Summary
Mutations in ribosomal proteins L4 and L22 confer resistance to erythromycin and other macrolide antibiotics in a variety of bacteria. L4 and L22 have elongated loops whose tips converge in the peptide exit tunnel near the macrolide binding site, and resistance mutations typically affect residues within these loops. Here, we use bacteriophage λ Red-mediated recombination, or “recombineering”, to uncover new L4 and L22 alleles that confer macrolide resistance in Escherichia coli. We randomized residues at the tips of the L4 and L22 loops using recombineered oligonucleotide libraries, and selected the mutagenized cells for erythromycin-resistant mutants. These experiments led to the identification of 341 different resistance mutations encoding 278 unique L4 and L22 proteins – the overwhelming majority of which are novel. Many resistance mutations were complex, involving multiple missense mutations, in-frame deletions, and insertions. Transfer of L4 and L22 mutations into wild-type cells by phage P1-mediated transduction demonstrated that each allele was sufficient to confer macrolide resistance. Although L4 and L22 mutants are typically resistant to most macrolides, selections carried out on different antibiotics revealed macrolide-specific resistance mutations. L22 Lys90Trp is one such allele, which confers resistance to erythromycin, but not tylosin or spiramycin. Purified L22 Lys90Trp ribosomes show reduced erythromycin binding, but have the same affinity for tylosin as wild-type ribosomes. Moreover, DMS methylation protection assays demonstrated that L22 Lys90Trp ribosomes bind tylosin more readily than erythromycin in vivo. This work underscores the exceptional functional plasticity of the L4 and L22 proteins, and highlights the utility of Red-mediated recombination in targeted genetic selections.
Keywords: antibiotic resistance, macrolide, recombineering, ribosomal proteins, mutagenesis
Introduction
Macrolide antibiotics – which include erythromycin and tylosin – are commonly used antibacterial agents that interfere with protein synthesis. These drugs bind to the large ribosomal subunit at the entrance of the nascent peptide exit tunnel, near the peptidyl transferase center, where they inhibit peptide bond formation and facilitate peptidyl-tRNA dissociation from the ribosome 1; 2; 3; 4. In addition, macrolides have been shown to interfere with ribosome assembly in several bacterial species 5; 6; 7. Resistance to macrolides is mediated by three general mechanisms: i) enzymatic inactivation of macrolides, ii) increased macrolide efflux, and iii) alteration of the macrolide-binding site 8; 9; 10. Modification of the macrolide-binding site is the most widespread resistance mechanism and is usually associated with the acquisition of Erm methyltransferases, which catalyze N6,N6-dimethylation of A2058 in 23S rRNA (Escherichia coli numbering is used throughout) 8; 11; 12. A2058 is critical for the binding of macrolides, lincosamides, and streptogramin B-type antibiotics (Fig. 1b), and its methylation confers resistance to all three classes of antibiotics 8; 9. Mutation of A2058, A2059, and other 23S rRNA residues within the macrolide-binding site can confer a similar resistance phenotype, but these mutations are typically limited to bacteria containing only one or two rRNA operons 10. Finally, the macrolide-binding site can be modified by mutations in rplD and rplV, which encode ribosomal proteins L4 and L22, respectively. L4 and L22 each have extended loops, which converge to form a narrowing in the exit tunnel adjacent to the macrolide-binding site (Fig. 1a and 1b) 1; 2; 3. Although less common than Erm-mediated resistance, L4 and L22 mutations are increasingly being identified in clinically relevant species such as Streptococcus pneumoniae, Haemophilus influenzae, and Staphylococcus aureus 13; 14; 15.
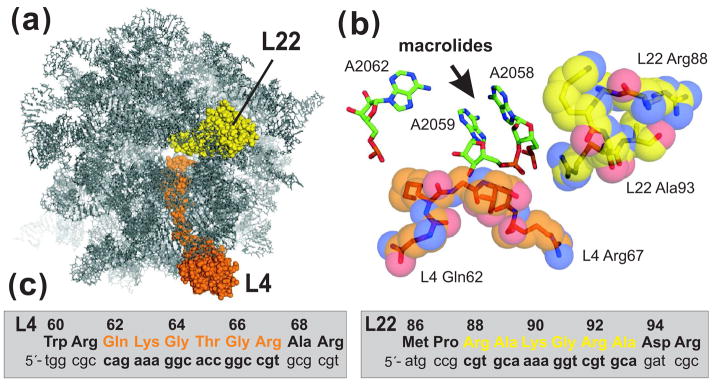
Figure 1.
L4 and L22 loops and the ribosomal macrolide-binding site. (a) The E. coli large ribosome subunit viewed from the cytoplasmic opening of the exit tunnel. All ribosomal proteins except L4 and L22 have been omitted from the rendering. (b) View of the macrolide-binding site. Residues A2058 and A2059 of 23S rRNA form a hydrophobic crevice into which macrolide antibiotics bind. Residue A2062 has been shown to form a reversible covalent bond with the aldehyde moiety of tylosin and spiramycin 1. The tips of the L4 and L22 loops are shown in orange and yellow, respectively. Structures were derived from PDB file 2AW4 45 and rendered using PyMol. (c) L4 and L22 nucleotide and protein sequences. L4 residues Gln62 to Arg67 (in orange) and L22 residues Arg88 to Ala93 (in yellow) were mutagenized by oligonucleotide library recombineering as described in the text.
Macrolide-resistance mutations in L4 and L22 are diverse, and include missense mutations, insertions and deletions 13; 15; 16; 17. Insertion and deletion mutations have been found at many positions along the L4 and L22 loops, but missense mutations tend to be localized to L4 Gln62 – Gly66 and L22 Arg88 – Ala93 13; 16; 18; 19; 20. These residues are closest to the macrolide-binding site (Fig. 1b), and are among the most highly conserved in eubacterial L4 and L22 proteins. Although L4 and L22 mutations have been identified in several bacterial species, only a handful of mutant ribosomes from E. coli have been characterized to date 21; 22; 23; 24. The L4 Lys63Glu mutation alters the structure of domain V within 23S rRNA and significantly decreases ribosome affinity for erythromycin 21; 22; 23; 25. Zengel and colleagues have recently characterized ten additional L4 and L22 macrolide-resistance alleles in E. coli, most of which also appear to reduce ribosome affinity for macrolides 24. In contrast, L22 containing a deletion of Met82-Lys83-Arg84 (L22 ΔMet82-Lys83-Arg84) confers macrolide resistance in E. coli and H. influenzae 17; 26, yet ribosomes from this mutant bind erythromycin with wild-type affinity 21; 23. Although macrolide resistance in the L22 ΔMet82-Lys83-Arg84 mutant is not yet fully understood, the mechanism is presumably distinct from that of L4 Lys63Glu and other mutations that prevent macrolide binding 23; 25; 27. Most other L4 and L22 mutations have been identified in clinical isolates, or in strains that have been selected in vitro by long-term culture in macrolide-containing media 13; 16; 28; 29; 30. Analysis of resistance alleles from these bacteria can be challenging because they often contain multiple L4 and L22 mutations and/or additional macrolide-resistance mutations in 23S rRNA 13; 16; 18; 19; 20. The ability to recapitulate these novel L4 and L22 alleles in a non-pathogenic model bacterium such as E. coli would facilitate the biochemical characterization of macrolide-resistant ribosomes.
In this report, we use bacteriophage λ Red-mediated recombination, or “recombineering”, to isolate new L4 and L22 mutations that confer macrolide resistance. The λ Red system is comprised of three proteins – gam, beta, and exo – which greatly enhance homologous recombination in E. coli 31. The beta protein facilitates the hybridization of single-stranded DNA at replication forks, allowing efficient recombination of synthetic oligonucleotides 32; 33. Court and colleagues have exploited oligonucleotide-based recombineering to introduce point mutations directly onto the E. coli chromosome 32. We reasoned that this methodology could be used to perform targeted mutagenesis of rplD and rplV for genetic selections. Using oligonucleotide libraries, we randomized individual codons corresponding to residues at the L4 and L22 loop tips and selected for macrolide-resistant mutants. These selections resulted in the isolation of several hundred rplD and rplV macrolide-resistance mutations, encoding 278 unique L4 and L22 proteins. Although L4 and L22 mutations typically confer resistance to multiple macrolides 9, we identified L22 Lys90 mutants that were resistant to erythromycin, but not tylosin or spiramycin. This resistance phenotype resulted from a specific decrease in ribosome affinity for erythromycin. These results demonstrate that an extraordinary number of L4 and L22 mutations confer macrolide resistance, and highlight the power of recombineering to generate novel proteins for structure-function analysis.
Results
Selection and screening of erythromycin-resistant mutants
To systematically identify L4 and L22 mutations that confer macrolide resistance, we performed a series of targeted mutagenesis experiments using bacteriophage λ Red-mediated recombination in E. coli 32; 34. We focused on L4 residues 62 – 67 and L22 residues 88 – 93 (Fig. 1b & 1c), because resistance mutations affect these residues most frequently 15. Twelve oligonucleotide libraries were designed for recombineering, each containing a randomized codon corresponding to one of the target residues in the L4 or L22 loops. Each oligonucleotide library was introduced into cells expressing the phage λ Red proteins, followed by selection for erythromycin-resistant mutants. A total of 1,731 erythromycin-resistant colonies were isolated from the twelve libraries, of which 1,398 survived secondary selection on fresh erythromycin plates (Table 1). Mutants that survived secondary erythromycin selection were subjected to 3′-mismatch PCR screening to detect isolates containing wild-type L4 and L22 (Table 1) 35. The rplD and rplV genes were sequenced from 903 of the screened mutants, resulting in the identification of 305 distinct mutations, several of which were independently isolated multiple times (for comprehensive analysis of identified mutations, see Supplementary Tables II & III). Wild-type rplD and rplV genes were found in 224 of the erythromycin-resistant isolates, corresponding to a false-positive rate of ~25% for the PCR screen (Table 1). Presumably, cells with wild-type L4 and L22 carry other unidentified mutations responsible for macrolide resistance.
Table 1.
L4 and L22 mutant selection statistics
| Macrolide | Mutagenized codona | Colonies isolated | Survived 2° selection | Passed PCR screen | Clones sequenced | L4/L22 mutantsb |
|---|---|---|---|---|---|---|
| Erythromycin | ||||||
| L4 | ||||||
| Gln62 | 136 | 117 | 98 | 96 | 69 | |
| Lys63 | 130 | 121 | 110 | 98 | 96 | |
| Gly64 | 159 | 149 | 117 | 96 | 96 | |
| Thr65 | 129 | 121 | 89 | 89 | 65 | |
| Gly66 | 170 | 167 | 99 | 96 | 56 | |
| Arg67 | 246 | 172 | 81 | 81 | 76 | |
| Gln62Phe | 33 | 31 | NPc | 31 | 20 | |
| Gln62Lys | 33 | 19 | NPc | 19 | 0 | |
| Thr65Pro | 31 | 31 | 31 | 24 | 24 | |
| Thr65Ser | 46 | 37 | 29 | 24 | 21 | |
| L22 | ||||||
| Arg88 | 201 | 82 | 49 | 48 | 33 | |
| Ala89 | 167 | 117 | 100 | 96 | 45 | |
| Lys90 | 98 | 98 | 96 | 96 | 96 | |
| Gly91 | 79 | 79 | 12 | 6 | 4 | |
| Arg92 | 135 | 94 | 61 | 48 | 30 | |
| Ala93 | 81 | 81 | 53 | 53 | 13 | |
| Spiramycin | ||||||
| L4 | ||||||
| Gln62 | 98 | 81 | 68 | 48 | 46 | |
| Lys63 | 97 | 93 | 62 | 48 | 48 | |
| Gly66 | 49 | 49 | 48 | 48 | 48 | |
| Tylosin | ||||||
| L22 | ||||||
| Lys90 | 97 | 88 | 57 | 48 | 47 | |
Individual codons were randomized using oligonucleotide libraries except where specific mutations are indicated.
Indicates that L4 or L22 mutations were identified within and/or adjacent to the target codon. Several erythromycin-resistant isolates contained wild-type L4 and L22, and presumably these cells contain unidentified macrolide-resistance mutations.
3′-mismatch PCR screening was not performed (NP) on mutants from these selections.
Although each oligonucleotide library was designed to introduce single missense mutations, a large proportion (~44%) of the L4 and L22 mutants contained mutations in multiple codons (Sup. Tables II & III). Generally, these complex mutations were comprised of target codon missense mutations, plus secondary point mutations or in-frame deletions affecting one or more neighboring codons (Sup. Tables II & III). We classified mutants according to the presence and type of secondary mutation, which revealed that each library had a tendency to produce mutations of a particular subclass (Table 2). For example, the L22 Ala89 library produced mostly missense mutations in the target codon coupled to neighboring in-frame deletions, whereas most L4 Thr65 mutants contained secondary point mutations (Table 2 and Sup. Tables II & III). Complex mutations were isolated exclusively from five libraries – L22 Ala89, L22 Gly91, L22 Arg92, L22 Ala93 and L4 Arg67 (Table 2). In contrast, target codon missense mutations were isolated primarily from the L4 Gln62, L4 Lys63, L4 Gly66 and L22 Lys90 library selections (Table 2). The mutations isolated from the twelve libraries are predicted to encode 248 unique L4 and L22 proteins (Tables 3 & 4). Each L4 and L22 allele was transferred into wild-type E. coli by phage P1-mediated transduction using a linked kanamycin resistance marker. All kanamycin-resistant transductants were also resistant to erythromycin (data not shown), strongly suggesting that the L4 and L22 alleles are sufficient to confer macrolide resistance.
Table 2.
Classification of L4 and L22 mutationsa
| Macrolide | Mutagenized codon | Simple missense | Missense + 2° point | Missense + 2° deletion | Deletion + 2° other | Insertions/duplication | Other/not in target codon | Total mutations |
|---|---|---|---|---|---|---|---|---|
| Erythromycin | ||||||||
| L4 | ||||||||
| Gln62 | 68 | 1 | 0 | 0 | 0 | 0 | 69 | |
| Lys63 | 94 | 0 | 1 | 1 | 0 | 0 | 96 | |
| Gly64 | 67 | 18 | 7 | 4 | 0 | 0 | 96 | |
| Thr65 | 2 | 44 | 11 | 7 | 1 | 0 | 65 | |
| Gly66 | 49 | 2 | 0 | 5 | 0 | 0 | 56 | |
| Arg67 | 0 | 11 | 30 | 31 | 0 | 4 | 76 | |
| Thr65Pro | 0 | 23 | 0 | 1 | 0 | 0 | 24 | |
| Thr65Ser | 0 | 5 | 9 | 7 | 0 | 0 | 21 | |
| L22 | ||||||||
| Arg88 | 8 | 0 | 12 | 13 | 0 | 0 | 33 | |
| Ala89 | 0 | 3 | 29 | 5 | 5 | 3 | 45 | |
| Lys90 | 93 | 3 | 0 | 0 | 0 | 0 | 96 | |
| Gly91 | 0 | 0 | 1 | 3 | 0 | 0 | 4 | |
| Arg92 | 0 | 0 | 18 | 5 | 3 | 4 | 30 | |
| Ala93 | 0 | 0 | 8 | 4 | 0 | 1 | 13 | |
| Spiramycin | ||||||||
| L4 | ||||||||
| Gln62 | 42 | 0 | 3 | 0 | 0 | 1 | 46 | |
| Lys63 | 48 | 0 | 0 | 0 | 0 | 0 | 48 | |
| Gly66 | 45 | 1 | 1 | 1 | 0 | 0 | 48 | |
| Tylosin | ||||||||
| L22 | ||||||||
| Lys90 | 0 | 46 | 1 | 0 | 0 | 0 | 47 | |
L4 and L22 mutations were classified with respect to presence and type of secondary mutation. The column labeled Mutagenized codon indicates the target codon for mutagenesis experiment. Simple missense mutations affected only target codons, with no other mutations identified in either L4 or L22. Target codon missense mutations coupled to secondary point mutations or secondary in-frame deletions are indicated as Missense + 2° point and Missense + 2° deletion, respectively. Deletions of target codons with additional secondary mutations in neighboring codons are indicated as Deletion + 2° other. Insertions arising from sequence duplications and mutations that did affect the target codon are indicated as such.
Table 3.
Predicted L4 proteins from macrolide-resistant E. coli mutantsa
| Gln62 | Lys63 | Gly64 | Thr65 | Gly66 | Arg67 |
|---|---|---|---|---|---|
| Q62D | K63A | G64I | T65D | G66A | Δ63-65, R67C |
| Q62E | K63C | G64L | T65C, G66C | G66C | Δ65-66, R67C |
| Q62F | K63D | G64P | T65C, G66D | G66D | Δ63-64, R67E |
| Q62I | K63E | Q62H, G64C | T65C, G66S | G66H | Δ65-66, R67F |
| Q62K | K63G | Δ61, G64F | G64V, T65C | G66Q | G64A, Δ65, R67F |
| Q62L | K63Ib | Δ63, G64I | K63Q, T65C | G66S | G64C, R67G |
| Q62M | K63L | Q62H, G64K | T65F, G66S | G66T | Q62H, R67G |
| Q62R | K63M | G64L, T65N | Δ60-61, T65F | T65I, G66A | Δ65-66, R67H |
| Q62V | K63P | G64L, R67G | T65G, G66S | G66D, R67C | Δ65, R67I |
| Q62W | K63Qc | W60C, G64L | T65H, G66S | Δ66-67 | G66C, R67K |
| K63R | Δ61, G64L | Δ63, G64N, T65I | Δ65-66, R67S | G66D, R67K | |
| (Spiramycin) | K63S | Q62H, G64L | K63Q, T65K | Δ64, R67K | |
| Q62Y | K63V | Δ62, G64M | T65L, G66C | (Spiramycin) | Δ65-66, R67K |
| Q62P, Δ64-65 | K63E, Δ64-65 | G64P, T65N | T65L, G66S | G66K | G64V, R67L |
| Q62S, Δ64-65 | Δ63-64 | G64P, T65S | Δ63, T65L | G66R | Δ65, R67L |
| Q62V, Δ64-65 | Δ61-63, G64R | Δ63, G64S, T65L | Δ65, G66S, Δ67 | Δ65, R67M | |
| (Spiramycin) | Q62H, G64S | Δ63, T65N | Δ65-66, R67M | ||
| K63H | Q62R, G64S | Δ64, T65N | G64A, Δ65, R67M | ||
| Δ63, G64S | G64C, T65P | Δ65, R67N | |||
| G64V, T65N | G64V, T65P | Δ65-66, R67N | |||
| G64V, T65S | W60L, T65P | Δ64, R67S | |||
| Δ63, G64V | K58N, T65Pd | Δ65, R67S | |||
| G64W, R67G | Δ62-63, T65P | K63N, Δ64-65, R67S | |||
| Δ61, Δ64 | Δ62-63, G64H, T65P | W60L, R67V | |||
| Δ64, T65S | T65Q, G66S | Q62H, R67V | |||
| Q62R, Δ63-64 | Q62L, T65Q | K63R, R67V | |||
| K63N, Δ64, T65Q | R67V, S70F | ||||
| T65R, G66D | R67V, S72F | ||||
| T65R, G66S | Δ65-66, R67V | ||||
| T65S, G66Ce | Δ62-63, G64R, R67V | ||||
| K58N, T65Se | Δ67-68 | ||||
| Δ61-62, K63Q, T65Se | Δ68-69, S70A | ||||
| Q62H, T65Se | T65N, Δ66-67 | ||||
| Δ63, T65Se | Δ64 | ||||
| Δ63, G64S, T65Se | Δ65 | ||||
| Δ63-64, T65Se | K63N, Δ64-65 | ||||
| T65V, G66A | |||||
| T65V, G66S | |||||
| Q62H, T65V | |||||
| T65W, G66S | |||||
| T65Y, G66S | |||||
| K63N, Δ64, T65Y | |||||
| Δ64-65 | |||||
| Δ65-66 | |||||
| Δ65-67 | |||||
| Δ65-66, R67G | |||||
| Q62E, K63R, Δ64-65 | |||||
| Ins65 [IEG]66 |
Unless otherwise indicated, all L4 mutations were isolated by selection for erythromycin-resistant mutants after codon randomization. Boldfaced alleles indicate a missense mutation within the target codon. Mutations isolated on spiramycin are indicated and italicized. Inserted (Ins) residues are enclosed within brackets, with subscripted numbers indicating the wild-type residues flanking the insertion. One letter amino acid code is used throughout.
Isolated from L4 Lys63Ile oligonucleotide mutagenesis and selection on erythromycin.
Isolated from L4 Lys63Gln oligonucleotide mutagenesis and selection on erythromycin.
Isolated from L4 Thr65Pro oligonucleotide mutagenesis and selection on erythromycin.
Isolated from L4 Thr65Ser oligonucleotide mutagenesis and selection on erythromycin.
Table 4.
Predicted L22 proteins from macrolide-resistant E. coli mutantsa
| Arg88 | Ala89 | Lys90 | Gly91 | Arg92 | Ala93 |
|---|---|---|---|---|---|
| R88P | A89D, G91N | K90F | A89E, Δ90, G91S | Ins91[AADRIAKG]92, R92C | A93C, Δ95-97 |
| R88D, Δ89, K90E | A89D, Δ90 | K90G | Δ91-92, A93Q | Δ90, R92E | A93D, Δ94-96, L97V |
| R88E, Δ90-91 | A89D, Δ92 | K90L | Δ91-92, A93S | R92E, Δ95-97, K98Q | A93F, Δ95-96 |
| R88E, Δ90-91, R92S | A89D, Δ90-91 | K90M | Δ89, K90E, R92E | Δ91, A93K | |
| R88E, Δ90, G91N | A89D, Δ90-92 | K90P | Δ89, K90E, R92E, I96L | Δ89-91, A93P | |
| Δ82-83, R84S, R88G | A89D, Δ91-92 | K90V | Δ89-90, R92F | A93W, D94G, Δ95 | |
| R88K, Δ91-92 | Δ87, A89D | K90W | Δ89, K90E, R92F | A93Y, R95Q, Δ96-98 | |
| Δ85-86, R88L | A89D, K90N, Δ91 | K90Y | A89E, Δ90, R92G | G91V, Δ92-93 | |
| R88N, Δ89, K90E | A89D, Δ90-91, R92I | A89E, K90V | Δ90, G91S, R92I | G91Y, Δ92-93 | |
| Δ85-87, R88P | A89D, R92P, Δ93 | A89E, K90W | Δ90, R92I, I96L | Δ93, D94N | |
| R88S, A89E, Δ90-91 | Ins88[IKGRA]89, A89D | Δ88-89, K90Q, R92K | Δ93-96, L97P | ||
| Δ88, A89D | Δ85-87, A89E | (Tylosin) | Δ90, G91S, R92L | ||
| Δ88, A89E | A89E, Δ90-91 R92N | A89E, K90D | Δ87, R88P, R92L, I96L | ||
| Δ88, A89P | R88P, A89F | A89E, K90P | R92P, Δ93-97 | ||
| Δ88, A89K, K90E | Δ87, A89F | K90P, G91S | Ins91[IAG]92, R92P | ||
| Δ88, A89P, K90E | A89H, Δ90-91, R92S | K90P, G91V | R92S, Δ94-96, L97V | ||
| Δ88, A89T, K90E | A89I, Δ90, G91S | K90P, Δ92 | Δ90-91, R92S, I96L | ||
| Δ88-89, K90D | Δ85-87, A89L | K90S, Δ91, R92V | |||
| Δ88-89, K90E | Ins84[KR]85, A89L | R92V, A93D, Δ94-96 | |||
| Δ88-89, K90Q | Δ87, A89N | Δ91-92 | |||
| K90N, Δ91-92 | Δ84-86, A89P | Δ95-96 | |||
| A89P, Ins90[GRE]91 | Δ96-98 | ||||
| Δ84-86, A89R | A89E, Δ90-92 | ||||
| Δ87, A89S | Ins91[NAD]92 | ||||
| Δ84, I85L, A89S | Δ91-93, I96L | ||||
| A89S, Ins89[KGRADRA]90 | Δ92, A93K, I96L | ||||
| Δ84, A89V | K90E, Δ91-92, I96L | ||||
| A89V, Δ90-92 | A89E, G91V, R92K, A93Q, D94I | ||||
| Ins88[R]89, A89V | |||||
| A89W, Δ91, R92G | |||||
| R88P, A89Y | |||||
| A89Y, Δ91-92 | |||||
| 86, P87T, A89Y | |||||
| Δ89-90, G91C | |||||
| Δ89-90, G91S | |||||
| Δ89-90, G91W | |||||
| Δ89-90, G91Y, R92I | |||||
| Δ84, I85L | |||||
| Δ90-91 |
Unless otherwise indicated, all L22 mutations were isolated by selection for erythromycin-resistant mutants after codon randomization. Boldfaced alleles indicate a missense mutation within the target codon. Mutations isolated on tylosin are indicated and italicized. Inserted (Ins) residues are enclosed within brackets, with subscripted numbers indicating the wild-type residues flanking the insertion. One letter amino acid code is used throughout.
Simple missense alleles
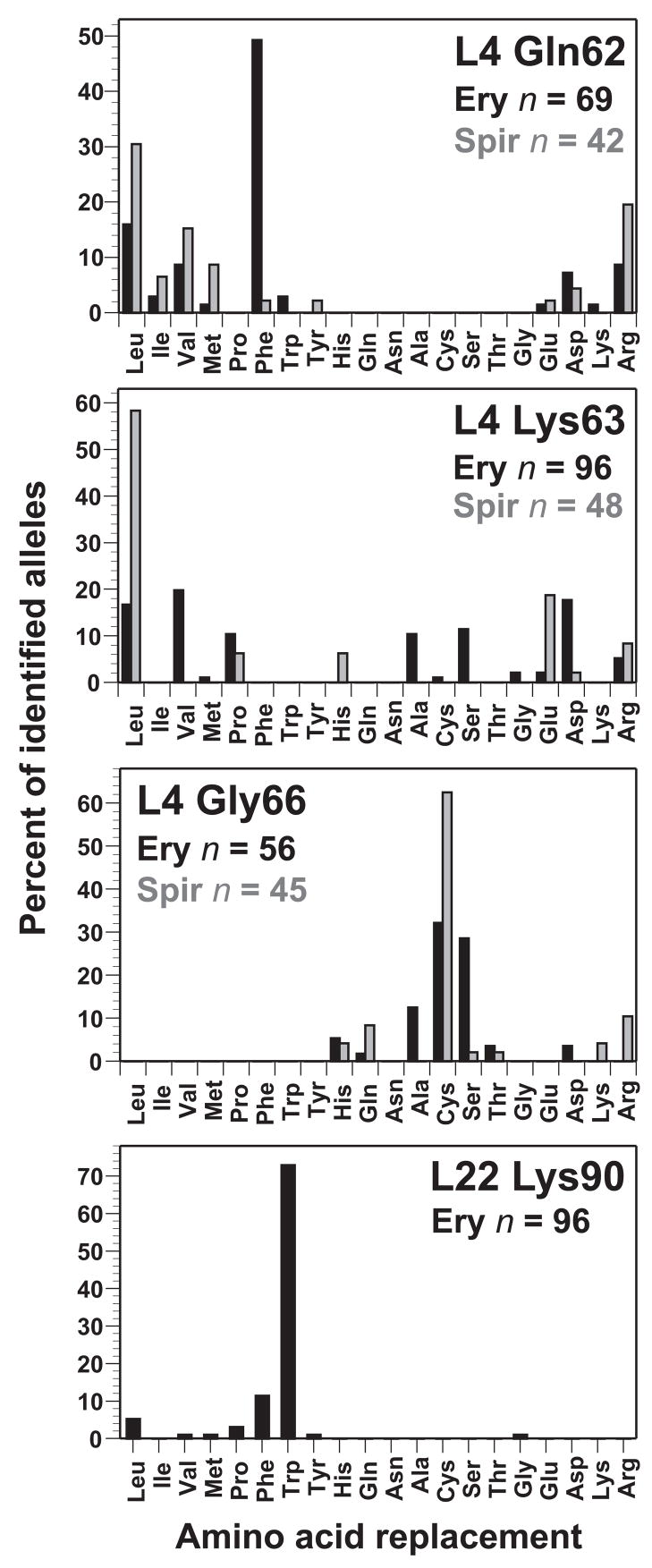
Because single missense mutations were identified primarily from the L4 Gln62, L4 Lys63, L4 Gly66 and L22 Lys90 libraries, allele isolation frequencies could be determined in a straightforward manner for these positions. Allele isolation frequency was expressed as the percent of total mutants identified for each position, and the resulting frequency distributions were plotted as histograms (Fig. 2). In general, each library produced a distinct allele distribution. Mutations isolated from the L4 Gln62 library resulted in changes to residues with hydrophobic and charged side chains (Fig. 2). Strikingly, L4 Gln62Phe mutations were identified in almost 50% of the L4 Gln62 mutants isolated by erythromycin selection (Fig. 2). Remarkable allele diversity was obtained from the L4 Lys63 library, in which 11 different amino acid changes were identified (Fig. 2). L4 Gly66 resistance alleles were more limited, with mutations to Ala, Cys and Ser residues identified most frequently (Fig. 2). Finally, the L22 Lys90 selection was dominated by the L22 Lys90Trp allele, which was identified in 70 of the 96 characterized mutants (Fig. 2).
Figure 2.
Isolation frequencies of L4 Gln62, L4 Lys63, L4 Gly66, and L22 Lys90 missense alleles. Frequencies are expressed as the percent of total mutants isolated from each selection. The total number of mutants identified for each position is indicated. L4 Gln62, L4 Lys63, and L4 Gly66 mutants isolated by spiramycin selection are shown in grey. Complex mutations isolated from these libraries are not shown in the histograms, but were included in the calculation of allele frequencies. See Supplemental Tables II & III for a complete enumeration of all isolated mutations.
The erythromycin selection protocol ensured that each mutant arose from an individual recombination event. Therefore, L4 and L22 alleles conferring the greatest erythromycin resistance were expected to be isolated at higher frequencies. We examined the erythromycin resistance properties of several L4 Gln62 mutants to determine whether a correlation exists between allele isolation frequency and antibiotic resistance. Because growth in the presence of erythromycin can select for other mutations that contribute to macrolide resistance, we used a linked kanamycin resistance marker to transfer L4 Gln62 mutations into wild-type cells by phage P1-mediated transduction prior to analysis. All of the analyzed L4 Gln62 mutants had similar macrolide resistance properties, although the L4 Gln62Glu mutant appeared to have a slight growth advantage in the presence of erythromycin (Table 5). In contrast, L4 Gln62Phe cells grew at a slower rate comparable to other L4 Gln62 mutants (Table 5). Because L4 Gln62Glu was isolated only once from the L4 Gln62 library selection compared to 34 independent isolates of L4 Gln62Phe (Fig. 2 and Sup. Table II), these data suggest that factors other than antibiotic resistance contribute to allele isolation frequency.
Table 5.
L4 mutant growth rates and macrolide sensitivitya
| L4 allele | Doubling time (min) | ERY doubling time (min) | ERY CDIC (μg/mL) | SPIR doubling time (min) | SPIR CDIC (μg/mL) |
|---|---|---|---|---|---|
| wild-type | 31 | 151 | 200 | 149 | 750 |
| Gln62Phe | 47 | 63 | 1000 | 79 | 2500 |
| Gln62Leu | 44 | 63 | 1125 | 78 | 2500 |
| Gln62Met | 41 | 56 | 1125 | 60 | 1875 |
| Gln62Asp | 43 | 58 | 1000 | 75 | 2500 |
| Gln62Glu | 39 | 55 | 1000 | 78 | 1875 |
| Gln62Lys | 42 | 64 | 1125 | 66 | 1875 |
| Gln62Arg | 43 | 63 | 1125 | 75 | 2500 |
| Lys63Glu | 44 | 61 | 1250 | 68 | 1750 |
| Lys63Asp | 44 | 69 | 1000 | 104 | 1000 |
| Lys63Leu | 36 | 65 | 750 | 77 | 2000 |
| Lys63Ala | 34 | 58 | 500 | 90 | 1000 |
| Lys63Val | 34 | 59 | 500 | 92 | 875 |
| Lys63His | 37 | 59 | 1000 | 53 | 2000 |
| Lys63Ile | 46 | 68 | 1250 | NDb | NDb |
| Lys63Gln | 37 | 59 | 750 | NDb | NDb |
| Gly66Cys | 38 | 57 | 1250 | 56 | 2500 |
| Gly66Ser | 33 | 55 | 1000 | 55 | 1750 |
| Gly66Ala | 34 | 55 | 875 | 74 | 1250 |
| Gly66Asp | 32 | 56 | 875 | 69 | 1500 |
| Gly66His | 39 | 56 | 1000 | 54 | 1750 |
| Gly66Arg | 42 | 61 | 1250 | 56 | 2250 |
Strains were grown in LB media at 37 °C with aeration. Growth rates were determined in the presence of erythromycin (ERY) at 200 μg/mL and spiramycin (SPIR) at 750 μg/mL, and expressed as the time required for cell mass to double during logarithmic growth. Macrolide cell-doubling inhibitory concentrations (CDIC) were determined as described in Materials and Methods.
Not determined.
Several factors could potentially influence allele isolation frequency, including: i) codon bias in libraries; ii) recombination efficiency; iii) colony selection bias; and iv) 3′-mismatch PCR screening, in which multiple nucleotide changes are presumably more likely to pass the screen than point mutations. To evaluate these factors, we conducted two recombination experiments using oligonucleotides encoding L4 Gln62Phe (coded as TTT) and L4 Gln62Lys (coded as AAA). The L4 Gln62Phe and L4 Gln62Lys mutants grew at essentially the same rate in media containing erythromycin (Table 5), yet these alleles were isolated at very different frequencies from the L4 Gln62 library (Fig. 2 and Sup. Table II). An equal number of erythromycin-resistant clones were randomly isolated from each transformation and subjected to secondary erythromycin selection (Table 1). Almost all of the L4 Gln62Phe transformants survived secondary erythromycin selection, compared to ~ 67% of the L4 Gln62Lys transformants (Table 1). Because L4 Gln62Lys mutants may be less likely to pass 3′-mismatch PCR screening due to the similarity between Gln and Lys codons, rplD was sequenced from all erythromycin-resistant transformants (Table 1). Most L4 Gln62Phe transformants contained the oligonucleotide-encoded mutation, whereas all of the L4 Gln62Lys transformants had wild-type L4 (Table 1). Thus, the L4 Gln62Phe oligonucleotide was significantly more effective in mutagenesis than the L4 Gln62Lys oligonucleotide. These results are consistent with previous work showing Red-mediated recombination frequencies can vary dramatically depending on the identity of the oligonucleotide-encoded mutation 32; 36.
Although a variety of alleles were isolated from the L4 Lys63 library, we were surprised that L4 Lys63Gln mutants were not isolated, because this mutation confers macrolide-resistance in Streptococcus pneumoniae and Haemophilus influenzae 13; 37. Additionally, the lack of L4 Lys63Ile mutants seemed unusual given that L4 Lys63Leu and L4 Lys63Val mutants were isolated readily (Fig. 2 and Sup. Table II). To determine whether L4 Lys63Gln and L4 Lys63Ile mutations confer erythromycin resistance to E. coli, we introduced oligonucleotides encoding these mutations into cells and selected for erythromycin-resistant mutants. L4 Lys63Gln and L4 Lys63Ile mutants were readily isolated from these transformations, and these mutants exhibited erythromycin resistance comparable to other L4 Lys63 mutants (Table 5). Moreover, the L4 Lys63Gln and L4 Lys63Ile mutants passed the 3′-mismatch PCR screen, indicating that they were probably not systematically discarded during the original L4 Lys63 library screen. Because a number of alleles were isolated only once or twice from the L4 Lys63 library (Fig. 2 and Sup. Table II), L4 Lys63Gln and L4 Lys63Ile mutants may have been identified if more mutants had been characterized from the library.
Complex mutations
Several libraries produced primarily complex mutations, suggesting that simple missense changes at these codons are not sufficient to confer erythromycin resistance. We examined the role of secondary mutations in erythromycin resistance using L4 Thr65 as a model position. Most L4 Thr65 mutants contained secondary point mutations in neighboring codons (Table 2 and Sup. Table II), and in some instances, the secondary mutations appeared to be sufficient for resistance. For example, L4 Gly66Ser was commonly identified as a secondary mutation when L4 Thr65 was changed to hydrophobic residues (Sup. Table II). L4 Gly66Ser was also frequently isolated from the L4 Gly66 library and this single mutation confers erythromycin resistance (Table 5 and Sup. Table II). In contrast, the L4 Gly64Cys and L4 Gly64Val secondary mutations associated with L4 Thr65Pro were not isolated from the L4 Gly64 library (Sup. Table II), suggesting they do not confer resistance as single mutations. To examine whether secondary mutations are required for resistance in the context of the L4 Thr65Pro mutation, we introduced an oligonucleotide encoding L4 Thr65Pro (coded as CCC) into cells and selected for erythromycin-resistant mutants. Twenty-four of the resulting mutants were sequenced, and none contained the simple L4 Thr65Pro missense mutation (Table 2 and Sup. Table II). Instead, nearly all L4 Thr65Pro mutants contained additional L4 Gly64Cys (71%) or L4 Gly64Val (21%) point mutations (Table 2 and Sup. Table II).
Because an L4 Thr65Pro mutant was not isolated by recombineering, we constructed this mutation within a plasmid-borne copy of the E. coli S10 operon, which encodes L4, L22 and several other ribosomal proteins. The chromosomal S10 locus was then deleted so that the mutated S10 plasmid was the only source of L4 protein 38. Cells expressing L4 Thr65Pro were viable and grew at the same rate as cells carrying a wild-type version of the S10 plasmid (Table 6). Additionally, L4 Thr65Pro cells were almost as sensitive to erythromycin as wild-type cells, consistent with our inability to isolate these mutants by erythromycin selection. We next generated L4 Gly64Cys and L4 Gly64Val mutants using the S10 plasmid, and compared their resistance properties to those of mutants carrying S10 plasmid versions of the originally isolated L4 Gly64Cys-Thr65Pro and L4 Gly64Val-Thr65Pro mutations. The L4 Gly64Cys mutant was as sensitive to erythromycin as wild-type cells (Table 6). The L4 Gly64Val mutant was significantly more resistant than wild-type cells, yet it grew relatively slowly in the presence of erythromycin (Table 6). In both instances, the complex L4 Gly64Cys-Thr65Pro and L4 Gly64Val-Thr65Pro mutations provided a higher level of erythromycin resistance than the corresponding single L4 Gly64Cys and L4 Gly64Val mutations (Table 6). These results demonstrate that secondary mutations significantly enhance the erythromycin resistance of L4 Thr65Pro mutants.
Table 6.
Plasmid-borne L4 mutant growth rates and erythromycin sensitivitya
| L4 allele | Doubling time (min) | ERY doubling time (min) | ERY CDIC (μg/mL) |
|---|---|---|---|
| pS10(wild-type) | 33 | 127 | 200 |
| pS10(Thr65Pro) | 32 | 95 | 250 |
| pS10(Gly64Cys) | 34 | 115 | 200 |
| pS10(Gly64Val) | 36 | 83 | 750 |
| pS10(Gly64Ser) | 34 | 128 | 200 |
| pS10(Gly64Cys-Thr65Pro) | 35 | 43 | 1500 |
| pS10(Gly64Val-Thr65Pro) | 39 | 62 | 1000 |
| pS10(Gly64Ala-Thr65Pro) | 33 | 43 | 2000 |
| pS10(Gly64Ser-Thr65Pro) | 34 | 45 | 2000 |
| pS10(Gly64Arg-Thr65Pro) | 39 | 56 | 1500 |
| pS10(Gly64Asp-Thr65Pro) | 37 | 48 | 2000 |
Strains were grown in LB media at 37 °C with aeration. Growth rates were determined in the absence and presence of erythromycin (200 μg/mL), and expressed as the time required for cell mass to double during logarithmic growth. Erythromycin cell-doubling inhibitory concentrations (CDIC) were determined as described in Materials and Methods.
Strikingly, most of the L4 Thr65Pro-associated secondary mutations were either L4 Gly64Cys or Gly64Val point mutations (Sup. Table II), implying that these particular L4 Gly64 secondary mutations are best able to confer resistance in combination with L4 Thr65Pro. However, the combination of L4 Gly64Ala and L4 Thr65Pro mutations confers high-level macrolide resistance in Staphylococcus aureus 14. Using the S10 plasmid system, we tested whether the other four possible L4 Gly64 point mutations (L4 Gly64Ala, L4 Gly64Asp, L4 Gly64Arg and L4 Gly64Ser) also confer erythromycin resistance in combination with L4 Thr65Pro. All of the resulting mutants were as resistant as cells carrying plasmid-borne L4 Gly64Cys-Thr65Pro or L4 Gly64Val-Thr65Pro mutations (Table 6). In fact, the L4 Gly64Ala-Thr65Pro, L4 Gly64Asp-Thr65Pro and L4 Gly64Ser-Thr65Pro mutants were more resistant to erythromycin than any other L4 or L22 mutant examined in this study (Tables 5, 6 and 7). We also constructed the L4 Gly64Ser single mutation on the S10 plasmid and the resulting mutant was indistinguishable from wild-type with respect to erythromycin resistance (Table 6). Taken together, these results suggest that secondary mutagenesis is a non-random process favoring specific base changes. The L4 Gly64Cys and L4 Gly64Val secondary mutations are both G-toT transversions, suggesting this change occurs at a higher frequency than other point mutations. However, the most common secondary mutation from the L4 Thr65 library was a G-to-A transition producing L4 Gly66Ser, which was associated with other L4 Thr65 mutations (Sup. Table II). Because secondary mutations were typically found adjacent to target codons, perhaps the sequence context of the primary mutation influences secondary mutagenesis.
Table 7.
L22 mutant growth rates and macrolide sensitivitya
| L22 allele | Doubling time (min) | ERY select (%) | ERY doubling time (min) | ERY CDIC (μg/mL) | TYL select (%) | TYL doubling time (min) | TYL CDIC (μg/mL) |
|---|---|---|---|---|---|---|---|
| wild-type | 31 | 0 | 151 | 200 | 0 | 93 | 1250 |
| Lys90Trp | 33 | 73 | 48 | 750 | 0 | 145 | 1125 |
| Lys90Phe | 34 | 11 | 96 | 750 | 0 | 217 | 1250 |
| Lys90Leu | 33 | 6 | 79 | 375 | 0 | 167 | 1000 |
| Lys90Pro | 35 | 3 | 71 | 500 | 0 | 68 | 2125 |
| Ala89Glu-Lys90Val | 36 | 2 | 47 | 1125 | 19 | 43 | 3125 |
| Ala89Glu-Lys90Pro | 37 | 0 | 44 | 1250 | 57 | 44 | 3250 |
| Ala89Glu-Lys90Asp | 36 | 0 | 44 | 1250 | 13 | 43 | 3750 |
| Ala89Glu-Lys90Trp | 38 | 1 | 44 | 1250 | 2 | 43 | 3375 |
| Lys90Pro-ΔArg92 | 40 | 0 | 51 | 1000 | 2 | 50 | 3250 |
| Lys90Pro-Gly91Val | 40 | 0 | 50 | 875 | 2 | 47 | 3250 |
| Lys90Pro-Gly91Ser | 39 | 0 | 54 | 750 | 4 | 44 | 3000 |
Strains were grown in LB media at 37 °C with aeration. Growth rates were determined in the presence of erythromycin (ERY) at 200 μg/mL and tylosin (TYL) at 1.25 mg/mL, and expressed as the time required for cell mass to double during logarithmic growth. Macrolide cell-doubling inhibitory concentrations (CDIC) were determined as described in Materials and Methods. Allele isolation frequencies from erythromycin (n = 96) and tylosin (n = 47) selections are expressed as percents.
Library recombineering effectively produced a diverse set of missense mutations for genetic selection. However, comprehensive identification of these mutations was difficult, in part, because of the complexity added by secondary mutations. This was readily apparent in the L22 libraries, in which most alleles were isolated only once (Sup. Table III). In contrast, although L4 Thr65 mutants were dominated by complex mutations, several alleles were isolated independently at least twice (Sup. Table II). Therefore, we decided to examine the coverage of complex mutations using L4 Thr65 as a model position. The original library selection showed that L4 Thr65 could be mutated to 15 different amino acid residues, almost always in combination with secondary mutations (Table 2 and Sup. Table II). Complex mutations containing L4 Thr65Ser were not identified from the L4 Thr65 library, even though the structurally related L4 Thr65Cys mutation was identified as a component of five different complex mutations (Table 3 and Sup. Table II). However, L4 Thr65Ser mutations were identified as secondary point mutations from the L4 Gly64 library (Table 3 and Sup. Table II). To examine whether L4 Thr65Ser mutants were missed during the L4 Thr65 library selection, we introduced an oligonucleotide encoding this mutation (as AGC) into cells and selected for erythromycin-resistant mutants. We sequenced rplD from 24 of the resulting transformants and identified nine different mutations, seven of which had not previously been identified in any of the previous selections (Sup. Table II). Most of these mutants contained L4 Thr65Ser missense mutations coupled to other neighboring mutations, but there were also a significant number of complex deletion mutations (Table 2 and Sup. Table II). Thus, there are clearly more erythromycin-resistance alleles that were not identified during the original library selections. Indeed, there may be several hundred additional L4 and L22 macrolide-resistance alleles, whose comprehensive identification would probably require the isolation and characterization of several thousand mutants.
Selection on spiramycin and tylosin
The macrolactone rings of all macrolide antibiotics have similar, overlapping binding sites on the large ribosomal subunit 1; 27. However, the appended sugar residues of some macrolides make unique contacts to the ribosome. Spiramycin contains a forosamine residue that appears to contact L4, whereas tylosin has a mycanose sugar that tilts in the opposite direction and makes contact with L22 1. To determine whether these unique interactions could be exploited to identify macrolide-specific resistance alleles, we repeated selections using the libraries that produced predominately missense mutations. The L4 Gln62, L4 Lys63, and L4 Gly66 libraries were selected for spiramycin-resistant mutants, and the L22 Lys90 library was selected for tylosin-resistant mutants. In general, the distributions of L4 alleles isolated on spiramycin were similar, but not identical to those isolated on erythromycin (Fig. 2). In some instances, changes in allele isolation frequency could be explained by the resistance properties of a given mutant. For example, L4 Gly66Ala, which was isolated several times on erythromycin but never on spiramycin, had a growth rate similar to other L4 Gly66 mutants on erythromycin, but grew significantly more slowly in spiramycin (Fig. 2 and Table 5). Additionally, the L4 Lys63Glu, L4 Lys63Leu, and L4 Lys63His mutants grew more rapidly in spiramycin than other L4 Lys63 mutants (Table 5), consistent with their higher isolation frequency in spiramycin versus erythromycin (Fig. 2). However, other differences in allele isolation frequency could not be explained in terms of macrolide resistance. It is possible that some differences in allele frequency observed between the selections were due to the relatively small number of characterized mutants. We note that Zengel and colleagues have reported significantly slower growth rates for some of the same mutants characterized here, particularly L4 Gly66Cys and L4 Gly66Arg 24. These discrepancies may reflect differences in genetic background. Additionally, we transduced all L4 and L22 alleles into wild-type cells prior to analysis, whereas the selected mutants were characterized directly in Zaman et al. 24.
In contrast to the spiramycin selections, L22 Lys90 alleles isolated on tylosin were largely distinct from those obtained on erythromycin (Table 7 and Sup. Table III). L22 Lys90Trp was the dominant mutation isolated on erythromycin (70 out of 96 mutants), but it was not identified in any of the mutants isolated on tylosin (Table 7 and Sup. Table III). Instead, tylosin-selected mutants commonly had L22 Ala89Glu secondary point mutations in addition to changes in L22 Lys90 (Table 7 and Sup. Table III). The most common tylosin-selected mutation was L22 Ala89Glu-Lys90Pro, which was identified in 27 of the 47 tylosin-selected mutants (Table 7 and Sup. Table III). We examined the macrolide resistance properties of all the tylosin-selected mutants and compared them with L22 Lys90Trp, L22 Lys90Phe, L22 Lys90Leu, and L22 Lys90Pro cells, which together accounted for 93% of the erythromycin-selected L22 Lys90 mutants. All of the alleles isolated by tylosin selection were able to confer resistance to both erythromycin and tylosin (Table 7). In contrast, the L22 Lys90Trp, L22 Lys90Phe and L22 Lys90Leu alleles isolated on erythromycin did not confer resistance to tylosin or spiramycin (Table 7 and data not shown). In fact, the L22 Lys90Trp and L22 Lys90Leu mutants were more sensitive to tylosin than wild-type cells (Table 7).
Characterization of L22 Lys90Trp and L22 Ala89Glu-Lys90Pro ribosomes
We next sought to characterize the basis of erythromycin-specific resistance observed with the L22 Lys90Trp mutant. Binding constants for erythromycin were determined using purified ribosomes from wild-type, L22 Lys90Trp and L22 Ala89Glu-Lys90Pro cells. The dissociation constant (KD) for erythromycin binding to wild-type ribosomes was ~10 nM (Table 8), consistent with previously reported values 39; 40; 41. L22 Ala89Glu-Lys90Pro ribosomes had essentially the same erythromycin affinity as wild-type ribosomes, but L22 Lys90Trp ribosomes showed significantly lower affinity for erythromycin (Table 8). Tylosin binding affinity was determined by a competitive IC50 analysis, in which tylosin was titrated to displace ribosome-bound, radiolabeled erythromycin 40. This analysis determined that wild-type ribosomes bind tylosin and erythromycin with similar affinity (Table 8). In contrast to the erythromycin binding studies, L22 Lys90Trp ribosomes bound tylosin with wild-type affinity, whereas L22 Ala89Glu-Lys90Pro ribosomes exhibited lower affinity for tylosin (Table 8).
Table 8.
Macrolide binding constantsa
| Ribosomes | Erythromycin KD (nM) | Tylosin IC50 (nM) |
|---|---|---|
| Wild-type | 11 (± 2.2) | 11 (± 4.2) |
| L22 Lys90Trp | 170 (± 48) | 12 (± 9.4) |
| L22 Ala89Glu-Lys90Pro | 12 (± 2.2) | 85 (± 13) |
Binding experiments were conducted with hydrophobic interaction chromatography-purified ribosomes and [3H]- or [14C]-labeled erythromycin as described 40. The IC50 value is the concentration of tylosin required to displace 50% of the radiolabeled erythromycin from the ribosome. Reported values are the average ± standard deviation from three independent binding titrations.
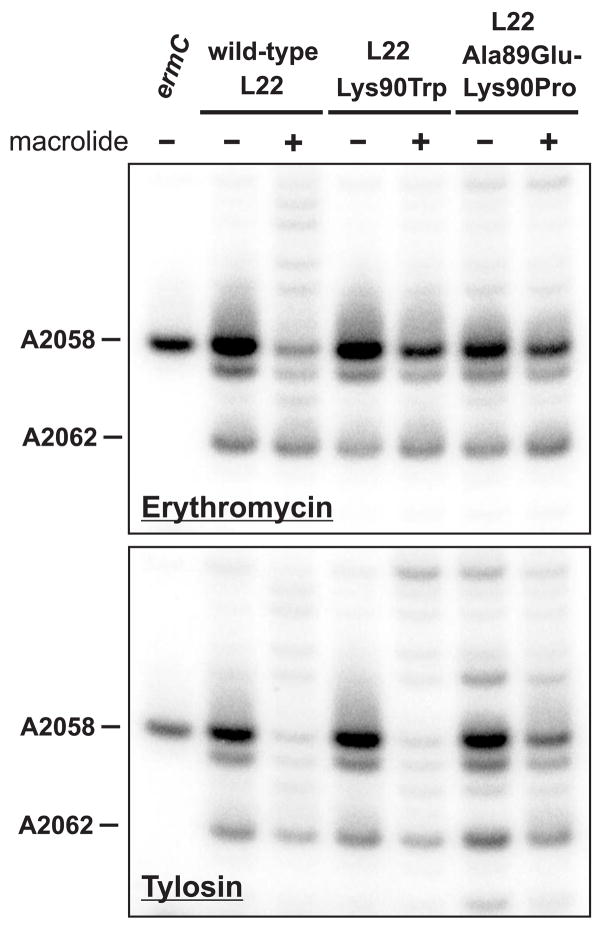
Although L22 Lys90Trp ribosomes have reduced affinity for erythromycin, the in vitro binding constant indicates that ribosomes would still be saturated at low micromolar concentrations of antibiotic. In addition, L22 Ala89Glu-Lys90Pro cells are highly resistant to erythromycin, yet their ribosomes bind the antibiotic with wild-type affinity in vitro. To further explore the basis of resistance in these mutants, we monitored macrolide binding in vivo using dimethyl sulfate (DMS) methylation protection assays. Ribosome-bound macrolides protect 23S rRNA residue A2058 from DMS-mediated methylation 42; 43. Cells were treated with DMS in the absence and presence of macrolides, and A2058 methylation was assessed by primer extension analysis. At concentrations slightly below the cell-doubling inhibitory concentration, both macrolides protected A2058 against methylation in wild-type ribosomes (Fig. 3). Significantly less A2058 protection was observed in the L22 Lys90Trp mutant treated with erythromycin, although tylosin protection was indistinguishable from that observed in wild-type cells (Fig. 3). These results indicate that erythromycin has a reduced residence time on L22 Lys90Trp ribosomes, whereas tylosin binding is apparently unaffected by this mutation. In contrast, methylation protection assays conducted with L22 Ala89Glu-Lys90Pro cells showed a reduction in the binding of both erythromycin and tylosin (Fig. 3). Thus, macrolide resistance in these L22 mutants appears to be the consequence of impaired antibiotic-ribosome binding in vivo.
Figure 3.
In vivo methylation protection assays. Wild-type and mutant cells were treated with dimethyl sulfate (DMS) in the presence and absence of macrolide antibiotics (150 μg/mL erythromycin and 1 mg/mL tylosin). Binding of macrolides to the ribosome inhibits DMS-mediated methylation of 23S rRNA residue A2058. Methylation was monitored by primer extension analysis as described in Materials and Methods. Primer extension products corresponding to DMS-mediated N1-methylation of A2058 and A2062 are indicated. Erythromycin had a greater inhibitory effect on DMS-mediated A2058 methylation in wild-type cells compared with the two L22 mutants. Tylosin significantly inhibited DMS-mediated A2058 methylation in wild-type and L22 Lys90Trp cells, but had less of an effect in L22 Ala89Glu-Lys90Pro mutant. A control reaction from cells overproducing the ErmC methyltransferase was included to serve as a marker for A2058 methylation.
Discussion
The results presented here show that an extraordinary number of ribosomal protein L4 and L22 variants confer resistance to macrolide antibiotics. Although several L4 and L22 mutants have been identified from a variety of bacterial species, to our knowledge, only 11 of the alleles isolated here have been previously reported 13; 14; 17; 21; 24; 37; 44. The remaining 267 L4 and L22 proteins described here appear to be novel. Much of this diversity can be attributed to unanticipated secondary mutations that accompanied many target codon mutations. It seems likely that these secondary mutations play an important role in macrolide resistance. This was confirmed for mutants containing the L4 Thr65Pro mutation, which was always associated with secondary point mutations or deletions. The L4 Thr65Pro mutation is unable to confer macrolide resistance by itself, but the combination of L4 Thr65Pro with any of the six L4 Gly64 point mutations confers high-level resistance to erythromycin. In this regard, secondary mutagenesis was beneficial because it produced many more macrolide resistance alleles for identification and characterization. However, secondary mutagenesis appears to be a non-random process that is context dependent. For example, G-to-T mutations affecting L4 Gly64 were isolated almost exclusively from the L4 Thr65Pro oligonucleotide selection, even though other secondary point mutations, such as the G-to-A mutation generating L4 Gly64Ser, result in even greater erythromycin resistance. The lack of L4 Gly64Ser secondary mutations in the L4 Thr65Pro selection experiment is not due to an intrinsically slow G-to-A mutation rate, because this transition was commonly identified as L4 Gly66Ser secondary mutations when L4 Thr65 was mutated to hydrophobic residues (Sup. Table II). Although the mechanism is unclear, these observations suggest that secondary mutagenesis depends not only upon recombineering, but also on the identity of the primary mutation encoded by the recombined oligonucleotide. We suspect that each library produced secondary mutations at very low frequencies, but these complex mutations were only isolated when the more abundant simple missense mutations were insufficient for macrolide resistance.
The remarkable number of macrolide resistance mutations suggests that almost any perturbation of the L4 or L22 loops is sufficient to modulate, or interfere with antibiotic binding. This appears to be the case for L4 Lys63, whose side-chain amino group coordinates the 2′-hydroxyl of A2060 and the phosphate of G2061 in 23S rRNA 45. Mutation of L4 Lys63 to almost any other residue is predicted to disrupt this coordination and could conceivably change the conformation of residues A2058 and A2059, whose unstacked bases form a hydrophobic crevice into which all macrolides bind (Fig. 1b) 1. Indeed, ribosomes containing L4 Lys63Glu have significantly lower affinity for erythromycin 21; 23. In addition, we find that substituting L4 Lys63 with Leu, Ile, Val, Met, Pro, Ala, Ser, Cys, Gly, Gln, Asp, Arg or His also confers macrolide resistance in E. coli. In fact, it appears that all residues, except those with large aromatic side chains, are able to functionally replace L4 Lys63 (E.J. Diner and C.S. Hayes, unpublished results). Several different L4 Gln62 and L4 Gly66 macrolide resistance alleles were also isolated. However, there are simple missense perturbations of the L4 loop (and presumably the L22 loop), which do not confer macrolide resistance (e.g. L4 Thr65Pro). Moreover, specific types of complex mutations were isolated from many of the libraries, implying that only a subset of L4 and L22 loop geometries are able to confer macrolide resistance.
Although the number of macrolide-resistance alleles is extraordinary, it is perhaps more remarkable that many of the mutants described appear to be quite healthy under the growth conditions studied here. L4 and L22 bind to 23S rRNA early during ribosome biogenesis and both are required for proper ribosome assembly and peptidyl transferase activity 46. In fact, some insertion mutations within the L4 and L22 loops have been shown to adversely affect ribosome assembly 24; 47. L4 also functions as a transcriptional and translational repressor of the S10 operon, ensuring that the encoded ribosomal proteins are synthesized in an equimolar ratio with rRNA 48; 49. Additionally, the L4 and L22 loops play a role in gene regulation by helping to mediate regulated ribosome pausing in E. coli 50; 51. Programmed ribosome pausing regulates the synthesis of the SecA general secretion ATPase in E. coli, and L22 mutations are known to modulate this essential regulation 51; 52. Therefore, there is ample opportunity for L4 and L22 mutations to adversely affect E. coli physiology. However, most of the mutants we characterized grew at rates very close to wild-type in the absence of macrolides, suggesting that ribosome assembly and function are near normal. Because L4 Gln62, L4 Lys63, L4 Gly66 and L22 Gly91 are completely conserved across all major divisions of eubacteria, we had anticipated that many macrolide-resistance mutations would significantly impair protein synthesis and lead to slow growth phenotypes. Thus, the highly conserved sequences of the L4 and L22 loops belie a striking structural and functional plasticity.
This work highlights a powerful application for Red-mediated recombination in genetic screens and selections. This methodology is particularly well suited for the mutagenesis of ribosomal proteins involved in antibiotic resistance. Using oligonucleotide library recombineering, we have mutagenized the rpsL and rpsE genes to obtain novel ribosomal S12 and S5 proteins that confer streptomycin and spectinomycin resistance, respectively (E.J. Diner, L.E. Holberger and C.S. Hayes, unpublished results). One obvious advantage of this approach is that mutagenesis is targeted to specific codons. This allows mutations of interest to be obtained at a much higher frequency than with spontaneous or chemical induced mutagenesis. Moreover, the oligonucleotide library approach permits routine isolation of atypical missense mutations such as Lys (AAA) to Phe (TTT). Because all three nucleotides must be mutated for this particular amino acid change, this missense mutation is virtually impossible to isolate though spontaneous mutagenesis. However, we find that not all mutations are recombineered with equal efficiency, and therefore some alleles are probably significantly underrepresented in the mutant population. Court and colleagues have previously reported this same phenomenon, and have convincingly demonstrated that methyl-directed mismatch DNA repair is responsible for the differences in recombination efficiency 32; 36. In an attempt to eliminate this bias from our library selections, we conducted several experiments using a mutS strain, which is deficient for mismatch DNA repair. However, macrolide-resistant mutants arose spontaneously at a high frequency in the mutS background. Moreover, many of the mutants isolated from these selections expressed wild-type L4 and L22 proteins from genes that contained dozens of silent mutations (B. D. Janssen and C. S. Hayes, unpublished results). Given that recombineering uncovered over 250 novel macrolide-resistance alleles, we feel that differential recombination efficiency is a minor barrier to allele diversity.
Genetic recombineering uncovered some unusual macrolide-specific resistance alleles. Typically, ribosomal mutations that confer resistance to one macrolide confer resistance not only to other macrolides, but also to lincosamides and streptogramins, which have overlapping binding-sites on the ribosome 9; 10. However, the L22 Lys90Trp, L22 Lys90Phe and L22 Lys90Leu mutations confer resistance to erythromycin, but not tylosin or spiramycin. Although the C2611U mutation in 23S rRNA exhibits a similar phenotype 53, we are unaware of other L22 or L4 mutations that confer resistance to erythromycin specifically. The basis of this phenotype appears to be an erythromycin-binding defect, which was observed both in vitro and in vivo. Modeling of the L22 Lys90Trp mutation predicts a dramatic van der Waals clash with A751 in domain II of 23S rRNA, suggesting that significant structural rearrangements accompany this amino acid change. Intriguingly, these structural perturbations appear more likely to interfere with the binding of tylosin, which makes contacts with L22 and 23S rRNA residue G748 1. In fact, methylation of G748 is sufficient to confer resistance to tylosin, but not erythromycin 54. How these L22 Lys90 mutations perturb ribosome binding of erythromycin, but not tylosin, is unclear at present. Additionally, it was somewhat surprising that we observed reduced erythromycin binding to L22 Ala89Glu-Lys90Pro ribosomes in vivo, given that these ribosomes bind erythromycin with wild-type affinity in vitro. Of course, binding studies conducted with non-translating ribosomes do not recapitulate in vivo conditions. Nascent peptides in the lumen of the exit tunnel inhibit macrolide binding 55; 56, and perhaps the L22 mutation significantly reduces the affinity of actively translating ribosomes for erythromycin. A similar discrepancy between erythromycin binding in vitro and in vivo has been reported for L22 ΔMet82-Lys83-Arg84 ribosomes 21; 57. Moore & Sauer have recently shown that L22 ΔMet82-Lys83-Arg84 ribosomes are as sensitive to erythromycin as wild-type ribosomes using in vitro translation reactions 57. They propose that the L22 ΔMet82-Lys83-Arg84 mutation confers resistance by reducing intracellular macrolide concentrations, perhaps by modulating the activity of TolC-dependent efflux pumps 57. Although the mechanism is unclear, these results challenge the prevailing models of macrolide resistance for this mutant. Clearly, further experimentation will be required to elucidate the basis of resistance in these intriguing L22 mutants.
Materials and Methods
Selection of macrolide-resistant mutants
All E. coli strains were derivatives of strain X90 58. Strain X90 ΔpioO::kan was constructed by P1 mediated transduction from strain JW3284 59. Plasmid pSIM5 60 was generously provided by Don Court (NCI-Frederick, MD). Genetic recombineering was performed essentially as described 34. Briefly, 50 – 100 μL of competent cells were transformed with 5 pmol of single-stranded oligonucleotide by electroporation (complete recombineering oligonucleotide sequences are presented in Supplementary Table I). Transformed cells were immediately suspended in 1.0 mL of Luria broth (LB) medium and 100 μL aliquots plated onto sterile nitrocellulose filters on LB agar plates containing no antibiotic. Antibiotic-free plates were incubated at 30 °C for 3 h, then the nitrocellulose filters were transferred onto LB agar plates containing one of the following macrolide antibiotics: erythromycin (100 μg/mL), spiramycin (550 μg/mL) or tylosin (1.25 mg/mL). Macrolide plates were further incubated at 37 °C for up to 36 h to select resistant colonies. Selections for each randomized position were performed independently at least three times. Isolated colonies were streaked onto fresh LB agar plates containing the appropriate macrolide antibiotic. Clones that survived the second macrolide selection were screened by mismatch mutation PCR 35, using primers listed in Supplemental Table I. Clones that produced little or no PCR product (i.e. likely to contain mutations) were selected for sequencing of rplD or rplV. The rplD gene was PCR amplified using primers rplD-for, (5′ - TGC TGC TGG TTA AAG GTG CTG TCC C) and rplD-rev, (5′ - CGT GCG GTG CAC GCA GCA CCT TCA GCA GAC G); and the resulting products sequenced using primer rplD-seq, (5′ - GCG ACC TGA TCG TTA AAC CAG CTG TGA AGG). The rplV gene was PCR amplified using primers rplV-for, (5′ - GGT GAA TTC GCA CCG ACT CGT ACT TAT CG) and rplV-rev, (5′ - GAG TTC CAT GGT TTT ACA ATA CCC AGG); and the resulting products sequenced using primer rplV-seq, (5′ - TAG GAG GAA CAT ATG GAA ACT ATC GCT AAA C). Each L4 and L22 allele conferred erythromycin resistance to wild-type cells after transfer by phage P1-mediated generalized transduction using the linked pioO::kan kanamycin resistance marker. To confirm allele transfer, both rplD and rplV genes were sequenced from each transductant.
Plasmid constructions
Plasmid pS10 (generously provided by S. Moore, University of Central Florida) is a ColA plasmid derivative carrying the entire S10 operon 38. QuickChange PCR (Stratagene) was used to remove the NcoI restriction site in rpsC, then rplD Pro59 was synonymously recoded to introduce an NcoI site, generating plasmid pS10-2. All plasmid-borne rplD mutations were made by PCR using reverse primer rplD-ApaLI, (5′- CGT GCG GTG CAC GCA GCA CCT TCA GCA GAC G), in combination with the following mutagenic forward primers: rplD(T65P)-NcoI, (5′- AAA CCA TGG CGC CAG AAA GGC CCC GGC CGT GCG CG); rplD(G64C)-NcoI (5′ - AAA CCA TGG CGC CAG AAA TGC ACC GGC CG); rplD(G64S)-NcoI, (5′ - AAA CCA TGG CGC CAG AAA AGC ACC GGC CG); rplD(G64V)-NcoI, (5′ - AAA CCA TGG CGC CAG AAA GTC ACC GGC CG); rplD(G64C-T65P)-NcoI, (5′ -AAA CCA TGG CGC CAG AAA TGC CCC GGC CG); rplD(G64V-T65P)-NcoI,(5′ - AAA CCA TGG CGC CAG AAA GTC CCC GGC CG); rplD(G64A-T65P)-NcoI, (5′- AAA CCA TGG CGC CAG AAA GCC CCC GGC CGT GCG CG); rplD(G64D-T65P)-NcoI, (5′- AAA CCA TGG CGC CAG AAA GAC CCC GGC CGT GCG CG); rplD(G64R-T65P)-NcoI, (5′- AAA CCA TGG CGC CAG AAA CGC CCC GGC CGT GCG CG); and rplD(G64S-T65P)-NcoI, (5′ - AAA CCA TGG CGC CAG AAA AGC CCC GGC CGT GCG CG); followed by ligation into plasmid pS10-2 using NcoI and ApaLI restriction sites (restriction sites are underlined). After constructs were confirmed by DNA sequencing, the chromosomal S10 operon was deleted as described 38, leaving the mutant pS10 plasmids as the sole source of L4 protein. The ermC gene from plasmid pHB201 61 was amplified using primers: ermC-Eco, (5′ - TTT GAA TTC ACC ATG AAC GAG AAA AAT ATA AAA CAC AG), and ermC-Pst, (5′ - TTT CTG CAG CCC TTA ACTY TAC TTA TTA AAT AAT TTA TAG C); digested with EcoRI and PstI, and ligated to plasmid pBAD24 to create an arabinose-inducible ErmC expression plasmid.
Macrolide sensitivity and growth rate determinations
Wild-type E. coli is intrinsically resistant to macrolide antibiotics. The minimum inhibitory concentration (MIC) of erythromycin is 625 μg/mL for our wild-type E. coli strain. Because erythromycin MICs for the selected mutants are even higher (L22 Lys90Trp MIC = 1.75 mg/mL; and L22 Ala89Glu-Lys90Pro MIC = 2.25 mg/mL), we assessed macrolide sensitivity using a modified MIC procedure to avoid very high concentrations of antibiotics. Mid-log phase cells were diluted to OD600 = 0.05 into a series of LB cultures containing increasing concentrations of macrolide, and grown at 37 °C with aeration until cultures without antibiotic had attained OD600 ~ 1.0. The macrolide concentration that prevented one OD600 doubling was taken as the cell-doubling inhibitory concentration (CDIC) for the antibiotic. Growth rates were determined by diluting mid-log phase to OD600 = 0.05 in fresh, pre-warmed LB media, with or without macrolide antibiotics (200 μg/mL erythromycin, 750 μg/mL spiramycin, or 1.25 mg/mL tylosin) followed by incubation at 37 °C with aeration. Cell growth was monitored by OD600 as a function of time and the doubling times calculated from log2 plots of growth curves.
Ribosome purification
E. coli cultures (1 L) were grown to early log phase, harvested onto crushed ice, followed by centrifugation to collect cells. Cells were resuspended and washed once in ice-cold HT-1 buffer [20 mM potassium HEPES (pH 7.5) – 100 mM potassium glutamate – 0.1 mM EDTA – 1 mM magnesium acetate] and collected by centrifugation. Cell pellets were then resuspended in ice-cold HT-1 buffer supplemented with 14 mM β-mercaptoethanol and 0.01% Tween-20, and cells broken by French press at 20,000 psi. Lysates were clarified by centrifugation at 30,000 × g for 10 min at 4 °C, and the supernatants layered onto 1.1 M sucrose cushions prepared in HT-1 buffer supplemented with 14 mM β-mercaptoethanol and 0.01% Tween-20. Sucrose cushions were centrifuged at 45,000 rpm (160,000 × g) in a Beckman MLS-50 rotor for 1 hr at 4 °C. The crude ribosome pellet was gently dissolved into HT-6 buffer (supplemented to 6 mM magnesium acetate) containing 14 mM β-mercaptoethanol and 0.01% Tween-20, followed by centrifugation at 20,000 × g to remove insoluble material. Magnesium acetate and ammonium sulfate were added to final concentrations of 10 mM and 1.5 M, respectively, followed by incubation on ice for 20 min to facilitate precipitation. Precipitate was removed by centrifugation at 20,000 × g, and the supernatant applied to a HiTrap™ butyl sepharose hydrophobic interaction column (GE Healthcare) pre-equilibrated in column wash buffer [20 mM potassium HEPES (pH 7.5) – 10 mM magnesium acetate – 1.5 M ammonium sulfate]. The column was washed with 5.0 mL of ice-cold column wash buffer, followed by a 5.0 mL wash of wash buffer containing 1.2 M ammonium sulfate. Ribosomes were eluted with 15.0 mL of 20 mM potassium HEPES (pH 7.5) – 10 mM magnesium acetate – 0.6 M ammonium sulfate. Purified ribosomes were concentrated and buffer exchanged into with HT-6 buffer using Amicon Ultra 30K centrifugal concentrators (Millipore). Purified ribosomes were quantified by absorption at 260 nm using an extinction coefficient of 3.91 × 107 M−1 cm−1.
Macrolide-ribosome binding analysis
Erythromycin binding constants for wild-type and L22 Ala89Glu-Lys90Pro ribosomes were determined by titrations of 6 nM purified ribosomes with [3H]-erythromycin (80 Ci/mmol, American Radiolabeled Chemicals). L22 Lys90Trp ribosome binding studies were performed with 100 nM purified ribosomes and [14C]-erythromycin (51.3 mCi/mmol, Perkin Elmer-New England Nuclear). All macrolide binding titrations were performed in HT-6 buffer supplemented with 14 mM β-mercaptoethanol and 0.01% Tween-20. Binding reactions were incubated at ambient temperature for 2 hr and then passed through a spin column containing Sephadex superfine G-25 (GE Healthcare) pre-equilibrated in binding buffer. The void volume was collected, subjected to scintillation counting, and specific binding calculated by subtracting background cpm obtained from reactions lacking purified ribosomes. A quadratic binding equation was fit to the experimental data by iterative non-linear regression as described 40, using the DeltaGraph 5.6 software package (Red Rock Software). Tylosin binding was assessed by competitive binding and quantified as IC50, the concentration of tylosin required to displace 50% of ribosome-bound radiolabeled erythromycin. IC50 values were determined by three-parameter iterative fits as described 40. All binding experiments were conducted at least three times and the average and standard deviations are reported.
Dimethyl sulfate (DMS) modification mapping
E. coli strains were grown in 25 mL of LB medium at 37 °C with aeration to an OD600 of 0.3, then macrolide antibiotics (150 μg/mL erythromycin or 1 mg/mL tylosin) were added, followed by further culture for 2 hr. Cells were harvested by centrifugation, and resuspended in 1.5 mL of pre-warmed LB medium containing the appropriate antibiotic and incubated at 37 °C with shaking for 10 min. Dimethyl sulfate (DMS) was added to a final concentration of 42 mM for 15 min, then reactions quenched by addition to 15 mL of stop solution [100 mM Tris-HCl (pH 7.5) – 50 mM sodium chloride – 2 mM magnesium acetate – 1 M β-mercaptoethanol]. Quenched cell suspensions were extracted with 15 mL of isoamyl alcohol, cells collected and resuspended in 15 mL of stop solution, followed by addition of 15 mL ice-cold methanol. Control reactions, in which stop solution was added to cells prior to DMS, demonstrated that quenching prevented modification during RNA isolation (data not shown). Total cellular RNA was extracted as described 62.
Primer extension analysis was performed to detect methylated adenosine residues. The 5′-radiolabeled oligonucleotide mac-DMS, (5′- CAG TGT CAA GCT ATA GTA AAG GTT CAC) was incubated with 3 μg of total RNA at 94 °C for 2 min, followed by annealing at 50 °C. Reverse transcriptase (Superscript® III, Invitrogen) and deoxynucleotides were added and incubated for 30 min at 50 °C. Reactions were quenched by addition of an equal volume of denaturing gel loading buffer, samples were heated to 95° C and run on 1 TBE – 8 M urea –10% polyacrylamide gels. Gels were exposed and developed by phosphorimaging using BioRad Quantity One software.
Supplementary Material
Acknowledgments
We thank Fernando Garza-Sánchez, Brian Janssen, Sean Moore and Zach Ruhe for helpful discussions. Additional thanks to Sean Moore for sharing unpublished data and generously providing the pS10 plasmid, and to Brian Janssen for conducting preliminary experiments. This work was supported by grant GM078634 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hansen JL, Ippolito JA, Ban N, Nissen P, Moore PB, Steitz TA. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol Cell. 2002;10:117–128. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 2.Schlunzen F, Harms JM, Franceschi F, Hansen HA, Bartels H, Zarivach R, Yonath A. Structural basis for the antibiotic activity of ketolides and azalides. Structure. 2003;11:329–338. doi: 10.1016/s0969-2126(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 3.Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 4.Tenson T, Lovmar M, Ehrenberg M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J Mol Biol. 2003;330:1005–1014. doi: 10.1016/s0022-2836(03)00662-4. [DOI] [PubMed] [Google Scholar]
- 5.Champney WS. Bacterial ribosomal subunit synthesis: a novel antibiotic target. Curr Drug Targets Infect Disord. 2001;1:19–36. doi: 10.2174/1568005013343281. [DOI] [PubMed] [Google Scholar]
- 6.Champney WS, Tober CL. Specific inhibition of 50S ribosomal subunit formation in Staphylococcus aureus cells by 16-membered macrolide, lincosamide, and streptogramin B antibiotics. Curr Microbiol. 2000;41:126–135. doi: 10.1007/s002840010106. [DOI] [PubMed] [Google Scholar]
- 7.Chittum HS, Champney WS. Erythromycin inhibits the assembly of the large ribosomal subunit in growing Escherichia coli cells. Curr Microbiol. 1995;30:273–279. doi: 10.1007/BF00295501. [DOI] [PubMed] [Google Scholar]
- 8.Nakajima Y. Mechanisms of bacterial resistance to macrolide antibiotics. J Infect Chemother. 1999;5:61–74. doi: 10.1007/s101560050011. [DOI] [PubMed] [Google Scholar]
- 9.Roberts MC. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol Lett. 2008;282:147–159. doi: 10.1111/j.1574-6968.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- 10.Vester B, Douthwaite S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother. 2001;45:1–12. doi: 10.1128/AAC.45.1.1-12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westh H, Hougaard DM, Vuust J, Rosdahl VT. Prevalence of erm gene classes in erythromycin-resistant Staphylococcus aureus strains isolated between 1959 and 1988. Antimicrob Agents Chemother. 1995;39:369–373. doi: 10.1128/aac.39.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peric M, Bozdogan B, Jacobs MR, Appelbaum PC. Effects of an efflux mechanism and ribosomal mutations on macrolide susceptibility of Haemophilus influenzae clinical isolates. Antimicrob Agents Chemother. 2003;47:1017–1022. doi: 10.1128/AAC.47.3.1017-1022.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prunier AL, Trong HN, Tande D, Segond C, Leclercq R. Mutation of L4 ribosomal protein conferring unusual macrolide resistance in two independent clinical isolates of Staphylococcus aureus. Microb Drug Resist. 2005;11:18–20. doi: 10.1089/mdr.2005.11.18. [DOI] [PubMed] [Google Scholar]
- 15.Franceschi F, Kanyo Z, Sherer EC, Sutcliffe J. Macrolide resistance from the ribosome perspective. Curr Drug Targets Infect Disord. 2004;4:177–191. doi: 10.2174/1568005043340740. [DOI] [PubMed] [Google Scholar]
- 16.Bogdanovich T, Bozdogan B, Appelbaum PC. Effect of efflux on telithromycin and macrolide susceptibility in Haemophilus influenzae. Antimicrob Agents Chemother. 2006;50:893–898. doi: 10.1128/AAC.50.3.893-898.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark C, Bozdogan B, Peric M, Dewasse B, Jacobs MR, Appelbaum PC. In vitro selection of resistance in Haemophilus influenzae by amoxicillin-clavulanate, cefpodoxime, cefprozil, azithromycin, and clarithromycin. Antimicrob Agents Chemother. 2002;46:2956–2962. doi: 10.1128/AAC.46.9.2956-2962.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canu A, Malbruny B, Coquemont M, Davies TA, Appelbaum PC, Leclercq R. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2002;46:125–131. doi: 10.1128/AAC.46.1.125-131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corcoran D, Quinn T, Cotter L, Fanning S. An investigation of the molecular mechanisms contributing to high-level erythromycin resistance in Campylobacter. Int J Antimicrob Agents. 2006;27:40–45. doi: 10.1016/j.ijantimicag.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 20.Cagliero C, Mouline C, Cloeckaert A, Payot S. Synergy between efflux pump CmeABC and modifications in ribosomal proteins L4 and L22 in conferring macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother. 2006;50:3893–3896. doi: 10.1128/AAC.00616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chittum HS, Champney WS. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J Bacteriol. 1994;176:6192–6198. doi: 10.1128/jb.176.20.6192-6198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregory ST, Dahlberg AE. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23 S ribosomal RNA. J Mol Biol. 1999;289:827–834. doi: 10.1006/jmbi.1999.2839. [DOI] [PubMed] [Google Scholar]
- 23.Wittmann HG, Stoffler G, Apirion D, Rosen L, Tanaka K, Tamaki M, Takata R, Dekio S, Otaka E. Biochemical and genetic studies on two different types of erythromycin resistant mutants of Escherichia coli with altered ribosomal proteins. Mol Gen Genet. 1973;127:175–189. doi: 10.1007/BF00333665. [DOI] [PubMed] [Google Scholar]
- 24.Zaman S, Fitzpatrick M, Lindahl L, Zengel J. Novel mutations in ribosomal proteins L4 and L22 that confer erythromycin resistance in Escherichia coli. Mol Microbiol. 2007;66:1039–1050. doi: 10.1111/j.1365-2958.2007.05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabashvili IS, Gregory ST, Valle M, Grassucci R, Worbs M, Wahl MC, Dahlberg AE, Frank J. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol Cell. 2001;8:181–188. doi: 10.1016/s1097-2765(01)00293-3. [DOI] [PubMed] [Google Scholar]
- 26.Apirion D. Three genes that affect Escherichia coli ribosomes. J Mol Biol. 1967;30:255–275. [PubMed] [Google Scholar]
- 27.Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Pereyre S, Guyot C, Renaudin H, Charron A, Bebear C, Bebear CM. In vitro selection and characterization of resistance to macrolides and related antibiotics in Mycoplasma pneumoniae. Antimicrob Agents Chemother. 2004;48:460–465. doi: 10.1128/AAC.48.2.460-465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pereyre S, Metifiot M, Cazanave C, Renaudin H, Charron A, Bebear C, Bebear CM. Characterisation of in vitro-selected mutants of Ureaplasma parvum resistant to macrolides and related antibiotics. Int J Antimicrob Agents. 2007;29:207–211. doi: 10.1016/j.ijantimicag.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Tait-Kamradt A, Davies T, Cronan M, Jacobs MR, Appelbaum PC, Sutcliffe J. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob Agents Chemother. 2000;44:2118–2125. doi: 10.1128/aac.44.8.2118-2125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu Rev Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 32.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci U S A. 2001;98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kmiec E, Holloman WK. Beta protein of bacteriophage lambda promotes renaturation of DNA. J Biol Chem. 1981;256:12636–12639. [PubMed] [Google Scholar]
- 34.Thomason L, Court DL, Bubunenko M, Costantino N, Wilson H, Datta S, Oppenheim A. Recombineering: genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol. 2007 doi: 10.1002/0471142727.mb0116s78. Chapter 1, Unit 1 16. [DOI] [PubMed] [Google Scholar]
- 35.Cha RS, Zarbl H, Keohavong P, Thilly WG. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl. 1992;2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- 36.Costantino N, Court DL. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc Natl Acad Sci U S A. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farrell DJ, Morrissey I, Bakker S, Buckridge S, Felmingham D. In vitro activities of telithromycin, linezolid, and quinupristin-dalfopristin against Streptococcus pneumoniae with macrolide resistance due to ribosomal mutations. Antimicrob Agents Chemother. 2004;48:3169–3171. doi: 10.1128/AAC.48.8.3169-3171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore SD, Baker TA, Sauer RT. Forced extraction of targeted components from complex macromolecular assemblies. Proc Natl Acad Sci U S A. 2008;105:11685–11690. doi: 10.1073/pnas.0805633105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pestka S. Binding of [14C]erythromycin to Escherichia coli ribosomes. Antimicrob Agents Chemother. 1974;6:474–478. doi: 10.1128/aac.6.4.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan K, Hunt E, Berge J, May E, Copeland RA, Gontarek RR. Fluorescence polarization method to characterize macrolide-ribosome interactions. Antimicrob Agents Chemother. 2005;49:3367–3372. doi: 10.1128/AAC.49.8.3367-3372.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovmar M, Tenson T, Ehrenberg M. Kinetics of macrolide action: the josamycin and erythromycin cases. J Biol Chem. 2004;279:53506–53515. doi: 10.1074/jbc.M401625200. [DOI] [PubMed] [Google Scholar]
- 42.Moazed D, Noller HF. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987;69:879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- 43.Xiong L, Korkhin Y, Mankin AS. Binding site of the bridged macrolides in the Escherichia coli ribosome. Antimicrob Agents Chemother. 2005;49:281–288. doi: 10.1128/AAC.49.1.281-288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matic V, Kosowska K, Bozdogan B, Kelly LM, Smith K, Ednie LM, Lin G, Credito KL, Clark CL, McGhee P, Pankuch GA, Jacobs MR, Appelbaum PC. Antipneumococcal activities of two novel macrolides, GW 773546 and GW 708408, compared with those of erythromycin, azithromycin, clarithromycin, clindamycin, and telithromycin. Antimicrob Agents Chemother. 2004;48:4103–4112. doi: 10.1128/AAC.48.11.4103-4112.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–34. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 46.Rohl R, Nierhaus KH. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 1982;79:729–733. doi: 10.1073/pnas.79.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zengel JM, Jerauld A, Walker A, Wahl MC, Lindahl L. The extended loops of ribosomal proteins L4 and L22 are not required for ribosome assembly or L4-mediated autogenous control. RNA. 2003;9:1188–1197. doi: 10.1261/rna.5400703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dean D, Yates JL, Nomura M. Escherichia coli ribosomal protein S8 feedback regulates part of spc operon. Nature. 1981;289:89–91. doi: 10.1038/289089a0. [DOI] [PubMed] [Google Scholar]
- 49.Zengel JM, Lindahl L. Ribosomal protein L4 of Escherichia coli: in vitro analysis of L4-mediated attenuation control. Biochimie. 1991;73:719–727. doi: 10.1016/0300-9084(91)90052-3. [DOI] [PubMed] [Google Scholar]
- 50.Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol Cell. 2005;19:333–343. doi: 10.1016/j.molcel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 51.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 52.Nakatogawa H, Ito K. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol Cell. 2001;7:185–92. doi: 10.1016/s1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]
- 53.Vannuffel P, Di Giambattista M, Morgan EA, Cocito C. Identification of a single base change in ribosomal RNA leading to erythromycin resistance. J Biol Chem. 1992;267:8377–8382. [PubMed] [Google Scholar]
- 54.Liu M, Kirpekar F, Van Wezel GP, Douthwaite S. The tylosin resistance gene tlrB of Streptomyces fradiae encodes a methyltransferase that targets G748 in 23S rRNA. Mol Microbiol. 2000;37:811–820. doi: 10.1046/j.1365-2958.2000.02046.x. [DOI] [PubMed] [Google Scholar]
- 55.Odom OW, Picking WD, Tsalkova T, Hardesty B. The synthesis of polyphenylalanine on ribosomes to which erythromycin is bound. Eur J Biochem. 1991;198:713–722. doi: 10.1111/j.1432-1033.1991.tb16071.x. [DOI] [PubMed] [Google Scholar]
- 56.Tai PC, Wallace BJ, Davis BD. Selective action of erythromycin on initiating ribosomes. Biochemistry. 1974;13:4653–4659. doi: 10.1021/bi00719a029. [DOI] [PubMed] [Google Scholar]
- 57.Moore SD, Sauer RT. Revisiting the mechanism of macrolide-antibiotic resistance mediated by ribosomal protein L22. Proc Natl Acad Sci U S A. 2008;105:18261–18266. doi: 10.1073/pnas.0810357105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J Biol Chem. 2002;277:33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- 59.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Datta S, Costantino N, Court DL. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 61.Bron S, Bolhuis A, Tjalsma H, Holsappel S, Venema G, van Dijl JM. Protein secretion and possible roles for multiple signal peptidases for precursor processing in bacilli. J Biotechnol. 1998;64:3–13. doi: 10.1016/s0168-1656(98)00099-6. [DOI] [PubMed] [Google Scholar]
- 62.Hayes CS, Sauer RT. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol Cell. 2003;12:903–911. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.