Abstract
Often fatal, acute lung injury has a complicated etiology. Previous studies from our laboratory in mice have demonstrated that survival during acute lung injury is a complex trait governed by multiple loci. We also found that the increase in metallothionein (MT) is one of the greatest noted in transcriptome-wide analyses of gene expression. To assess the role of MT in nickel-induced acute lung injury, the survival of Mt-transgenic, Mt1/2(+/+), and Mt1/2(+/+) mice was compared. Pulmonary inflammation and global gene expression were compared in Mt1/2(+/+) and Mt1/2(+/+) mice. Gene-targeted Mt1/2(+/+) mice were more susceptible than Mt1/2(+/+) mice to nickel-induced inflammation, surfactant-associated protein B transcript loss, and lethality. Similarly, Mt-transgenic mice exhibited increased survival. MAPPFinder analyses also noted significant decreases in genes involved in protein processing (e.g., ubiquitination, folding), which were greater in Mt1/2(+/+) mice as compared with Mt1/2(+/+) mice early in the progression of acute lung injury, possibly due to a zinc-mediated transcript destabilization. In contrast, transcript levels of genes associated with the inflammatory response, extracellular matrix regulation, and coagulation/fibrinolysis were increased more in Mt1/2(+/+) mice as compared with Mt1/2(+/+) mice late in the development of acute lung injury. Thus, MT ultimately improves survival in the progression of acute lung injury in mice. Transcriptome-wide analysis suggests that this survival may be mediated through changes in the destabilization of transcripts associated with protein processing, the subsequent augmentation of transcripts controlling inflammation, extracellular matrix regulation, coagulation/fibrinolysis, and disruption of surfactant homeostasis.
Keywords: microarray, surfactant, inflammation, fibrinolysis, extracellular matrix
Acute lung injury is a severe clinical syndrome that occurs from multiple causes, including infection, trauma, and inhalation of irritants. Estimates of the incidence of acute lung injury range from 3 to 75 cases/100,000 individuals in the population per year. Pathologic conditions associated with the development of acute lung injury include diffuse alveolar damage, inflammatory cell influx and activation, pulmonary edema and hemorrhage, alteration of surfactant production, and insufficient gas exchange (1, 2). Resolution of acute lung injury is often problematic, as the mortality rate is 20–40% (1). Patients that do survive often have lasting adverse pulmonary difficulties, such as interstitial fibrosis and reduced lung compliance.
Previous studies from our laboratory have assessed aspects of the molecular mechanisms involved in the pathogenesis of acute lung injury in mice using inhaled nickel. The cumulative adverse effects of inflammation, extracellular matrix alterations, fibrinolysis, and disruption in surfactant homeostasis characterize lung injury in this and other models. In our mouse model of acute lung injury, one key event from nickel exposure is a decrease in the expression of lung surfactant–associated protein B (Sftpb) (3–6), which is essential for normal pulmonary function (7, 8).
In mice, as acute lung injury progresses, increases in metallothionein (MT)-1 transcript levels were identified to be one of the greatest increases noted in our transcriptome-wide analyses of gene expression after exposure to nickel (3) or hyperoxia (9). Metallothioneins are cysteine-rich, low molecular weight proteins that are primarily controlled at the transcriptional level (10). MT protein exists intracellularly, bound predominantly with zinc and, to a lesser extent, copper, and has been widely studied for its role in the detoxification and homeostasis of metals (11). In addition, the redox capacity of the metal-thiolate bonds allows MT to guard against reactive oxygen species (12, 13). Of the four mouse Mt genes, Mt1 and Mt2 are expressed in nearly all organs, and are induced by metals, glucocorticoids, and cytokines (14, 15).
Previously, MT was shown to be protective against LPS-induced lung inflammation and edema (16), although the molecular changes that differ between Mt1/2(+/+) and Mt1/2(+/+) wild-type mice during the development of lung injury are not well understood. The purpose of this study was to investigate the role of MT in the protection from nickel-induced acute lung injury using Mt-transgenic, Mt1/2(+/+), and Mt1/2(+/+) mice.
MATERIALS AND METHODS
Mice and Exposure Protocol
Mt1/2(+/+) mice (129S7/SvEvBrd-Mt1tm1Bri Mt2tm1Bri/J), with their respective 129S1/SvImJ Mt1/2(+/+) wild-type mice, as well as Mt-transgenic (C57BL/6J-Tg(Mt1)174Bri/J), with their respective C57BL/6J wild-type mice (males and females, aged 7–10 wk) were purchased from The Jackson Laboratory (Bar Harbor, ME). The Mt-transgenic mice used in this study were originally characterized by Palmiter and colleagues (17). Although equipped with 56 copies of the Mt1 transgene driven by the Mt1 promoter, these Mt-transgenic mice have approximately twice the level of basal lung MT protein as compared with controls. However, known Mt inducers can markedly increase transcript and protein levels of MT protein in the lungs of Mt-transgenic mice (18). All mice were housed in our animal facilities ⩾ 1 wk before exposure. Nickel aerosol was generated from 50 mM NiSO4•6H2O (Sigma, St. Louis, MO), and monitored as described previously (19). Mice were exposed to 150 ± 15 μg Ni2+/m3 in a 0.32-m3 stainless-steel inhalation chamber. For survival analyses, mice were continuously exposed, and survival time was recorded. All experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Cincinnati Medical Center.
Real-Time Quantitative PCR for Sftpb mRNA
Total RNA from lungs of Mt1/2(+/+) and Mt1/2(+/+) mice exposed to nickel for 72 h was reverse-transcribed into cDNA using the following reaction mixture: 200 ng total RNA from each sample in 10 μl RNase-free water (Invitrogen, Carlsbad, CA), 1.0 μl of oligo dT-15 (Promega, Madison, WI), 1.0 μl of 10 mM deoxynucleotide triphosphate mix (Invitrogen). The reaction mixture was incubated at 65°C for 5 min, and then chilled (4°C). First-strand buffer (5×, 4.0 μl), 2.0 μl 0.1 M DTT, 1.0 μl SuperScript II (Invitrogen,), and 1.0 μl RNasin (Promega) were then added to the reaction and further incubated at 42°C for 1 h. The reaction was terminated by heating the mixture at 70°C for 5 min, and then stored at 4°C. cDNA (2.0 μl) was used in the subsequent real-time quantitative PCR reaction using SYBR Green in a 50-μl reaction mixture containing 0.2 μM of each primer mixture and 25 μl of QuantiTech SYBR Green 2× PCR buffer (Qiagen, Valencia, CA). Prevalidated primer mixtures for Sftpb (Cat. No. PPM29254A) and the hypoxanthine guanine phosphoribosyl transferase 1 (Hprt1) housekeeping gene (Cat. No. PPM03559A) were purchased from SuperArray (Frederick, MD). Quantitative PCR was performed on an Applied Biosystems 7900HT System (Foster City, CA) with the following conditions: 95°C for 15 min, followed by 40 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s, followed by a dissociation curve analysis. For quantitation of data, the comparative Δ (ΔCT) method was used. ΔCT = CT (SFTPB) − CT (HPRT1), and this value was calculated for each sample, where CT = cycle number threshold. The ΔCT calculation involved finding the difference between each sample's ΔCT and its mean control ΔCT. These values were then transformed to absolute values using the formula where comparative expression level = 2 −Δ (ΔCT).
Bronchoalveolar Lavage
Bronchoalveolar lavage (BAL) fluid was collected from Mt1/2(+/+) and Mt1/2(+/+) mice (n = 4–6) exposed to nickel for 24, 48, or 72 h and compared with strain-matched, nonexposed control animals (n = 5). Mice were anesthetized with an intraperitoneal injection of 50 mg/kg of sodium pentobarbital, the diaphragm punctured, and the lungs lavaged with two 1-ml aliquots of Hanks' balanced salt solution without Ca2+ and Mg2+ (pH 7.2, 37°C; Invitrogen). Recovered BAL fluid samples were immediately placed on ice. Aliquots (200 μl) of lavage fluid were cytocentrifuged (Cytospin 3; Shandon Scientific Ltd., Astmoor, Runcorn, UK), and the cells were stained with Hemacolor (EM Science, Gibbstown, NJ) for differential cell analysis. Differential cell counts were performed by identifying at least 300 cells according to standard cytological procedures (20). Total cell counts were performed with a hemacytometer using trypan blue (Invitrogen). To assess the amount of hemoglobin in BAL, 900 μl of distilled, deionized H2O was added to 100 μl aliquots of BAL fluid, centrifuged at 3,000 rpm for 10 min, and then absorbance at 412 nm was measured (21). The remaining BAL fluid samples were centrifuged (500 × g, 5 min, 4°C), and the cell-free supernatants were decanted and stored at −80°C. Total BAL protein was measured using a bicinchoninic acid assay, with BSA used as the standard (Pierce, Rockford, IL). Total protein concentration in BAL fluid was used as an indicator of lung injury and changes in lung permeability.
RNA Expression Analysis by Oligonucleotide Microarray Analysis
Mt1/2(+/+) and Mt1/2(+/+) mice were exposed to aerosolized nickel for 3, 8, 24, 48, and 72 h. After exposure, mice were killed with pentobarbital (followed by exsanguinations), and the lungs were removed, placed in liquid nitrogen, and stored at −80°C. Total cellular RNA was isolated from frozen lung tissue with TRIzol (Invitrogen), and quantity was assessed by A260/A280 spectrophotometric absorbance (SmartSpec 3000; Bio-Rad, Hercules, CA). RNA quality was assessed by separation with a denaturing formaldehyde/agarose/ethidium bromide gel, and quantified by analysis with an Agilent Bioanalyzer (Quantum Analytics, Foster City, CA). To examine differential gene expression of 13,443 70-mer oligonucleotides, a microarray was fabricated by the Genomic and Microarray Laboratory, Center for Environmental Genetics, University of Cincinnati (http://microarray.uc.edu/), using a commercial library (Qiagen-Operon, Alameda, CA). Seventy-mer clones from the Operon Library were amplified by PCR and printed onto glass slides (Omnigrid Microarrayer; GeneMachines, San Carlos, CA). These clones consisted of 8,077 known genes, 5,017 RIKEN cDNAs, 210 nonannotated sequences/segments/IMAGE clones, and 139 genes for hypothetical proteins. Each exposure group and nonexposed control group consisted of nine mice. RNA samples from three mice per group were pooled for each microarray, and three separate microarrays per exposure group were compared with nonexposed controls using 20 μg total RNA per array. Each sample of mRNA was reverse-transcribed and randomly reciprocal-tagged with fluorescent Cyanine 3 (Cy3) or Cyanine 5 (Cy5) (e.g., Cy3 for nonexposed control and Cy5 for each exposure group). Cy3 and Cy5 samples were cohybridized with the printed 70-mers. After hybridization, slides were washed and scanned at 635 (Cy5) and 532 (Cy3) nm (GenePix 4000B; Axon Instruments, Inc., Union City, CA). Normalization of the data was performed in three steps for each microarray, as described previously (22).
Assessment of Microarray Data and Statistical Analysis
Statistical analysis of microarray data was performed by fitting the following mixed-effects linear model for each gene separately: Yijk = μ + Ai + Sj + Ck+ εijk, where: Yijk corresponds to the normalized log-intensity on the ith array (i = 1,..., 48), labeled with the kth dye (k = 1 for Cy5 and k = 2 for Cy3) and for the jth treatment condition; μ is the overall mean log-intensity; Ai is the effect of the ith array; Sj is the effect of the jth treatment condition; and Ck is the effect of the kth dye. Assumptions about model parameters were the same as described by Wolfinger and colleagues (23), with array effects assumed to be random, and treatment and dye effects assumed to be fixed. This model was fitted for each gene, and statistical significance of the differential expression between exposure groups after adjusting for the array and dye effects was assessed by calculating P values for corresponding linear contrasts. Multiple hypotheses testing adjustment was performed by calculating a false discovery rate (FDR) (24). The data normalization and the statistical analysis were performed using SAS statistical software (SAS Institute Inc., Cary, NC).
To further analyze the microarray dataset, we used MAPPFinder to dynamically link microarray data to the Gene Ontology Consortium hierarchy database (www.genmapp.org). MAPPFinder generates a gene expression profile at the level of biological processes, cellular components, and molecular functions that allows for identification of specific biological pathways that are being altered (25). We focused on the alteration of pathways that occur early (3 h) and late (72 h) in the progression of nickel-induced acute lung injury. The results, calculated using Fisher's Exact test, are expressed as a z score for a given pathway, and values ⩾ 2.0 were considered to be significant. GenMAPP was used to view microarray data on biological pathways (26).
BAL data and real-time quantitative PCR data are presented as means ± SEM. Significant differences among groups were identified by ANOVA. Individual comparisons between groups were confirmed by the two-tailed Student's t test. A P value < 0.05 was considered statistically significant.
RESULTS
Survival
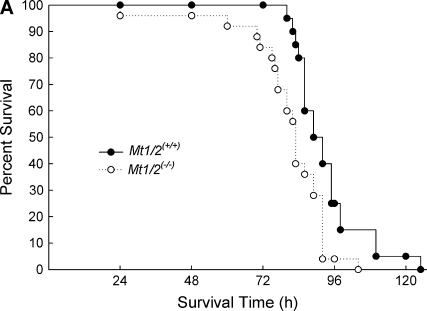
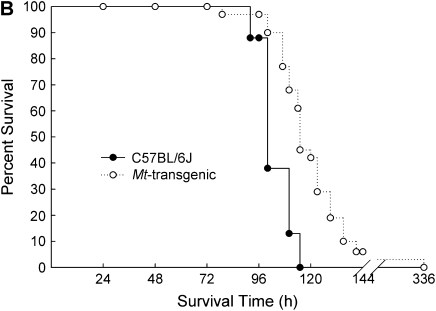
The absence or overexpression of Mt in mice had a significant influence on mean survival time (MST) after continuous exposure to nickel. In Figure 1A, Mt1/2(+/+) mice were more susceptible than strain-matched Mt1/2(+/+) 129S1/SvImJ mice (MST = 84.8 ± 3.3 h versus 96.8 ± 4.5 h, respectively). In contrast, Figure 1B shows that Mt-transgenic mice had a greater MST (124.8 ± 7.5 h) than strain-matched C57BL/6J mice (103.4 ± 2.7 h). The survival curves of Mt1/2(+/+) versus Mt1/2(+/+) mice and Mt-transgenic versus C57BL/6J mice were significantly different as determined by a Bayesian Weibull survival model (P < 0.05).
Figure 1.
Survival of Mt1/2(+/+) and Mt-transgenic mice exposed to nickel. (A) Increased susceptibility to nickel-induced lethality in the absence of Mt1 and Mt2. Mt1/2(+/+) (n = 10) and Mt1/2(+/+) (n = 10) mice were exposed to 150 ± 15 μg Ni2+/m3, and survival was assessed. (B) Protective effect of Mt1 overexpression on nickel-induced lethality. Mt-transgenic (n = 31) and C57BL/6J (n = 8) mice were exposed to 150 ± 15 μg Ni2+/m3, and survival times were recorded. The survival curves of Mt1/2(+/+) versus Mt1/2(+/+) mice and Mt-transgenic versus C57BL/BJ6 mice were significantly different as determined by a Bayesian Weibull survival model (P < 0.05).
SFTPB mRNA Levels
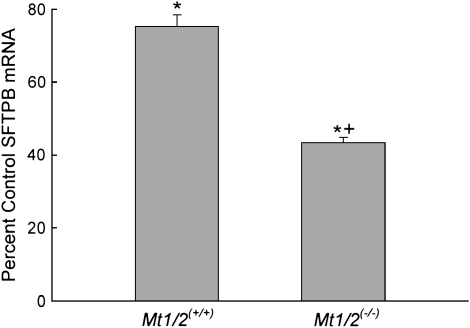
Previously, we reported that SFTPB transcript levels decrease in lungs of inbred mice exposed to nickel (3–6). In the present study, we determined by quantitative real-time RT-PCR that SFTPB mRNA levels in the lungs are decreased in Mt1/2(+/+) and Mt1/2(+/+) mice after a 72-h exposure compared with nonexposed, strain-matched control animals (Figure 2). The nickel-induced decrease of SFTPB mRNA is significantly greater in Mt1/2(+/+) mice (43% of control) than Mt1/2(+/+) mice (75% of control).
Figure 2.
Surfactant-associated protein B (SFTPB) lung mRNA levels in Mt1/2(+/+) and Mt1/2(+/+) mice exposed to 150 ± 15 μg Ni2+/m3 for 72 h. For quantitation of the data, the comparative cycle threshold Δ (ΔCT) method was used, where ΔCT = CT (SFTPB) − CT (HPRT1). The ΔCT calculation involved finding the difference between each sample's ΔCT and its corresponding mean nonexposed control ΔCT. Data are presented as percent of nonexposed, strain-matched control values (means ± SEM, n = 5 mice/group). *Significant difference from nonexposed control group, P < 0.05. +Significant difference from Mt1/2(+/+) group, P < 0.05.
BAL Measurements
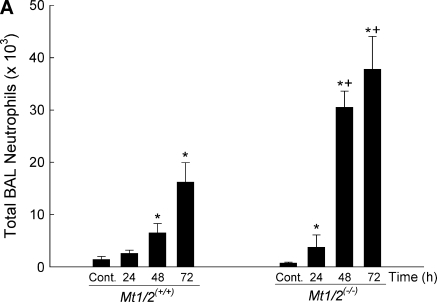
The absence of Mt also had an effect on inflammation (i.e., BAL neutrophils and macrophages) and lung permeability (i.e., total BAL protein) assessed in BAL fluid during nickel exposure. At 24 h, Mt1/2(+/+) mice had more BAL neutrophils compared with nonexposed control animals (Figure 3A). Both Mt1/2(+/+) and Mt1/2(+/+) mice had more BAL neutrophils after 48 and 72 h of exposure than did nonexposed control animals. However, Mt1/2(+/+) mice had more BAL neutrophils than Mt1/2(+/+) mice after 48 and 72 h of exposure. In addition, there was a suppression of macrophages in BAL, compared with nonexposed control animals, after NiSO4 exposure in both Mt1/2(+/+) and Mt1/2(+/+) mice, with Mt1/2(+/+) mice having more BAL macrophages than Mt1/2(+/+) mice after 72 h (7.05 ± 0.45 × 103 versus 2.56 ± 0.76 × 103; P < 0.05).
Figure 3.
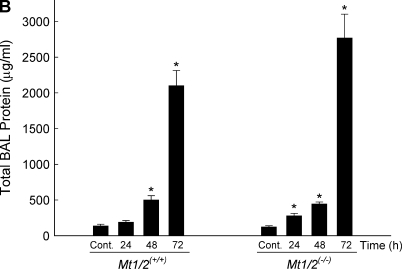
Total number of neutrophils (A), macrophages (B), and total protein (C) in bronchoalveolar lavage fluid of Mt1/2(+/+) and Mt1/2(+/+) mice exposed to 150 ± 15 μg Ni2+/m3 for 24, 48, or 72 h, or nonexposed control animals (n = 4–6 mice/group). Data are presented as means ± SEM. *Significant difference from nonexposed control group, P < 0.05. +Significant difference from Mt1/2(+/+) group, P < 0.05.
Total BAL protein was also increased in Mt1/2(+/+) and Mt1/2(+/+) mice after nickel exposure. Similar to BAL neutrophils, Mt1/2(+/+) mice had more total BAL protein after 24 h of exposure, and both Mt1/2(+/+) and Mt1/2(+/+) mice had more total BAL protein after 48 and 72 h of exposure compared with nonexposed control animals (Figure 3B). Nickel-exposed Mt1/2(+/+) mice also had more total BAL protein than nickel-exposed Mt1/2(+/+) mice at 24 h (278 ± 34 μg/ml versus 191 ± 21 μg/ml, respectively; P = 0.07) and 72 h (2,767 ± 336 μg/ml versus 2,101 ± 214 μg/ml, respectively; P = 0.13), although the difference was not statistically significant. Similarly, Mt1/2(+/+) mice had a greater amount of hemoglobin (an index of alveolar hemorrhage) in BAL fluid than did Mt1/2(+/+) mice after 72 h of nickel exposure, as assessed by measuring absorbance at 412 nm (0.029 ± 0.003 OD versus 0.014 ± 0.002 OD, respectively; P < 0.05).
Identification of Transcripts that Differed between Mt1/2(+/+) and Mt1/2(−/−) Mice
After nickel exposure, lung mRNA expression of Mt1/2(+/+) and Mt1/2(+/+) mice was analyzed using oligonucleotide microarrays at selected times. Total significant changes in transcripts (either increased or decreased levels as compared with nonexposed, strain-matched control animals, FDR ⩽ 0.1) at each time point were identified for the Mt1/2(+/+) and Mt1/2(+/+) strains.
Nonexposed Mt1/2(+/+) and Mt1/2(+/+) mice differed by only four transcripts. These differences included three transcription factors: CCAAT/enhancer binding protein delta (27), TSC22 domain family 3 (28), and Kruppel-like factor 9 (29), which were increased 2.2 ± 0.3–fold, 2.1 ± 0.3–fold, and 1.8 ± 0.3–fold in Mt1/2(+/+) mice compared with Mt1/2(+/+) mice, respectively.
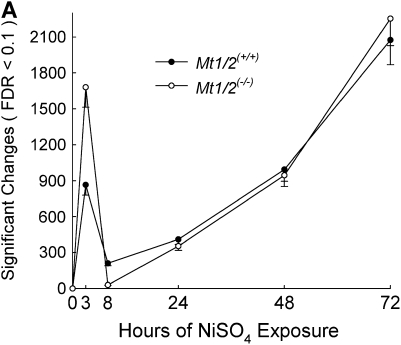
With the initiation of exposure, Mt1/2(+/+) mice were more responsive than Mt1/2(+/+) mice, and differences were greatest at the earlier times. For example, nearly twice as many significant changes in transcript levels in Mt1/2(+/+) mice than in Mt1/2(+/+) mice were noted at 3 h of nickel exposure (Figure 4A). At 8 h, however, more significant changes in transcript levels occurred in Mt1/2(+/+) mice than Mt1/2(+/+) mice. The number of significant transcript changes in Mt1/2(+/+) and Mt1/2(+/+) mice were nearly the same at 24, 48, and 72 h.
Figure 4.
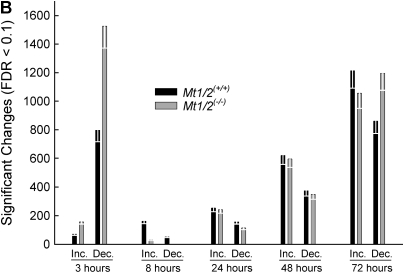
Identification of lung transcripts different between Mt1/2(+/+) and Mt1/2(+/+) mice after nickel exposure as assessed by oligonucleotide microarray analysis. Mt1/2(+/+) and Mt1/2(+/+) mice were exposed to 150 ± 15 μg Ni2+/m3 for 3, 8, 24, 48, and 72 h, and nonexposed (control), and lung mRNA was isolated and analyzed using oligonucleotide microarray (n = 3 arrays/time, 3 mice/array). (A) Total significant gene changes (false discovery rate [FDR] ⩽ 0.1). (B) Total increases and decreases (FDR ⩽ 0.1). In both (A) and (B), each white error bar represents the 0.1 FDR for each group.
Of the total significant changes in transcript levels in Mt1/2(+/+) and Mt1/2(+/+) mice at 3 h, ∼ 90% were decreases in both strains. However, twice as many transcripts decreased in Mt1/2(+/+) mice as compared with Mt1/2(+/+) mice at 3 h (Figure 4B). The number of increased and decreased transcripts was greater in Mt1/2(+/+) than in Mt1/2(+/+) mice at 8 h, but the response (as measured by the number of transcripts that changed) of each strain was similar at 24 and 48 h.
To further analyze the functional significance of the differences in gene expression, MAPPFinder was used to identify molecular pathways (using classification terms in the Gene Ontology Consortium categories of biological processes and molecular function) that contained an overrepresented number of altered transcript levels. After a 3-h exposure, several significantly altered molecular pathways (i.e., z score ⩾ 2.0 and at least 5 genes changed) were revealed with MAPPFinder and are presented in Table 1. Interestingly, many more molecular pathways significantly changed in Mt1/2(+/+) mice than in Mt1/2(+/+) mice (20 versus 2, respectively). Of those pathways that were altered in Mt1/2(+/+) mice and not in Mt1/2(+/+) mice, several were related to translational processes, such as protein biosynthesis (cytosolic ribosome, z score = 2.8; ribosome, z score = 2.4; ribosome biogenesis, z score = 2.1), ubiquitination (ubiquitin ligase complex, z score = 2.3), and transport (protein transporter activity, z score = 2.4). In addition, the molecular functional group of insulin-like growth factor binding genes was overrepresented (z score = 3.9) in Mt1/2(+/+) mice.
TABLE 1.
IDENTIFICATION OF MOLECULAR PATHWAYS ALTERED IN NICKEL-INDUCED ACUTE LUNG INJURY OF GENE-TARGETED MICE LACKING METALLOTHIONEIN 1 AND 2 [Mt1/2(−/−)] AND 129S1/SVIMJ STRAIN-MATCHED WILD-TYPE MICE [Mt1/2(+/+)] EXPOSED TO NICKEL FOR 3 h
|
Mt1/2(+/+)
|
Mt1/2(+/+)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Pathway/GO Name | Genes Changed | Genes Measured | z Score | Pathway/GO Name | Genes Changed | Genes Measured | z Score | |
| Regulation of cell growth | 11 | 21 | 4.1 | Interleukin receptor activity | 5 | 16 | 2.6 | |
| Insulin-like growth factor binding | 8 | 13 | 3.9 | Regulation of cell growth | 5 | 21 | 2.2 | |
| Growth factor binding | 7 | 11 | 3.9 | |||||
| Extracellular matrix structural constituent conferring tensile strength | 8 | 15 | 3.4 | |||||
| Nuclear pore | 7 | 13 | 3.2 | |||||
| Collagen | 8 | 17 | 3.0 | |||||
| Cytosolic ribosome | 11 | 26 | 2.8 | |||||
| Synaptosome | 5 | 10 | 2.5 | |||||
| Ribosome | 28 | 89 | 2.4 | |||||
| Protein transporter activity | 34 | 125 | 2.4 | |||||
| Transcription from Pol II promoter | 7 | 16 | 2.3 | |||||
| Lymph gland development | 5 | 11 | 2.3 | |||||
| Ubiquitin ligase complex | 5 | 8 | 2.3 | |||||
| Peroxisome | 12 | 36 | 2.2 | |||||
| DNA packaging | 6 | 10 | 2.2 | |||||
| Cytosol | 21 | 89 | 2.1 | |||||
| Chromatin | 8 | 22 | 2.1 | |||||
| Ribosome biogenesis | 14 | 38 | 2.1 | |||||
| Structural molecule activity | 21 | 90 | 2.0 | |||||
| Neuropeptide Y receptor activity | 7 | 19 | 2.0 | |||||
Definition of abbreviations: GO, Gene Ontology Consortium; Mt, metallothionein.
MAPPFinder was used to calculate the cumulative total of genes changed for a parent GO term and its children, and provides a statistical z Score to assess significance. z Score values ⩾ 2.0 were considered to be significant.
Based on the MAPPFinder results, and the observation that 90% of the total expression changes at 3 h were decreases in both Mt1/2(+/+) and Mt1/2(+/+) mice, we examined genes involved in pre- and posttranslational biological processes that were decreased in both strains. Thirty-five transcripts with an FDR < 0.1 were identified that were decreased more in Mt1/2(+/+) mice than in Mt1/2(+/+) mice (Table 2). Two transcripts, ubiquitin B and sorting nexin 12, were decreased in Mt1/2(+/+) mice but increased in Mt1/2(+/+) mice.
TABLE 2.
EXPRESSION DIFFERENCES IN GENES INVOLVED IN PRE- AND POST-TRANSLATIONAL PROCESSES IN THE LUNGS OF GENE-TARGETED MICE LACKING METALLOTHIONEIN 1 AND 2 [Mt1/2(−/−)] AND 129S1/SvImJ STRAIN-MATCHED WILD-TYPE MICE [Mt1/2(+/+)] EXPOSED TO NICKEL FOR 3 h
| Accession ID | Gene Name | Symbol | GO Biological Process | Mt1/2(+/+) Fold Change | Mt1/2(+/+) Fold Change | Mt1/2(−/−)/ Mt1/2(+/+) Ratio |
|---|---|---|---|---|---|---|
| NM_011664 | Ubiquitin B | Ubb | Protein modification | −2.3 ± 0.6 | +1.1 ± 0.3 | — |
| NM_018875 | Sorting nexin 12 | Snx12 | Protein transport | −2.2 ± 0.6 | +2.5 ± 1.4 | — |
| NM_011883 | Ring finger protein 13 | Rnf13 | Protein ubiquitination | −3.9 ± 1.2 | −1.5 ± 0.4 | 2.5 |
| NM_024267 | Importin 4 | Ipo4 | Protein transport | −4.1 ± 1.6 | −1.7 ± 0.8 | 2.4 |
| NM_026768 | Mitochondrial ribosomal protein S18A | Mrps18a | Protein biosynthesis | −2.6 ± 0.5 | −1.1 ± 0.2 | 2.3 |
| NM_018753 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | Ywhab | Protein targeting | −2.7 ± 0.9 | −1.1 ± 0.4 | 2.3 |
| NM_007597 | Calnexin | Canx | Protein folding | −2.7 ± 0.9 | −1.2 ± 0.3 | 2.2 |
| NM_013876 | Ring finger protein 11 | Rnf11 | Protein ubiquitination; ubiquitin-dependent protein catabolism | −2.4 ± 0.7 | −1.1 ± 0.3 | 2.2 |
| NM_145486 | Membrane-associated ring finger (C3HC4) 2 | March2 | Protein ubiquitination | −2.4 ± 0.5 | −1.1 ± 0.2 | 2.1 |
| NM_020600 | Ribosomal protein S14 | Rps14 | Protein biosynthesis; ribosome biogenesis | −2.2 ± 0.5 | −1.1 ± 0.2 | 2.0 |
| NM_019865 | Ribosomal protein L36a | Rpl36a | Protein biosynthesis; ribosome biogenesis | −3.8 ± 1.7 | −1.9 ± 0.9 | 2.0 |
| NM_030743 | Zinc finger protein 313 | Zfp313 | Protein ubiquitination | −2.5 ± 0.8 | −1.3 ± 0.4 | 1.9 |
| NM_013923 | Ring finger protein (C3HC4 type) 19 | Rnf19 | Protein ubiquitination | −2.3 ± 0.6 | −1.2 ± 0.3 | 1.9 |
| NM_013787 | S-phase kinase-associated protein 2 (p45) | Skp2 | Ubiquitin cycle | −6.3 ± 2.2 | −3.3 ± 1.7 | 1.9 |
| NM_023651 | Peroxisomal biogenesis factor 13 | Pex13 | Intracellular protein transport | −2.1 ± 0.5 | −1.2 ± 0.3 | 1.8 |
| NM_022997 | Vacuolar protein sorting 35 | Vps35 | Protein transport | −2.2 ± 0.6 | −1.3 ± 0.4 | 1.8 |
| NM_025586 | Ribosomal protein L15 | Rpl15 | Protein biosynthesis | −2.2 ± 0.5 | −1.3 ± 0.3 | 1.7 |
| AF100956 | Zinc finger protein 297 | Zfp297 | Protein folding | −2.0 ± 0.6 | −1.2 ± 0.3 | 1.7 |
| NM_024478 | GrpE-like 1, mitochondrial | Grpel1 | Protein folding | −2.3 ± 0.5 | −1.4 ± 0.3 | 1.7 |
| NM_009078 | Ribosomal protein L19 | Rpl19 | Protein biosynthesis | −2.5 ± 0.6 | −1.5 ± 0.4 | 1.7 |
| NM_138946 | Ribosomal protein S18 | Rps18 | Protein biosynthesis | −2.3 ± 0.4 | −1.4 ± 0.3 | 1.6 |
| NM_011187 | Proteasome (prosome, macropain) subunit, beta type 7 | Psmb7 | Ubiquitin-dependent protein catabolism | −2.1 ± 0.4 | −1.3 ± 0.3 | 1.6 |
| NM_007475 | Acidic ribosomal phosphoprotein P0 | Arbp | Ribosome biogenesis and assembly | −2.7 ± 0.7 | −1.7 ± 0.4 | 1.6 |
| NM_026693 | Gamma-aminobutyric acid (GABA-A) receptor-associated protein-like 2 | Gabarapl2 | Intracellular protein transport | −3.6 ± 1.1 | −2.4 ± 0.8 | 1.5 |
| NM_019727 | Sorting nexin 1 | Snx1 | Intracellular protein transport | −2.9 ± 0.9 | −1.8 ± 0.7 | 1.5 |
| NM_021433 | Syntaxin 6 | Stx6 | Intracellular protein transport | −2.1 ± 0.5 | −1.4 ± 0.5 | 1.5 |
| NM_025481 | SMAD specific E3 ubiquitin protein ligase 2 | Smurf2 | Ubiquitin cycle | −2.0 ± 0.5 | −1.3 ± 0.3 | 1.5 |
| NM_013911 | F-box and leucine-rich repeat protein 12 | Fbxl12 | Ubiquitin cycle | −2.4 ± 0.6 | −1.7 ± 0.4 | 1.5 |
| NM_009082 | Ribosomal protein L29 | Rpl29 | Protein biosynthesis; ribosome biogenesis | −2.0 ± 0.6 | −1.3 ± 0.3 | 1.5 |
| NM_012052 | Ribosomal protein S3 | Rps3 | Protein biosynthesis | −2.2 ± 0.5 | −1.6 ± 0.4 | 1.4 |
| NM_011300 | Ribosomal protein S7 | Rps7 | Protein biosynthesis | −2.0 ± 0.4 | −1.5 ± 0.6 | 1.4 |
| NM_026405 | RAB32, member RAS oncogene family | Rab32 | Protein transport | −2.4 ± 0.4 | −1.9 ± 0.4 | 1.3 |
| NM_017478 | Coatomer protein complex, subunit gamma 2 | Copg2 | Protein transport | −2.3 ± 0.6 | −2.0 ± 0.9 | 1.2 |
| NM_030559 | Vacuolar protein sorting 16 (yeast) | Vps16 | Intracellular protein transport | −2.1 ± 0.7 | −1.7 ± 0.6 | 1.2 |
| NM_029782 | Calreticulin 3 | Calr3 | Protein folding | −2.2 ± 0.5 | −1.9 ± 0.6 | 1.2 |
| NM_025592 | Ribosomal protein L35 | Rpl35 | Protein biosynthesis | −2.0 ± 0.6 | −1.8 ± 0.6 | 1.1 |
| NM_024212 | Ribosomal protein L4 | Rpl4 | Protein biosynthesis | −2.2 ± 0.5 | −2.0 ± 0.6 | 1.1 |
Definition of abbreviations: GO, Gene Ontology Consortium; Mt, metallothionein.
Values are mean fold changes compared to corresponding nonexposed control values ± SEM, with an Mt1/2(+/+) to Mt1/2(+/+) expression ratio. Genes were considered statistically significant if average intensity > 300 and false discovery rate ⩽ 0.1. Gene ontology biological processes were derived from the Gene Ontology Consortium (MGI).
MAPPFinder analysis of oligonucleotide microarray gene expression data from lungs of Mt1/2(+/+) and Mt1/2(+/+) mice exposed to nickel for 72 h demonstrated that there were few differences in the number and type of changed pathways. However, within pathways that changed in both strains (e.g., cytokine activity, chemotaxis, extracellular matrix, and coagulation/fibrinolysis), transcripts of several genes that could contribute to lung inflammation and injury were increased more in Mt1/2(+/+) mice than in Mt1/2(+/+) mice (Table 3).
TABLE 3.
EXPRESSION DIFFERENCES IN GENES INVOLVED IN THE INFLAMMATORY RESPONSE, CHEMOTAXIS, EXTRACELLULAR MATRIX REGULATION, AND COAGULATION AND FIBRINOLYSIS IN THE LUNGS OF GENE-TARGETED MICE LACKING METALLOTHIONEIN 1 AND 2 [Mt1/2(−/−)] AND 129S1/SvImJ STRAIN-MATCHED WILD-TYPE MICE [Mt1/2(+/+)]EXPOSED TO NICKEL FOR 72 h
| Accession ID | Gene Name | Symbol | Mt1/2(+/+) Fold Change | Mt1/2(+/+) Fold Change | Mt1/2(−/−)/Mt1/2(+/+) Ratio | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Inflammatory Response/Chemotaxis | ||||||||||
| NM_011330 | Small chemokine (C-C motif) ligand 11 | Ccl11 | 6.9 ± 1.7 | 2.4 ± 0.6 | 2.9 | |||||
| NM_009704 | Amphiregulin | Areg | 7.9 ± 2.4 | 3.6 ± 1.0 | 2.2 | |||||
| NM_011332 | Chemokine (C-C motif) ligand 17 | Ccl17 | 8.3 ± 1.8 | 3.9 ± 0.7 | 2.1 | |||||
| NM_007577 | Complement component 5, receptor 1 | C5r1 | 3.4 ± 0.7 | 1.7 ± 0.4 | 2.0 | |||||
| NM_009140 | Chemokine (C-X-C motif) ligand 2 | Cxcl2 | 5.2 ± 2.0 | 2.7 ± 1.0 | 1.9 | |||||
| AI323594 | Chemokine (C-C motif) ligand 2 | Ccl2 | 23.5 ± 12.4 | 12.8 ± 6.3 | 1.8 | |||||
| NM_020509 | Resistin-like α | Retnla | 11.9 ± 3.0 | 7.9 ± 1.8 | 1.7 | |||||
| NM_008361 | IL-1β | Il1b | 6.8 ± 2.0 | 4.5 ± 1.4 | 1.5 | |||||
| NM_015783 | INF-α–inducible protein | G1p2 | 2.7 ± 1.0 | 2.0 ± 0.7 | 1.4 | |||||
| NM_010798 | Macrophage migration inhibitory factor | Mif | 2.5 ± 0.8 | 1.8 ± 0.5 | 1.4 | |||||
| NM_013611 | Nodal | Nodal | 2.2 ± 0.5 | 1.6 ± 0.4 | 1.4 | |||||
| NM_019568 | Chemokine (C-X-C motif) ligand 14 | Cxcl14 | 3.3 ± 0.6 | 2.5 ± 0.4 | 1.3 | |||||
| NM_009402 | Peptidoglycan recognition protein 1 | Pglyrp1 | 3.0 ± 0.6 | 2.3 ± 0.5 | 1.3 | |||||
| NM_008336 | INF-α family, gene B | Ifnab | 2.2 ± 0.4 | 2.0 ± 0.4 | 1.2 | |||||
| NM_008334 | INF-α family, gene 7 | Ifna7 | 2.1 ± 0.5 | 1.7 ± 0.4 | 1.2 | |||||
| M58004 | Chemokine (C-C motif) ligand 6 | Ccl6 | 2.0 ± 0.5 | 1.7 ± 0.4 | 1.2 | |||||
| Extracellular Matrix | ||||||||||
| NM_010217 | Connective tissue growth factor | Ctgf | 4.3 ± 1.2 | 1.7 ± 0.5 | 2.5 | |||||
| X56304 | Tenascin C | Tnc | 31.2 ± 11.0 | 15.1 ± 5.3 | 2.1 | |||||
| NM_013599 | Matrix metalloproteinase 9 | Mmp9 | 3.1 ± 0.6 | 1.6 ± 0.3 | 1.9 | |||||
| NM_011593 | Tissue inhibitor of metalloproteinase 1 | Timp1 | 29.2 ± 6.0 | 16.3 ± 3.3 | 1.8 | |||||
| NM_009928 | Procollagen, type XV | Col15a1 | 2.2 ± 0.3 | 1.5 ± 0.2 | 1.5 | |||||
| NM_011580 | Thrombospondin 1 | Thbs1 | 26.7 ± 8.6 | 19.7 ± 6.4 | 1.4 | |||||
| D28599 | Chondroitin sulfate proteoglycan 2 | Cspg2 | 9.9 ± 2.5 | 7.5 ± 1.9 | 1.3 | |||||
| NM_009369 | Transforming growth factor β–induced | Tgfbi | 2.9 ± 0.5 | 2.2 ± 0.4 | 1.3 | |||||
| NM_008608 | Matrix metalloproteinase 14 (membrane-inserted) | Mmp14 | 4.1 ± 0.7 | 3.3 ± 0.6 | 1.2 | |||||
| NM_008485 | Laminin γ2 | Lamc2 | 2.9 ± 0.5 | 2.4 ± 0.4 | 1.2 | |||||
| NM_009614 | A disintegrin and metalloproteinase domain 15 (metargidin) | Adam15 | 2.0 ± 0.4 | 1.6 ± 0.3 | 1.2 | |||||
| NM_007925 | Elastin | Eln | 4.0 ± 0.9 | 3.6 ± 0.9 | 1.1 | |||||
| NM_007993 | Fibrillin 1 | Fbn1 | 2.1 ± 0.4 | 2.0 ± 0.4 | 1.1 | |||||
| Coagulation and Fibrinolysis | ||||||||||
| NM_008871 | Serine (or cysteine) proteinase inhibitor, clade E, member 1 | Serpine1 | 14.1 ± 7.6 | 4.4 ± 1.8 | 3.2 | |||||
| NM_008872 | Plasminogen activator, tissue | Plat | 8.7 ± 2.0 | 4.9 ± 1.2 | 1.8 | |||||
| NM_011113 | Urokinase plasminogen activator receptor | Plaur | 3.8 ± 0.8 | 2.8 ± 0.6 | 1.4 | |||||
Definition of abbreviation: Mt, metallothionein.
Values are mean fold changes over corresponding nonexposed control values ± SEM, with a Mt1/2(+/+) to Mt1/2(+/+) expression ratio. Genes were considered statistically significant if average intensity > 300 and false discovery rate (FDR) ⩽ 0.1.
Lung MT2 mRNA expression was significantly induced in a time-dependent manner in Mt1/2(+/+) mice after nickel exposure (increased 4.4 ± 1.3–fold at 3 h, 10.1 ± 2.6–fold at 8 h, 15.4 ± 4.0–fold at 24 h, 16.9 ± 4.3–fold at 48 h, and 33.6 ± 8.6–fold at 72 h). Although an oligonucleotide for MT1 was not on the microarray, because MT1 and MT2 transcripts are coordinately expressed from the same gene locus, it is likely that MT1 followed the same trend as we observed previously in our model of acute lung injury (3).
DISCUSSION
MT Induction Leads to Protection during Acute Lung Injury
Based on the observation that the increase in MT mRNA is one of the largest among all mRNAs assayed after nickel-induced acute lung injury, and because of its putative role in protection from several cellular stressors, we tested and determined that MT is protective against nickel-induced acute lung injury. Aside from its known role of binding and sequestering certain metals, the mechanism by which MT elicits protection after treatment of cells and animals to different and chemically diverse cellular stressors, is still controversial, although studied extensively (10). MT can efficiently scavenge hydroxyl radicals in vitro (13). It may be argued, however, that because MT concentrations are low relative to the concentration of other scavengers (e.g., reduced glutathione and enzymes that handle hydroxyl radicals), this cellular role for MT may be modest. Although MT can also protect against other reactive oxygen species, such as superoxide and hydrogen peroxide (30), Zn-MT, the major cellular form of MT in vivo, may not function as effectively as MT in scavenging reactive oxygen species. Nickel has been shown to cause mild oxidative stress at high concentrations (31), but there is little evidence that ionic nickel (Ni2+) directly participates in redox cycling that could generate reactive oxygen species in the lung (32). It is also unlikely that the protective effects of MT involve the direct sequestration of nickel by MT because MT has a low affinity for nickel as compared with other metals (33).
A more reasonable explanation for MT protection throughout nickel-induced acute lung injury may be indirect and related to the capability of MT to bind and release copper and zinc. Although labile, metal ions do not typically exist as free metals (e.g., free copper or free zinc), but delivery of metals to metalloproteins is assisted by metallochaperones that protect by guiding metals to specific targeted compartments and proteins (34). About one-third of structurally characterized proteins are metalloproteins, requiring metal cofactors for function. The assembly and activity of these proteins is a complex process requiring protein-protein interactions, often involving several accessory and helper proteins.
MT is known to bind Cu1+ with high affinity, and protects against copper-induced toxicity (35). Little is known regarding the effect of nickel-on-copper sequestration or copper-catalyzed oxidative damage, but it is clear that Cu2+ must remain tightly associated with proteins within cells. Free Cu2+ reacts with cellular reductants, such as ascorbate to produce Cu1+, which is redox-labile, and may produce hydroxyl radicals. Although nickel is unlikely to participate in redox reaction under physiologic conditions, it is likely that mice experienced oxidative stress during acute lung injury, because numerous transcripts regulated by oxidative stress (including glutamate-cysteine ligase, catalytic subunit, glutathione S-transferase α 1, thioredoxin reductase 1, and heme oxygenase 1) progressively increased during acute lung injury. The accompanying activation of inflammatory cells can also initiate reactive oxygen species formation. The resultant oxidative stress can lead to release of copper from MT, and thereby enhance the formation of reactive oxygen species and potentiate cellular damage (36).
In addition, MT plays an essential role in zinc homeostasis (sequestration and presentation), and Zn-MT may be viewed as a labile pool of cellular zinc for multiple zinc-containing metalloproteins. Physiologic evidence supporting this hypothesis is indirect, but can be derived from MT's capacity for preserving pregnancy under conditions of zinc deficiency (37, 38). Thus, the induction of additional MT could represent an attempt to chaperone zinc, thereby assisting cells in combating cellular stresses by providing zinc for cellular processes, including furnishing zinc to metalloproteins. Although, the mechanism for this function of MT is untested, the relationship of Mt gene expression to zinc homeostasis is evident. The Mt1 and Mt2 genes are regulated by metal response element binding transcription factor (MTF)-1, which is a zinc-sensing transcription factor (39). The strong induction of Mt gene expression in nickel-induced acute lung injury is likely to have resulted from zinc-mediated activation of MTF1. In addition, hypoxia can develop during respiratory insufficiency and can augment MTF1 activation (40). Nickel, like cobalt, mimics hypoxia, and can lead to stabilization of hypoxia-induced factor-1α (41–43). The activation of MTF1 may be from zinc that is displaced from labile intracellular binding sites, or altered transport into or out of the cell. In the presence of adequate MT levels, or when transgenic MT is induced, MT can quickly sequester zinc, and homeostasis is maintained. It should also be noted that the source of zinc is unlikely to be due to displacement from Zn-MT by nickel because nickel does not bind to MT.
Initial Decrease in Transcript Levels Is Greater in Mt1/2(−/−) than in Mt1/2(+/+) Mice after Nickel Exposure
Alterations in specific molecular pathways during acute lung injury, identified using oligonucleotide microarray and MAPPFinder analyses, differed between Mt1/2(+/+) and Mt1/2(+/+) mice after nickel exposure. A major advantage of this type of analysis is that it creates an understanding of the biology occurring within the dataset, in contrast to hierarchical clustering or self-organizing maps that arrange genes according to similarity in pattern of gene expression (44). It was interesting to note that before exposure, lung mRNA levels for all but four transcripts in Mt1/2(+/+) mice were comparable to those of Mt1/2(+/+) mice. This is consistent with previous studies that demonstrate little overt pulmonary phenotype in Mt1/2(+/+) mice when compared with Mt1/2(+/+) mice (45).
In contrast, the magnitude and dimension of the transcripts that differed between Mt1/2(+/+) and Mt1/2(+/+) mice was more evident during acute lung injury. Of the pathways that initially decreased more in Mt1/2(+/+) mice than in Mt1/2(+/+) mice, many were related to protein processing. Protein processing is critical to proper assembly and maturation of biologically active forms of peptides, especially secreted proteins such as SFTPB (46). Interestingly, 90% of the total significant gene changes in Mt1/2(+/+) and Mt1/2(+/+) mice at 3 h were mRNA decreases. This large-scale decrease in mRNA levels as an early response to stress is similar to that observed previously with viral infection of lung cells in vitro (47). Furthermore, twice as many decreases in different transcripts were observed in Mt1/2(+/+) mice as compared with Mt1/2(+/+) mice at 3 h, suggesting that, in the absence of Mt, this early injury involves factors responsible for normal transcriptional processing. Rapid large-scale decreases in global mRNA levels are likely a result of transcript degradation rather than diminished transcription or elongation of transcripts. Regulation of eukaryotic mRNA stability is often controlled by AU-rich elements, which are regulatory motifs in the 3′ untranslated region of many transcripts that serve as targets for RNA-binding proteins. Many of the identified RNA-binding proteins (e.g., zinc finger 36 or tristetraprolin) contain zinc-binding domains, and represent a mechanism for the zinc stabilitization of AU-rich elements containing mRNAs (48). Moreover, although less is known about the mechanism of regulated mRNA stability in mammalian cells, mRNA decay in yeast during nonpermissive heat stress is selective and is enriched in genes involved in protein biosynthesis and ribosomal biogenesis (49, 50). The repertoire of selectively decreased mRNAs found in yeast under heat stress was similar to our finding in the mouse lung during acute lung injury, and support genome-wide mRNA stability as a possible mechanism by which MT could participate in the overall surveillance of the transcriptome.
Development of Transcript Differences between Mt1/2(+/+) and Mt1/2(−/−) Mice as Lung Injury Progressed
In contrast to the initial destabilization and massive decrease in transcript levels, the increase in levels of transcripts associated with the regulation of inflammation was augmented more in Mt1/2(+/+) mice than in Mt1/2(+/+) mice as lung injury progressed. Several of these transcripts have previously been associated with acute lung injury (Table 3), and include chemokine ligand 2 (a.k.a. MIP-2α), chemokine (C-C motif) ligand 2 (a.k.a. MCP-1), CCL6 (a.k.a. C10), CCL11 (a.k.a. eotaxin); CCL17 (a.k.a TARC), complement component 5 receptor 1, IL-1β, and macrophage migration inhibitory factor (5, 51–55). Additional inflammatory mediators not previously associated with acute lung injury increased more in Mt1/2(+/+) mice than in Mt1/2(+/+) mice included resistin-like α (a.k.a. FIZZ1), which is a mediator of allergic pulmonary inflammation (56), and chemokine ligand 14 (a.k.a. BRAK or MIP-2γ), which is a mediator of neutrophil chemotaxis (57). The differences in inflammatory mediators was first noted at 8–24 h, and became maximal at 72 h, paralleling the differences noted in BAL neutrophils (Figure 3A) between mouse strains. These findings are consistent with those of LPS-induced lung inflammation and edema (16), and support the protective role identified for MT.
Transcript levels of specific extracellular matrix proteins also were increased more in Mt1/2(+/+) mice than Mt1/2(+/+) mice at 72 h during acute lung injury. Connective tissue growth factor (also known as IGFBP-rP) is a fibroblast mitogen and promoter of collagen deposition that is increased in oxygen- (9) and bleomycin-induced lung injury (58), and in the BAL fluid of patients with pulmonary sarcoidosis (59). Tissue inhibitor of metalloproteinase 1 can mediate inflammation and repair processes during acute lung injury through stabilization of matrix components (60), and tenascin C is an extracellular matrix glycoprotein that may play a role similar to tissue inhibitor of metalloproteinase 1 (61). Increased gelatinolytic and collagenolytic activities via induction of matrix metalloproteinase 9 (a.k.a. gelatinase B) contribute to the pathogenesis of acute lung injury (62, 63). Thrombospondin 1 is a potent activator of latent TGF-β (64) that can play a role in the development of acute lung injury through alteration of fibroproliferative responses, lung permeability, and inflammatory cell influx (4, 65).
Additionally, acute lung injury is characterized by alteration of fibrinolytic activity within the lung (66). In Mt1/2(+/+) mice, transcript levels for proteins involved in the modulation of fibrinolysis were greater than in Mt1/2(+/+) mice and included serine (or cysteine) proteinase inhibitor, clade E, member 1 (also known as PAI-1), plasminogen activator tissue, and urokinase plasminogen activator receptor. In particular, serine (or cysteine) proteinase inhibitor, clade E, member 1 is the major inhibitor of fibrinolytic activity in the alveolar space (66). The differences in transcript levels of proteins involved in the modulation of the extracellular matrix and fibrinolysis agreed with the difference in the magnitude of lung injury observed between Mt1/2(+/+) and Mt1/2(+/+) mice.
Metallothionein Protects against Nickel-induced Lethality and Diminution of SFTPB Transcript Levels
Previous investigations from our laboratory suggested Mt1 and Mt2 to be possible candidate genes for nickel-induced acute lung injury because they are harbored together near a suggestive quantitative-trait locus on chromosome 8 that was identified for genetic susceptibility in mice (67), and MT1 was one of the most induced transcripts in the lungs of mice after nickel exposure (3). In the present study, Mt-transgenic mice survived longer than strain-matched C57BL/6J control mice after continuous nickel exposure. This may be due to the elevated basal level of MT mRNA and protein in the lungs of Mt-transgenic mice as compared with controls, as well as the likely larger increase in levels obtained after induction of lung injury. In Mt1/2(+/+) mice, MT2 mRNA levels also significantly increased throughout nickel exposure (exceeding 30-fold at 72 h). Lung MT1 and MT2 mRNA is notably increased in rodents by zinc (68) and several other inhaled toxicants (9, 51, 69–71). We established that Mt1/2(+/+) mice were more susceptible to lethality and increased lung inflammation (BAL neutrophils) and permeability (BAL hemoglobin and protein) after nickel exposure. These data are consistent with those of previous studies, in which Mt1/2(+/+) mice were more susceptible to the lethal effects of mercury vapor (72), as well as increased lung inflammation and edema induced by intratracheal challenge with LPS (16) and ovalbumin (73). With respect to survival, the magnitude of protection we observed was small but consistent with our previous genetic analysis that demonstrated that survival is a complex trait under the control of at least five chromosomal regions (67). The region on chromosome 8 (near where Mt1 and Mt2 are located) was predicted to explain 10–15% of the phenotypic variance.
Produced principally by the alveolar type II epithelial cells, SFTPB is unequivocally necessary for normal pulmonary function (7, 8). A small hydrophobic peptide, SFTPB enhances surfactant adsorption and spreading, and is necessary for the surface tension–reducing properties of pulmonary surfactant. Mice lacking Sftpb succumb to respiratory failure shortly after birth, whereas heterozygous Sftpb mice (containing 50% of wild-type SFTPB levels) survive, suggesting that a 50% loss of SFTPB can be endured under normoxic conditions (74). We have previously demonstrated that exposure to nickel decreases lung SFTPB transcript levels in mice (3–6). Furthermore, preservation of lung SFTPB levels may protect against nickel-induced acute lung injury (4, 5). In the present study, SFTPB mRNA is decreased more in Mt1/2(+/+) mice than in Mt1/2(+/+) mice after nickel exposure for 72 h, which likely contributes to the decreased survival time observed in Mt1/2(+/+) mice.
In summary, Mt has a protective role in the development of nickel-induced acute lung injury as assessed by survival and pulmonary inflammation. Additionally, SFTPB transcript levels were decreased more in Mt1/2(+/+) mice than in Mt1/2(+/+) mice. Oligonucleotide microarray analysis revealed an early (3 h) wholesale decrease in transcripts involved in protein processing and ribosomal biogenesis that was greater in Mt1/2(+/+) mice and suggests a novel role of MT in selective transcript stabilization during stress. Subsequently, MT deficiency lead to augmentation of the increase in transcripts associated with the inflammation, extracellular matrix regulation, and coagulation and fibrinolysis with the progression of acute lung injury. These data should stimulate further investigation of the protective role of MT in the pathogenesis of acute lung injury.
This work was supported by National Institutes of Health grants ES10562, ES06096, HL65612, ES07250, and ES012463.
Originally Published in Press as DOI: 10.1165/rcmb.2005-0248OC on September 15, 2005
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med 2000;342:1334–1349. [DOI] [PubMed] [Google Scholar]
- 2.Chollet-Martin S. Polymorphonuclear neutrophil activation during the acute respiratory distress syndrome. Intensive Care Med 2000;26:1575–1577. [DOI] [PubMed] [Google Scholar]
- 3.McDowell SA, Gammon K, Bachurski CJ, Wiest JS, Leikauf JE, Prows DR, Leikauf GD. Differential gene expression in the initiation and progression of nickel-induced acute lung injury. Am J Respir Cell Mol Biol 2000;23:466–474. [DOI] [PubMed] [Google Scholar]
- 4.Hardie WD, Prows DR, Piljan-Gentle A, Dunlavy MR, Wesselkamper SC, Leikauf GD, Korfhagen TR. Dose-related protection from nickel-induced lung injury in transgenic mice expressing human transforming growth factor-α. Am J Respir Cell Mol Biol 2002;26:430–437. [DOI] [PubMed] [Google Scholar]
- 5.McDowell SA, Gammon K, Zingarelli B, Bachurski CJ, Aronow BJ, Prows DR, Leikauf GD. Inhibition of nitric oxide restores surfactant gene expression following nickel-induced acute lung injury. Am J Respir Cell Mol Biol 2003;28:188–198. [DOI] [PubMed] [Google Scholar]
- 6.Wesselkamper SC, Case LM, Henning LN, Borchers MT, Tichelaar JW, Mason JM, Dragin N, Medvedovic M, Sartor MA, Tomlinson CR, et al. Gene expression changes during the development of acute lung injury: role of TGF-β. Am J Respir Crit Care Med (In press) [DOI] [PMC free article] [PubMed]
- 7.Ingenito EP, Mora R, Cullivan M, Marzan Y, Haley K, Mark L, Sonna LA. Decreased surfactant protein-B expression and surfactant dysfunction in a murine model of acute lung injury. Am J Respir Cell Mol Biol 2001;25:35–44. [DOI] [PubMed] [Google Scholar]
- 8.Melton KR, Nesslein LL, Ikegami M, Tichelaar JW, Clark JC, Whitsett JA, Weaver TE. SP-B deficiency causes respiratory failure in adult mice. Am J Physiol Lung Cell Mol Physiol 2003;285:L543–L549. [DOI] [PubMed] [Google Scholar]
- 9.Perkowski S, Sun J, Singhal S, Santiago J, Leikauf GD, Albelda SM. Gene expression profiling of the early pulmonary response to hyperoxia in mice. Am J Respir Cell Mol Biol 2003;28:682–696. [DOI] [PubMed] [Google Scholar]
- 10.Palmiter RD. The elusive function of metallothioneins. Proc Natl Acad Sci USA 1998;95:8428–8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klaassen CD, Liu J, Choudhuri S. Metallothionein: an intracellular protein to protect against cadmium toxicity. Annu Rev Pharmacol Toxicol 1999;39:267–294. [DOI] [PubMed] [Google Scholar]
- 12.Klaassen CD, Cagen SZ. Metallothionein as a trap for reactive organic intermediates. Adv Exp Med Biol 1981;136(Pt A):633–646. [DOI] [PubMed] [Google Scholar]
- 13.Thornalley PJ, Vasak M. Possible role for metallothionein in protection against radiation-induced oxidative stress: kinetics and mechanism of its reaction with superoxide and hydroxyl radicals. Biochim Biophys Acta 1985;827:36–44. [DOI] [PubMed] [Google Scholar]
- 14.Karin M, Slater EP, Herschman HR. Regulation of metallothionein synthesis in HeLa cells by heavy metals and glucocorticoids. J Cell Physiol 1981;106:63–74. [DOI] [PubMed] [Google Scholar]
- 15.De SK, McMaster MT, Andrews GK. Endotoxin induction of murine metallothionein gene expression. J Biol Chem 1990;265:15267–15274. [PubMed] [Google Scholar]
- 16.Takano H, Inoue K, Yanagisawa R, Sato M, Shimada A, Morita T, Sawada M, Nakamura K, Sanbongi C, Yoshikawa T. Protective role of metallothionein in acute lung injury induced by bacterial endotoxin. Thorax 2004;59:1057–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmiter RD, Sandgren EP, Koeller DM, Brinster RL. Distal regulatory elements from the mouse metallothionein locus stimulate gene expression in transgenic mice. Mol Cell Biol 1993;13:5266–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iszard MB, Liu J, Liu Y, Dalton T, Andrews GK, Palmiter RD, Klaassen CD. Characterization of metallothionein-I–transgenic mice. Toxicol Appl Pharmacol 1995;133:305–312. [DOI] [PubMed] [Google Scholar]
- 19.Wesselkamper SC, Prows DR, Biswas P, Willeke K, Bingham E, Leikauf GD. Genetic susceptibility to irritant-induced acute lung injury in mice. Am J Physiol Lung Cell Mol Physiol 2000;279:L575–L582. [DOI] [PubMed] [Google Scholar]
- 20.Saltini C, Hance AJ, Ferrans VJ, Basset F, Bitterman PB, Crystal RG. Accurate quantification of cells recovered by bronchoalveolar lavage. Am Rev Respir Dis 1984;130:650–658. [DOI] [PubMed] [Google Scholar]
- 21.Regal JF, Fraser DG, Weeks CE, Greenberg NA. Dietary phytoestrogens have anti-inflammatory activity in a guinea pig model of asthma. Proc Soc Exp Biol Med 2000;223:372–378. [DOI] [PubMed] [Google Scholar]
- 22.Karyala S, Guo J, Sartor M, Medvedovic M, Kann S, Puga A, Ryan P, Tomlinson CR. Different global gene expression profiles in benzo[a] pyrene- and dioxin-treated vascular smooth muscle cells of AHR-knockout and wild-type mice. Cardiovasc Toxicol 2004;4:47–73. [DOI] [PubMed] [Google Scholar]
- 23.Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS. Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 2001;8: 625–637. [DOI] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc [Ser A] 1995;B57:289–300. [Google Scholar]
- 25.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using gene ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol 2003;4:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet 2002;31:19–20. [DOI] [PubMed] [Google Scholar]
- 27.Choi AM, Sylvester S, Otterbein L, Holbrook NJ. Molecular responses to hyperoxia in vivo: relationship to increased tolerance in aged rats. Am J Respir Cell Mol Biol 1995;13:74–82. [DOI] [PubMed] [Google Scholar]
- 28.Berrebi D, Bruscoli S, Cohen N, Foussat A, Migliorati G, Bouchet-Delbos L, Maillot MC, Portier A, Couderc J, Galanaud P, et al. Synthesis of glucocorticoid-induced leucine zipper (GILZ) by macrophages: an anti-inflammatory and immunosuppressive mechanism shared by glucocorticoids and IL-10. Blood 2003;101:729–738. [DOI] [PubMed] [Google Scholar]
- 29.Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics 2005;85:551–556. [DOI] [PubMed] [Google Scholar]
- 30.Tamai KT, Gralla EB, Ellerby LM, Valentine JS, Thiele DJ. Yeast and mammalian metallothioneins functionally substitute for yeast copper-zinc superoxide dismutase. Proc Natl Acad Sci USA 1993;90:8013–8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang X, Frenkel K, Klein CB, Costa M. Nickel induces increased oxidants in intact cultured mammalian cells as detected by dichlorofluorescein fluorescence. Toxicol Appl Pharmacol 1993;120:29–36. [DOI] [PubMed] [Google Scholar]
- 32.Zilbermann I, Maimon E, Cohen H, Meyerstein D. Redox chemistry of nickel complexes in aqueous solutions. Chem Rev 2005;105:2609–2625. [DOI] [PubMed] [Google Scholar]
- 33.Waalkes MP, Harvey MJ, Klaassen CD. Relative in vitro affinity of hepatic metallothionein for metals. Toxicol Lett 1984;20:33–39. [DOI] [PubMed] [Google Scholar]
- 34.Finney LA, O'Halloran TV. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 2003;300: 931–936. [DOI] [PubMed] [Google Scholar]
- 35.Kawai K, Liu SX, Tyurin VA, Tyurina YY, Borisenko GG, Jiang JF, St Croix CM, Fabisiak JP, Pitt BR, Kagan VE. Antioxidant and antiapoptotic function of metallothioneins in HL-60 cells challenged with copper nitrilotriacetate. Chem Res Toxicol 2000;13:1275–1286. [DOI] [PubMed] [Google Scholar]
- 36.Fabisiak JP, Tyurin VA, Tyurina YY, Borisenko GG, Korotaeva A, Pitt BR, Lazo JS, Kagan VE. Redox regulation of copper-metallothionein. Arch Biochem Biophys 1999;363:171–181. [DOI] [PubMed] [Google Scholar]
- 37.Dalton T, Fu K, Palmiter RD, Andrews GK. Transgenic mice that overexpress metallothionein-I resist dietary zinc deficiency. J Nutr 1996;126: 825–833. [DOI] [PubMed] [Google Scholar]
- 38.Andrews GK, Geiser J. Expression of the mouse metallothionein-I and -II genes provides a reproductive advantage during maternal dietary zinc deficiency. J Nutr 1999;129:1643–1648. [DOI] [PubMed] [Google Scholar]
- 39.Andrews GK. Cellular zinc sensors: MTF-1 regulation of gene expression. Biometals 2001;14:223–237. [DOI] [PubMed] [Google Scholar]
- 40.Murphy BJ, Andrews GK, Bittel D, Discher DJ, McCue J, Green CJ, Yanovsky M, Giaccia A, Sutherland RM, Laderoute KR, et al. Activation of metallothionein gene expression by hypoxia involves metal response elements and metal transcription factor-1. Cancer Res 1999;59:1315–1322. [PubMed] [Google Scholar]
- 41.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG Jr. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 2001;292:464–468. [DOI] [PubMed] [Google Scholar]
- 42.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001;292:468–472. [DOI] [PubMed] [Google Scholar]
- 43.Salnikow K, Davidson T, Costa M. The role of hypoxia-inducible signaling pathway in nickel carcinogenesis. Environ Health Perspect 2002;110:831–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998;95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beattie JH, Wood AM, Newman AM, Bremner I, Choo KH, Michalska AE, Duncan JS, Trayhurn P. Obesity and hyperleptinemia in metallothionein (-I and -II) null mice. Proc Natl Acad Sci USA 1998;95:358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ueno T, Linder S, Na CL, Rice WR, Johansson J, Weaver TE. Processing of pulmonary surfactant protein B by napsin and cathepsin H. J Biol Chem 2004;279:16178–16184. [DOI] [PubMed] [Google Scholar]
- 47.Brum LM, Lopez MC, Varela JC, Baker HV, Moyer RW. Microarray analysis of A549 cells infected with rabbitpox virus (RPV): a comparison of wild-type RPV and RPV deleted for the host range gene, SPI-1. Virology 2003;315:322–334. [DOI] [PubMed] [Google Scholar]
- 48.Worthington MT, Pelo JW, Sachedina MA, Applegate JL, Arseneau KO, Pizarro TT. RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/tristetraprolin protein. J Biol Chem 2002;277:48558–48564. [DOI] [PubMed] [Google Scholar]
- 49.Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol Cell Biol 2004;24:5534–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duttagupta R, Tian B, Wilusz CJ, Khounh DT, Soteropoulos P, Ouyang M, Dougherty JP, Peltz SW. Global analysis of Pub1p targets reveals a coordinate control of gene expression through modulation of binding and stability. Mol Cell Biol 2005;25:5499–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnston CJ, Finkelstein JN, Gelein R, Oberdorster G. Pulmonary cytokine and chemokine mRNA levels after inhalation of lipopolysaccharide in C57BL/6 mice. Toxicol Sci 1998;46:300–307. [DOI] [PubMed] [Google Scholar]
- 52.Belperio JA, Dy M, Burdick MD, Xue YY, Li K, Elias JA, Keane MP. Interaction of IL-13 and C10 in the pathogenesis of bleomycin-induced pulmonary fibrosis. Am J Respir Cell Mol Biol 2002;27:419–427. [DOI] [PubMed] [Google Scholar]
- 53.Inoue T, Fujishima S, Ikeda E, Yoshie O, Tsukamoto N, Aiso S, Aikawa N, Kubo A, Matsushima K, Yamaguchi K. CCL22 and CCL17 in rat radiation pneumonitis and in human idiopathic pulmonary fibrosis. Eur Respir J 2004;24:49–56. [DOI] [PubMed] [Google Scholar]
- 54.Melendez AJ, Ibrahim FB. Antisense knockdown of sphingosine kinase 1 in human macrophages inhibits C5a receptor-dependent signal transduction, Ca2+ signals, enzyme release, cytokine production, and chemotaxis. J Immunol 2004;173:1596–1603. [DOI] [PubMed] [Google Scholar]
- 55.Lai KN, Leung JC, Metz CN, Lai FM, Bucala R, Lan HY. Role for macrophage migration inhibitory factor in acute respiratory distress syndrome. J Pathol 2003;199:496–508. [DOI] [PubMed] [Google Scholar]
- 56.Holcomb IN, Kabakoff RC, Chan B, Baker TW, Gurney A, Henzel W, Nelson C, Lowman HB, Wright BD, Skelton NJ, et al. FIZZ1, a novel cysteine-rich secreted protein associated with pulmonary inflammation, defines a new gene family. EMBO J 2000;19:4046–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao X, Zhang W, Wan T, He L, Chen T, Yuan Z, Ma S, Yu Y, Chen G. Molecular cloning and characterization of a novel CXC chemokine macrophage inflammatory protein-2 gamma chemoattractant for human neutrophils and dendritic cells. J Immunol 2000;165:2588–2595. [DOI] [PubMed] [Google Scholar]
- 58.Lasky JA, Ortiz LA, Tonthat B, Hoyle GW, Corti M, Athas G, Lungarella G, Brody A, Friedman M. Connective tissue growth factor mRNA expression is upregulated in bleomycin-induced lung fibrosis. Am J Physiol Lung Cell Mol Physiol 1998;275:L365–L371. [DOI] [PubMed] [Google Scholar]
- 59.Allen JT, Knight RA, Bloor CA, Spiteri MA. Enhanced insulin-like growth factor binding protein-related protein 2 (connective tissue growth factor) expression in patients with idiopathic pulmonary fibrosis and pulmonary sarcoidosis. Am J Respir Cell Mol Biol 1999;21:693–700. [DOI] [PubMed] [Google Scholar]
- 60.Madtes DK, Elston AL, Kaback LA, Clark JG. Selective induction of tissue inhibitor of metalloproteinase-1 in bleomycin-induced pulmonary fibrosis. Am J Respir Cell Mol Biol 2001;24:599–607. [DOI] [PubMed] [Google Scholar]
- 61.Zhao Y, Young SL, McIntosh JC. Induction of tenascin in rat lungs undergoing bleomycin-induced pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 1998;274:L1049–L1057. [DOI] [PubMed] [Google Scholar]
- 62.Ricou B, Nicod L, Lacraz S, Welgus HG, Suter PM, Dayer JM. Matrix metalloproteinases and TIMP in acute respiratory distress syndrome. Am J Respir Crit Care Med 1996;154:346–352. [DOI] [PubMed] [Google Scholar]
- 63.Warner RL, Beltran L, Younkin EM, Lewis CS, Weiss SJ, Varani J, Johnson KJ. Role of stromelysin 1 and gelatinase B in experimental acute lung injury. Am J Respir Cell Mol Biol 2001;24:537–544. [DOI] [PubMed] [Google Scholar]
- 64.Crawford SE, Stellmach V, Murphy-Ullrich JE, Ribeiro SM, Lawler J, Hynes RO, Boivin GP, Bouck N. Thrombospondin-1 is a major activator of TGF-beta1 in vivo. Cell 1998;93:1159–1170. [DOI] [PubMed] [Google Scholar]
- 65.Pittet JF, Griffiths MJ, Geiser T, Kaminski N, Dalton SL, Huang X, Brown LA, Gotwals PJ, Koteliansky VE, Matthay MA, et al. TGF-beta is a critical mediator of acute lung injury. J Clin Invest 2001;107:1537–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Idell S, Peters J, James KK, Fair DS, Coalson JJ. Local abnormalities of coagulation and fibrinolytic pathways that promote alveolar fibrin deposition in the lungs of baboons with diffuse alveolar damage. J Clin Invest 1989;84:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prows DR, Leikauf GD. Quantitative trait analysis of nickel-induced acute lung injury in mice. Am J Respir Cell Mol Biol 2001;24:740–746. [DOI] [PubMed] [Google Scholar]
- 68.Wesselkamper SC, Chen LC, Gordon T. Development of pulmonary tolerance in mice exposed to zinc oxide fumes. Toxicol Sci 2001;60:144–151. [DOI] [PubMed] [Google Scholar]
- 69.Hart BA. Cellular and biochemical response of the rat lung to repeated inhalation of cadmium. Toxicol Appl Pharmacol 1986;82:281–291. [DOI] [PubMed] [Google Scholar]
- 70.Liu J, Lei D, Waalkes MP, Beliles RP, Morgan DL. Genomic analysis of the rat lung following elemental mercury vapor exposure. Toxicol Sci 2003;74:174–181. [DOI] [PubMed] [Google Scholar]
- 71.Johnston CJ, Stripp BR, Reynolds SD, Avissar NE, Reed CK, Finkelstein JN. Inflammatory and antioxidant gene expression in C57BL/6J mice after lethal and sublethal ozone exposures. Exp Lung Res 1999;25:81–97. [DOI] [PubMed] [Google Scholar]
- 72.Yoshida M, Satoh M, Shimada A, Yasutake A, Sumi Y, Tohyama C. Pulmonary toxicity caused by acute exposure to mercury vapor is enhanced in metallothionein-null mice. Life Sci 1999;64:1861–1867. [DOI] [PubMed] [Google Scholar]
- 73.Inoue K, Takano H, Yanagisawa R, Sakurai M, Ichinose T, Sadakane K, Hiyoshi K, Sato M, Shimada A, Inoue M, et al. Role of metallothionein in antigen-related airway inflammation. Exp Biol Med (Maywood) 2005;230:75–81. [DOI] [PubMed]
- 74.Clark JC, Weaver TE, Iwamoto HS, Ikegami M, Jobe AH, Hull WM, Whitsett JA. Decreased lung compliance and air trapping in heterozygous SP-B–deficient mice. Am J Respir Cell Mol Biol 1997;16:46–52. [DOI] [PubMed] [Google Scholar]