 promoter region reveals shared and unique properties in comparison with other C-signal-dependent promoters. J Bacteriol. 2004;186:3766–3776. doi: 10.1128/JB.186.12.3766-3776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viswanathan K, Viswanathan P, Kroos L. Mutational analysis of the Myxococcus xanthus Ω4406 promoter region reveals an upstream negative regulatory element that mediates C-signal dependence. J Bacteriol. 2006;188:515–524. doi: 10.1128/JB.188.2.515-524.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueki T, Inouye S. Identification of an activator protein required for the induction of fruA, a gene essential for fruiting body development in Myxococcus xanthus. Proc Natl Acad Sci USA. 2003;100:8782–8787. doi: 10.1073/pnas.1533026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 26.Majdalani N, Gottesman S. The Rcs phosphorelay: A complex signal transduction system. Annu Rev Microbiol. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 27.Pristovsek P, et al. Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box. J Biol Chem. 2003;278:17752–17759. doi: 10.1074/jbc.M301328200. [DOI] [PubMed] [Google Scholar]
- 28.Mouslim C, Latifi T, Groisman EA. Signal-dependent requirement for the coactivator protein RcsA in transcription of the RcsB-regulated ugd gene. J Biol Chem. 2003;278:50588–50595. doi: 10.1074/jbc.M309433200. [DOI] [PubMed] [Google Scholar]
- 29.Barnard A, Wolfe A, Busby S. Regulation at complex bacterial promoters: How bacteria use different promoter organizations to produce different regulatory outcomes. Curr Opin Microbiol. 2004;7:102–108. doi: 10.1016/j.mib.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Tullius T. Homeodomains: Together again for the first time. Structure. 1995;3:1143–1145. doi: 10.1016/s0969-2126(01)00250-7. [DOI] [PubMed] [Google Scholar]
- 31.Fried M, Crothers DM. Equilibria and kinetics of lac repressor–operator interactions by polyacrylamide gel electrophoresis. Nucl Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schar J, Sickmann A, Beier D. Phosphorylation-independent activity of atypical response regulators of Helicobacter pylori. J Bacteriol. 2005;187:3100–3109. doi: 10.1128/JB.187.9.3100-3109.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otten SL, Olano C, Hutchinson CR. The dnrO gene encodes a DNA-binding protein that regulates daunorubicin production in Streptomyces peucetius by controlling expression of the dnrN pseudo response regulator gene. Microbiology. 2000;146:1457–1468. doi: 10.1099/00221287-146-6-1457. [DOI] [PubMed] [Google Scholar]
- 34.Guthrie EP, et al. A response-regulator-like activator of antibiotic synthesis from Streptomyces coelicolor A3(2) with an amino-terminal domain that lacks a phosphorylation pocket. Microbiology. 1998;144(3):727–738. doi: 10.1099/00221287-144-3-727. [DOI] [PubMed] [Google Scholar]
- 35.Mutsuda M, Michel KP, Zhang X, Montgomery BL, Golden SS. Biochemical properties of CikA, an unusual phytochrome-like histidine protein kinase that resets the circadian clock in Synechococcus elongatus PCC 7942. J Biol Chem. 2003;278:19102–19110. doi: 10.1074/jbc.M213255200. [DOI] [PubMed] [Google Scholar]
- 36.Williams SB, Vakonakis I, Golden SS, LiWang AC. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: A potential clock input mechanism. Proc Natl Acad Sci USA. 2002;99:15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strayer C, et al. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- 38.Mangan S, Zaslaver A, Alon U. The coherent feed-forward loop serves as a sign-sensitive delay element in transcription networks. J Mol Biol. 2003;334:197–204. doi: 10.1016/j.jmb.2003.09.049. [DOI] [PubMed] [Google Scholar]
- 39.Shimkets LJ, Kaiser D. Murein components rescue developmental sporulation of Myxococcus xanthus. J Bacteriol. 1982;152:462–470. doi: 10.1128/jb.152.1.462-470.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higgs PI, Jagadeesan S, Mann P, Zusman DR. EspA, an orphan hybrid histidine protein kinase, regulates the timing of expression of key developmental proteins of Myxococcus xanthus. J Bacteriol. 2008;190:4416–4426. doi: 10.1128/JB.00265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kroos L. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Ann Rev Genet. 2007;41:13–39. doi: 10.1146/annurev.genet.41.110306.130400. [DOI] [PubMed] [Google Scholar]
- 42.Viswanathan P, Ueki T, Inouye S, Kroos L. Combinatorial regulation of genes essential for Myxococcus xanthus development involves a response regulator and a LysR-type regulator. Proc Natl Acad Sci USA. 2007;104:7969–7974. doi: 10.1073/pnas.0701569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY; Woodbury, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.