Abstract
Cells have evolved biomolecular networks that process and respond to changing chemical environments. Understanding how complex protein interactions give rise to emergent network properties requires time-resolved analysis of cellular response under a large number of genetic perturbations and chemical environments. To date, the lack of technologies for scalable cell analysis under well-controlled and time-varying conditions has made such global studies either impossible or impractical. To address this need, we have developed a high-throughput microfluidic imaging platform for single-cell studies of network response under hundreds of combined genetic perturbations and time-varying stimulant sequences. Our platform combines programmable on-chip mixing and perfusion with high-throughput image acquisition and processing to perform 256 simultaneous time-lapse live-cell imaging experiments. Nonadherent cells are captured in an array of 2,048 microfluidic cell traps to allow for the imaging of eight different genotypes over 12 h and in response to 32 unique sequences of stimulation, generating a total of 49,000 images per run. Using 12 devices, we carried out >3,000 live-cell imaging experiments to investigate the mating pheromone response in Saccharomyces cerevisiae under combined genetic perturbations and changing environmental conditions. Comprehensive analysis of 11 deletion mutants reveals both distinct thresholds for morphological switching and new dynamic phenotypes that are not observed in static conditions. For example, kss1Δ, fus3Δ, msg5Δ, and ptp2Δ mutants exhibit distinctive stimulus-frequency-dependent signaling phenotypes, implicating their role in filtering and network memory. The combination of parallel microfluidic control with high-throughput imaging provides a powerful tool for systems-level studies of single-cell decision making.
Keywords: yeast, systems biology, microscopy, live-cell imaging
Cellular processes are governed by complex protein signaling networks that function as robust and dynamic control systems, ensuring appropriate responses to sustained and transient stimuli. These networks feature emergent properties, including bistability, adaptation, and memory that make their behavior inherently dependent on previous stimulation and current cell states. As examples, system bistability provides a selective advantage by allowing populations of cells to test the responses of alternative states to a given condition (1, 2); network adaptation to a sustained change in stimulant concentration limits the metabolic cost of a sustained response (3, 4); and network memory allows more rapid accommodation of recurrent stimulations (5, 6). Because of experimental tractability, these emergent properties were first studied in model systems and have recently been uncovered in key mammalian regulatory networks, including those dysregulated in disease (7).
Because of the facility of genetic manipulations and availability of reporters, yeast has emerged as the prototypical model of cell signaling. In particular, the pheromone response pathway in Saccharomyces cerevisiae is arguably the best-characterized mitogen-activated protein kinase (MAPK) signaling network and has been a particularly fruitful model of eukaryotic signaling. MAPK signaling is of central importance to a wide range of cellular decision-making processes, responding to a staggering range of stimuli, including growth factors, cytokines, hormones, cellular adhesion, stress, and nutrient conditions (8). Regulated signaling governs cellular growth and differentiation whereas deviations from normal MAPK regulation are implicated in the onset of disease, including cancer (9).
The yeast pheromone response is initiated by the binding of a mating peptide, either α-factor or a-factor, to a membrane-localized G protein-coupled receptor, either Ste2 or Ste3 on MATa or MATα cells, respectively. Pheromone signaling is communicated through a MAPK signaling cascade that ultimately results in the phosphorylation of key substrates, including the cyclin-dependent kinase inhibitor Far1, which initiates growth arrest, and the transcription factor Ste12, which activates a program of gene expression involving >200 genes (8, 10–13). Despite the wealth of detailed information gleaned from 30 years of biochemical and genetic studies, the vast majority of these data are qualitative and derived from measurements of cellular response under conditions of constant α-factor levels. More recently, genome-wide analysis of transcription, protein expression, and protein interactions has been applied to systems-level studies of the pheromone response, delineating the tapestry of protein–protein interactions that mediate signaling (14, 15). However, these studies are limited by poor temporal and spatial resolution, making it difficult to probe the dynamics of network function. Perhaps most importantly, these methods require the study of large populations of cells and are completely blind to cell differences that arise from a combination of desynchronization, bistability, and stochastic variation in gene expression.
The combination of fluorescent reporters and quantitative microscopy has recently been used to address cell–cell variability in the yeast mating response under constant environmental conditions (2, 16). Such techniques are scalable to high-throughput formats in multiwell plates but provide only crude control over the microenvironment and are poorly suited to the study of response dynamics or history effects. Indeed, little is known regarding cellular regulation in dynamically changing environments. This dearth of understanding is largely because of the technological challenges involved in precisely controlling time-varying conditions and limitations in throughput. Achieving a quantitative understanding of protein network function requires new tools for high-throughput studies under a large number of genetic perturbations and changing chemical environments and with single-cell resolution (17).
In particular, microfluidics offers the combined advantages of precision fluid control necessary for exchange of media conditions around cells and scalability for parallel analysis of multiple conditions on a single device. In yeast, the precise microfluidic control of conditions has been applied to investigations of modest number of genotypes or chemical conditions under both constant (2, 18) or changing (19–21) media conditions. More scalable devices have been applied to the culture and analysis of mammalian cells on-chip (22, 23) although these have to date be focused primarily on adherent cell types (22, 24, 25). In particular, microfluidic large-scale integration (26, 27) of devices having hundreds to thousands of valves has proven a powerful technique for simultaneously realizing the advantages of temporal control over media conditions and scalability of culture. Here, we further extend the throughput and functionality of this approach in the development of a microfluidic high-throughput single-cell analysis platform optimized for live-cell imaging studies of yeast. This system features a throughput of 256 simultaneous perfusion experiments with nonadherent yeast, integrated on-chip mixing, and control software for programmable control of media conditions and image-processing algorithms and computational infrastructure for large-scale data analysis.
We use our platform to investigate the role of signaling genes in network memory and the filtering of transient stimulation. Recent studies have demonstrated the capacity for memory in cellular circuits, including the pheromone pathway (28), but were limited to long-time scales (5) or in very few conditions (6, 28). Using our microfluidic platform we examined cellular memory by performing >3,000 experiments investigating the combined effect of gene deletions and changing stimulant conditions on the mating response. These studies show that the mating system depends strongly on the frequency of stimulation and identifies genes that play a dominant role in regulating memory.
Results
High-Throughput Microfluidic Live-Cell Imaging Platform.
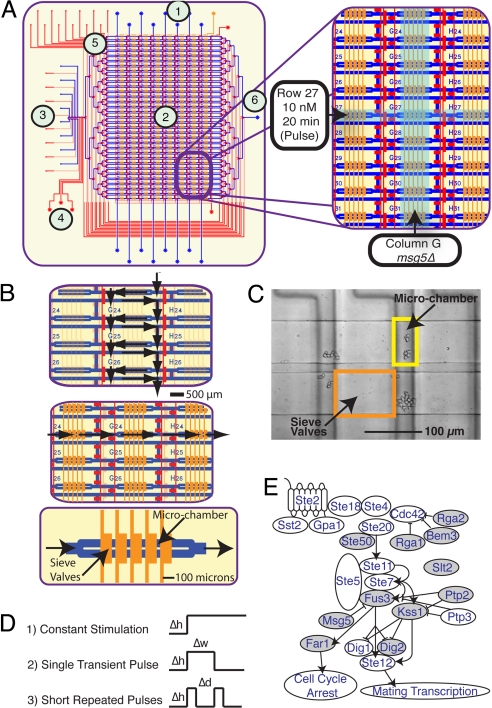
To test the combined effect of genetic perturbations and chemical sequences on mating response we developed a microfluidic live-cell imaging matrix in which eight yeast strains are tested against a total of 32 stimulant-concentration sequences for a total of 256 simultaneous experiments (Fig. 1A). Unique genetic and chemical conditions are created along the matrix columns and rows, respectively. During cell loading, all columns are isolated by actuation of row-valves (top to bottom) (Fig. 1B Top), confining each yeast strain to a unique column without cross-contamination. At each vertex of the matrix, the loaded cells were trapped in a perfusion chamber formed by a series of cell traps designed to immobilize yeast while allowing for the rapid and complete exchange of surrounding media (Fig. 1 B Bottom and C). Each cell trap consists of a partially closed valve that, when actuated, creates a cell filter to allow media exchange while retaining the cells [supporting information (SI) Fig. S1]. Cells are loaded through each column unimpeded and then trapped stochastically upon hydraulic actuation of the trap valves at ≈120 kPa of pressure. At each of the 256 perfusion chambers, five sieve valves are actuated over a doubled flow line to create eight cell traps (2,048 per device). The size of these chambers is easily adjusted and in the current experiments was designed to accommodate ≈600 cells before reaching confluence.
Fig. 1.
Schematic of the microfluidic device. (A) Layout of microfluidic live-cell imaging matrix. Device features two layers of channels including a flow structure (blue), in which cells and reagents are introduced, and a control structure (red) for pneumatic valves. Regions of the device are indicated including (1) cell-loading ports, (2) experiment matrix, (3) chemical inputs and control, (4) peristaltic pump, (5) fluidic multiplexer, and (6) waste outlet. Each column of imaging matrix corresponds to a single yeast genotype. Each row corresponds to a single experimental condition. (B) Control architecture for cell loading and perfusion. Dark red and orange or transparent red and orange lines indicate actuated and nonactuated control lines, respectively. (Top) Valve actuation isolated columns to direct flow through matrix during cell loading. (Middle and Bottom) During perfusion (Middle) rows are isolated, and flow is directed horizontally across the matrix perfusion chambers (Bottom) formed by arrays of pneumatically actuated cell traps (dark orange). (C) Differential interference contrast image of cell yeast cells in perfusion chambers. (D) Stimulation conditions. (1) Step function. The cells are stimulated with constant α-factor solutions, with different pulse heights (Δh) (α-factor concentrations). (2) Pulse function. The cells are stimulated with a transient α-factor solution, with different pulse heights (Δh) and pulse widths (Δw) (duration of stimulus) analyzed. (3) Short repeated pulses. The cells are stimulated with short repeated pulses of α-factor with different pulse heights (Δh) and different delays between pulses (Δd). (E) Pheromone pathway. Gray nodes indicate genes deleted in this work. Only selected interactions are displayed for clarity. Arrowheads represent activating interactions; blunted lines represent repressive interactions.
Perfusion of immobilized yeast allows for studies under well-defined and time-varying chemical conditions. Our device features fluidic elements for the periodic programmable mixing and delivery of chemical formulations to each row of the matrix to generate arbitrary chemical sequences of nutrient and stimulant concentrations in time. On-chip generation of programmable chemical conditions is accomplished by a peristaltic pump that precisely meters varying proportions of eight stock reagents to enable accurate and continuous control of stimulant concentration. Sequences of varying numbers of 120-pL aliquots of the input reagents are mixed in line by Taylor dispersion as they are transported from the mixing element to the array (Fig. 2E). A single mixing element controls all rows of the matrix by using a time-division multiplexing strategy in which each row is sequentially addressed by using a fluidic multiplexer (26). Between sequential perfusions the entire fluidic path connecting the mixer and matrix is purged through wash channels located between every row, thereby eliminating cross-contamination. Experiments using a fluorescent tracer show that contamination between rows is <1 part in 10,000, which was the detection limit of our detector. Automated perfusion of each row is performed periodically during experiments at ≈100-s intervals to maintain nutrient levels, remove metabolites, and change conditions, thereby enabling long-term study of response under constant (chemostatic) or changing (chemodynamic) conditions. The measured generation time for yeast grown in SCD medium at room temperature was 220 min and was found to be independent of seeding density <200 cells per chamber, resulting in an ≈10× increase in cell number over a 12-h experiment. This growth rate is consistent with off-chip measurements in bulk culture (≈240 min).
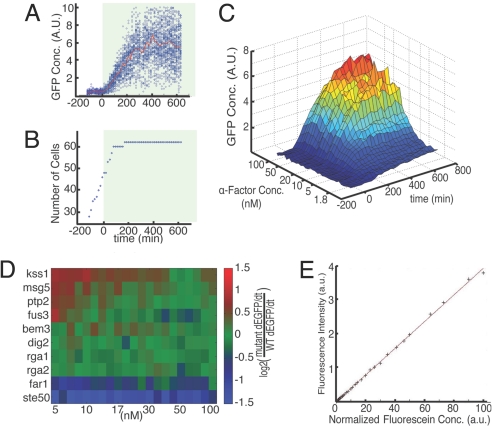
Fig. 2.
Mating response to persistent α-factor stimulation. (A and B) WT time course data showing mean and variation of response to constant stimulation with 20 nM α-factor. Stimulated with α-factor at t = 0 is indicated by shading. (A) Measured GFP concentration, reporting mating-specific gene expression, of each cell, with mean of population indicated in red. (B) Number of cells in the chamber as a function of time showing arrest under α-factor stimulation. (C) Time course and dose response of mean GFP concentration in WT cells for all α-factor concentrations. (D) Strain comparison of signaling under constant stimulation. Initial dGFP/dt for all strains at the given concentrations relative to WT is shown (see SI Text). Initial dGFP/dt is calculated as the slope of a line fitted to the population averaged GFP concentrations between 30 and 180 min. (E) Performance of on-chip chemical formulation. Fluorescent measurements of 32 concentrations generated on-chip as detailed in SI Text and Table S1 are shown.
Throughout each experiment, the cells are confined in the vertical direction by 3.5-μm height of the perfusion chambers, restricting them to a monolayer of cells in a single focal plane and allowing for long-term imaging over multiple generations (Fig. S2). In each experiment, high-resolution brightfield (Nomarski) and fluorescence images of all 256 chambers were taken with 15-min time resolution over the entire length of each experiment (12.5 h). Two fields of view are required for complete imaging of each chamber so that a single experimental run generates >50,000 images capturing millions of single-cell measurements. To handle the volume of raw image data, we developed an image analysis pipeline to record single-cell data, including cell number, cell size, cell morphology, and concentration of a fluorescent gene expression reporter molecule [green fluorescent protein (GFP)] (Fig. S3).
Imaging Studies of Pheromone Response Pathway.
Microfluidic parallelization allows for the simultaneous collection of unified datasets in a single experiment, thereby allowing for sensitive comparisons of wild-type (WT) with multiple mutant responses under a wide array of changing chemical conditions. We investigated the signaling response of WT cells and a panel of 11 mutants having deletions of mating signaling genes (DIG2, RGA1, RGA2, SLT2, MSG5, PTP2, FUS3, KSS1, STE50, FAR1, and BEM3) that are reported to have subtle or complex mutant phenotypes under constant α-factor stimulation (10, 13, 29–32). Mutant response was screened against a wide range of static and time-varying (Fig. 1D) conditions, including (i) constant stimulation under finely varied concentrations to measure dose–response of pathway activation and morphological variability; (ii) transient pulses of varying concentration and duration to measure pathway deactivation and adaptation; and (iii) repeated short pulses of varying concentration and frequency to measure cellular memory of transient stimulation (Fig. S4). Mating-specific gene expression was reported by using an enhanced GFP gene under the control of a minimal promoter including the tandem pheromone-response elements of the PRM1 promoter (33). The BAR1 gene, encoding a secreted α-factor protease, was deleted from all strains to focus on the roles of intracellular elements. Details of strain construction are included in the online SI Text.
Response Under Chemostatic Conditions.
Frequent medium exchange allows for precise control of chemical conditions over long times to perform highly resolved studies of the dose–response of signaling output (Fig. S5). Using this control, we validated our platform in the high-throughput analysis of all 12 genotypes under static conditions of finely varied α-factor concentrations. Using five identical devices, we tested 8 strains per device with at least three replicates for each of the 12 strains. This analysis rapidly and faithfully reproduced a broad range of observations collated from previous studies (2, 16) and further extended these results in terms of the number of chemical conditions, range of genetic perturbations, and temporal resolution. The signaling response of all strains was measured across 32 exponentially distributed α-factor concentrations, ranging from 1 to 100 nM. Gene expression was detectable over the full range of concentrations and showed a 15-fold increase at saturating concentrations of 30–100 nM α-factor (16). A representative dataset for one of the 256 experiments, showing the distribution of single-cell GFP expression and growth rate for WT cells <20 nM α-factor stimulation, is shown in Fig. 2 A and B. The signaling response was mapped for each mutant as high-resolution GFP expression surfaces, showing the interplay among stimulation strength, time, and GFP concentration (Fig. 2C for WT). The simultaneous testing of identical stimulation conditions in multiple strains allows for precise comparative analysis by normalization of expression to WT response (Fig. 2D). Under constant stimulation, we identified hyperresponders (kss1Δ, msg5Δ, and ptp2Δ), WT-like responders (fus3Δ, slt2Δ, dig2Δ, rga1Δ, rga2Δ, and bem3), and hyporesponders (far1Δ and ste50Δ) (Fig. 2D). Generally, the degree of differential expression was found to be concentration-dependent, with hyper- and hyporesponding phenotypes exhibited most strongly at low nonsaturating α-factor concentrations, highlighting the context-specific effect of nonessential genetic perturbations to network output (2, 16).
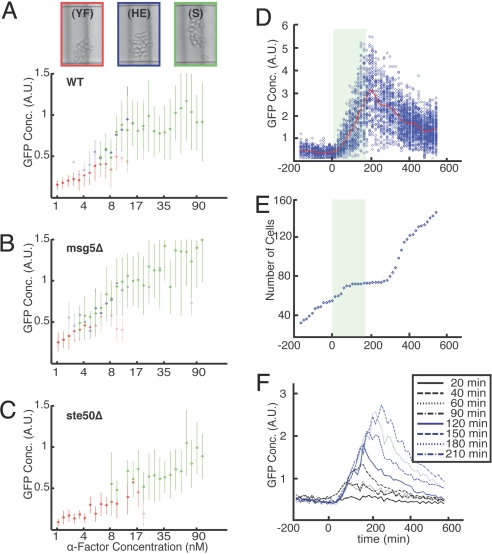
High-throughput imaging allows for the direct comparison of morphological transitions across varying genetic backgrounds as a function of α-factor dose. Gene expression, cell cycle arrest, and cell morphology changes are tightly coupled in the mating response. However, in contrast to mating pathway-dependent gene expression, which increased continuously with increasing α-factor concentration, cell cycle arrest and morphological transitions were found to exhibit defined thresholds. Analysis of single-cell morphology after 6 h under varying α-factor concentrations reveals three distinct cell types: proliferating ovoid cells at very low concentrations (<4 nM), highly elongated cells at intermediate concentrations (4–20 nM), and cells with shmoos (mating projections) at high concentrations (>20 nM). At intermediate α-factor concentrations (4–20 nM), we find the coexistence of all three morphological types (2, 34–37) with characteristic levels of transcriptional output, a phenomenon that has been attributed to network bistability (2). Fig. 3A depicts average WT gene expression in each morphological cluster after 6 h. Interestingly, some mutant strains were found to undergo morphological transitions at different thresholds of α-factor concentration and to support the coexistence of phenotypes over differing concentration ranges (Fig. S6). For example, the morphological switch in msg5Δ mutants is more sensitive, exhibiting elongated morphologies at lower α-factor concentrations than WT (Fig. 3B). In contrast, ste50Δ presents no elongated morphology at any concentrations tested at 6 h (Fig. 3C). Interestingly, fus3Δ displayed a delayed morphological response, with no observable elongation or shmooing until 10 h.
Fig. 3.
Morphological and transient stimulation responses. (A–C) Morphological response of WT (A), msg5Δ (B), ste50Δ (C) across all α-factor concentrations under constant stimulation. Color code for morphology shows representative images of yeast form cells (YF) at 3.2 nM (red); hyperelongated cells (HE) at 17 nM (blue), and shmoo cells (S) at 90 nM (green). The figures presents the mean GFP concentration, reporting mating-specific gene expression as a function of α-factor concentration for each classification, with dot opacity indicating the fraction of cells with that morphology. Error bars represent SD of measured GFP expression for cells of each morphology. Measurements were taken 360 min after exposure to pheromone. (D and E) WT time course response to a 180-min duration 50 nM α-factor pulse. Cells are stimulated with α-factor at t = 0; shading indicates the presence of α-factor. (D) GFP concentration, reporting mating-specific gene expression per cell with mean of population indicated in red with nearest-neighbor time point averaging used to smooth the mean curve. (E) Total number of cells vs. time showing transient growth arrest during α-factor pulse. Each blue diamond is the total number of cells in the microchamber array at a given time. (F) Representative data of population mean GFP concentration in response to transient α-factor pulse of varying duration. Data are shown for 20 nM α-factor condition.
Pathway Response Under Dynamic Stimulation.
Single-pulse experiments.
Microfluidics offers unique opportunities for measuring cellular response to precisely controlled time-varying stimulation and with high temporal resolution (19, 20, 38). We used this temporal control to investigate differences in network memory between mutants. In particular, the propagation of signal through the MAPK network results in changes in the dynamic state of the system, including alterations in protein expression, phosphorylation, localization, and complex assembly. These changes modulate the ability of the network to transmit signals (network capacity), giving rise to memory effects in which the cellular response is history-dependent (5).
We first measured cellular response to transient α-factor and tested whether cellular recovery depends on duration of stimulation. We stimulated yeast with single-transient pulses of α-factor across a broad range of both stimulation strength and pulse duration; all combinations of four α-factor concentration (5, 10, 20, and 50 nM) and eight pulse widths (20, 40, 60, 90, 120, 150, 180, and 210 min) were tested. Consistent with experiments under static conditions, we observed no threshold of response and measured expression in all conditions (Fig. 3F and SI Text).
Release from stimulation resulted in a characteristic decay time of 3.6 h, beginning ≈30 min after release, which was independent of pulse duration and the maximum level of GFP. This is consistent with reported GFP maturation times and dilution of GFP during cell growth, suggesting that the rapid deactivation of signaling output is independent of input dose (Fig. 3F). In contrast to the case of periodic stimulation (described below), single-pulse stimulation revealed no new differences between mutants, suggesting that any changes in network dynamics arise through transients with fast characteristic time scales or adaptation occurring at very long time scales. Similarly, analysis of cell cycle response (Fig. S7C) indicates that cell growth quickly resumes upon α-factor removal (Fig. 3E). No morphological changes were observed in any cells for pulses shorter than 90 min even at saturating α-factor concentrations, indicating that the emergence of a full mating response requires sustained stimulation. Directly probing signaling at faster time scales by using single-pulse experiments is limited by low expression and the long maturation time of GFP and will require future studies with faster reporters such as those using fluorescence resonance energy transfer, photoactivatable GFP (39), or mRNA tagging (40).
Response to periodic stimulation.
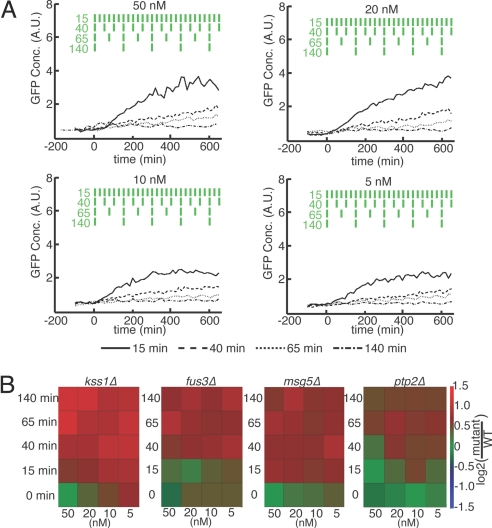
Under constant stimulation, different deletion mutants may exhibit phenotypes that are indistinguishable, thus making it difficult to assign unique functions to these genes. These genes may nonetheless have distinct roles in controlling short time-scale network dynamics and thus may be separated by analysis under varying stimulation. Although these differences are difficult to detect with fluorescent protein reporters, the monitoring of response under periodic stimulation allows for integration of the GFP output to amplify subtle differences across conditions and mutant genotypes. We used this strategy to investigate mutant variations in pathway memory by measuring transcriptional output to repeated 10-min pulses of pheromone of varying frequency. All strains were tested under repeated pulse conditions of varying concentrations (5, 10, 20, and 50 nM) and delay times (15, 40, 65, and 140 min) between pulses (Fig. 4A). Although the WT response was found qualitatively to increase with total time-averaged α-factor dose, there were notable deviations from this trend that suggest a more complicated dependence on the frequency response of signaling. Cells were found to respond comparatively more strongly to repeated intermittent pulses of low pheromone than would be expected under a model of response to simple time-averaged concentration. For instance, conditions of repeated 10-min pulses of 5 nM pheromone every 25 min, corresponding to a time-averaged dose of 2 nM, resulted in a gene expression response similar to conditions of 50 and 20 nM stimulation with pulse delays of 40 min (corresponding to time-averaged dose of 10 and 4 nM, respectively). Similarly, 10-min pulses of both 5 and 50 nM pheromone every 75 min, corresponding to 0.67 and 6.7 nM time-averaged doses, respectively, show a very similar response. Taken together, these observations under periodic stimulation indicate that pathway output depends strongly on cell history and the frequency of signal input.
Fig. 4.
Mating response to short repeated pulses of α-factor. (A) Population mean GFP concentration, reporting mating-specific gene expression, of WT cells for the 50, 20, 10, and 5 nM α-factor concentrations across four pulse delay lengths (15, 40, 60, and 140 min). In each condition, 10-min pulses of α-factor are used. The green rectangles depict the four pulse patterns of α-factor stimulation over time; values indicated delay time in min. (B) Sensitivity of kss1Δ, fus3Δ, msg5Δ, and ptp2Δ mutants under periodic stimulation under conditions of varying α-factor concentration (columns) and pulse delays (rows). Mean population GFP concentration over three experimental replicates are shown normalized to WT. Data are taken at t = 600 min.
Mutants Implicated in Dynamic Phenotypes.
Analysis under periodic stimulation allows for the classification of mutants on the basis of differing dynamic response. To test this idea, we compared the dynamic responses of all mutants (Fig. S8B) and found distinct patterns of hypersensitivity for mutants lacking two highly related kinases, Fus3 and Kss1 (11), and two phosphatases, Msg5 and Ptp2 (13). All four of these mutants exhibit WT-like mating phenotypes, are indistinguishable under saturating pheromone concentrations (50 nM) (16), and were found to be increasingly hypersensitive at low α-factor concentrations (Fig. 4B). This divergence from WT behavior was greatly amplified under low-frequency periodic stimulation across all concentrations tested. Moreover, the frequency response of mating pathway-dependent gene expression under varying pheromone concentrations was unique to each mutant.
The ability to stratify mutants unambiguously on the basis of response to time-varying stimulation provides a stringent test for the development and testing of quantitative network models and suggests new regulatory roles of for signaling proteins. Across all conditions, kss1Δ mutants exhibited the greatest divergence from WT. In addition to reported hypersensitivity at low pheromone concentrations, kss1Δ mutants display hypersensitivity under intermittent pheromone stimulation. This effect was evident for all transient conditions, even when pulses are de-layed by as little as 15 min, and was most pronounced for low-frequency stimulation with high-pheromone concentrations. By comparison, mutant fus3Δ cells, which show similar pathway output to WT under all constant stimulation conditions, exhibit hypersensitivity to transient stimulation only for pulse delays of 40 min or more. Also, whereas the degree of hypersensitivity for fus3Δ cells was found to depend primarily on the frequency of stimulation, the sensitivity of kss1Δ mutants exhibits both concentration and frequency dependence, being most pronounced for transient pulses of high concentration. Taken together, these results implicate Kss1 in a regulatory mechanism that acts to filter both weak and intermittent α-factor stimulation whereas Fus3 appears primarily to filter transient signals (Fig. 4B). Analysis of the phosphatase mutants reveals a similar trend in which the hypersensitivity of msg5Δ mutants is largely determined by frequency whereas ptp2Δ mutants exhibit hypersensitivity depending on both pulse frequency and pheromone concentration. Similarities in behavior were also noted between kinase and phosphatase deletion mutants. The sensitivity trend for fus3Δ mutants under dynamic stimulation, having a frequency threshold with little concentration dependence, is similar to that of msg5Δ mutants at pulse delays longer than 15 min. At short pulse delays (15 min), kss1Δ mutants exhibit hypersensitivity similar to that of msg5Δ mutants.
Discussion
Here, we have presented a system for high-throughput measurements of single-cell response over a broad array of precisely controlled and time-varying conditions. Further improvements will allow for both increased genotypic throughput and more refined analysis of single cells in time. Straightforward device modifications will allow for the parallel analysis of ≈40 strains, allowing for comprehensive network-scale analysis on a single device run. Additionally, an important component of single-cell analysis is the ability to track single-cell response and lineage through time, something that is difficult to automate in the current format because of the motion of cells within the traps during perfusion sequences. We are currently refining cell immobilization techniques, image processing algorithms, and data analysis methods to enable automated high-throughput tracking of tens of thousands of single cells, spanning multiple genotypes and stimulation conditions, through time.
Microfluidics provides a powerful method for high-throughput imaging analysis with programmable control over the chemical environment, offering a new temporal dimension to live-cell imaging studies. Our present work demonstrates that the analysis of cellular networks under static conditions or with coarse chemical resolution is insufficient to reveal the function of genes in regulating network response. Indeed, dynamic analysis of mutants compromised for genes known to be key players in the pheromone response, including Kss1, Fus3, Msg5, and Ptp2, reveals unique properties of network response that are invisible under constant stimulation and that suggest possible mechanisms of network regulation. For instance, the similarity in frequency threshold for the hypersensitivity of kss1Δ and msg5Δ mutants may be caused by increased Fus3 activity in both of these mutants. The Kss1 kinase competes with Fus3 (41), and Msg5 phosphatase acts on Fus3 (13).
Usually, the response to transient stimulus is discussed in the context of signal filtering to prevent unproductive response. Conversely, we speculate that frequency-dependent signal memory enables productive responses in natural environments. For instance, one might expect that the paracrine induction and autorepression of the mating response, including the secretion of pheromone and pheromone-degrading enzymes, coupled with hydrodynamics, varying cell density, and cell motion, create spatiotemporal variations in pheromone concentration and intermittent opportunities for successful mating. Pathway mechanisms selected to filter or remember stimulations over appropriate time scales could act to prime cells for more rapid response, thus increasing mating success. Testing of such hypotheses will ultimately require combined approaches based on quantitative modeling and experiment. We contend that high-throughput single-cell measurements of network dynamics will provide a stringent test for in silico models and are essential for ultimately developing a quantitative and predictive understanding of cellular decision making.
Methods
Cell Loading.
For each experiment, yeast cells were grown with aeration overnight in YPD (30C), diluted, and grown to log phase in synthetic complete medium with 2% dextrose (SCD), sonicated for 5 s, and concentrated to an A600 of 3. The chip was primed with SCD medium containing 20 mg/mL BSA for ≈3 h before cell loading. Strains used in this work are shown in Table S2.
Microfluidic Fabrication.
Fabrication of the microfluidic device was accomplished by using multilayer soft lithography (27, 42). Our chips used a three-layer design: The top layer was a “flow layer,” containing the cells and the chemical channels. The middle layer was a “control layer,” containing channels used for pneumatic valves. The bottom layer was a “blank layer,” used to tightly seal the control channels to the glass slide. All devices were made from polydimethylsiloxan (RTV615; General Electric).
Image Acquisition.
Microfluidic devices were mounted onto a Leica DMIRE2 fluorescent microscope modified with a custom LED brightfield source to increase acquisition speed. Cells were imaged with a 40× air objective (HCX, long working distance, FLUOTAR PL with correction collar, NA = 0.6) and 1,344- × 1,024-pixel cooled CCD camera (ORCA-ER; Hamamatsu Photonics, differential interference contrast), and fluorescent images were captured with a 100-ms and 250-ms exposure, respectively.
Supplementary Material
Acknowledgments.
We thank G. Carter, P. Hieter, I. Barrett, S. Ben-Aroya, and J. Stoepel for helpful discussions and M. Homenuke, M. Despotovic, and A. Carter for technical assistance. This work was supported by National Institutes of Health Grants R21 EB005757-01 and P50-GM076547 (to R.J.T., A.N., S.A.R., I.S., T.G., and C.L.H.), the Michael Smith Foundation for Health Research (R.J.T. and C.L.H.), the Academy of Finland (A.N.), the Canadian Institute for Health Research, and the Swiss National Science Foundation (D.F.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813416106/DCSupplemental.
References
- 1.Ingolia NT, Murray AW. Positive-feedback loops as a flexible biological module. Curr Biol. 2007;17:668–677. doi: 10.1016/j.cub.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paliwal S, et al. MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature. 2007;446:46–51. doi: 10.1038/nature05561. [DOI] [PubMed] [Google Scholar]
- 3.Zhou J, Arora M, Stone DE. The yeast pheromone-responsive G α protein stimulates recovery from chronic pheromone treatment by two mechanisms that are activated at distinct levels of stimulus. Cell Biochem Biophys. 1999;30:193–212. doi: 10.1007/BF02738067. [DOI] [PubMed] [Google Scholar]
- 4.Doi K, et al. MSG5, a novel protein phosphatase promotes adaptation to pheromone response in S. cerevisiae. EMBO J. 1994;13:61–70. doi: 10.1002/j.1460-2075.1994.tb06235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- 6.Sigal A, et al. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- 7.Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb-E2F switch underlies the restriction point. Nat Cell Biol. 2008;10:476–482. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]
- 8.Chen RE, Thorner J. Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1773:1311–1340. doi: 10.1016/j.bbamcr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 10.Sabbagh W, Jr, Flatauer LJ, Bardwell AJ, Bardwell L. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol Cell. 2001;8:683–691. doi: 10.1016/s1097-2765(01)00322-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madhani HD, Styles CA, Fink GR. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 12.Choi KY, Satterberg B, Lyons DM, Elion EA. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 13.Zhan XL, Deschenes RJ, Guan KL. Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev. 1997;11:1690–1702. doi: 10.1101/gad.11.13.1690. [DOI] [PubMed] [Google Scholar]
- 14.Roberts CJ, et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 15.Flory MR, et al. Quantitative proteomic analysis of the budding yeast cell cycle using acid-cleavable isotope-coded affinity tag reagents. Proteomics. 2006;6:6146–6157. doi: 10.1002/pmic.200600159. [DOI] [PubMed] [Google Scholar]
- 16.Colman-Lerner A, et al. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- 17.Zamir E, Bastiaens PI. Reverse engineering intracellular biochemical networks. Nat Chem Biol. 2008;4:643–647. doi: 10.1038/nchembio1108-643. [DOI] [PubMed] [Google Scholar]
- 18.Groisman A, et al. A microfluidic chemostat for experiments with bacterial and yeast cells. Nat Methods. 2005;2:685–689. doi: 10.1038/nmeth784. [DOI] [PubMed] [Google Scholar]
- 19.Bennett MR, et al. Metabolic gene regulation in a dynamically changing environment. Nature. 2008;454:1119–1122. doi: 10.1038/nature07211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hersen P, McClean MN, Mahadevan L, Ramanathan S. Signal processing by the HOG MAP kinase pathway. Proc Natl Acad Sci USA. 2008;105:7165–7170. doi: 10.1073/pnas.0710770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mettetal JT, Muzzey D, Gomez-Uribe C, van Oudenaarden A. The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science. 2008;319:482–484. doi: 10.1126/science.1151582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King KR, et al. A high-throughput microfluidic real-time gene expression living cell array. Lab Chip. 2007;7:77–85. doi: 10.1039/b612516f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung PJ, Lee PJ, Sabounchi P, Lin R, Lee LP. Continuous perfusion microfluidic cell culture array for high-throughput cell-based assays. Biotechnol Bioeng. 2005;89:1–8. doi: 10.1002/bit.20289. [DOI] [PubMed] [Google Scholar]
- 24.Gomez-Sjoberg R, Leyrat AA, Pirone DM, Chen CS, Quake SR. Versatile, fully automated, microfluidic cell culture system. Anal Chem. 2007;79:8557–8563. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- 25.Cheong R, Wang CJ, Levchenko A. High-content cell screening in a microfluidic device. Mol Cell Proteomics. 2008 Oct; doi: 10.1074/mcpM8000291-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 27.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 28.McClean MN, Mody A, Broach JR, Ramanathan S. Cross-talk and decision making in MAP kinase pathways. Nat Genet. 2007;39:409–414. doi: 10.1038/ng1957. [DOI] [PubMed] [Google Scholar]
- 29.Narayanaswamy R, et al. Systematic profiling of cellular phenotypes with spotted cell microarrays reveals mating-pheromone response genes. Genome Biol. 2006;7:R6. doi: 10.1186/gb-2006-7-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rad MR, Xu G, Hollenberg CP. STE50, a novel gene required for activation of conjugation at an early step in mating in Saccharomyces cerevisiae. Mol Gen Genet. 1992;236:145–154. doi: 10.1007/BF00279653. [DOI] [PubMed] [Google Scholar]
- 31.Yu RC, et al. Negative feedback that improves information transmission in yeast signalling. Nature. 2008;456:755–761. doi: 10.1038/nature07513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breitkreutz A, Boucher L, Tyers M. MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr Biol. 2001;11:1266–1271. doi: 10.1016/s0960-9822(01)00370-0. [DOI] [PubMed] [Google Scholar]
- 33.Heiman MG, Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J Cell Biol. 2000;151:719–730. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hao N, et al. Regulation of cell signaling dynamics by the protein kinase-scaffold Ste5. Mol Cell. 2008;30:649–656. doi: 10.1016/j.molcel.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erdman S, Snyder M. A filamentous growth response mediated by the yeast mating pathway. Genetics. 2001;159:919–928. doi: 10.1093/genetics/159.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farley FW, Satterberg B, Goldsmith EJ, Elion EA. Relative dependence of different outputs of the Saccharomyces cerevisiae pheromone response pathway on the MAP kinase Fus3p. Genetics. 1999;151:1425–1444. doi: 10.1093/genetics/151.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levi JD. Mating reaction in yeast. Nature. 1956;177:2. [Google Scholar]
- 38.Charvin G, Cross FR, Siggia ED. A microfluidic device for temporally controlled gene expression and long-term fluorescent imaging in unperturbed dividing yeast cells. PLoS ONE. 2008;3:e1468. doi: 10.1371/journal.pone.0001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 40.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz MA, Madhani HD. Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu Rev Genet. 2004;38:725–748. doi: 10.1146/annurev.genet.39.073003.112634. [DOI] [PubMed] [Google Scholar]
- 42.Hansen CL, Sommer MO, Quake SR. Systematic investigation of protein phase behavior with a microfluidic formulator. Proc Natl Acad Sci USA. 2004;101:14431–14436. doi: 10.1073/pnas.0405847101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.