Abstract
Receptor tyrosine kinases (RTKs) process extracellular cues by activating a broad array of signaling proteins. Paradoxically, they often use the same proteins to elicit diverse and even opposing phenotypic responses. Binary, ‘on–off' wiring diagrams are therefore inadequate to explain their differences. Here, we show that when six diverse RTKs are placed in the same cellular background, they activate many of the same proteins, but to different quantitative degrees. Additionally, we find that the relative phosphorylation levels of upstream signaling proteins can be accurately predicted using linear models that rely on combinations of receptor-docking affinities and that the docking sites for phosphoinositide 3-kinase (PI3K) and Shc1 provide much of the predictive information. In contrast, we find that the phosphorylation levels of downstream proteins cannot be predicted using linear models. Taken together, these results show that information processing by RTKs can be segmented into discrete upstream and downstream steps, suggesting that the challenging task of constructing mathematical models of RTK signaling can be parsed into separate and more manageable layers.
Keywords: partial least-squares regression, protein microarray, PTB domain, receptor tyrosine kinase, SH2 domain
Introduction
Receptor tyrosine kinases (RTKs) constitute a large family of single-spanning membrane proteins found only in Metazoans (Robinson et al, 2000). Their primary role is to mediate intercellular communication by recognizing extracellular ligands and translating that information into an appropriate cellular response (Schlessinger, 2000). The intracellular region of an RTK contains a tyrosine kinase domain as well as several tyrosine residues that are phosphorylated upon receptor activation. These phosphotyrosines act as relays for information transmission, and the sequences surrounding these sites define signal specificity. Intracellular signaling proteins bind to these sites of tyrosine phosphorylation through Src homology 2 (SH2) or phosphotyrosine-binding (PTB) domains, initiating a variety of signaling cascades within the cell.
RTKs can elicit diverse and even opposing phenotypic responses, ranging from adhesion to migration, differentiation to proliferation, and survival to apoptosis (Schlessinger, 2000; Yarden and Sliwkowski, 2001). Although no two receptors feature identical sequences surrounding their pTyr sites, there is considerable qualitative overlap in the pathways they activate (Fambrough et al, 1999; Simon, 2000). The ability of RTKs to signal through common pathways, yet to induce diverse phenotypic responses, has largely been attributed to differences in cellular context, as signaling proteins are differentially expressed in different cell types (Jordan et al, 2000; Simon, 2000). For example, fibroblast growth factor receptor 1 (FGFR1) induces differentiation in neuronal cells, but induces proliferation in fibroblasts (Marshall, 1995; Lin et al, 1996). When expressed in the same cellular background, however, different RTKs have also been shown to elicit different phenotypic responses. For example, activation of epidermal growth factor receptor (EGFR) induces proliferation in PC12 neuronal cells, whereas activation of FGFR1 induces differentiation (Pollock et al, 1990; Lin et al, 1996). How, then, are intrinsic differences between RTKs manifested within the same cell type? Where does the information reside that defines these differences? How is that information processed?
To address these questions, we expressed six diverse RTKs in the same cellular background and monitored their signaling properties by quantitative immunoblotting. We found that although they activated many of the same signaling proteins, they did so to different degrees. We then used protein microarrays to define a quantitative interaction map for each receptor by measuring the affinity of almost every human SH2 and PTB domain for phosphopeptides representing pTyr sites on the receptors. Using partial least-squares regression (PLSR), we found that the relative phosphorylation levels of upstream signaling proteins could be accurately predicted using linear combinations of receptor-docking affinities, and that much of the predictive information resides in the docking sites for two central signaling proteins: phosphoinositide 3-kinase (PI3K) and Shc1. We also found that the relative phosphorylation levels of downstream proteins could not be predicted using linear models, suggesting that RTK signaling can be segmented into discrete upstream and downstream layers.
Results and discussion
To determine at a quantitative level how different RTKs behave when placed in the same cellular background, we selected six well-studied and phylogenetically diverse RTKs: EGFR, FGFR1, insulin-like growth factor 1 receptor (IGF1R), hepatocyte growth factor receptor (MET), neurotrophic tyrosine kinase receptor type 2 (NTRK2), and platelet-derived growth factor receptor β (PDGFRβ). Six stable cell lines were generated by transfecting the full-length coding region for each receptor into human embryonic kidney Flp-In-293 cells, which do not normally express these receptors at appreciable levels (Figure 1A). The resulting cell lines grew normally and, in each case, the receptor was produced at ∼105 copies per cell and activated by its cognate ligand in a dose-dependent manner (Supplementary Figure S1).
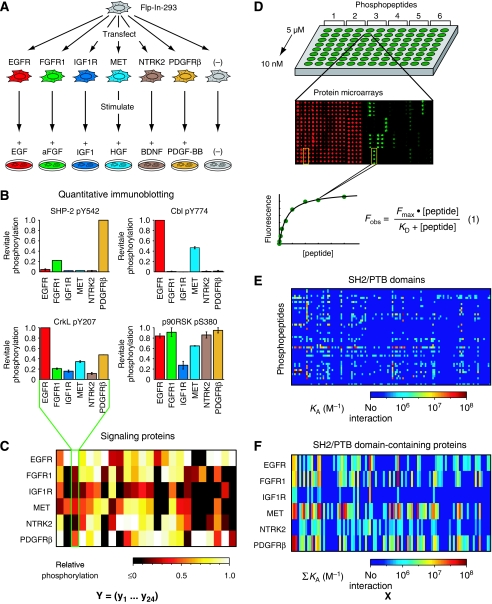
Figure 1.
Measurement of the intrinsic differences among six receptor tyrosine kinases. (A) The full-length coding regions for six RTKs were introduced into Flp-In-293 cells to generate stable cell lines. Each cell line was serum-starved for 24 h and stimulated for 5 min with a saturating concentration of the indicated growth factor. (B) Cell lysates were analyzed by quantitative immunoblotting to determine the relative levels of 24 phosphorylation sites on 23 signaling proteins across the six cell lines. Representative results are shown for four phosphorylation sites. Error bars indicate the range of biological duplicates. The other 20 bar graphs are provided in Supplementary Figure S2. (C) Heat map illustrating the relative levels of the 24 phosphorylation sites across the six cell lines. The columns of this matrix, Y, constitute relative phosphorylation vectors for each signaling event. (D) Protein microarrays comprising almost every human SH2 and PTB domain were printed in individual wells of 96-well microtiter plates and probed with eight concentrations of each phosphopeptide, ranging from 10 nM to 5 μM. Phosphopeptides were derived from established sites of tyrosine phosphorylation on the six RTKs. For each domain–peptide interaction, a saturation-binding curve was obtained and the observed fluorescence, Fobs, was fit to equation (1) to obtain an equilibrium dissociation constant, KD. (E) KD values were converted to KA values (KD=1/KA) and each phosphopeptide was represented as a vector of KA values. (F) Each receptor vector was defined as the sum of its constituent phosphopeptide vectors. The receptor-docking affinity matrix, X, comprises the six receptor vectors. Source data is available for this figure at www.nature.com/msb.
To obtain a broad and quantitative view of how each receptor activates intracellular signaling proteins, the six cell lines were serum-starved for 24 h and stimulated for 5 min with saturating levels of the appropriate ligand. This early time point was chosen because many signaling proteins peak in their phosphorylation levels within the first 10 min of stimulation and because we wanted to capture immediate, receptor-dependent signaling events without additional complications arising from feedback loops and other forms of network regulation. Quantitative immunoblotting was then used to measure the relative phosphorylation levels of a wide range of proteins that have previously been implicated in RTK signaling (Figure 1B and C). In total, we queried 65 sites of phosphorylation on 57 proteins and observed growth factor-induced phosphorylation of 24 sites on 23 proteins (Supplementary Table SI). To compare the phosphorylation levels of a given protein across the six cell lines, lysate concentrations were normalized, basal phosphorylation was subtracted, and each level was calculated relative to the maximum observed level for that site (Figure 1B; Supplementary Figure S2; Supplementary information). Duplicate experiments were in close agreement (r=0.91; Supplementary Figure S3A). Interestingly, each receptor induced a distinct pattern of phosphorylation. Some proteins, such as SHP-2 and Cbl, were phosphorylated in as few as two of the cell lines, while others, such as CrkL and p90RSK, were phosphorylated in all six (Figure 1B). For every site of phosphorylation, quantitative differences were observed across the six cell lines and the rank order varied depending on the site. Thus, although these six receptors have previously been shown to activate many of the same pathways, they do so to different degrees when placed in the same cellular context. What, then, accounts for these differences? As RTKs initiate signaling by recruiting proteins to sites of tyrosine phosphorylation (Schlessinger, 2000), we asked whether there was information in the recruitment properties of the pTyr sites on these receptors that could explain the observed differences.
Sites of tyrosine phosphorylation are recognized by either SH2 (Sadowski et al, 1986) or PTB domains (Kavanaugh and Williams, 1994). To obtain a genome-wide, unbiased, and quantitative measure of the recruitment potential of each receptor, we prepared protein microarrays comprising nearly every SH2 and PTB domain encoded in the human genome (Figure 1D; Supplementary Table SII) (Jones et al, 2006). We then probed these arrays with fluorescently labeled, 18-residue phosphopeptides with sequences derived from every known site of tyrosine phosphorylation on each of the six receptors (Supplementary Table SIII). Equilibrium dissociation constants (KD values) were obtained by probing the arrays with eight concentrations of each peptide and fitting the resulting fluorescence data (Supplementary information) to an equation that describes saturation binding (Figure 1D) (Jones et al, 2006). In total, we queried 96 SH2 domains and 37 PTB domains with 47 phosphopeptides and observed 652 interactions with KD⩽2 μM (Supplementary Table SIV). Weaker interactions could not be quantified using this approach. When we repeated this process, duplicate KD measurements were in close agreement (r=0.85; Supplementary Figure S3B) and the mean KD was used for subsequent analyses.
Of the 131 domains, 112 domains representing 74 different proteins bound at least one phosphopeptide. As anticipated, there was considerable qualitative overlap between the six receptors: 50 of these proteins recognized peptides from at least three receptors and 21 of them recognized peptides from at least five receptors (Supplementary Figure S4). In general, the domains that recognized the most receptors are those found in well-studied signaling proteins, including the lipid-modifying enzyme PI3K; the transcription factors Stat1 and Stat2; the non-receptor tyrosine kinases Src and Abl1; the guanine nucleotide exchange factor Vav2; the adaptor proteins Crk, CrkL, and Nck; and the scaffold proteins Shc1 and Grb7. Thus, if viewed in strictly binary terms, these phylogenetically diverse receptors differ very little in their recruitment properties with respect to these core signaling proteins. At the quantitative level, however, they differ substantially. For example, although five of the six receptors feature docking sites for the regulatory subunit of PI3K, there is only one low-affinity site (KD=590 nM) on IGF1R, but there are five sites, including one high-affinity site (KD=10 nM), on PDGFRβ. Quantitative differences in both the number of docking sites and the binding affinities at these sites may therefore explain the observed differences in signaling elicited by each receptor.
To test this hypothesis, we represented each phosphopeptide as a row vector of association constants, KA, with each element in the vector corresponding to a different SH2 or PTB domain-containing protein (Figure 1E). For proteins that contained two domains that bound the same peptide, the larger KA was used. In addition, the three isoforms of the regulatory subunit of PI3K were treated as a single protein as their SH2 domains behaved similarly. The binding vector for a given receptor was then defined as the sum of its phosphopeptide vectors to take into account the number of docking sites, as well as the affinities at each site. The implicit assumption in adding the phosphopeptide vectors is that multiple docking sites for the same protein within a given receptor act independently of each other. While this is probably not always true, it is the simplest way to combine the data and is a reasonable approximation. In addition, the phosphopeptide vectors were all weighted equally as the relative stoichiometry of phosphorylation at each pTyr site was not known. Thus, the intrinsic signaling capabilities of the six receptors was captured in the matrix X, which comprises six rows, one for each receptor, and 74 columns, one for each SH2 or PTB domain-containing protein (Figure 1F). In a similar manner, the cellular activity of the RTKs was captured in the matrix Y, which comprises six rows, one for each receptor, and 24 columns (y1…y24), one for each phosphorylation site that was monitored by immunoblotting (Figure 1C).
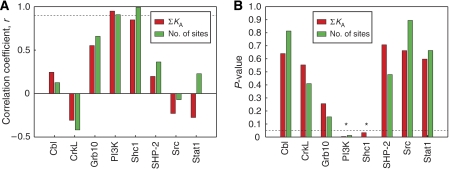
The simplest connection between the in vitro binding data and the cellular phosphorylation data is a one-to-one relationship in which the degree to which an SH2/PTB-containing protein is phosphorylated correlates linearly with its docking affinities. Of the eight proteins for which both microarray and immunoblotting data were obtained, significant correlations were observed for two: Shc1 (r=0.82, P=0.045) and PI3K (r=0.94, P=0.0059) (Figure 2). These correlations depend heavily on the number of Shc1- and PI3K-docking sites on each receptor. If the number of docking sites is taken into account but the affinities are ignored, the correlation actually improves for Shc1 (r=0.99, P=0.0001), but gets slightly worse for PI3K (r=0.91, P=0.013) (Figure 2). If the quantitative information is ignored and the interactions are treated as binary, correlations become meaningless as each protein recognized five of the six receptors. These results are consistent with a model in which Shc1 and PI3K interact directly with the activated receptors and are not influenced substantially by other docking proteins. For these two proteins, information processing is approximately linear and univariate.
Figure 2.
Correlations between docking affinities and relative phosphorylation levels for SH2 and PTB domain-containing proteins. (A) Correlation coefficients, r, were determined for the eight SH2 or PTB domain-containing proteins for which both docking affinities and relative phosphorylation levels were measured. Red bars show correlations using docking affinities (sum of KA values); green bars show correlations using only the number of docking sites with KD⩽2 μM. The dotted line indicates a correlation coefficient of 0.9. (B) P-values for the correlations shown in (A). The dotted line indicates a P-value of 0.05. P-values less than 0.05 are marked with an asterisk.
The same is not true, however, for the other SH2/PTB-containing proteins that were monitored by immunoblotting; significant correlations were not observed (Figure 2). For these proteins, the reductionist assumption that they bind directly to the receptor and act independently is too simplistic. Some proteins that contain SH2 or PTB domains have been shown to compete with each other for the same pTyr sites (Zhang et al, 2003), and many have been shown to interact with each other and with components of the cell membrane (Schlessinger and Lemmon, 2003). Thus, it is likely that they are inextricably interconnected. Are their relationships complex and nonlinear, or can they be approximated using relatively simple models that depend on combinations of docking affinities, rather than on single affinities alone?
The simplest multivariate model is one in which the phosphorylation levels of a given protein, yi, can be predicted using a linear combination of docking affinities. As the number of variables (docking affinities) exceeds the number of observations (RTKs), we used PLSR to regress each yi against X. PLSR reduces the dimensionality of X by decomposing it into a small number of orthogonal components that capture most of the covariance between X and yi. Each component is a linear combination of docking affinities, weighted by how much they contribute to predicting each immunoblot (yi). We found that four components were sufficient to capture ∼90% of the covariance with each yi. To guard against overfitting and to assess the predictive value of the docking affinities, we built our models using leave-one-out cross-validation: each model was trained using data from five receptors and then used to predict the immunoblotting data for the sixth receptor based on its docking affinities. This procedure was performed in all six combinations and the cross-validated residual between these predictions and the observed data, Q2, was calculated. To assess the significance of these predictions, we repeated our calculations 2000 times for each phosphorylation site using randomized X matrices and then calculated P-values for each model. Models were built using all of the microarray data, as well as subsets of the data that included only the SH2/PTB-containing proteins that bound at least two receptors, at least three receptors, at least four receptors, or at least five receptors. Similar results were obtained in every case, but the significance of the results increased as the number of variables was reduced (Supplementary Figure S5). Most of the information content in X resides in the 21 SH2/PTB-containing proteins that recognize at least five receptors and hence the results presented below are based on these data alone.
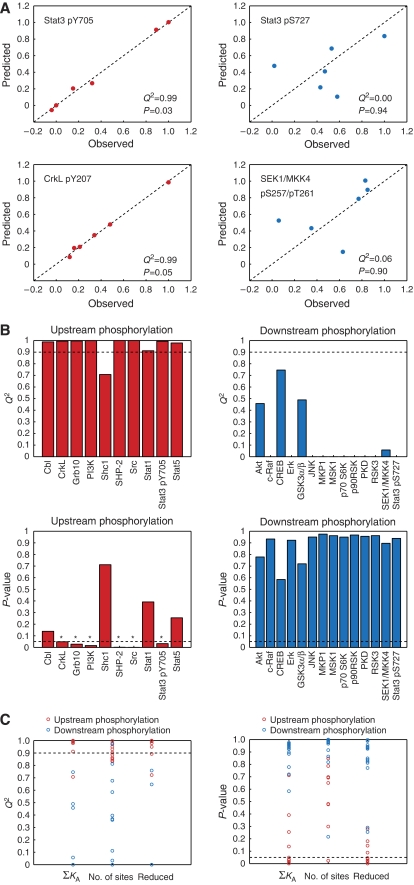
Of the 24 phosphorylation sites that we monitored by immunoblotting, nine were accurately predicted using linear combinations of docking affinities (Q2⩾0.9; Figure 3A and B; Supplementary Figure S6). Of these, six passed significance testing (P⩽0.05 and false discovery rate ⩽0.1). Interestingly, all nine of these sites are found on proteins that contain SH2 or PTB domains and therefore represent upstream signaling events (Figure 3B; Supplementary Figure S6). Moreover, only two phosphorylation sites that occur on SH2/PTB-containing proteins had a Q2 value less than 0.9: pTyr239/240 of Shc1 and pSer727 of Stat3. As noted earlier, the relative phosphorylation levels of Shc1 can be explained using only the number of Shc1-docking sites on each receptor (Figure 2); combinations of docking affinities are not required. More interesting is pSer727 of Stat3 (Q2=−0.33; P=0.94; Figure 3A). This serine residue is phosphorylated in a protein kinase C-dependent fashion and so represents a downstream signaling event (Aziz et al, 2007). In contrast, Tyr705 of Stat3 can be phosphorylated by the RTK itself and so represents an upstream event (Hwang et al, 2003); its phosphorylation is accurately predicted (Q2=0.99; P=0.03; Figure 3A). Similar to pSer727 of Stat3, the other 13 downstream signaling events could not be predicted using linear models (Figure 3B; Supplementary Figure S6). Thus, the phosphorylation sites that we monitored by immunoblotting naturally segregate into two groups: upstream phosphorylation events that are accurately predicted and downstream phosphorylation events that are not. From this, we submit that information processing by RTK signaling networks can be segmented into an upstream layer comprising proteins that are activated in an approximately linear manner through combinations of receptor-docking affinities and a downstream layer comprising proteins that are activated in a nonlinear manner. We note, however, that this result does not prove that the upstream step is linear mechanistically, but rather that this step can be approximated using relatively simple linear models.
Figure 3.
Linear combinations of docking affinities can predict upstream, but not downstream, signaling events. PLSR models were tested using leave-one-out cross-validation. (A) Predicted relative phosphorylation levels were plotted as a function of observed relative phosphorylation levels for each signaling event. Representative plots are shown for two upstream events (Stat3 pY705 and CrkL pY207) and two downstream events (Stat3 pS727 and SEK1/MKK4 pS257/pT261). The cross-validated residual, Q2, and the P-value are shown for each model. Plots for the other 20 models are provided in Supplementary Figure S6. (B) Q2 and P-values for all 24 PLSR models. Q2 values below zero were set to zero for display purposes. (C) Q2 and P-values for PLSR models built using: (1) the number of docking sites and the docking affinities (ΣKA); (2) only the number of docking sites (no. of sites); and (3) only the four variables with the highest VIP scores (reduced). Red circles represent upstream signaling events and blue circles represent downstream signaling events.
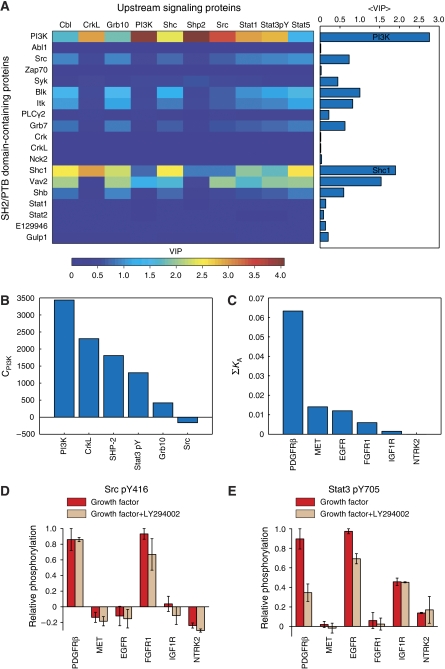
Both the number of docking sites and the docking affinities are important for predicting upstream signaling events. If only the number of docking sites is taken into account, the models perform less well and the results are less significant (Figure 3C). If only binary information is used (proteins are described either to interact or not interact with a receptor), all predictions fail as, at this level, the receptors are very similar (X is close to singular). To determine where most of the predictive information resides, we assessed the contribution of each SH2/PTB-containing protein to each PLSR model by calculating their variable importance in the projection (VIP; see Materials and methods). Reduced models were then prepared using only the most important variables. For all of the upstream signaling events, reduced models that included only the four most important variables performed almost as well as the full PLSR models (Figure 3C). On average, the two most important variables were PI3K and Shc1 (Figure 4A). In other words, much of the information needed to predict the relative phosphorylation levels of upstream signaling proteins resides in the number and affinity of PI3K- and Shc1-docking sites on the RTK.
Figure 4.
Contribution of SH2 and PTB domains in predicting the relative phosphorylation levels of upstream signaling proteins. (A) Heat map showing the variable importance in the projection (VIP) for each SH2 or PTB domain-containing protein in each PLSR model of upstream signaling events. The average across all 10 models is shown to the right. (B) Bar graph showing the coefficients for PI3K, CPI3K, in the six statistically significant PLSR models. (C) Bar graph showing the sum of PI3K-docking affinities for each RTK. (D, E) Relative phosphorylation levels for (D) Src pY416 and (E) Stat3 pY705 across the six cell lines, with and without PI3K inhibitor LY294002 (100 μM). Bar graphs for the other eight upstream signaling events are provided in Supplementary Figure 7. Source data is available for this figure at www.nature.com/msb
As these models are statistical in nature, this observation does not necessarily mean that PI3K and Shc1 have a causative function in determining the strength of signaling through other upstream proteins. PI3K- and Shc1-binding sites may have co-evolved with some other feature of RTKs that determines their ability to activate upstream proteins, such as kinase specificity or localization of the receptors to different membrane microdomains. Nevertheless, it is possible that these proteins do have a causative function in determining the extent to which other signaling proteins are activated.
This hypothesis cannot be tested by altering the abundance of PI3K or Shc1, as altering the composition of the cell would change the parameter values in the models. It is possible, however, to alter the catalytic activity of PI3K without altering its abundance using the small molecule inhibitor LY294002. If PI3K activity has a causative function in determining the degree to which upstream proteins are phosphorylated, we would expect LY294002 treatment to have the largest effect on proteins that have high PLSR coefficients for PI3K (Figure 4B), and on receptors with the strongest recruitment potential for PI3K (Figure 4C). We therefore stimulated all six cell lines in the presence or absence of LY294002 and assessed the relative phosphorylation levels of the upstream signaling proteins by immunoblotting (Figure 4D and E; Supplementary Figure S7; Supplementary information). Interestingly, the relative phosphorylation levels of Src, which has a low coefficient for PI3K (Figure 4B), were minimally affected by LY294002 treatment (Figure 4D), whereas the relative phosphorylation levels of Stat3, which has a high positive coefficient for PI3K (Figure 4B), were affected in a manner consistent with the number and affinity of PI3K-docking sites on the six RTKs (Figure 4E). This result suggests that, at least for Stat3, the contribution of PI3K in the PLSR model is, in part, dependent on its kinase activity. The same result was not observed, however, for all of the upstream signaling proteins (Supplementary Figure S7). The Stat3 result is not easily explained based on our current RTK wiring diagrams and a mechanistic understanding of this observation will require further investigation.
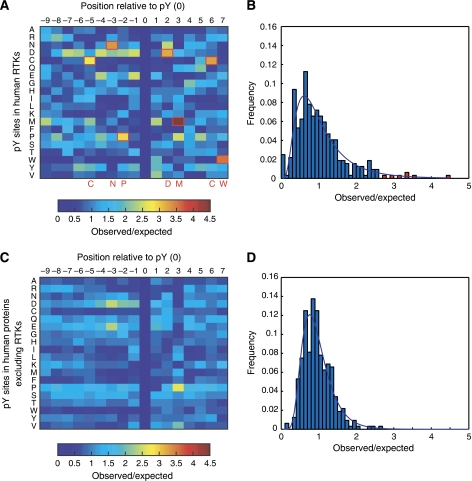
The overall importance of PI3K and Shc1 in RTK signaling was recently highlighted in a comprehensive map of the ErbB network, which revealed that a large fraction of information converges on a small number of signaling molecules, all of which can be modulated by PI3K and Shc1 (Oda et al, 2005). Interestingly, when we examined the sequences surrounding all known sites of tyrosine phosphorylation on human RTKs as reported in the Phospho. ELM database (Diella et al, 2008), we observed a distinct and significant (P<0.05) bias for sites that feature the consensus binding sequences for the PTB domain of Shc1 (NPXpY) (Songyang et al, 1995) and the SH2 domains of PI3K (pYXXM) (Songyang et al, 1993; Yaffe et al, 2001) (Figure 5A and B). This bias is not observed in known sites of tyrosine phosphorylation derived from all other human proteins (Figure 5C and D). Thus, we find that, despite activating many of the same proteins, intrinsic differences between RTKs are manifested in the degree to which they activate upstream signaling proteins and that much of this information resides in the number and affinity of docking sites for PI3K and Shc1. As these proteins lie upstream of the Akt and MAP kinase signaling pathways, and as these two pathways have been found repeatedly to have a central function in RTK biology, it is likely that our observations are not specific to HEK Flp-In-293 cells, but extend to more physiological settings as well.
Figure 5.
Bias for PI3K- and Shc1-binding sites in human RTKs. (A) Heat map showing the bias for each of the 20 amino acids at each position relative to sites of tyrosine phosphorylation in human RTKs. (B) Histogram of the observed/expected frequencies in (A). The line is a log-normal fit to the data. The red bars indicate significant deviations (P<0.05) and reflect biases (red squares in (A)). The biases for Cys at positions −5 and +6 and for Trp at position +7 are likely due to the conservation of structurally important residues at these locations relative to the conserved phosphotyrosine residue in the activation loop of the kinase domain. The biases for Asn at position −3 and Pro at position −2 match the consensus recognition sequence for the PTB domain of Shc1 (NPXpY). The bias for Met at position +3 matches the consensus recognition sequence for the SH2 domains of PI3K (pYXXM) and we frequently observe Asp at position +2 in phosphopeptides that are recognized by these domains. (C) Same as for (A), but using sites of tyrosine phosphorylation on all human proteins excluding RTKs. (D) Same as for (B), but using sites of tyrosine phosphorylation on all human proteins excluding RTKs.
Recently, Miller-Jensen et al (2007) showed that the phenotypic response of cells to external stimuli can be predicted using models that rely on linear combinations of a common set of downstream signaling proteins. Coupled with our results, this suggests that different RTKs may be able to elicit different phenotypic responses in the same cell type by activating a common set of signaling proteins, but to different quantitative degrees. In addition, our study, coupled with that of Miller-Jensen et al, supports a model in which information processing by RTK signaling networks can be segmented into three discrete layers: an upstream layer comprising proteins that are activated in a linear manner through combinations of receptor-docking affinities; an intermediate layer in which these signals are processed in a nonlinear manner; and a downstream layer in which integrators of signaling combine in a linear manner to determine cellular outcome. We submit that the difficult task of constructing mathematical models of RTK signaling can be parsed into discrete problems and that our greatest challenge lies in dissecting the middle layer.
Materials and methods
Cell culture, immunoblotting, ELISA, and protein microarray experiments
Stable cell lines were generated by co-transfecting Flp-In-293 cells (Invitrogen, Carlsbad, CA) with the plasmid pEF5/FRT/V5-DEST bearing the open reading frame for each RTK and the accessory plasmid pOG44 according to the manufacturer's directions (Invitrogen). Cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum, 2 mM glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 150 μg/ml hygromycin B. All cell culture and immunoblotting experiments were performed using standard procedures. Rabbit-derived primary antibodies were from Cell Signaling Technologies (Beverly, MA; Supplementary Table SI). For quantitative immunoblots, bands were detected with IRDye 680-labeled goat–anti-rabbit IgG (LI-COR Biosciences, Lincoln, NE) and imaged using an Odyssey Infrared Imaging System (LI-COR Biosciences). Expression levels of the RTKs were determined using ELISA kits from Invitrogen for EGFR and MET, and from R&D Systems (Minneapolis, MN) for NTRK2 and PDGFRβ. All protein microarray experiments were performed as described earlier (Jones et al, 2006; Kaushansky et al, 2008).
PLSR
To define the receptor-docking affinity matrix, X, the matrix of KD values (Supplementary Table SIV) was converted to a matrix of KA values (KA=1/KD). If a protein contained two domains that bound the same peptide, the higher KA value was used. Each phosphopeptide was then expressed as a row vector of KA values:
and each receptor was defined as the sum of its constituent phosphopeptide vectors:
The raw receptor-docking affinity matrix, Xraw, was then assembled from the six receptor vectors:
 |
The raw matrix was adjusted such that every SH2/PTB-containing protein (i.e. every column) was mean-centred and weighted according to its average affinity. This yielded the final receptor-docking affinity matrix, X. For the models presented in Figure 3, proteins that bound fewer than five receptors were removed from the matrix. Models obtained using all of the data or increasingly smaller subsets are shown in Supplementary Figure S5.
Relative phosphorylation levels of signaling proteins, as measured by immunoblotting, were calculated by first subtracting the level observed in the mock-treated, parental Flp-In-293 cell line and then dividing each value by the maximum observed value for that site across the six cell lines. Each phosphorylation site was treated as a separate vector, y, and each y was mean-centred and variance-normalized. A PLSR (Geladi and Kowalski, 1986) was then performed separately on each y. For cross-validation, each receptor (row ‘i') was removed once from both X and y, the regression was performed, and the resulting model was used to predict the value of yi. The residual, Q2, of this prediction was then compared with residuals generated from randomly shuffling X 2000 times. The distribution of these residuals was used to calculate the P-value of the observed Q2.
The weighted sum of squares (also known as the VIP) for each variable, k, was calculated according to equation (5):
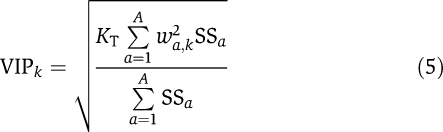 |
where KT is the total number of variables, a is the principal component, and SSa is the sum of squares for that component.
Amino-acid frequencies near sites of tyrosine phosphorylation
Experimentally determined sites of tyrosine phosphorylation in human proteins were acquired from the Phospho.ELM database (Diella et al, 2008). Of the 1397 identified sites, 196 were in RTKs and 1201 were in proteins other than RTKs. The amino-acid frequencies at positions upstream and downstream of pTyr sites were calculated and then normalized to the expected frequency of each amino acid in all human proteins (Echols et al, 2002). The resulting histograms of observed/expected frequencies were fit to a log-normal distribution from which P-values were calculated. All analyses were performed using MATLAB 7.4. (The MathWorks, Inc., Natick, MA). More detailed protocols are provided in Supplementary information.
Supplementary Material
Supplementary information: This supplementary file includes: Supplementary Methods; Supplementary References; Supplementary Figures 6, 7, 8, 9, 10, 11, 12; Supplementary Tables 1, 2, 3
Supplementary Table 4: Equilibrium dissociation constants for the binding of phosphopeptides to human SH2 or PTB domains.
Acknowledgments
We thank Jiunn-Ren Chen for helpful suggestions with the modeling and Jeffrey Knott, Susan Rogers, and Colleen Hunter for peptide synthesis. This study was supported by awards from the WM Keck Foundation, the Arnold and Mabel Beckman Foundation, and the Camille and Henry Dreyfus Foundation and by grants from the National Institutes of Health (R33 CA128726 and R21 CA126720). AG is the recipient of an NSF Graduate Research Fellowship, JAK is the recipient of a Howard Hughes Medical Institute Predoctoral Fellowship in the Biological Sciences, EMB and AK were supported in part by the NIH Molecular, Cellular, and Chemical Biology Training Grant (5 T32 GM07598-25), MS is the recipient of an Alfred and Isabel Bader fellowship and a Jacques-Émile Dubois fellowship, and JR is an employee of Cell Signaling Technology Inc.
Author contributions GM, AG, JAK, and EMB designed the study; AW-Y and MS generated the cell lines; JAK and EMB performed the cell-based experiments; JR supervised the peptide synthesis; AG and AK performed the protein microarray experiments; BHC contributed to the microarray experiments; AG performed the modeling experiments; AG, JAK, EMB, and GM interpreted the data; AG and GM wrote the paper, with contributions from JAK and EMB; and GM supervised the research.
Footnotes
John Rush is an employee of and stockholder in Cell Signaling Technologies Inc. (Danvers, MA) and Gavin MacBeath is an advisor for and stockholder in Merrimack Pharmaceuticals Inc. (Cambridge, MA) and Makoto Life Sciences Inc. (Bedford, MA).
References
- Aziz MH, Manoharan HT, Sand JM, Verma AK (2007) Protein kinase Cepsilon interacts with Stat3 and regulates its activation that is essential for the development of skin cancer. Mol Carcinog 46: 646–653 [DOI] [PubMed] [Google Scholar]
- Diella F, Gould CM, Chica C, Via A, Gibson TJ (2008) Phospho.ELM: a database of phosphorylation sites—update 2008. Nucleic Acids Res 36: D240–D244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols N, Harrison P, Balasubramanian S, Luscombe NM, Bertone P, Zhang Z, Gerstein M (2002) Comprehensive analysis of amino acid and nucleotide composition in eukaryotic genomes, comparing genes and pseudogenes. Nucleic Acids Res 30: 2515–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fambrough D, McClure K, Kazlauskas A, Lander ES (1999) Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell 97: 727–741 [DOI] [PubMed] [Google Scholar]
- Geladi P, Kowalski BR (1986) Partial least-squares regression: a tutorial. Anal Chim Acta 185: 1–17 [Google Scholar]
- Hwang JH, Kim DW, Suh JM, Kim H, Song JH, Hwang ES, Park KC, Chung HK, Kim JM, Lee TH, Yu DY, Shong M (2003) Activation of signal transducer and activator of transcription 3 by oncogenic RET/PTC (rearranged in transformation/papillary thyroid carcinoma) tyrosine kinase: roles in specific gene regulation and cellular transformation. Mol Endocrinol 17: 1155–1166 [DOI] [PubMed] [Google Scholar]
- Jones RB, Gordus A, Krall JA, MacBeath G (2006) A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature 439: 168–174 [DOI] [PubMed] [Google Scholar]
- Jordan JD, Landau EM, Iyengar R (2000) Signaling networks: the origins of cellular multitasking. Cell 103: 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushansky A, Gordus A, Chang B, Rush J, Macbeath G (2008) A quantitative study of the recruitment potential of all intracellular tyrosine residues on EGFR, FGFR1 and IGF1R. Mol Biosyst 4: 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh WM, Williams LT (1994) An alternative to SH2 domains for binding tyrosine-phosphorylated proteins. Science 266: 1862–1865 [DOI] [PubMed] [Google Scholar]
- Lin HY, Xu J, Ornitz DM, Halegoua S, Hayman MJ (1996) The fibroblast growth factor receptor-1 is necessary for the induction of neurite outgrowth in PC12 cells by aFGF. J Neurosci 16: 4579–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CJ (1995) Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80: 179–185 [DOI] [PubMed] [Google Scholar]
- Miller-Jensen K, Janes KA, Brugge JS, Lauffenburger DA (2007) Common effector processing mediates cell-specific responses to stimuli. Nature 448: 604–608 [DOI] [PubMed] [Google Scholar]
- Oda K, Matsuoka Y, Funahashi A, Kitano H (2005) A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol 1: 2005.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock JD, Krempin M, Rudy B (1990) Differential effects of NGF, FGF, EGF, cAMP, and dexamethasone on neurite outgrowth and sodium channel expression in PC12 cells. J Neurosci 10: 2626–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, Wu YM, Lin SF (2000) The protein tyrosine kinase family of the human genome. Oncogene 19: 5548–5557 [DOI] [PubMed] [Google Scholar]
- Sadowski I, Stone JC, Pawson T (1986) A noncatalytic domain conserved among cytoplasmic protein-tyrosine kinases modifies the kinase function and transforming activity of Fujinami sarcoma virus P130gag-fps. Mol Cell Biol 6: 4396–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J (2000) Cell signaling by receptor tyrosine kinases. Cell 103: 211–225 [DOI] [PubMed] [Google Scholar]
- Schlessinger J, Lemmon MA (2003) SH2 and PTB domains in tyrosine kinase signaling. Sci STKE 2003: RE12. [DOI] [PubMed] [Google Scholar]
- Simon MA (2000) Receptor tyrosine kinases: specific outcomes from general signals. Cell 103: 13–15 [DOI] [PubMed] [Google Scholar]
- Songyang Z, Margolis B, Chaudhuri M, Shoelson SE, Cantley LC (1995) The phosphotyrosine interaction domain of SHC recognizes tyrosine-phosphorylated NPXY motif. J Biol Chem 270: 14863–14866 [DOI] [PubMed] [Google Scholar]
- Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, Neel BG, Birge RB, Fajardo JE, Chou MM, Hanafusa H, Schaffhausen B, Cantley LC (1993) SH2 domains recognize specific phosphopeptide sequences. Cell 72: 767–778 [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Leparc GG, Lai J, Obata T, Volinia S, Cantley LC (2001) A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat Biotechnol 19: 348–353 [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX (2001) Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2: 127–137 [DOI] [PubMed] [Google Scholar]
- Zhang T, Ma J, Cao X (2003) Grb2 regulates Stat3 activation negatively in epidermal growth factor signalling. Biochem J 376: 457–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information: This supplementary file includes: Supplementary Methods; Supplementary References; Supplementary Figures 6, 7, 8, 9, 10, 11, 12; Supplementary Tables 1, 2, 3
Supplementary Table 4: Equilibrium dissociation constants for the binding of phosphopeptides to human SH2 or PTB domains.