SUMMARY
In mitosis, the spindle checkpoint detects a single unattached kinetochore, inhibits the anaphase-promoting complex or cyclosome (APC/C), and prevents premature sister-chromatid separation. The checkpoint kinase Bub1 contributes to checkpoint sensitivity through phosphorylating the APC/C activator, Cdc20, and inhibiting APC/C catalytically. We report here the crystal structure of the kinase domain of Bub1, revealing the requirement of an N-terminal extension for its kinase activity. Though the activation segment of Bub1 is ordered and has structural features indicative of active kinases, the C-terminal portion of this segment sterically restricts substrate access to the active site. Bub1 uses docking motifs, so-called KEN boxes, outside its kinase domain to recruit Cdc20, one of two known KEN-box receptors. The KEN boxes of Bub1 are required for the spindle checkpoint in human cells. Therefore, its unusual active-site conformation and mode of substrate recruitment suggest that Bub1 has an exquisitely tuned specificity for Cdc20.
INTRODUCTION
In mitosis, the spindle microtubules attach to the kinetochores of sister chromatids asynchronously (Tanaka and Desai, 2008). A single unattached inetochore can be sensed by the spindle checkpoint (Musacchio and Salmon, 2007; Rieder et al., 1995), which delays the onset of anaphase until all sister chromatids achieve bi-orientation. The unattached kinetochores need to emit diffusible checkpoint signals to block the premature separation of the attached ones. The nature of the diffusible checkpoint signals has not been definitively established. It is, however, clear that these signals ultimately block the ubiquitin ligase activity of a large molecular machine called the anaphase-promoting complex or cyclosome (APC/C) (Yu, 2002, 2007). Inhibition of APC/C stabilizes securin and cyclin B, which prevent anaphase onset and mitotic exit.
APC/C substrates contain small peptide motifs or degrons that are required for their efficient ubiquitination, such as the destruction box (D box) and KEN box (Peters, 2006; Yu, 2007). The mitotic activator of APC/C, Cdc20, binds directly to these degrons and recruits substrates to APC/C for ubiquitination (Peters, 2006; Yu, 2007). Multiple spindle checkpoint proteins, including Mad2, BubR1, and Bub1, collaborate to inhibit the APC/C–Cdc20 complex (APC/CCdc20) in mitosis by blocking the function of Cdc20 (Yu, 2007). Mad2 and BubR1 bind to Cdc20 and inhibit APC/CCdc20 stoichiometrically (Fang et al., 1998; Tang et al., 2001). By contrast, Bub1 phosphorylates Cdc20 and inhibits APC/CCdc20 catalytically (Tang et al., 2004). The kinase activity of Bub1 toward Cdc20 is enhanced in mitosis (Tang et al., 2004). Furthermore, chromosome-bound Bub1 is hyperactive (Chen, 2004). The kinase activity of Bub1 is required for the nocodazole-triggered metaphase arrest in Xenopus egg extracts supplemented with low concentrations of sperm nuclei (Chen, 2004). These results suggest that phosphorylation of Cdc20 by Bub1 contributes to the sensitivity of the spindle checkpoint. Although the mechanisms by which Mad2 and Bub1 inhibit APC/CCdc20 are unclear, two recent studies have shown that Mad3 (the yeast ortholog of BubR1) contains KEN boxes and binding of the KEN boxes of Mad3 to Cdc20 competitively blocks substrate binding, thus inhibiting the activity of APC/CCdc20 (Burton and Solomon, 2007; Diaz-Martinez and Yu, 2007; King et al., 2007).
In this study, we show that the kinase activity of Bub1 is required for the spindle checkpoint in human cells. We have determined the crystal structure of the C-terminal domain of Bub1, consisting of its kinase domain and a 60-residue fragment N-terminal to it. The structure reveals that the N-terminal extension organizes the ATP-binding pocket and the activation segment of Bub1 in a way analogous to the activation of cyclin-dependent kinases (Cdk) by cyclins. Mutations of the N-terminal extension disrupt the kinase activity of Bub1. Compared to other kinases, Bub1 has an extended substrate recognition loop which blocks the active site of Bub1 and limits the access of nonspecific substrates. Bub1 contains two KEN boxes in its central region which mediates its degradation APC/CCdh1 in the G1 phase of the cell cycle (Qi and Yu, 2007). We show here that the KEN boxes of Bub1 are also required for Cdc20 binding, efficient phosphorylation of Cdc20 by Bub1, and the spindle checkpoint. Because Cdc20 is one of only two known KEN-box receptors, Bub1 has a well-tuned specificity towards Cdc20. These findings provide insight into how multiple APC/C-inhibitory mechanisms are coordinated during checkpoint signaling.
RESULTS AND DISCUSSION
Structure Determination of the Kinase Domain of Bub1
Bub1 contains an N-terminal tetratricopeptide repeat (TPR) domain that is required for its kinetochore localization and a C-terminal serine/threonine (S/T) kinase domain (Figure 1A) (Bharadwaj and Yu, 2004; Kiyomitsu et al., 2007). Phosphorylation of Cdc20 by Bub1 inhibits APC/CCdc20 (Tang et al., 2004). Ectopic expression of a non-phosphorylatable mutant of Cdc20 compromises the spindle checkpoint in human cells (Tang et al., 2004). These findings implicate a role for the kinase activity of Bub1 in the spindle checkpoint. We thus formally tested whether the Bub1 kinase activity was required for the spindle checkpoint in human cells. We found that HeLa cells depleted of Bub1 by RNA interference (RNAi) failed to undergo mitotic arrest in the presence of nocodazole, as indicated by their lower mitotic index and decreased levels of securing and phospho-histone H3 (Figure S1). Ectopic expression of RNAi-resistant wild-type Myc-Bub1 in Bub1 RNAi cells restored the Bub1 protein to its endogenous levels and rescued the mitotic arrest deficiency of these cells. By contrast, expression of a kinase-dead (KD) mutant of Myc-Bub1 was less effective in rescuing the defect of Bub1 RNAi cells. These results indicate that the kinase activity of Bub1 is required for the spindle checkpoint in human cells.
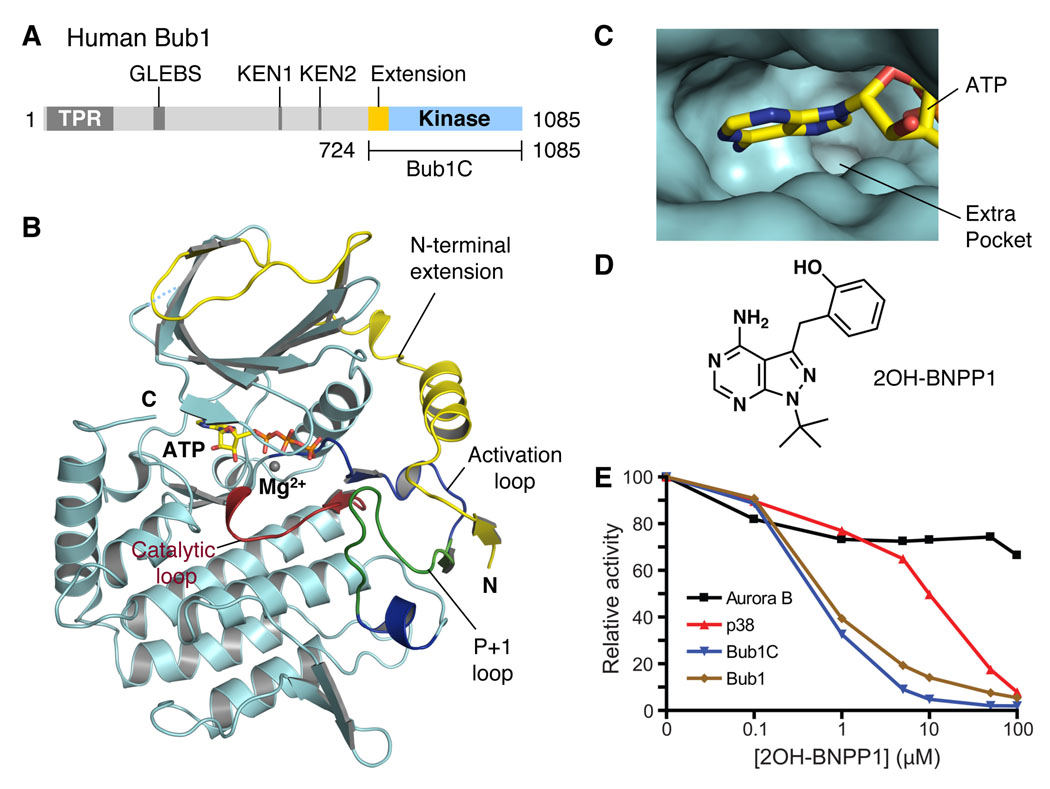
Figure 1. Structure and Inhibitor of the Extended Kinase Domain of Bub1.
(A) Domain architecture of human Bub1. TPR, tetratricopeptide repeat; GLEBS, Gle2-binding sequence; KEN, lysine-glutamate-asparagine.
(B) Ribbon drawing of the crystal structure of the extended kinase domain of Bub1. The N-terminal extension is colored yellow. The substrate-binding P+1 loop is shown in green while the rest of the activation segment is in blue. The catalytic loop is shown in red. ATP is shown as sticks. The Mg2+ ion is shown as a gray sphere. The N- and C-termini are indicated. All structure figures were generated with PyMOL (http://pymol.sourceforge.net/).
(C) The ATP-binding site of Bub1. ATP is shown as sticks. The extra pocket is indicated.
(D) The chemical structure of 2OH-BNPP1.
(E) Determination of the IC50 values of 2OH-BNPP1 against Aurora B, p38, Bub1C, and Bub1.
We next crystallized a C-terminal fragment of human Bub1 (referred to as Bub1C) containing residues 724−1085 in the presence of ATP and determined its crystal structure using the single-wavelength anomalous diffraction (SAD) method using data to a resolution of 2.5 Å (Figure 1B and Table S1). Bub1C consists of the kinase domain and an N-terminal extension (Figure 1A, B). The kinase domain contains residues 784-1085 and adopts a canonical kinase fold with two lobes. ATP and the active site are located at the interface of the two lobes. The N-terminal extension contains three β strands and an α helix and wraps around the N-lobe of the kinase domain.
Identification of a Chemical Inhibitor of Bub1
Chemical inhibitors of mitotic regulators, such as monastrol and blebbistatin, are powerful tools for studying rapid processes in mitosis (Mayer et al., 1999; Straight et al., 2003). Highly specific inhibitors of single members of large protein families, such as protein kinases, are difficult to obtain, however. Mutations of large residues (so-called “gatekeeper” residues) in the ATP-binding pocket of kinases to glycines or alanines enlarge the active sites of kinases and enable their inhibition by adenine analogs (Alaimo et al., 2001; Bishop et al., 2000). Introduction of these mutant kinases into cells that lack the corresponding wild-type kinases then sensitizes these cells to the inhibition by these orthogonal inhibitors, allowing the dissection of the cellular functions of kinases (Alaimo et al., 2001). The vertebrate Bub1 proteins contain a glycine (G866 in human Bub1) at the C-terminal end of strand β5 (Figure S2), a position occupied by a large hydrophobic gatekeeper residue in most other kinases. As a result, the active site of Bub1 has an unusual additional pocket that is not fully occupied by ATP, suggesting that inhibitors designed to inhibit engineered kinases with glycine gatekeepers might be good inhibitors of wild-type Bub1 (Figure 1C). We thus tested whether the wild-type Bub1 kinase was inhibited by inhibitor analogs designed to inhibit mutant kinases with glycine gatekeeper residues. Among 49 candidate inhibitors tested, 2OH-BNPP1 is the most potent inhibitor of both Bub1C and the full-length Bub1 with IC50s around 250 nM (Figure 1D, 1E, and S3). As controls, 2OH-BNPP1 did not appreciably inhibit another mitotic kinase, Aurora B, at concentrations up to 100 µM and inhibited p38MAPK with a much larger IC50 (about 10 µM). Incubation of human cells with 2OH-BNPP1 produces phenotypes that are consistent with Bub1 inhibition (data not shown). 2OH-BNPP1 is thus a specific inhibitor of Bub1 and a useful tool for studying mitosis of human cells. A broader screen for Bub1 inhibitors may reveal more selective inhibitors for this kinase because of its unusual glycine gatekeeper residue.
Mutations of the gatekeeper residues to glycines in several kinases sensitize these kinases to the inhibition by bulky adenine analogs, such as 1NM-PP1 (Bishop et al., 2000; Jaeschke et al., 2006; Papa et al., 2003). 1NM-PP1, however, did not inhibit Bub1 (data not shown). A closer inspection of the ATP-binding pocket of Bub1 revealed that M850 might occlude the binding of 1NM-PP1 and other bulky adenine analogs commonly used to inhibit mutant kinases with glycine gatekeeper residues.
The N-terminal Extension Acts as a “Mini-Cyclin” to Activate Bub1
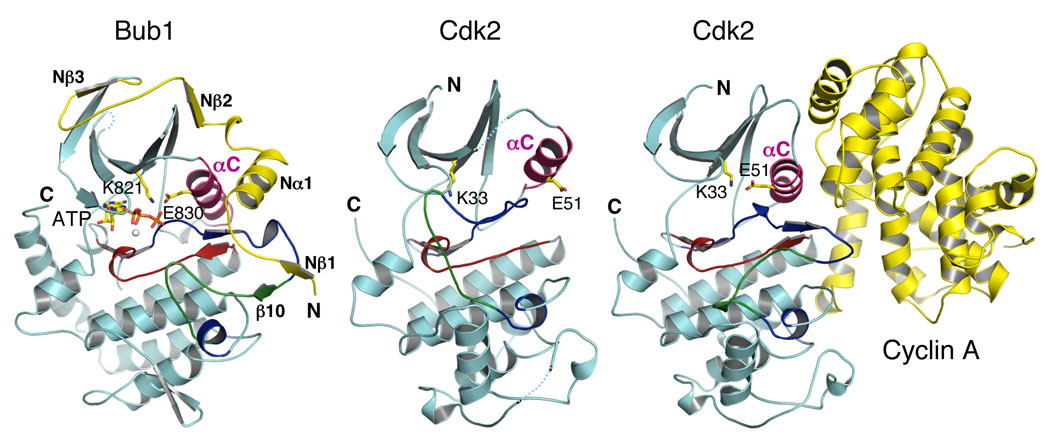
The sequence of the N-terminal extension is highly conserved among Bub1 proteins from different organisms (Figure S2), suggesting that it is functionally important. The N-terminal extension makes extensive contacts with the N-lobe and the activation segment of the Bub1 kinase domain (Figure 2). A segment of the N-terminal extension consisting of Nβ1, NαA, and Nβ2 restrains the motion of the αC helix of Bub1 (Figure 2). This arrangement allows E830 on αC to engage in favorable electrostatic interactions with K821, positioning it for ATP binding. Nβ1, P741, and W742 form extensive hydrophobic interactions with residues of the activation loop and serve as the N-terminal anchor point for the N-terminal extension (Figure 3A and 3B). Nβ1 forms a parallel β sheet with β10, which is a part of the substrate-binding loop (often referred to as the P+1 loop) in other kinases. Thus, the N-terminal extension also helps to stabilize the conformation of the activation segment of Bub1. These functions of the N-terminal extension of Bub1 are reminiscent of the ways in which cyclins activate Cdks (Figure 2) (Pavletich, 1999). First, cyclins restrain the movement of αC and enable ATP binding by facilitating the interaction between a glutamate on αC and the conserved ATP-binding lysine. Second, cyclins contact the activation loop of Cdks and stabilize its active conformation. Both activating mechanisms of cyclins are likely shared by the N-terminal extension of Bub1 in promoting the kinase activity of Bub1.
Figure 2. The N-terminal Extension Activates Bub1 with a Cyclin-like Mechanism.
Ribbon diagrams of Bub1C, Cdk2, and Cdk2–Cyclin A. The N- and C-termini are indicated. The color schemes are as described in Figure 1B except that αC is colored magenta. The ATP-binding lysine residue and the conserved glutamate residue in αC are shown as sticks. Cyclin A is shown in yellow.
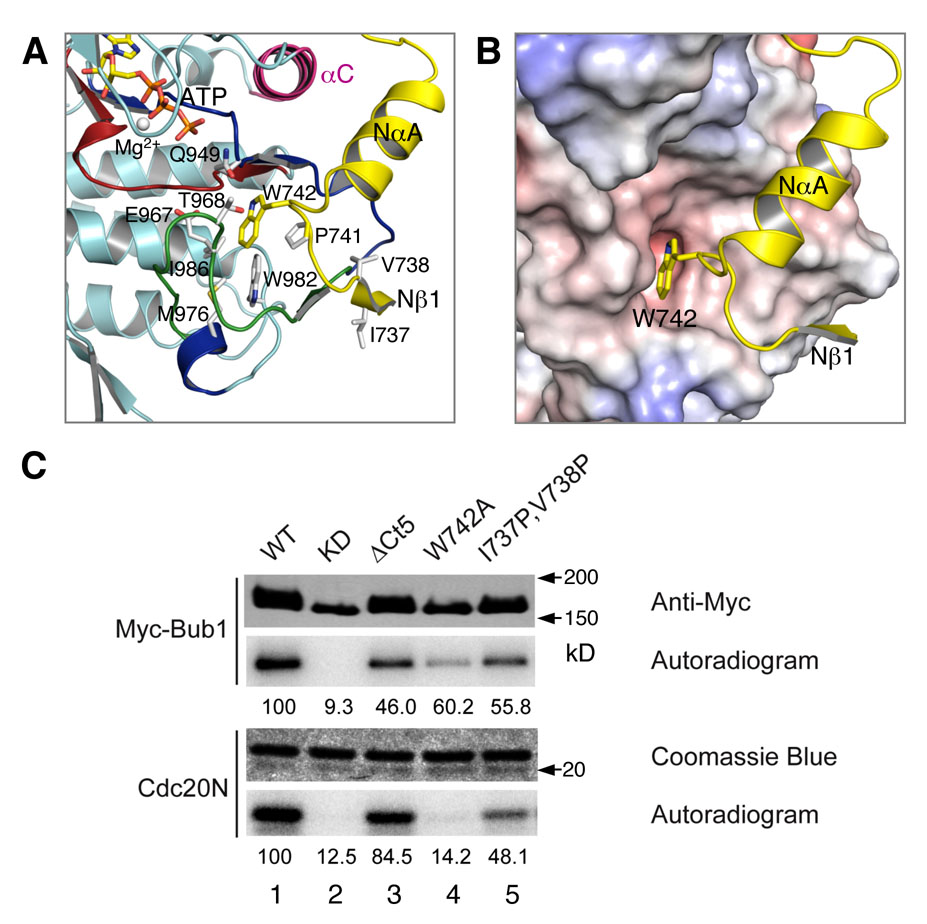
Figure 3. The N-terminal Extension Is Required for the Kinase Activity of Bub1.
(A) The N-terminal anchor of the N-terminal extension. The color schemes are as described in Figure 2. W742 and its neighboring residues are shown in sticks and labeled.
(B) Ribbon diagram of the N-terminal extension superposed with the surface of the rest of the protein. W742 is shown in sticks.
(C) In vitro kinase assays of full-length Bub1 wild-type (WT) and mutants. Myc-Bub1 WT and the indicated mutants were expressed in mitotic HeLa cells, immunoprecipitated with anti-Myc, and assayed for their auto-kinase activity and kinase activity using the N-terminal fragment of Cdc20 (Cdc20N) as a substrate. Anti-Myc blot of the Myc-Bub1 proteins and Coomassie staining of the Cdc20N protein are also shown. The kinase activities are quantified, normalized to the amount of kinase, and presented as relative values directly below each lane.
We next used site-directed mutagenesis to study the function of the N-terminal extension of Bub1. As discussed above, W742 and residues in Nβ1 form extensive hydrophobic interactions with the activation segment of Bub1. Consistently, Bub1C W742A was inactive in phosphorylating an N-terminal fragment of Cdc20 (Cdc20N) (data not shown). We then tested whether the same mutation also affected the activity of full-length Bub1. Myc-Bub1 WT, KD, and W742A were immunoprecipitated from lysates of mitotic HeLa cells transfected with plasmids encoding these proteins and subjected to kinase assays. Bub1 W742A was inactive in phosphorylating Cdc20N (Figure 3C). Mutations that are designed to disrupt Nβ1, such as I737P/V738P, also diminished the kinase activity of Bub1. Our results indicate that the N-terminal extension is required for the kinase activity of full-length Bub1.
Finally, several conserved C-terminal basic residues of Bub1 are near the ATP-binding pocket (Figure S2). Similar basic residues in TAO2 are required for its kinase activity (Zhou et al., 2004). Deletion of the C-terminal five residues of Bub1 (Bub1 ΔCt5) only slightly reduced its kinase activity towards Cdc20N (Figure 3C), indicating that these residues do not play a major role in ATP binding of Bub1.
The Unusual Conformation of the Activation Segment of Bub1
Kinases in their inactive states typically have disordered activation segments whereas active kinases generally have ordered ones (Nolen et al., 2004). In many kinases, the transition between disordered and ordered conformations of the activation segment is regulated by phosphorylation of the activation loop, binding of cofactors, or both. Our crystal structure of the kinase domain of Bub1 has features characteristic of active kinases. First, the activation segment of Bub1 is ordered in our crystal structure (Figure 3A). The N-terminal end of the relatively short activation loop of Bub1 folds into a 310 helix. Several conserved residues in the activation loop engage in extensive hydrophobic interactions with residues in the N-terminal extension. These elaborate hydrophobic interactions maintain the integrity of the active site, as disruption of these interactions by the W742A mutation inactivates Bub1 (Figure 3C). Second, the pairing between β6 preceding the catalytic loop and β9 at the start of the activation segment is a hallmark of active kinases (Figure 4A) (Nolen et al., 2004). This pairing is observed in Bub1 (Figure 4B).
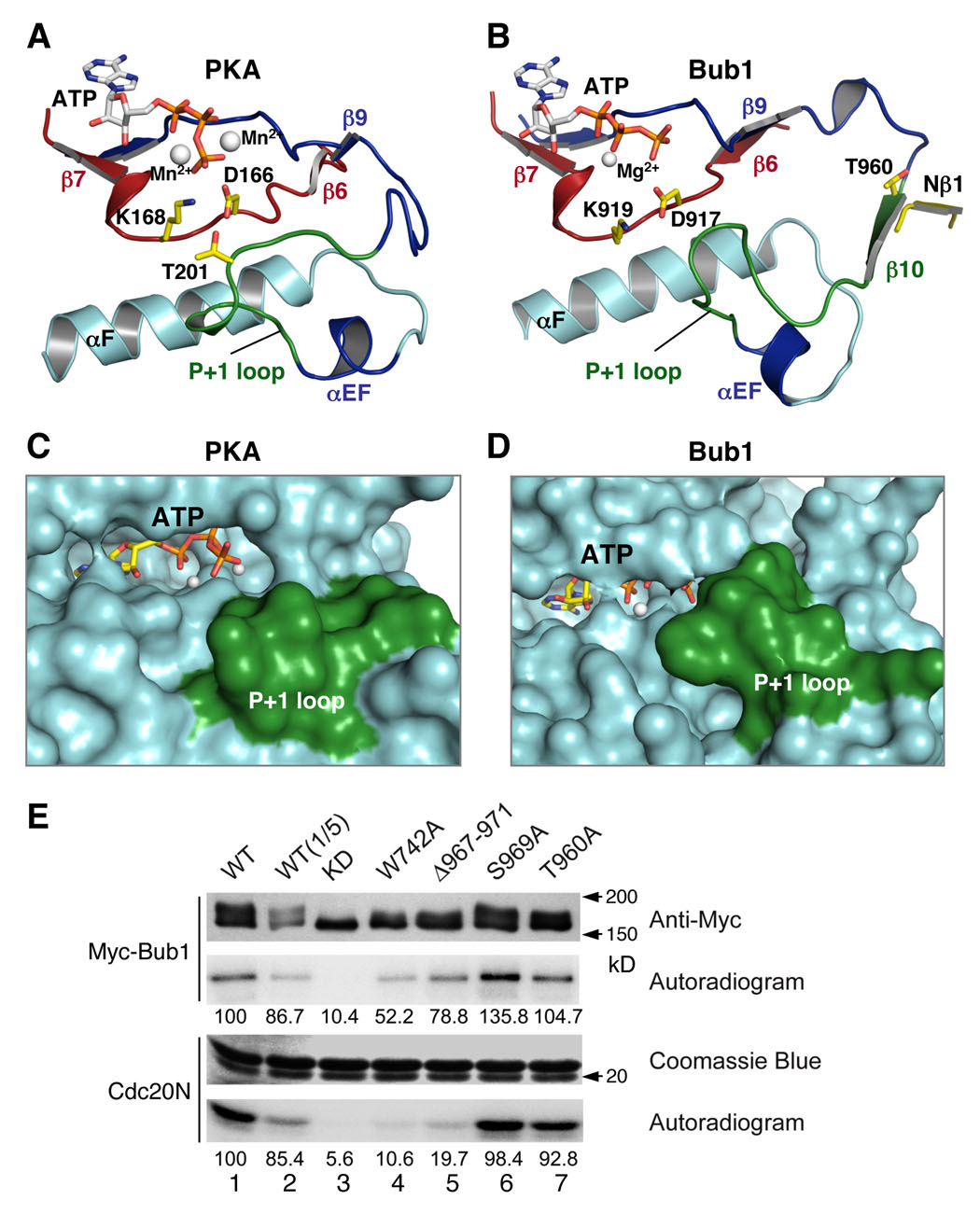
Figure 4. The C-terminal Region of the P+1 Loop in Bub1 Restricts Substrate Access and Confers Specificity.
(A) The active conformation of the activation segment of PKA.
(B) The conformation of the activation segment of Bub1.
(C) Surface diagram of PKA with the P+1 loop colored green and ATP shown as sticks.
(D) Surface diagram of Bub1 with the P+1 loop colored green and ATP shown as sticks.
(E) In vitro kinase assays of full-length Bub1 wild-type (WT) and mutants. The assays were performed as described in Figure 3C. Lane 2 contained one-fifth of the reaction mixture in lane 1. The kinase activities are quantified, normalized to the amount of kinase, and presented as relative values.
On the other hand, in our structure, Bub1 has an extended P+1 loop that adopts a conformation not found in active kinases. A characteristic GT dipeptide motif marks the start of the P+1 loop in many S/T kinases, including Bub1 and PKA (Figure S2) (Nolen et al., 2004). In the active state of PKA and other S/T kinases, the hydroxyl group of the threonine in this GT motif (T201 in PKA) forms a hydrogen bond with the catalytic aspartate (D166 in PKA). The C-terminal part of the P+1 loop forms a 310 helix. These two structural features allow much of the P+1 loop to lie parallel to the catalytic loop and to form the foundation for substrate binding (Figure 4A and 4C). In our structure of Bub1, T960 of the GT motif is located at the start of β10, which pairs with Nβ1 (Figure 4B). The pairing between β10 and Nβ1 places T960 about 19 Å away from the catalytic aspartate residue, D917. The C-terminal half of the P+1 loop in Bub1 adopts a hairpin-like structure that covers the catalytic loop and restricts substrate access to ATP (Figure 4D). Therefore, the P+1 loop of Bub1 adopts a conformation that is sub-optimal for substrate binding. Significant structural rearrangement of the C-terminal half of the P+1 loop (residues 965–972) has to occur during catalysis to allow efficient phosphorylation of substrates by Bub1.
We next tested whether this part of the P+1 loop in Bub1 was required for efficient phosphorylation of Cdc20N. Deletion of residues 967–971 (Bub1 Δ967–971) reduced the kinase activity of Bub1 towards Cdc20N about 5 fold (Figure 4E, compare lanes 1 and 5), whereas the same mutant retained about 80% of its autokinase activity. Similar results were obtained with mutations of V974, which is located in helix αEF and just C-terminal to the P+1 loop (data not shown). Therefore, mutations in the C-terminal portion of the P+1 loop in Bub1 affects its kinase activity towards different substrates to varying degrees, suggesting that this region might be directly involved in substrate binding. We cannot, however, rule out the possibility that perturbation of the P+1 loop disrupts the structural integrity of the active site of Bub1, thus indirectly affecting substrate binding.
Taken together, Bub1 in our crystal structure is likely in the active state, but it is not optimized for substrate binding. How can the P+1 loop of Bub1 adopt a conformation suitable for substrate binding? We envision two possible mechanisms: (1) pre-organization and (2) induced fit. In the pre-organization mechanism, posttranslational modifications of Bub1 or binding of other proteins pre-organize the P+1 loop into an optimal conformation for substrate binding. In the induced fit mechanism, binding of substrate induces a conformational change in the P+1 loop. The vertebrate Bub1 proteins contain only three conserved phosphorylatable residues in the activation segment: T960, T968, and S969 (Figure S2). Mutations of T960 and S969 to alanines did not affect the kinase activity of Bub1 towards Cdc20N (Figure 4E). By contrast, the T968A mutation greatly diminished the kinase activity of Bub1 towards Cdc20N (data not shown). Using mass spectrometry analysis, we have mapped multiple phosphorylation sites on the endogenous Bub1 protein isolated from mitotic HeLa cell lysates (J. Kang and H. Yu, unpublished results). T968 was not one of the phosphorylation acceptor residues identified in Bub1. In fact, none of the phosphorylation sites are located in the kinase domain. It is thus unclear which mechanism Bub1 uses to organize its P+1 loop. Regardless of the mechanism, the unusual conformation of the P+1 loop in Bub1 is expected to restrict the access of nonspecific substrates to its active site and enhance the substrate specificity of Bub1.
The KEN Boxes of Bub1 Bind to Cdc20
The sub-optimal conformation of the P+1 loop in Bub1 prompted us to search for additional mechanisms that might promote substrate binding to Bub1. Cdc20 is the only known physiologically relevant substrate of Bub1 (Tang et al., 2004). To examine whether the N-terminal non-kinase region of Bub1 enhanced phosphorylation of Cdc20, we performed kinase assays with Bub1C or the full-length Bub1 as the kinase and Cdc20N or full-length Cdc20 as the substrate. Full-length recombinant Bub1 phosphorylated both full-length Cdc20 and Cdc20N efficiently (Figure 5A). In contrast, Bub1C only phosphorylated Cdc20N, but not full-length Cdc20, suggesting that the N-terminal region of Bub1 was required for the efficient phosphorylation of full-length Cdc20. This finding further suggests that the Bub1-phosphorylation site(s) are masked in full-length Cdc20, but are accessible in Cdc20N.
Figure 5. The KEN Boxes of Bub1 Recruit Cdc20 for Efficient Phosphorylation at a Critical Site.
(A) In vitro kinase assays of full-length or C-terminal Bub1. Recombinant full-length or C-terminal Bub1 were assayed with the full-length (Cdc20) or N-terminal fragment of Cdc20 (Cdc20N) as substrates. Anti-Bub1 blot of the Bub1 proteins and Coomassie staining of the Cdc20 proteins are also shown. The kinase activities are quantified, normalized to the amount of kinase, and presented as relative values.
(B) Lysates of nocodazole-treated HeLa cells transfected with the indicated Myc-Bub1 plasmids were immunoprecipitated with anti-Myc. The lysates and anti-Myc immunoprecipitates (IP) were blotted with anti-Myc (top panel) and anti-Cdc20 (bottom panel).
(C) IP-kinase assays of Myc-Bub1 wild-type (WT), kinase-dead (KD), and ΔKEN. Both Cdc20N and full-length Cdc20 were used as substrates. The Cdc20 reactions were also blotted with an antibody against phospho-S153 (pS153) in Cdc20. The kinase activities are quantified, normalized to the amount of kinase, and presented as relative values (± standard deviations).
(D) Inhibition of Cdc20 phosphorylation by the Bub1520–653 fragment. Full-length Bub1 was assayed with the full-length (Cdc20) or N-terminal fragment of Cdc20 (Cdc20N) as the substrate. Synthetic Bub1 KEN-box peptides or recombinant Bub1520–653 were included in the kinase reactions. The KEN1/2-WT lane contained a mixture of the Bub1 KEN1 (residues 530–548) and KEN2 (residues 620–638) peptides each at a concentration of 140 µM. The KEN1/2-control lane contained two mutant peptides, KEN1-AAA with the KEN motif mutated to alanines and KEN2-rev with the reverse sequence of KEN2, each at a concentration of 140 µM. The Bub1520–653 lane contained 4 µM of purified recombinant His6-Bub1520–653. The kinase activities are quantified, normalized to the amount of kinase, and presented as relative values.
(E) Model for the recruitment of Cdc20 by the KEN boxes of Bub1. A structural model of the WD40 repeats of Cdc20 is shown in the ribbon diagram. The C-terminal IR box required for the binding of Cdc20 to APC/C is also indicated.
We next constructed plasmids encoding three Myc-tagged Bub1 fragments that spanned its non-kinase region (Figure S4A). HeLa cells were transfected with these plasmids and treated with nocodazole. The cell lysates were immunoprecipitated with anti-Myc. Both lysates and immunoprecipitates were blotted with anti-Cdc20. Only Bub1482–723 interacted with the endogenous Cdc20 (Figure S4B). Two recent studies have established that KEN boxes of Mad3 bind to Cdc20, block substrate binding to Cdc20, and inhibit APC/CCdc20 (Burton and Solomon, 2007; King et al., 2007). We have confirmed that binding of BubR1 (the human ortholog of Mad3) to Cdc20 requires the KEN boxes of BubR1 (B. Li and H. Yu, unpublished results). Intriguingly, Bub1 also contains two KEN boxes, which mediate its ubiquitination by APC/CCdh1 (Qi and Yu, 2007). Bub1482–723 contains both KEN boxes of Bub1 (Figure 5B). We next tested whether the KEN boxes of Bub1 were required for Cdc20 binding. Deletion of either KEN box in Bub1482–723 weakened its binding to Cdc20 whereas deletion of both KEN boxes abolished Cdc20 binding (Figure 5A). Our results thus indicate that Bub1 contains KEN boxes in its non-kinase region, which mediate Cdc20 binding. Although binding between the endogenous BubR1 and Cdc20 proteins can be readily detected in HeLa cell lysates, we have failed to detect binding between endogenous Bub1 and Cdc20 (data not shown), suggesting that the Bub1–Cdc20 interaction is transient.
The KEN Boxes of Bub1 Are Required for Phosphorylation of a Critical Site in Cdc20
Are the KEN boxes of Bub1 required for efficient phosphorylation of Cdc20? To answer this question, we first determined the major Bub1 phosphorylation site(s) in Cdc20. We had previously mapped six Bub1 phosphorylation sites at the N-terminal region of Cdc20 (Figure S5A) (Tang et al., 2004). Mutation of all six sites to alanines (Cdc20N6A) abolished phosphorylation by Bub1C (Figure S5B). Re-introduction of the S153 site into Cdc20N6A (Cdc205A S153) restored phosphorylation by Bub1C (Figure S5B). This finding indicates that S153 is a major Bub1 phosphorylation site in Cdc20. We next introduced the S153A mutation into full-length Cdc20 and tested whether this mutant was refractory to Bub1 inhibition in an APC/C ubiquitination assay. The ubiquitin ligase activity of APC/C in complex with wild-type (WT) Cdc20 was greatly reduced by Bub1 (Figure S5D, compare lanes 2 and 3). By contrast, Cdc20 S153A was refractory to Bub1 inhibition (Figure S5D, compare lanes 4 and 5). Therefore, S153 is a critical Bub1 phosphorylation site in Cdc20 in vitro. The functional importance of the S153 site in the spindle checkpoint remains to be established, although ectopic expression of a Cdc20 mutant with S153 and five other N-terminal phosphorylation sites mutated to alanines causes defects in the spindle checkpoint (Tang et al., 2004).
We then raised an antibody that selectively detected phospho-S153 of Cdc20 in vitro (Figure S5C). Bub1 WT phosphorylated Cdc20N and full-length Cdc20 efficiently at S153, based on 32P incorporation and anti-pS153 blots (Figure 5C). By contrast, Bub1 ΔKEN only weakly phosphorylated S153 in full-length Cdc20 (Figure 5C). As controls, Bub1 ΔKEN phosphorylated itself as efficiently as did Bub1 WT. In addition, phosphorylation of Cdc20N (which lacked the ability to bind to KEN boxes) by Bub1 ΔKEN was only slightly reduced, as compared to Bub1 WT. This result implicates a role of the KEN boxes of Bub1 in efficient phosphorylation of the full-length Cdc20 at a site critical for APC/C inhibition.
To better demonstrate the role of KEN boxes in mediating the Bub1–Cdc20 interaction, we tested whether phosphorylation of Cdc20 by Bub1 was inhibited by synthetic peptides containing the KEN boxes of Bub1 (Figure 5D). Addition of both KEN-box peptides did not inhibit Bub1-mediated phosphoryation of Cdc20 or Cdc20N. However, addition of a 134-residue fragment of Bub1 (Bub1520–653) containing both KEN boxes greatly diminished the phosphorylation of full-length Cdc20, but not Cdc20N (Figure 5D). This finding supports a role of this KEN-box-containing region of Bub1 in Cdc20 recruitment. The failure of isolated KEN-box peptides to inhibit Bub1 could be due to the fact that Bub1 contains two KEN boxes, which might engage in multivalent binding to Cdc20. Alternatively, additional motifs near the KEN boxes are required for efficient Cdc20 binding by Bub1.
Our findings are consistent with the following mode of interaction between Bub1 and Cdc20 (Figure 5E), which suggests the regulation of the specificity of Bub1 at two levels. First, the P+1 loop in the kinase domain of Bub1 interacts specifically with residues adjacent to the phosphorylation site, such as S153 in Cdc20, and determines the sequence specificity of Bub1 phosphorylation at the local level. Second, the KEN boxes of Bub1 and possibly its surrounding residues bind to the WD40 repeats of Cdc20 and recruit Cdc20 to Bub1. The close proximity between the active site of Bub1 and the N-terminal Bub1 phosphorylation motif in Cdc20 enables efficient phosphorylation of S153 in Cdc20.
The KEN Boxes of Bub1 Are Required for the Spindle Checkpoint
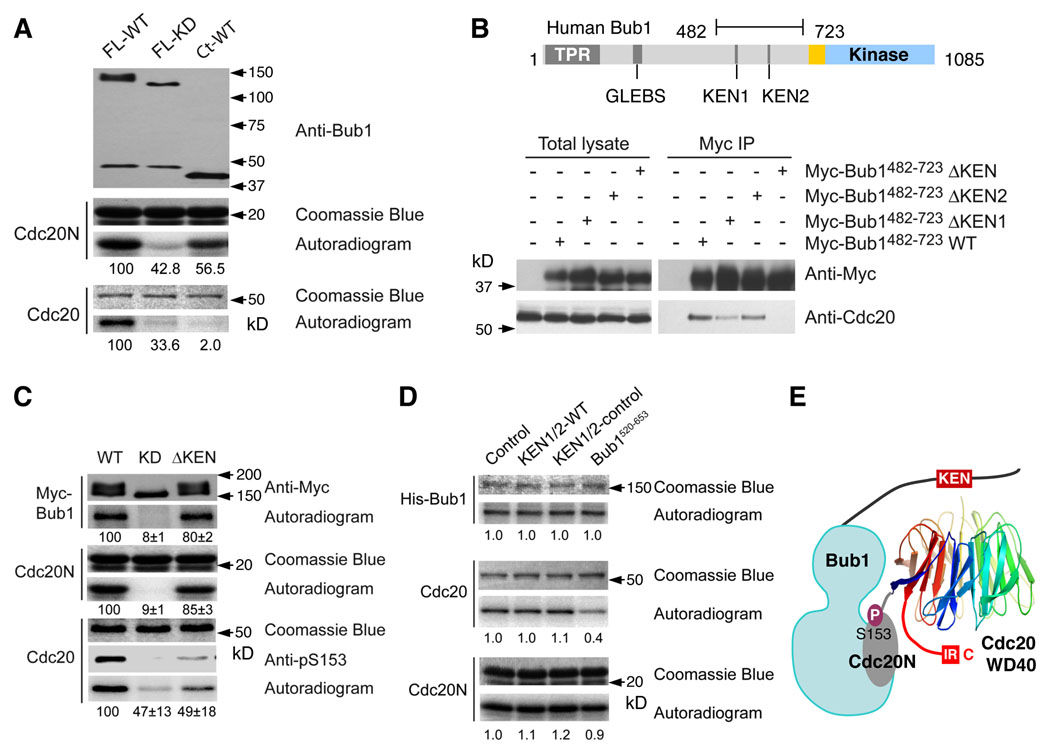
To determine the functional importance of the KEN boxes of Bub1, we generated HeLa cell lines stably expressing RNAi-resistant mCherry-Bub1 WT or ΔKEN, depleted the endogenous Bub1 protein using RNAi, and treated these cells with Taxol (Figure 6A). Ectopic expression of mCherry-Bub1 WT largely rescued the mitotic arrest deficiency of Bub1 RNAi cells in the presence of Taxol (Figure 6B and 6C). mCherry-Bub1 ΔKEN was significantly less effective in restoring Taxol-triggered mitotic arrest in these cells. Therefore, the KEN boxes of Bub1 are required for the spindle checkpoint in human cells.
Figure 6. The KEN Boxes of Bub1 Are Required for the Spindle Checkpoint.
(A) HeLa tet-on cells stably expressing RNAi-resistant mCherry-Bub1 wild-type (WT) or ΔKEN were transfected with Bub1 siRNA and then treated with Taxol. The cell lysates were blotted with the indicated antibodies. The positions of mCherry-Bub1 and the endogenous (Endo.) Bub1 are indicated.
(B) Cells described in (A) were incubated with Hoechst 33342 for 30 min and visualized using an inverted fluorescence microscope. The DNA morphology is shown in the left panels. The overlay of DNA in cyan and differential interference contrast (DIC) in red is shown in the right panels.
(C) The mitotic indices of the cells described in (A) and (B). About 1000 cells from three independent experiments were counted. The averages and standard deviations are shown.
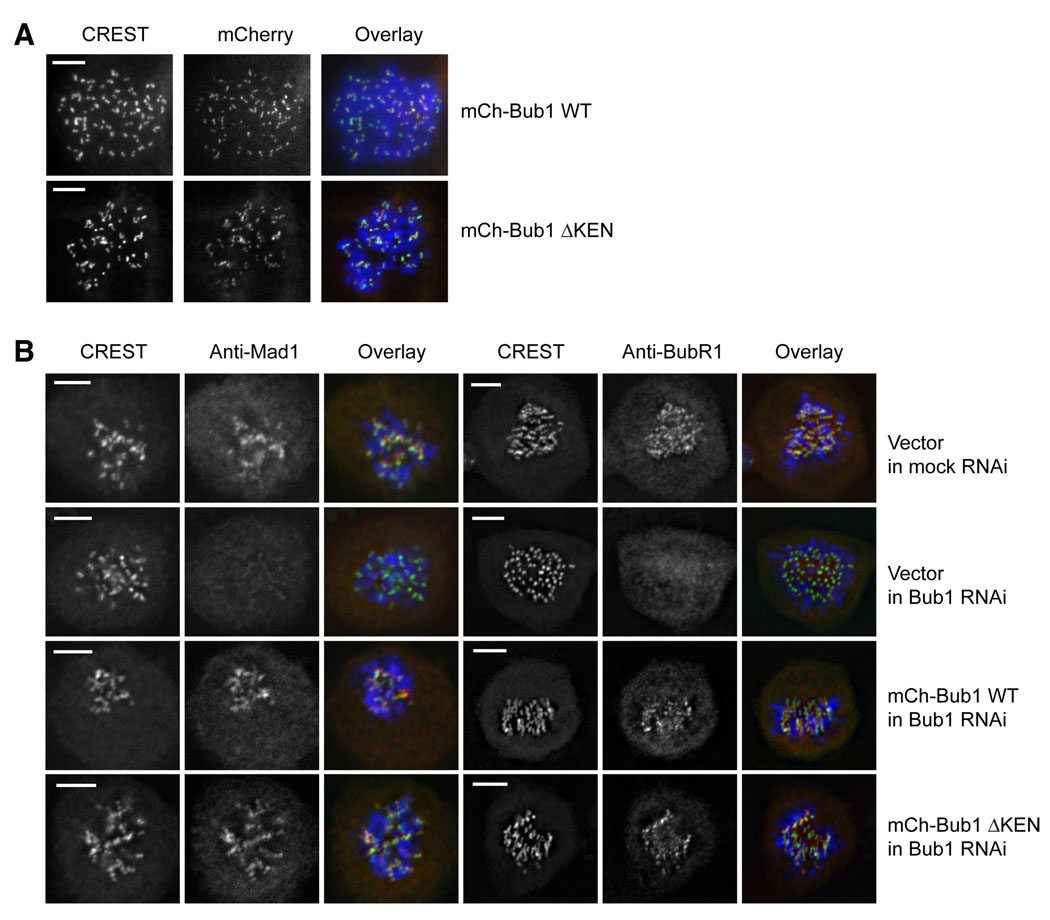
Bub1 has two known functions in the spindle checkpoint: (1) direct inhibition of APC/CCdc20 through phosphorylation of Cdc20 (Tang et al., 2004); and (2) recruitment of other checkpoint proteins such as BubR1, Mad1, and Mad2 to kinetochores (Qi et al., 2006; Sharp-Baker and Chen, 2001; Vigneron et al., 2004; Yu and Tang, 2005). Though our antibody against Cdc20 phospho-S153 could detect this phosphorylation event on purified recombinant Cdc20, it failed to reliably detect the endogenous Cdc20 phosphorylated at S153, presumably due to its low avidity. This technical difficulty prevented us from testing directly whether phosphorylation of Cdc20 at S153 was defective in mCherry-Bub1 ΔKEN-expressing cells. On the other hand, similar to mCherry-Bub1 WT, mCherry-Bub1 ΔKEN retained its kinetochore localization in mitosis (Figure 7A). Expression of mCherry-Bub1 ΔKEN in Bub1 RNAi cells restored the kinetochore localization of Mad1 and BubR1 (Figure 7B). Therefore, mutation of the Bub1 KEN boxes does not affect the kinetochore targeting of itself and other downstream checkpoint components. These findings further suggest that the checkpoint defect in mCherry-Bub1 ΔKEN-expressing cells is caused by the inefficient phosphorylation of Cdc20 by this Bub1 mutant.
Figure 7. The KEN Boxes of Bub1 Are Not Required for the Kinetochore Localiation of BubR1 and Mad1.
(A) HeLa tet-on cells stably expressing mCherry-Bub1 wild-type (WT) or ΔKEN were fixed and stained with CREST (green in overlay) and DAPI (blue in overlay). The scale bars indicate 5µm.
(B) HeLa tet-on cells stably expressing RNAi-resistant mCherry-Bub1 wild-type (WT) or ΔKEN were transfected with Bub1 siRNA, fixed, and stained with anti-Mad1 (three left panels, red in overlay) or anti-BubR1 (three right panels, red in overlay). The cells were co-stained with CREST (green in overlay) and DAPI (blue in overlay). The scale bars indicate 5µm.
Docking Interactions between Kinases and Substrates
Docking interactions between non-kinase regions of kinases and their protein substrates are common and promote phosphorylation of substrates by increasing the local concentration of phosphoacceptor residues, among other mechanisms (Figure S6) (Remenyi et al., 2006). For example, the Src family tyrosine kinases contain SH3 and SH2 domains that bind to proline-rich and phospho-tyrosine-containing docking motifs on certain substrates, respectively (Lim, 2002). The polo-like kinase (Plk1) uses its polo-box domain (PBD) to recognize phospho-S/T motifs that are created by priming kinases (Lowery et al., 2005). This interaction promotes efficient phosphorylation of Plk1 substrates at other sites. We have now shown that efficient phosphorylation of Cdc20 by Bub1 requires a docking interaction between Cdc20 and the KEN boxes of Bub1. Therefore, the Cdc20–KEN box interaction likely promotes Cdc20 phosphorylation by Bub1, in ways analogous to the binding between SH3 and proline-rich peptides.
There is an important difference between the docking interaction involving Bub1 and Cdc20 and the docking interactions of other kinases (Figure S6). In Src, Plk1, and numerous other cases, the kinases contain the receptors for the docking motifs, which are located in the substrates. In the case of Bub1–Cdc20 interaction, the kinase Bub1 contains the docking motifs (KEN boxes), whereas the substrate Cdc20 is the receptor for KEN boxes. Obviously, both types of arrangements are functionally equivalent in promoting substrate phosphorylation. The unusual configuration of the Bub1–Cdc20 docking event does, however, provide important clues about the substrate specificity of Bub1. To engage the KEN boxes of Bub1, protein substrates of Bub1 need to contain domains that can serve as KEN-box receptors. While many proteins may contain short peptide motifs that can bind to SH3, PBD, and other docking motif receptors, very few proteins are expected to contain receptors for KEN boxes. In fact, only Cdc20 and Cdh1 are known KEN-box receptors (Yu, 2007). Cdh1 is not efficiently phosphorylated by Bub1 (Tang et al., 2004), presumably due to the lack of an optimal phosphoacceptor residue in Cdh1 that can interact with the P+1 loop of the kinase domain of Bub1. Although we cannot rule out the possibility that Bub1 uses different mechanisms to promote phosphorylation of other substrates, it is clear that Bub1 is exquisitely tuned to phosphorylate Cdc20 through the docking interaction between Cdc20 and the KEN boxes in Bub1.
Implications for Spindle Checkpoint Signaling
Multiple checkpoint mechanisms inhibit APC/CCdc20, including binding of Mad2 to Cdc20, binding of Mad3/BubR1 to Cdc20, and phosphorylation of Cdc20 by Bub1 (Yu, 2007). Although each of the three proteins on its own is able to inhibit APC/CCdc20 in vitro, Mad2, BubR1, and Bub1 must cooperate with each other in human cells because depletion of each protein alone by RNAi causes checkpoint defects (Meraldi et al., 2004; Tang et al., 2004). The spatiotemporal coordination of these APC/C-inhibitory checkpoint mechanisms in vivo remains unclear.
Recently, it has been shown that Mad3 contains KEN boxes, which bind to Cdc20 and inhibit APC/CCdc20 through competing with substrate binding (Burton and Solomon, 2007; King et al., 2007). We have shown herein that Bub1 uses its KEN boxes to recruit Cdc20 for efficient phosphorylation. This finding suggests that binding of BubR1 to Cdc20 and phosphorylation of Cdc20 by Bub1 are competing events and need to be coordinated in cells. Two possible ways of coordination can be envisioned. In one way, phosphorylation of Cdc20 by Bub1 may need to occur prior to BubR1 binding to Cdc20, because BubR1 likely has a higher affinity towards Cdc20. Bub1-mediated phosphorylation of Cdc20 may then promote binding of BubR1 and Mad2 to Cdc20. Alternatively, because Bub1 can more efficiently phosphorylate Cdc20 that is not bound to BubR1, Bub1 and BubR1 may inhibit different pools of Cdc20. Experiments aimed at testing these hypotheses will advance our understanding of the spindle checkpoint.
CONCLUSION
In this study, we have determined the structure of the extended kinase domain of Bub1, identified a chemical inhibitor of Bub1, and uncovered a role of the KEN boxes of Bub1 as docking motifs for Cdc20. Our combined structural, biochemical, and functional analyses indicate that Bub1 is a specific kinase optimized for Cdc20 phosphoryation. The expected competition between Bub1 and BubR1 in Cdc20 recognition further suggests testable models for the spatiotemporal coordination of multiple APC/C-inhibitory mechanisms during spindle checkpoint signaling in living cells.
EXPRIMENTAL PROCEDURES
Protein Expression and Purification
Recombinant baculoviruses encoding an N-terminal His6-tagged C-terminal fragment of human Bub1 (Bub1C) containing residues 724–1085 were constructed using the Bac-to-Bac system (Invitrogen) according to manufacturer’s protocols. Sf9 cells were infected with the Bub1C baculovirus and harvested about 50 hrs post-infection. Cells were resuspended in lysis buffer (25 mM phosphate pH 7.4, 150 mM KCl, 20 mM imidazole, 0.1% (v/v) Triton X-100, 1 mM PMSF, 5 mM DTT, and a protease inhibitor cocktail). After sonication and centrifugation, the supernatant was filtered through 0.45 µm filters and passed through a 5-ml HisTrap Chelating HP column (GE Healthcare). After washing with the wash buffer (25 mM phosphate pH 7.4, 150 mM KCl, 20 mM imidazole, 1 mM PMSF, and 5 mM DTT), the bound proteins were eluted with a linear gradient (0–70%) of the elution buffer (25 mM phosphate pH 7.4, 300 mM KCl, 300 mM imidazole, 1 mM PMSF, 5 mM DTT). The His6-Bub1C-containing fractions were pooled, desalted and purified using ion exchange chromatography (resource S 5/5 and resource Q 5/5, GE Healthcare). His6-Bub1C was incubated with tobacco etch virus (TEV) protease and passed through a HisTrap Chelating HP column (GE Healthcare). Bub1C was further purified by a gel filtration column (Superdex 200, GE Healthcare), concentrated to 15 mg/ml in the purification buffer (50 mM Tris pH 7.7, 100 mM KCl, 5 mM MgCl2, 5 mM DTT, and 1 mM PMSF), flash frozen in liquid nitrogen, and stored at −80°C for crystallization or kinase assays.
The seleno-methionine variant of Bub1C protein was expressed in Sf9 cells as described (Zhou et al., 2004). Briefly, Sf9 cells cultured in the EX-420 medium (JRH Bioscience) were infected with the Bub1C baculovirus. The cells were harvested at 20 hrs post-infection, washed twice with the EX-420 medium without L-methionine, and resuspended into this medium. The cells were incubated for 6 hrs to deplete the remaining L-methionine, harvested again, and resuspended into the EX-420 medium supplied with 100 mg/l seleno-methionine. After an additional 24–30 hrs, the cells were harvested and the seleno-methionine Bub1C protein was purified as described above.
Crystallization, Data Collection, and Structure Determination
The Bub1C protein was thawed in ice-cold water, and ATP was added to a final concentration of 10 mM. Crystals were grown at 20° C by the vapor diffusion method in sitting drop mode by mixing 1.5 µl protein with 1.5 µl crystallization solution (20% (w/v) polyethylene glycol 3350, 0.1 M sodium formate (pH 6.29), and 25 mM DTT) and equilibrating against 200 µl of reservoir solution. Large single crystals were obtained by repeated seeding. The crystals were cryo-protected in reservoir solution supplemented with 18% (v/v) glycerol and then flash-cooled in liquid propane. The crystals exhibit the symmetry of space group C2221 with cell dimensions of a = 109 Å, b = 148 Å, c = 47 Å, and contained one molecule of Bub1C in the asymmetric unit with a solvent content of 46%. The seleno-methionine-derivatized Bub1C crystals were obtained in the same way and had similar cell parameters.
Diffraction data were collected at beamline 19-ID (SBC-CAT) of the Advanced Photon Source (Argonne National Laboratory, Argonne, Illinois, USA) and processed with HKL2000 (Otwinowski and Minor, 1997). Native and seleno-methionine-derivatized crystals diffracted to a minimum Bragg spacing of about 2.3 Å and about 3.5 Å, respectively (Table S1). All crystals showed significant anisotropic diffraction. Single-wavelength anomalous diffraction data on the seleno-methionine Bub1C crystals were collected at 3.2 Å with weak anomalous signal present to about 3.5 Å.
Phases for the seleno-methionine variant were obtained from a single anomalous dispersion (SAD) experiment. Using data to 3.5 Å, twelve of the 15 expected seleno-methionine sites were identified using SHELX (Schneider and Sheldrick, 2002; Sheldrick, 2002) and refined using SHARP (Bricogne et al., 2003). The resulting electron density map clearly showed several α-helices, which allowed placing the similar structures of the kinase domains of c-Jun, CK2, and twitchin (PDB codes: 1jnk, 1daw, and 1kob, respectively). Using these structures, the phases from the selenium-SAD experiment, and the known positions of twelve selenium atoms as guides, a model was constructed manually in an iterative fashion using the program XtalView and automatically rebuilt and refined with the programs ARP/wARP and Refmac5 of the CCP4 package (Consortium, 1994; Murshudov et al., 1997; Perrakis et al., 1999). Completion of the model and final refinement were then carried out using the native data to 2.32 Å. The final model contains residues 735–806, 814–931, 939–1080 of Bub1C, one molecule Mg2+-ATP, two chloride ions and 20 water molecules. After final refinement, the model had an Rwork of 23.2% and an Rfree of 30.2%. The model has good geometry, except for three residues that are outliers in a Ramachandran plot as defined by MolProbity (Davis et al., 2007). One of them, D946, is a direct ligand of the bound ATP. Its electron density is well defined. The other two residues, P776 and T778, are located in a surface loop with weak electron density.
Cell Culture and Transfection
HeLa tet-on cells were cultured in DMEM (Invitrogen) supplemented with fetal bovine serum. The Bub1 ΔKEN1 (with K535, E536, and N537 mutated to alanines), Bub1 ΔKEN2 (with K625, E626, and N627 mutated to alanines), and Bub1 ΔKEN (with both KEN motifs mutated to alanines) mutants were constructed using the QuikChange mutagenesis kit (Qiagen). Plasmid transfections were performed using the Effectene reagent (Qiagen). To establish stable cell lines, HeLa tet-on cells were transfected with the pIRES-mCherry-Bub1 WT or ΔKEN plasmids and incubated with 0.5 µg/ml puromycin. Surviving clones that expressed mCherry-Bub1 WT or ΔKEN at similar levels were transfected with Bub1 siRNA for 24 hr and treated with 100 nM Taxol for 18 hr. After a 30-min incubation with Hoechst 33342, the cells were analyzed using an inverted fluorescence microscope. Round cells with condensed chromosomes were counted as mitotic cells.
Kinase Assays
To determine the IC50 values of 2OH-BNPP1 for Bub1, Bub1C, Aurora B and p38, about 0.1 µg of various kinases were incubated with 2 µg of substrates for 30 min at RT in 25 µl of kinase buffer I (50 mM Tris, pH 7.5, 0.2 M NaCl, 1 mM DTT, 0.1 mM ATP, 0.1 µCi/µl γ-32P-ATP) containing various concentrations of 2OH-BNPP1. Reaction mixtures were quenched with SDS sample buffer, separated on SDS-PAGE, and analyzed using a phosphoimager. The Immunoprecipitation (IP) kinase assays were performed as described previously (Kang et al., 2007).
Immunofluorescence (IF)
For Bub1 localization, HeLa tet-on cells stably expressing mCherry-Bub1 WT or ΔKEN were grown in 6-well plates, treated with Taxol for 18 hr, and sedimented on slides using Shandon cytospin 4 (Thermo electron). The cells were fixed in 4% (w/v) paraformaldehyde for 10 min and incubated with the appropriate primary antibodies (1 µg/ml) in IF buffer I (PBS with 0.1% (v/v) Triton X-100) containing 3% (w/v) BSA for 2 hr. The cells were further washed with IF buffer I, incubated with the appropriate FITC-, Cy3-, or Cy5-conjugated secondary antibodies (4 µg/ml, Molecular Probes) in IF buffer I containing 3% (w/v) BSA for 1 hr, washed again with IF buffer I containing 1 µg/ml DAPI, and analyzed using a 63X objective on a Zeiss Axiovert 200M inverted fluorescence microscope. The images were acquired with the Intelligent Imaging software and processed with Photoshop.
For Mad1 and BubR1 localization, HeLa tet-on cells were grown in 4-well chamber slides, permeabilized with IF buffer II (60 mM PIPES, pH 7.0, 20 mM HEPES, pH 7.5, 5 mM EGTA, pH 8.0, 2 mM MgCl2, and 4 M glycerol) containing 0.2% (v/v) Triton, fixed in IF buffer II containing 4% (w/v) paraformaldehyde and 0.05% (v/v) glutaraldehyde for 10 min, and incubated with the appropriate primary antibodies (1 µg/ml) in IF buffer I containing 3% (w/v) BSA for 2 hr. The cells were then processed and analyzed as described above.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Sheng Ye for assistance with structure determination. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by University of Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research. This work is supported by the National Institutes of Health (GM61542 and GM76481 to H.Y. and AI44009 to K.M.S.), the W. M. Keck Foundation, the March of Dimes Foundation, the Welch Foundation, and the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The atomic coordinates of Bub1C have been deposited in the Protein Data Bank with the accession code 3E7E.
REFERENCES
- Alaimo PJ, Shogren-Knaak MA, Shokat KM. Chemical genetic approaches for the elucidation of signaling pathways. Curr. Opin. Chem. Biol. 2001;5:360–367. doi: 10.1016/s1367-5931(00)00215-5. [DOI] [PubMed] [Google Scholar]
- Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23:2016–2027. doi: 10.1038/sj.onc.1207374. [DOI] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH. Phosphorylation and activation of Bub1 on unattached chromosomes facilitate the spindle checkpoint. EMBO J. 2004;23:3113–3121. doi: 10.1038/sj.emboj.7600308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TC. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Martinez LA, Yu H. Running on a treadmill: dynamic inhibition of APC/C by the spindle checkpoint. Cell Div. 2007;2:23. doi: 10.1186/1747-1028-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeschke A, Karasarides M, Ventura JJ, Ehrhardt A, Zhang C, Flavell RA, Shokat KM, Davis RJ. JNK2 is a positive regulator of the cJun transcription factor. Mol. Cell. 2006;23:899–911. doi: 10.1016/j.molcel.2006.07.028. [DOI] [PubMed] [Google Scholar]
- Kang J, Chen Y, Zhao Y, Yu H. Autophosphorylation-dependent activation of human Mps1 is required for the spindle checkpoint. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20232–20237. doi: 10.1073/pnas.0710519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EM, van der Sar SJ, Hardwick KG. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS ONE. 2007;2:e342. doi: 10.1371/journal.pone.0000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Obuse C, Yanagida M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 2007;13:663–676. doi: 10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Lim WA. The modular logic of signaling proteins: building allosteric switches from simple binding domains. Curr. Opin. Struct. Biol. 2002;12:61–68. doi: 10.1016/s0959-440x(02)00290-7. [DOI] [PubMed] [Google Scholar]
- Lowery DM, Lim D, Yaffe MB. Structure and function of Polo-like kinases. Oncogene. 2005;24:248–259. doi: 10.1038/sj.onc.1208280. [DOI] [PubMed] [Google Scholar]
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr.D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Papa FR, Zhang C, Shokat K, Walter P. Bypassing a kinase activity with an ATP-competitive drug. Science. 2003;302:1533–1537. doi: 10.1126/science.1090031. [DOI] [PubMed] [Google Scholar]
- Pavletich NP. Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J. Mol. Biol. 1999;287:821–828. doi: 10.1006/jmbi.1999.2640. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Qi W, Tang Z, Yu H. Phosphorylation- and polo-box-dependent binding of Plk1 to Bub1 is required for the kinetochore localization of Plk1. Mol. Biol. Cell. 2006;17:3705–3716. doi: 10.1091/mbc.E06-03-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Yu H. KEN-box-dependent degradation of the Bub1 spindle checkpoint kinase by the anaphase-promoting complex/cyclosome. J. Biol. Chem. 2007;282:3672–3679. doi: 10.1074/jbc.M609376200. [DOI] [PubMed] [Google Scholar]
- Remenyi A, Good MC, Lim WA. Docking interactions in protein kinase and phosphatase networks. Curr. Opin. Struct. Biol. 2006;16:676–685. doi: 10.1016/j.sbi.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Sharp-Baker H, Chen RH. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 2001;153:1239–1250. doi: 10.1083/jcb.153.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. Macromolecular phasing with SHELXE. Zeitschrift Fur Kristallographie. 2002;217:644–650. [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- Tanaka TU, Desai A. Kinetochore-microtubule interactions: the means to the end Curr. Opin. Cell Biol. 2008;20:53–63. doi: 10.1016/j.ceb.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- Tang Z, Shu H, Oncel D, Chen S, Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol. Cell. 2004;16:387–397. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Vigneron S, Prieto S, Bernis C, Labbe JC, Castro A, Lorca T. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol. Biol. Cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 2002;14:706–714. doi: 10.1016/s0955-0674(02)00382-4. [DOI] [PubMed] [Google Scholar]
- Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol. Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Yu H, Tang Z. Bub1 multitasking in mitosis. Cell Cycle. 2005;4:262–265. [PubMed] [Google Scholar]
- Zhou T, Raman M, Gao Y, Earnest S, Chen Z, Machius M, Cobb MH, Goldsmith EJ. Crystal structure of the TAO2 kinase domain: activation and specificity of a Ste20p MAP3K. Structure. 2004;12:1891–1900. doi: 10.1016/j.str.2004.07.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.