Abstract
The rosid clade (70,000 species) contains more than one-fourth of all angiosperm species and includes most lineages of extant temperate and tropical forest trees. Despite progress in elucidating relationships within the angiosperms, rosids remain the largest poorly resolved major clade; deep relationships within the rosids are particularly enigmatic. Based on parsimony and maximum likelihood (ML) analyses of separate and combined 12-gene (10 plastid genes, 2 nuclear; >18,000 bp) and plastid inverted repeat (IR; 24 genes and intervening spacers; >25,000 bp) datasets for >100 rosid species, we provide a greatly improved understanding of rosid phylogeny. Vitaceae are sister to all other rosids, which in turn form 2 large clades, each with a ML bootstrap value of 100%: (i) eurosids I (Fabidae) include the nitrogen-fixing clade, Celastrales, Huaceae, Zygophyllales, Malpighiales, and Oxalidales; and (ii) eurosids II (Malvidae) include Tapisciaceae, Brassicales, Malvales, Sapindales, Geraniales, Myrtales, Crossosomatales, and Picramniaceae. The rosid clade diversified rapidly into these major lineages, possibly over a period of <15 million years, and perhaps in as little as 4 to 5 million years. The timing of the inferred rapid radiation of rosids [108 to 91 million years ago (Mya) and 107–83 Mya for Fabidae and Malvidae, respectively] corresponds with the rapid rise of angiosperm-dominated forests and the concomitant diversification of other clades that inhabit these forests, including amphibians, ants, placental mammals, and ferns.
Keywords: community assembly, divergence time estimates, phylogeny, rapid radiation
Great progress has been made in elucidating deep-level angiosperm relationships during the past decade. The eudicot clade, with ≈75% of all angiosperm species, comprises several major subclades: rosids, asterids, Saxifragales, Santalales, and Caryophyllales (1–3). Investigations have converged on the branching pattern of the basalmost angiosperms, revealing that Amborellaceae, Nymphaeales [in the sense of APG II (3) and including Hydatellaceae (4)], and Austrobaileyales are successive sisters to all other extant angiosperms (reviewed in ref. 2). Analyses of complete plastid genome sequences have resolved other problematic deep-level relationships, suggesting that Chloranthaceae and magnoliids are sister to a clade of monocots and eudicots plus Ceratophyllaceae (5, 6). Likewise, progress has been made in clarifying relationships within the large monocot (7) and asterid (8) clades.
Despite these successes, the rosids stand out as the largest and least-resolved major clade of angiosperms; basal nodes within the clade have consistently received low internal support (1, 2, 9, 10). The rosid clade comprises ≈70,000 species and 140 families (2, 11). Containing more than a quarter of total angiosperm and ≈39% of eudicot species diversity, the rosid clade is broader in circumscription than the traditional Rosidae or Rosanae (e.g., 12; reviewed in ref. 2). The oldest fossil flowers conforming to the rosids are from the late Santonian to Turonian (≈84–89.5 Mya) (9, 11, 13, 14).
Rosids exhibit enormous heterogeneity in habit, habitat, and life form, comprising herbs, shrubs, trees, vines, aquatics, succulents, and parasites. The rosid clade also contains novel biochemical pathways, such as the machinery necessary for symbiosis with nitrogen-fixing bacteria (nitrogen-fixing clade) and defense mechanisms such as glucosinolate production (Brassicales) and cyanogenic glycosides (2). Many important crops, including legumes (Fabaceae) and fruit crops (Rosaceae), are rosids. Furthermore, 4 of the 5 published complete angiosperm nuclear genome sequences are rosids (with 2 other rosids, Manihot and Ricinus, well underway): Arabidopsis (Brassicaceae), Carica (Caricaceae), Populus (Salicaceae), and Vitis (Vitaceae, sister to other rosids). The variation in morphological, chemical, and ecological features and the importance of rosids as genetic and genomic models require a phylogenetic perspective for interpreting large-scale evolutionary patterns across the clade. Finally, most lineages of extant temperate and tropical forest trees are rosids (e.g., Betulaceae, Celtidaceae, Fabaceae, Fagaceae, Malvaceae, Sapindaceae, and Ulmaceae). In temperate North America ≈71% of the forest tree species are rosids (15). Similarly, rosids often constitute >50% of tropical tree species diversity. For example, they comprise 59% of the tree species on Barro Colorado Island, Panama (16), 60% of the tree flora of Paraguay (17), 60% on Puerto Rico and the Virgin Islands (18), and 63% of the tree species of Brazil (19). Analysis of tropical forest plots from around the world again suggests a major role of rosid trees (20). Half the species richness of the Neotropical plots is made up of just 11 families, 4 of which are rosids, with Fabaceae the most important (Moraceae, Meliaceae, and Euphorbiaceae are other major rosids in the Neotropics); Fabaceae are also the most important family of trees in Africa, but are replaced as number one by another rosid family, Dipterocarpaceae, in Southeast Asia (20). The important ecological role of many rosid families further argues for greater resolution of rosid phylogeny.
Although studies agree on the composition of the rosid clade, deep-level relationships within the rosids remain enigmatic. Vitaceae usually appear as sister to all other rosids, although support for this placement is generally low (reviewed in refs. 2 and 10). Most remaining rosids appear in 2 large subclades (1–3): eurosids I [Fabidae (21)] and eurosids II [Malvidae (21)], with jackknife support of 77% and 95%, respectively. Fabidae include (i) the nitrogen-fixing clade (Rosales, Fabales, Cucurbitales, and Fagales); (ii) Zygophyllales; and (iii) a weakly supported clade of Celastrales, Oxalidales, and Malpighiales [COM group (22)]. Malvidae include Brassicales, Malvales, Sapindales, and Tapisciaceae.
Some rosid clades (Crossosomatales, Geraniales, and Myrtales), however, do not fall into either Fabidae or Malvidae, and their relationships remain unclear. Myrtales are often resolved as sister to Fabidae, whereas Crossosomatales and Geraniales appear with Malvidae, but without strong support (1, 2, 10). Furthermore, the placements of several problematic families—Aphloiaceae, Apodanthaceae, Geissolomataceae, Huaceae, Ixerbaceae, Picramniaceae, and Strasburgeriaceae—also need to be ascertained.
Rosid diversification, like angiosperm phylogeny as a whole, is characterized by repeating patterns of radiation from deep to shallow levels (2). Lack of resolution at the base of the rosid phylogeny in previous studies suggests an early radiation of crown-group rosids into major clades. Similar polytomies are evident within both Fabidae and Malvidae and within clades further nested within Fabidae (i.e., Malpighiales). However, it is unclear whether the apparent radiations within the rosids are real or merely artifacts due to poorly resolved relationships. Improved analysis of phylogeny, coupled with dating of diversification events, could help to distinguish between these alternatives.
To resolve deep relationships within the rosids and evaluate hypotheses of repeated radiations, we constructed a dataset for >100 rosids using sequences of 12 genes (two nuclear and 10 plastid) representing >18,000 bp. We also constructed a dataset using the entire plastid inverted repeat (IR; 24 genes plus intervening spacers; > 25,000 bp), a region with great utility at deep levels in the angiosperms (23); these datasets were also combined (>43,000 bp). Our goals were to (i) elucidate deep relationships among the major lineages of rosids; (ii) determine the placement of large, problematic lineages (e.g., Crossosomatales, Geraniales, Myrtales), and enigmatic families (Aphloiaceae, Huaceae, Ixerbaceae, Picramniaceae, and Strasburgeriaceae) not placed in either Fabidae or Malvidae in previous studies; (iii) date the early diversification of the rosid clade; and (iv) reconsider the fossil record in light of this resolved phylogeny.
Results
Parsimony Analyses.
Maximum parsimony (MP) analysis of the total evidence dataset (12 targeted genes plus IR for 117 taxa) produced 9 shortest trees (Fig. S1) that differ mainly in the relationships recovered among some Malpighiales. There are no instances of strongly supported conflicting relationships (i.e., > 80% BS support) among trees from the individual genes (trees not shown; tree statistics in Table S1). Single-gene analyses generally recovered clades that correspond to angiosperm orders (in the sense of ref. 3), but, with few exceptions, failed to resolve interordinal relationships. MP analyses of the combined nuclear genes (18S and 26S rDNA; trees not shown) also provided limited resolution and support of relationships. In contrast, MP analyses of combined plastid and nuclear datasets (12 targeted genes) yielded trees with much greater resolution (Figs. S2–S5).
Maximum Likelihood Analyses/Comparison with Parsimony.
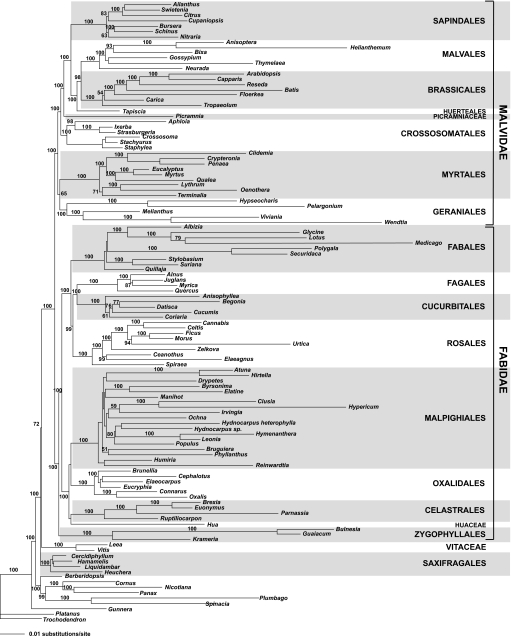
Maximum likelihood (ML) analyses of 10 targeted plastid genes and 10 plastid plus 2 nuclear genes (Figs. S2–S5) yielded topologies nearly identical to the ML total evidence tree for 117 taxa (Fig. 1). In MP analyses of the total evidence dataset (12 targeted genes plus IR), Zygophyllales were placed in Malvidae as sister to Geraniales, whereas with ML, Zygophyllales were sister to Fabidae (Fig. 1). A more minor difference is that in MP analyses, Fabales were sister to Rosales whereas in ML analyses Fabales were sister to a clade of Rosales, Cucurbitales, and Fagales. With both ML and MP, the slowly evolving IR region alone yielded a topology (Fig. S2) identical to that retrieved by ML analysis of the total evidence dataset (Fig. 1). The conflicts between MP and ML analyses are likely due to the inherent difficulties that long branches pose for parsimony (see below); indeed both Fabales and Zygophyllales contain lineages that are on long branches in these trees. Therefore, for the remainder of this article we will focus on the ML topology of the total evidence tree. In this tree (Fig. 1), Vitaceae are sister to all other rosids, which in turn form 2 large clades, Fabidae and an expanded Malvidae, each with a bootstrap value of 100%.
Fig. 1.
ML tree resulting from GARLI analysis of total evidence data set (2 nuclear genes, 10 plastid genes) for 117 members of the rosid clade and outgroups and the IR for 59 taxa. Numbers above branches are bootstrap values.
The approximately unbiased (AU) topology test indicated that the sister relationship of Zygophyllales to Malvidae observed in MP can be rejected (P < 0.001) in favor of the ML placement of Zygophyllales as sister to an expanded Fabidae using both the total evidence dataset and the 12 targeted genes dataset.
Divergence Time Estimates.
Cross-validation determined a smoothing value of 1,000 for each of the penalized likelihood (PL) analyses. In general, when the root of the tree was fixed to an age of 125 Mya, age estimates were an average of 5–10 million years older than when the root was constrained to a maximum age of 125 Mya (Table 1). Likewise, based on the bootstrap estimates of variation, the 2 different methods usually did not fall within one standard deviation of one another. Our estimates for the origin of crown group rosids from the 2 analyses ranged from 110 (± 6) to 93 (± 6) Mya, in the Early to Late Cretaceous, followed by rapid diversification into the Fabidae and expanded Malvidae clades ≈108 (± 6) to 91 (± 6) Mya and 107 (± 6) to 83 (± 7) Mya, respectively (Table 1).
Table 1.
Divergence time estimates
| Calde/group | Age |
||||
|---|---|---|---|---|---|
| PL-1 | PL-2 | BRC-1 | BRC-2 | Wikström et al. | |
| Rosid crown group | 108 (114–102) | 91 (97–85) | 114 (116–111) | 113 (115–110) | 95 (98–92) |
| Vitaceae/rosid split | 111 (115–109) | 92 (96–88) | 117 (119–113) | 116 (118–113) | 108 (112–104) |
| Brassicalescrown | 73 (76–70) | 60 (63–57) | 55 (63–50) | 55 (63–46)) | 71 (75–67) |
| Brassicalesstem | 89 (94–85) | 74 (80–68) | 86 (96–80) | 88 (90–84) | 85 (89–81) |
| Celastralescrown | 81 (87–75)) | 56 (62–50) | 91 (100–80) | 74 (83–68) | 85 (88–82) |
| Celastralesstem | 104 (108–100)) | 91 (95–87) | 109 (112–106) | 108 (110–105) | 89 (92–86) |
| Crossosomatalescrown | 88 (94–82)) | 51 (–45) | 88 (100–70) | 84 (91–79) | 56 (62–50) |
| Crossosomatalesstem | 105 (112–98) | 81 (88–74) | 103 (107–99) | 100 (105–97) | 91 (95–87) |
| Cucurbitalescrown | 78 (83–73) | 80 (85–75) | 74 (90–64) | 68 (78–61) | 66 (68–64) |
| Cucurbitalesstem | 103 (107–100) | 88 (92–84) | 105 (109–100) | 103 (106–101) | 84* |
| Fabalescrown | 87 (90–84) | 72 (75–69) | 93 (100–83) | 83 (92–77) | 74 (77–71) |
| Fabalesstem | 104 (109–99) | 89 (92–86) | 109 (112–104) | 107 (109–105) | 89 (91–87) |
| Fagalescrown | 90 (93–87) | 88 (92–84) | 96 (100–91) | 95 (98–92) | 61 (65–57) |
| Fagalesstem | 103 (107–99) | 88 (92–84) | 105 (109–100) | 103 (106–101) | 84* |
| Geranialescrown | 103 (109–97) | 68 (74–62) | 101 (109–91) | 94 (100–88) | 88 (92–84) |
| Geranialesstem | 107 (114–100) | 83 (90–76) | 109 (113–105) | 106 (110–102) | 99 (103–95) |
| Malpighialescrown | 92 (93–90) | 90 (91–89) | 102 (106–100) | 102 (104–100) | 77 (80–74) |
| Malpighialesstem | 103 (107–99) | 91 (95–87) | 105 (112–102) | 107 (109–105) | 88 (91–85) |
| Malvalescrown | 78 (80–76) | 74 (76–72) | 79 (88–75) | 77 (80–73) | 67 (71–63) |
| Malvalesstem | 89 (93–85) | 74 (78–70) | 86 (96–80) | 81 (84–79) | 80 (84–76) |
| Myrtalescrown | 85 (89–81) | 78 (82–74) | 94 (99–90) | 91 (96–86) | 78 (82–74) |
| Myrtalesstem | 106 (111–101) | 83 (86–80) | 107 (111–102) | 103 (107–99) | 100 (103–97) |
| Oxalidalescrown | 69 (74–64) | 62 (67–57) | 62 (70–55) | 63 (69–59) | 72 (75–69) |
| Oxalidalesstem | 102 (109–95) | 91 (98–84) | 105 (112–94) | 104 (108–98) | 88 (91–85) |
| Rosalescrown | 93 (96–90) | 88 (91–85) | 96 (103–88) | 97 (101–93) | 76 (79–73) |
| Rosalesstem | 105 (110–100) | 89 (94–84) | 107 (111–102) | 106 (108–104) | 88 (90–86) |
| Sapindalescrown | 63 (66–60) | 71 (73–69) | 70 (75–61) | 71 (76–66) | 57 (61–53) |
| Sapindalesstem | 96 (102–90) | 76 (80–72) | 96 (102–90) | 94 (97–90) | 80 (84–76) |
| Zygophyllalescrown | 79 (88–70) | 55 (64–46) | 89 (102–79) | 70 (85–55) | 64 (68–60) |
| Zygophyllalesstem | 108 (114–102) | 91 (97–85) | 112 (115–110) | 111 (113–109) | 95 (98–92) |
PL-1 represents an analysis where the root of the tree was fixed to an age of 125 mya. PL-2 represents an analysis where the root was constrained to be a maximum of 125 mya. Ranges on PL divergence time represent ± 1 standard deviation of the mean based on 100 bootstrap replicates. Each Bayesian relaxed clock (BRC) analysis partitioned data by genome, giving a different model of evolution and rate change to each genome (see Materials and Methods for more detail). The BRC-1 analysis treated priors on fossils as being drawn from a uniform distribution between the minimum age of the fossil and 125 million years. The BRC-2 analysis treated priors on fossils as being drawn from a lognormal distribution (see Table S4). The Wikström et al. (34) ages based on maximum likelihood calculations and standard error estimates. All ages are millions of years.
*Calibration point.
The divergence times estimated under the uncorrelated lognormal (UCLN) relaxed-clock model with the data partitioned by genome, treating fossils as drawn from a uniform distribution (UCLN-uniform), estimated a coefficient of variation of 0.815 [95% HPD (highest posterior density): 0.777, 0.863] and covariance of 0.136 (95% HPD: 5.62 × 10−2, 0.237) for the plastid partition and a coefficient of variation of 0.634 (95% HPD: 0.543, 0.736) and covariance of 0.067 (95% HPD: 1.62 × 10−2, 0.147) for the nuclear partition. Parameter values for the UCLN relaxed-clock model with fossils treated as being drawn from a lognormal distribution (UCLN-lognormal) were almost identical. Accordingly, these data appear suited to divergence-time estimation methods that assume a positive autocorrelation of substitution rate variation (e.g., refs. 24–28). However, for the Bayesian relaxed-clock models, the partitioned model with fossils drawn from a uniform distribution was determined to be the best-fitting model based on Bayes factors (mean for the log of posterior equaled −3.76 × 105 for the “uniform” model and −3.78 × 105 for the “lognormal” model). Our estimates for the origin of crown group rosids from the 2 fossil treatments ranged from 115 (119–112) to 113 (117–111) Mya, once again in the Early Cretaceous, followed by rapid diversification into the Fabidae and Malvidae crown groups ≈112 (115–110) to 110 (113–109) Mya and 109 (113–105) to 106 (111–102) Mya, respectively. In general, the variance on nodes tended to be smaller for the analysis that treated fossils as being drawn from a lognormal distribution.
Discussion
Phylogenetic Relationships.
MP and ML analyses of the total evidence dataset yielded similar topologies. However, the primary difference between the MP and ML trees is significant, with Zygophyllales appearing as either part of an expanded Malvidae with MP or sister to the expanded Fabidae with ML. MP bootstrap values for Fabidae and Malvidae (77% and 75%, respectively) are lower than ML bootstrap values (both 100%). The AU test results provide statistical support for the ML total evidence topology by rejecting the MP position of Zygophyllales as sister to Malvidae. IR data alone (below) also support the ML topology. In addition, previous studies (MP and Bayesian) of a 3-gene dataset with more rosid taxa also placed Zygophyllales in Fabidae, but in an unresolved position (2, 10). Zygophyllales are characterized by a long branch, which could affect the placement of the clade (particularly with parsimony); the branches within Zygophyllales (to Krameria, to Guaiacum, and to Bulnesia) are particularly long (Figs. S2–S5). Zygophyllales are highly enigmatic and share few non-DNA traits with any other rosid lineage (2).
With both ML and MP (Fig. S2), the IR dataset alone yielded a topology identical to that retrieved by ML analysis of the total evidence dataset (Fig. 1). These results parallel those reported for Saxifragales (23), illustrating the value of the slowly evolving IR in deep-level phylogeny reconstruction. Despite far fewer parsimony-informative sites than “fast” genes, the slowly evolving IR genes provide higher resolution and support for relationships, providing a topology identical to the total evidence ML tree.
We will base our discussion on the ML total evidence tree for 117 taxa (Fig. 1) for several reasons. Nodes in the ML tree receive high bootstrap support, whereas with MP, many crucial relationships (including placement of Zygophyllales) have low bootstrap values. Parsimony has also had difficulty resolving other deep-level phylogenetic problems in angiosperms that appear to be rapid radiations and that are similarly characterized by a combination of short and long branches (e.g., refs. 6 and 23). Furthermore, ML and Bayesian approaches are better than MP under heterogeneous evolution involving lineage- and gene-specific rate variation (29–31).
In previous studies, Fabidae were not well supported, and relationships were unresolved within the clade (1, 2, 10). Our analysis not only provides strong support for Fabidae (100% bootstrap with ML), but also indicates that Zygophyllales are sister to 2 major subclades within Fabidae (each with BS = 100%): the nitrogen-fixing clade (Cucurbitales, Fagales, Fabales, Rosales) and Celastrales, Oxalidales, and Malpighiales [COM group (22)], plus Huaceae.
Previous studies placed Tapisciaceae, Brassicales, Malvales, and Sapindales in Malvidae, but with little support for relationships among these clades (reviewed in refs. 2 and 10). Furthermore, Geraniales, Myrtales, and Crossosomatales were not placed in either Fabidae or Malvidae in previous analyses. We provide the first strong evidence that these 3 clades, and Picramniaceae [placed by some (12) within Simaroubaceae, Sapindales], are part of an expanded Malvidae (100% bootstrap with ML). We propose that Malvidae be formally expanded to include Geraniales, Myrtales, Crossosomatales, and Picramniaceae. Furthermore, Crossosomatales should be expanded to include Ixerbaceae and Strasburgeriaceae (2).
This hypothesis of rosid relationships is greatly improved compared with trees based on approximately ≈4,700 bp (8) or ≈8,400 bp (32). This study and other recent analyses (5, 6, 23) suggest that with enough data, many, if not most, remaining deep-level “radiations” in the flowering plants can be resolved. For example, a putative rapid radiation in Saxifragales was resolved, despite long-branch attraction problems, with >25,000 base pairs (23). Highly problematic deep-level relationships involving basal angiosperm lineages have similarly been resolved through analysis of ≈42,000 bp (6).
Divergence Times.
Our estimates for the origin of crown group rosids ranged from 115 to 93 Mya (late Aptian to early Turonian), in the Early to Late Cretaceous, followed by rapid diversification into the Fabidae and Malvidae crown groups ≈112 to 91 Mya (Albian to Coniacian), and 109 to 83 Mya (Cenomanian to Santonian), respectively (Table 1). With the exception of the PL analysis that constrained the root to be a maximum age of 125 Mya, age estimates were quite consistent. For the most part, associated errors in age estimates overlapped across all nodes in the tree so that we cannot precisely resolve the actual timing and rates of these successive divergences.
The oldest confirmed fossils of Vitaceae are Paleocene, ≈60 Mya (33), which is anomalously young in view of the mid- to late-Cretaceous ages known for members of Fabidae and Malvidae. We predict, based on divergence time estimates presented above, that Vitaceae should have a Cretaceous fossil history. However, the distinctive seeds of Vitaceae have not been detected in the pre-Tertiary fossil record.
Our estimate for the time of origin of the crown group rosid clade is similar to those of both Wikström et al. (34) (117–108 Mya, their node 15) and Davies et al. (35) (≈115–110 Mya). Inspection of the ages of rosid lineages obtained here revealed no general pattern to these node ages being older or younger, when compared with Wikström et al. (34). Regardless of analyses, the rosid clade appears to have diversified rapidly into several major lineages, over a period of <15 million years, and perhaps as quickly as 4 to 5 million years. To place this in perspective, this narrow window of initial diversification of 4–5 million years represents a timeframe comparable to the rapid radiation of the Hawaiian silversword alliance (Asteraceae), which arose from a North American ancestor 5 Mya (36). Saxifragales (23), Malpighiales (34, 37), and Mesangiospermae (6) also each appear to have arisen and diversified over a very short period.
Rise of Angiosperm-Dominated Forests.
We hypothesize that the bursts in diversification in the rosids correspond to the rapid rise of angiosperm-dominated forests (38, 39). Fossil leaf assemblages of late Albian to middle Cenomanian age (≈104–97 Mya) have been interpreted as “an explosive increase in the structural diversity of flowering plants” (ref. 39, p. 259), not only magnoliids and platanoids, but also new types of rosids with “pinnately compound leaves and simple leaves showing evidence for derivation from a compound-leaved ancestor” (ref. 39, p. 259). This interval partially coincides with our inferred dates of the divergence for the rosid crown group and of the Fabidae and Malvidae. The results of these diversifications are already well-marked in the Late Cretaceous, by which time taxa related to Saxifragales, Rosales, Malpighiales, and Fagales are common, as confirmed by well-preserved charcoalified floral remains (37, 40, 41).
Our estimates of the timing of the rapid diversification of these rosid lineages are comparable to published values (34, 35), and those provided in more focused analyses within rosid clades. For example, Davis et al. (37) concluded that Malpighiales are very old and began to diversify during the mid-Cretaceous (≈112–94 Mya), perhaps as small to mid-sized trees in tropical rain forest understories. A similar window of diversification is seen in woody species in clades other than the rosids, such as the woody Saxifragales [e.g., Altingiaceae (23)]. Within asterids, Cornales also appear to have diversified during this same window of time (e.g., 35), as did Ericales (34, 35).
The diversification of rosids also corresponds to that of several major insect groups. For example, the diversification of major ant lineages is attributed to the “rise in angiosperm-dominated forests” (42) and corresponds to the time period estimated here for the rosid radiation. This period also corresponds to the radiation of other major herbivores, such as beetles and hemipterans (43, 44). Similarly, the “principal splits” underlying the diversification of the extant lineages of placental mammals occurred in a similar timeframe, from 100 to 85 Mya (45). Diversification in amphibians occurred slightly later (80–85 Mya) and is similarly attributed to the rise of angiosperm forests (46), especially given that 82% of amphibian species live in these forests. The majority of extant ferns similarly resulted from a Cretaceous diversification (initiated ≈100 Mya) coupled with the rise of angiosperm forests; divergence time estimates suggest that ferns diversified “in the shadow of the angiosperms” (47). The rise of all of these lineages appears to have closely tracked the rise of angiosperm-dominated forests, and most of these key forest lineages occur within the rosid clade. Hence, the radiations we have detected in rosids largely represent the rapid rise of angiosperm-dominated forests and associated codiversification events.
Materials and Methods
Taxon Sampling.
A total of 117 species (including 104 species of rosids) was sampled for 12 genes (see DNA Amplification and Sequencing); a subset of 59 taxa was sampled for the plastid IR. We sampled broadly across the rosids, including several exemplars of most orders (SI Appendix). For larger orders, multiple families spanning the phylogenetic diversity of the clade were included. We also broadly sampled orders and families whose placements remain uncertain, including Picramniaceae, Huaceae, Aphloiaceae, Ixerbaceae, and Strasburgeriaceae (3).
For most taxa, the same DNA was used throughout the study. For a few genera, sequences for 1 or 2 of the genes sequenced in earlier studies represent a different congeneric species from that used here. We selected non-rosids representing the other major lineages of eudicots as outgroups (Fig. 1). Species names and voucher/collection information are given in SI Appendix.
DNA Amplification and Sequencing.
We targeted 10 plastid [rbcL, atpB, matK, the psbBTNH region (= 4 genes), rpoC2, ndhF, and rps4] and 2 nuclear genes (18S and 26S rDNA) for all taxa. Primers for amplification and sequencing are provided in Table S2. PCR protocols follow ref. 23. We sequenced the entire IR (see ref. 23) for 59 of the rosids used in our 12-gene analyses (due to expense, we did not sequence the IR for all taxa). We also extracted IR sequences for all publicly available complete plastid genome sequences for rosids, and for several outgroups (Fig. 1 and SI Appendix). Sequences were generated on an ABI 3730 XL DNA sequencer.
Alignment.
Preliminary alignments of the 12 targeted genes were obtained independently for each locus using Clustal X (48) and adjusted manually. To assist in the alignment of protein-coding regions, sequences were also aligned by amino acid. Alignment of coding sequences for all genes except ndhF and rpoC2 was straightforward; the latter 2 genes were more problematic because they possess many indels and some regions of high sequence variability. Regions that were difficult to align, and short incomplete regions at the beginnings and ends of genes, were excluded from the analyses. The entire IR was aligned as in ref. 23. The aligned, analyzed lengths of the genes included are given in Table S3.
Phylogenetic Analysis.
We analyzed 4 datasets: (i) 12 targeted genes for 117 taxa; (ii) the IR for 59 taxa; (iii) 12 targeted genes plus IR for 117 taxa (with those taxa not sequenced for the IR scored as having missing data); and (iv) 12 targeted genes plus IR for those taxa sequenced for both (59 taxa). We also analyzed each of the 12 targeted genes individually.
Gaps were treated as missing data. MP and ML analyses were used to infer the phylogeny. With the exception of the nuclear data partition, all MP analyses were conducted using heuristic searches with 1,000 replicates of random taxon addition with tree-bisection-reconnection (TBR) branch swapping, saving all shortest trees per replicate. For the nuclear partition, MP heuristic searches involved 3,000 random addition replicates, saving no more than 1,000 trees per replicate. Bootstrap support was estimated following the methods of ref. 6 except for the nuclear partition, in which only 10,000 shortest trees per replicate were saved for the bootstrap analysis.
For ML analyses we used GARLI v. 0.942 (49). MrModelTest 2.2 (people.scs.fsu.edu/∼nylander/mrmodeltest2/mrmodeltest2.html) and the Akaike information criterion was used to determine the appropriate evolutionary model (GTR + I + Γ). GARLI runs, including ML bootstrap analyses, were conducted following ref. 6.
To test the statistical significance of the differing positions of Zygophyllales in MP and ML analyses, we performed AU tests of topology (50) using the total evidence (all taxa) and the 12 targeted genes datasets. For both datasets we tested the total evidence ML topology against the topology with the highest ML score constrained to have Zygophyllales form a monophyletic group with Malvidae. To find the latter tree topology, we performed ML searches of both datasets using GARLI v. 0.951 (49) with the topological constraint of Zygophyllales + Malvidae enforced and a GTR + I + Γ model. The AU test was performed in CONSEL v. 0.1i (51).
Divergence Time Estimates.
Given the lack of rate constancy among lineages (based on a likelihood ratio test: P < 0.001), for all datasets divergence times were estimated under a relaxed molecular clock. We used both penalized likelihood (PL, in r8s v.1.70: 27, 53) and a Bayesian method (BEAST v.1.4.7) (53–55). For PL analyses the smoothing parameter (λ) was determined by cross-validation. The ML topology was used. The basal polytomy involving the outgroups (Fig. 1) was resolved by rooting the tree with Platanaceae. The tree was then imported into PAUP* (56), where branch lengths were reestimated under a GTR + I + Γ model for use in PL analyses. To quantify errors in divergence time estimates, we used a 3-step non-parametric bootstrap (36).
The uncorrelated lognormal (UCLN) model implemented in BEAST (53) was used to infer divergence times. The underlying model of molecular evolution was specified to be GTR + I + Γ. The estimation of absolute divergence times requires calibrating (or constraining) the age of 1 or more nodes. For each analysis, we initiated 4 independent MCMC analyses from starting trees with branch lengths that satisfied the respective priors on divergence times. Convergence of each chain to the target distribution was assessed using Tracer v.1.4 (57) and by plotting time series of the log posterior probability of sampled parameter values. After convergence was achieved, each chain was sampled every 1,000 steps until 5,000 samples were obtained. Model fit of the different UCLN relaxed clock models was assessed using Bayes factors (58) as implemented in Tracer v.1.4.
Rosids are abundant in the fossil record (11), but many of the Tertiary fossils are too young to be relevant in dating deep divergences. To provide insight into the timing of divergences for the topologies presented, we selected fossils representing the geologically oldest examples that could confidently be assigned to particular subclades. Those that were used to provide minimum age constraints are given in Table S4. We did not include the Rose Creek flower as a constraint, but its estimated age of ≈98 million years is consistent with the results presented. For the assignment of numerical ages to stages of the Cretaceous, we followed ref. 59. We treated the age of the root node in several ways (see ref. 6 and Table S4).
Supplementary Material
Acknowledgments.
We thank D. Swofford for access to computational resources (supported by National Science Foundation Grant EF-0331495); M. Gitzendanner for help with some of the phylogenetic analyses; and B. Moore, S. Magallón, and an anonymous reviewer for helpful comments on the manuscript. This work was supported by Assembling the Tree of Life (AToL) Grants EF-0431266 and EF-0431242 (National Science Foundation) and Deep Time Research Coordination Networks Grant NSF DEB-0090283.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU002149 to EU002556).
This article contains supporting information online at www.pnas.org/cgi/content/full/0813376106/DCSupplemental.
References
- 1.Judd WS, Olmstead RG. a survey of tricolpate (eudicot) phylogeny. Amer J Bot. 2004;91:1627–1644. doi: 10.3732/ajb.91.10.1627. [DOI] [PubMed] [Google Scholar]
- 2.Soltis DE, Soltis PS, Endress PK, Chase MW. Phylogeny and Evolution of Angiosperms. Sunderland, MA: Sinauer; 2005. [Google Scholar]
- 3.APG II. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. Bot J Linn Soc. 2003;141:399–436. [Google Scholar]
- 4.Saarela JM, et al. Hydatellaceae identified as a new branch near the base of the angiosperm phylogenetic tree. Nature. 2007;446:312–315. doi: 10.1038/nature05612. [DOI] [PubMed] [Google Scholar]
- 5.Jansen RK, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome scale-data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci USA. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham SW, et al. Robust inference of monocot deep phylogeny using an expanded multigene plastid data set. Aliso. 2006;22:3–20. [Google Scholar]
- 8.Bremer B, et al. Phylogenetics of asterids based on 3 coding and 3 non-coding chloroplast DNA markers and the utility of non-coding DNA at higher taxonomic levels. Mol Phylogenet Evol. 2002;24:274–301. doi: 10.1016/s1055-7903(02)00240-3. [DOI] [PubMed] [Google Scholar]
- 9.Schönenberger J, von Balthazar M. Reproductive structures and phylogenetic framework of the rosids - progress and prospects. Plant Syst Evol. 2006;260:87–106. [Google Scholar]
- 10.Soltis DE, Gitzendanner MA, Soltis PS. A 567-taxon data set for angiosperms: The challenges posed by Bayesian analyses of large data sets. Int J Plant Sci. 2007;168:137–157. [Google Scholar]
- 11.Magallón-Puebla S, Crane PR, Herendeen P. Phylogenetic pattern, diversity, and diversification of eudicots. Ann Mo Bot Gard. 1999;86:297–372. [Google Scholar]
- 12.Cronquist A. An Integrated System of Classification of Flowering Plants. New York: Columbia Univ Press; 1981. [Google Scholar]
- 13.Crepet WL, Nixon KC, Gandolfo MA. Fossil evidence and phylogeny: The age of major angiosperm clades based on mesofossil and macrofossil evidence from Cretaceous deposits. Amer J Bot. 2004;91:1683–1699. doi: 10.3732/ajb.91.10.1666. [DOI] [PubMed] [Google Scholar]
- 14.Basinger JF, Dilcher DL. Ancient bisexual flowers. Science. 1984;224:511–513. doi: 10.1126/science.224.4648.511. [DOI] [PubMed] [Google Scholar]
- 15.Elias T. The Complete Trees of North America: Field Guide and Natural History. NY: van Nostrand Reinhold; 1980. [Google Scholar]
- 16.Croat TB. Flora of Barro Colorado Island. Palo Alto, CA: Stanford Univ Press; 1978. [Google Scholar]
- 17.Lopez AJ, Little EL, Ritz GF, Rombold JS, Hahn WJ. Arboles communes del Paraguay: Ñande yvyra mata kuera. Lopez, Paraguay: Cuerpo de Paz; 1987. [Google Scholar]
- 18.Little EL, Jr., Wadsworth FH. Common trees of Puerto Rico and the Virgin Islands. Washington, DC: U.S. Department of Agriculture; 1964. [Google Scholar]
- 19.Lorenzi H. Brazilian Trees. Avenida, Brazil: RR Donnelley; 2000. [Google Scholar]
- 20.Gentry AH. Changes in plant community diversity and floristic composition on environmental and geographical gradients. Ann Mo Bot Gard. 1988;75:1–34. [Google Scholar]
- 21.Cantino PD, et al. Towards a Phylogenetic Nomenclature of Tracheophyta. Taxon. 2007;56:822–846. [Google Scholar]
- 22.Endress PK, Matthews ML. Floral structure and systematics in four orders of rosids, including a broad survey of floral mucilage cells. Plant Syst Evol. 2006;260:223–251. [Google Scholar]
- 23.Jian S, et al. Resolving an ancient, rapid radiation in Saxifragales. Syst Biol. 2007;57:1–20. doi: 10.1080/10635150801888871. [DOI] [PubMed] [Google Scholar]
- 24.Thorne JL, Kishino H, Painter IS. Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- 25.Huelsenbeck JP, Larget B, Swofford DL. A compound Poisson process for relaxing the molecular clock. Genetics. 2000;154:1879–1892. doi: 10.1093/genetics/154.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kishino H, Thorne JL, Bruno WJ. Performance of a divergence time estimation method under a probabilistic model of rate evolution. Mol Biol Evol. 2001;18:352–361. doi: 10.1093/oxfordjournals.molbev.a003811. [DOI] [PubMed] [Google Scholar]
- 27.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 28.Aris-Brosou S, Yang Z. Bayesian models of episodic evolution support a late Precambrian explosive diversification of he Metazoa. Mol Biol Evol. 2003;20:1947–1954. doi: 10.1093/molbev/msg226. [DOI] [PubMed] [Google Scholar]
- 29.Gadagkar SR, Kumar S. Maximum-likelihood outperforms maximum parsimony even when evolutionary rates are heterotachous. Mol Biol Evol. 2005;22:2139–2141. doi: 10.1093/molbev/msi212. [DOI] [PubMed] [Google Scholar]
- 30.Whitfield JB, Lockhart PJ. Deciphering ancient rapid radiations. Trends Ecol Evol. 2007;22:258–265. doi: 10.1016/j.tree.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 31.Whitfield JB, Kjer KM. Ancient rapid radiations of insects: Challenges for phylogenetic analysis. Annu Rev Entomol. 2008;53:449–472. doi: 10.1146/annurev.ento.53.103106.093304. [DOI] [PubMed] [Google Scholar]
- 32.Soltis DE, et al. Gunnerales are sister to other core eudicots: Implications for the evolution of pentamery. Amer J Bot. 2003;90:461–470. doi: 10.3732/ajb.90.3.461. [DOI] [PubMed] [Google Scholar]
- 33.Chen I, Manchester SR. Seed morphology of modern and fossil Ampelocissus (Vitaceae) and implications for phytogeography. Amer J Bot. 2007;94:1534–1553. doi: 10.3732/ajb.94.9.1534. [DOI] [PubMed] [Google Scholar]
- 34.Wikström N, Savolainen V, Chase MW. Evolution of the angiosperms: Calibrating the family tree. Proc R Soc London Ser B. 2001;268:2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies TJ, et al. Darwin's abominable mystery: Insights from a supertree of the angiosperms. Proc Natl Acad Sci USA. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baldwin BG, Sanderson MJ. Age and rate of diversification of the Hawaiian silversword alliance (Compositae) Proc Natl Acad Sci. 1998;95:9402–9406. doi: 10.1073/pnas.95.16.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis CC, Webb CO, Wurdack KJ, Jaramillo CA, Donoghue MJ. Explosive radiation of Malpighiales supports a Mid-Cretaceous origin of modern tropical rain forests. Am Nat. 2005;165:E36–E65. doi: 10.1086/428296. [DOI] [PubMed] [Google Scholar]
- 38.Crane PR. In: The origins of angiosperms and their biological consequences. Friis EM, Chaloner WG, Crane PR, editors. Cambridge, UK: Cambridge Univ Press; 1987. pp. 107–144. [Google Scholar]
- 39.Upchurch GR, Wolfe JA. Cretaceous vegetation of the Western Interior and adjacent regions of North America. In: Caldwell WGE, Kauffman EG, editors. Evolution of the Western Interior Basin. Vol. 39. Geological Association of Canada; 1993. pp. 243–281. Special Paper. [Google Scholar]
- 40.Crane PR, Herendeen P, Friis EM. Fossils and plant phylogeny. Amer J Bot. 2004;91:1683–1699. doi: 10.3732/ajb.91.10.1683. [DOI] [PubMed] [Google Scholar]
- 41.Endress PK, Friis EM. Rosids—reproductive structures, fossil and extant, and their bearing on deep relationships. Plant Syst Evol. 2006;260:83–85. [Google Scholar]
- 42.Moreau CS, Bell CD, Vila R, Archibald SB, Pierce N. Phylogeny of the ants: Diversification in the age of angiosperms. Science. 2006;312:101–104. doi: 10.1126/science.1124891. [DOI] [PubMed] [Google Scholar]
- 43.Farrell BD. “Inordinate fondness” explained: Why are there so many beetles? Science. 1998;281:555. doi: 10.1126/science.281.5376.555. [DOI] [PubMed] [Google Scholar]
- 44.Wilf P, et al. Timing the radiations of leaf beetles: Hispines on gingers from latest Cretaceous to Recent. Science. 2000;289:291–294. doi: 10.1126/science.289.5477.291. [DOI] [PubMed] [Google Scholar]
- 45.Bininda-Emonds ORP, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 46.Roelants K, et al. Global patterns of diversification in the history of modern amphibians. Proc Natl Acad Sci USA. 2007;104:887–892. doi: 10.1073/pnas.0608378104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider H, et al. Ferns diversified in the shadow of angiosperms. Nature. 2004;428:553–557. doi: 10.1038/nature02361. [DOI] [PubMed] [Google Scholar]
- 48.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTALX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zwickl DJ. Austin, TX: University of Texas; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence data sets under the maximum likelihood criterion. Ph.D dissertation. [Google Scholar]
- 50.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 51.Shimodaira H, Hasegawa M. CONSEL: For assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- 52.Sanderson MJ. r8s: Inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 53.Drummond AJ, Rambaut A. BEAST v1.4.1. 2006 Available from http://beast.bio.ed.ac.uk/
- 54.Ho SYW, Phillips MJ, Drummond AJ, Cooper A. Accuracy of rate estimation using relaxed-clock models with a critical focus on the early metazoan radiation. Mol Biol Evol. 2005;22:1355–1363. doi: 10.1093/molbev/msi125. [DOI] [PubMed] [Google Scholar]
- 55.Moore BR, Donoghue MJ. Correlates of diversification in the plant clade Dipsacales: Geographic movement and evolutionary innovations. Amer Nat. 2007;170:S28–S55. doi: 10.1086/519460. [DOI] [PubMed] [Google Scholar]
- 56.Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2000. Ver 4. [Google Scholar]
- 57.Drummond AJ, Rambaut A. TRACER Version 1.3. 2003 Available at http://evolve.zoo.ox.ac.uk.
- 58.Nylander JAA. Uppsala, Sweden: Uppsala University; 2004. MrModeltest Version 2. Program distributed by the author. Available at http://people.scs.fsu.edu/nylander/mrmodeltest2/mrmodeltest2.html. [Google Scholar]
- 59.Gradstein FM, et al. A Geologic Time Scale 2004. Cambridge, UK: Cambridge Univ Press; 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.