Abstract
In this report, the authors compare and contrast medical informatics (MI) and bioinformatics (BI) and provide a viewpoint on their complementarities and potential for collaboration in various subfields. The authors compare MI and BI along several dimensions, including: (1) historical development of the disciplines, (2) their scientific foundations, (3) data quality and analysis, (4) integration of knowledge and databases, (5) informatics tools to support practice, (6) informatics methods to support research (signal processing, imaging and vision, and computational modeling, (7) professional and patient continuing education, and (8) education and training. It is pointed out that, while the two disciplines differ in their histories, scientific foundations, and methodologic approaches to research in various areas, they nevertheless share methods and tools, which provides a basis for exchange of experience in their different applications. MI expertise in developing health care applications and the strength of BI in biological “discovery science” complement each other well. The new field of biomedical informatics (BMI) holds great promise for developing informatics methods that will be crucial in the development of genomic medicine. The future of BMI will be influenced strongly by whether significant advances in clinical practice and biomedical research come about from separate efforts in MI and BI, or from emerging, hybrid informatics subdisciplines at their interface.
With the Human Genome Project (HGP) published in draft form,1 new challenges in functional genomics have followed hard on its heels, opening up a wide variety of medical applications. Altman,2 Sander,3 and others4,5,6,7,8 have recently emphasized the need for new genomic-based approaches in medicine, such as studying genome-related risk factors for various diseases, developing novel diagnostics tests, creating updated cancer cell classifications, or integrating genetic and medical data in clinical practice. Both bioinformatics (BI) and medical informatics (MI) are widely expected to have important roles in supporting these types of efforts, but whether they will do so together or apart is a matter of debate among researchers in both disciplines.4
Bioinformatics involves the development and application of novel informatics techniques in the biological (especially genomic) sciences. It is a young, successful discipline, which already has its own professional societies, meetings, and scientific journals focused on a clear research agenda, having contributed critically to the successes of the human and other genome projects. In contrast, MI is a more established field that has pioneered the development and introduction of informatics methods in clinical medicine and biomedical research but has recently found itself increasingly challenged by the emergence of BI.9
New approaches in MI have been called for to bridge the divide with BI in developing novel methods for the emerging joint field of biomedical informatics (BMI).10,11,12 This raises an interesting set of strategic issues for BI and MI, focused around the question: Is enough known already for BI to productively focus on developing methods and tools for enhancing the transfer of genomic results to medicine, and what novel clinically oriented MI methods will be needed to make such transfers a success?
Relationship of BI and MI to Medical Research and Practice
Beginning in the late 1950s, the introduction of computers into medical settings was followed by the implementation of clinical and bibliographic databases, computerized medical records (CPRs),13,14 and medical information systems (MISs)15 during the next two decades, contributing to the rapid development of MI.16,17 Pioneering medical consultation systems, based first on logical and statistical methods, and later on expert knowledge-based methods,18,19,20,21,22,23,24,25 attracted considerable attention, had high impact in scientific journal publications, and served as models for similar research and many successful applications in other disciplines. Large clinical databases26,27 and literature indexing and search technologies developed by medical informatics researchers led to significant shifts in medical research and practice.
It is now common for physicians to use systems such as Medline28 in conjunction with MISs and CPRs without thinking twice and referring to them simply as “data sources.” Taking for granted these results of MI research is clear evidence of their success, but it tends to hide and underestimate the fact that it took more than 30 years to reach such a level of “seamless” technology. Medline is not just a database with millions of records. Its developer, the U.S. National Library of Medicine, has supported research on medical vocabularies, information retrieval, and natural language processing for more than three decades to improve the capabilities of Medline.28,29 Similarly, CPRs are not ordinary databases but incorporate research results on cognitive studies of physician–patient interaction, human interfaces, knowledge representation, system interoperability, and coding standards, among others30; they are much more than simple “data sources.”
Something similar may already be happening to BI, given the responses recently reported in a Bioinformatics journal editorial31 in which professionals outside the field are cited as considering BI research to be easy and cheap, yielding free software, and producing rapid publication of easily verified predictions. While most BI researchers might disagree with such opinions, the fact that they are widely held is likely to influence the future of the field just as similar opinions have influenced MI in the past. Many of the BI programs are complex software systems that use a mix of mathematical models and expert heuristics, which are hard to evaluate out of context and in general, but whose benefits are obvious for specific problems of biological inquiry and analysis.32,33 While it is impossible to anticipate exactly how and when BI results will substantially affect the practice of medicine, a careful look at the experiences of MI might help anticipate some of the technological and scientific challenges for clinical applications of genomics.
MI and BI—Complementarities for Genomic Medicine
A comparison between BI and MI can yield useful insights for planning the future of both disciplines. Within BI, some professionals have suggested that researchers from other areas, such as physicists or statisticians, should mainly contribute to BI tasks, given their expertise in dealing with the massive quantities of experimental data.34 Yet, maybe the most fruitful collaborations lie with the more proximal discipline of MI,2,4,5,6,35 which has been dealing with problems of structuring complex and large quantities of data and knowledge in biomedicine for the last 30 years.
In this report, we compare the disciplines from a number of different perspectives. ▶ summarizes the main points of this comparison. Below we briefly expand on the contrasting features of MI and BI summarized in ▶.
Table 1.
Selected Similarities and Differences between Medical Informatics (MI) and Bioinformatics (BI)
| Feature Comparison and Contrast |
||
|---|---|---|
| Discipline Feature | Bioinformatics | Medical Informatics |
| 1. Academic discipline, development, and interdisciplinary links. | Gradual build-up, focusing on science-driven methods of analysis for data from emerging gene and protein technologies. Early connections with cybernetics, information, and automata theory gave an information-oriented focus to genomics. | Rapid launch in the 1960s based on computer technology opportunities in health care. Few connections of medicial practice with “preinformatics” disciplines (mathematical modeling, statistics, linguistics, AI, etc.)—though fundamental for public health epidemiology, laboratory medicine, and radiology. |
| 2. Scientific content and informatics goals. | Molecular biology and biochemistry—massive data analysis and management demands for biological scientific discovery. | Medicine—standardization, efficient data processing and specialized analysis and management tools for clinical practice, biomedical research, and education. |
| 3. Data quality and analysis: noise and uncertainty. Role of data mining. | Scientific data—controlled noise and uncertainty models and prospective studies allow relatively frequent direct application of data mining methods. | Clinical data—other than for controlled clinical trials, the incompleteness, subjectivity, and uncertainty of retrospective patient data make mining results difficult to replicate and transfer. |
| 4. Integrating data and knowledge: networks, databases, interoperability, standardization, and information retrieval. | Many specialized databases publicly available over the Web. Interoperability good; quality issues addressed by increased curation. | Pioneering networking (Medline, SUMEX-AIM), documentation standards, vocabularies, and coding (e.g., UMLS, SNOMED, HL7). Clinical databases mostly nonpublic. |
| 5. Tools to support medical practice. | Recent emphasis on ontologies, network models, etc. No involvement in clinical applications until gene expression and SNP studies—still in early stages. | Emphasis on information systems for patient care, hospital, and medical information (HIS, MIS), education, and telemedicine. Representation of medical knowledge and language (i.e., terminology, indexing, disease taxonomies), central for information retrieval and decision support. |
| 6. Tools to support research and practice: signal processing, imaging and visualization, computational modeling. | Signal processing biophysics-oriented. Three-dimensional graphic visualization of molecular structures for scientific research. Image analysis of functional data and network modeling and simulation. | Signal, image processing, and computational modeling bioengineering-oriented. Three-dimensional visualization of radiologic, histologic, other imaging for clinical and educational use. Related to clinically useful (logic, coding, text, and graphics) knowledge representations and systems architectures. |
| 7. Professional and health consumer education. | Widely available tools for scientific data analysis. Prevention and personalized diagnosis and treatment seen as future promises. | Widely available Web sites, multimedia tools for accessing wide range of biomedical information, analysis tools less widely disseminated. |
| 8. Education and training of MI and BI professionals: role as scientists, engineers, and information brokers. | Recent creation of training programs, rapidly expanding. | Training programs from the 1970s, firmly consolidated—much interdisciplinary experience. |
Development of MI and BI as Academic Informatics Disciplines
Even before the introduction of computers into medical settings and biology laboratories, one can note some interesting differences between MI's and BI's underlying fields of study. Cybernetics, information theory, and automata theory, emerging in the 1940s and 1950s, introduced ideas critical to the foundations of computer science (CS) and informatics, which usually is taken to embrace CS, information technology (IT), and the information/library sciences more generally. Pioneers such as Wiener, Shannon, and von Neumann, were involved in the study of biological issues and contributed to the thinking that led to breaking the genetic code.36 Cryptographers were also involved in these efforts, using computers to carry out complex calculations to check the feasibility of theoretical proposals for the genetic code, which, while not directly successful, introduced a set of concepts and metaphors into genetics, such as “information,” “message,” and “code,” which were decisive in establishing the centrality of information in genetics through the central dogma of molecular biology.37 The practice of medicine, in contrast, had fewer connections with these or other “preinformatics” disciplines such as mathematics, statistics, and linguistics, with exceptions involving mathematical modeling in neurology and immunology and statistical analyses of radiologic images.38,39,40
When MI developed as an academic field in the late 1960s and early 1970s, it concentrated more on practical issues of clinical documentation, information systems, and technology in which computers had been introduced to increase productivity of medical processes41,42 more than for basic biology studies. Scientific inquiry did, nevertheless, provide a focus for work on medical knowledge representation and language understanding, implemented in software as pioneering statistical and artificial intelligence (AI) consultation and decision support systems of the 1960s and 1970s. It also led to foundational work on terminologies, coding, medical records, and indexing for Medline, since carried forward through the Unified Medical Language System29 and related research. It has been said that many MI pioneers arrived to the field by chance, because they were working on issues related to specific informational needs in various clinical settings,42 although complemented by academic specialists from linguistics, mathematics, computer science, engineering, and other disciplines.41,42
Bioinformatics experienced a more gradual development evolving from the individual efforts of researchers who helped analyze early deoxyribonucleic acid (DNA) and protein sequence data and, later, the macromolecular structural and functional data needed for genomic discovery. BI was able to build on work in computational biology involving a large number of mathematically oriented investigators and theoreticians.43 When BI emerged in the 1990s, many of its practitioners had already been working on traditional scientific tasks for which informatics methods and software became a practical means to an end, rather than being the primary focus, as in MI.
Scientific Content and Informatics Goals of MI and BI
While this topic has already been introduced above in contrasting the academic disciplines, it can be made more specific with the following observations. Biology has been transformed from a largely descriptive science to one grounded scientifically on theories and principles such as Darwin's theory of evolution, Mendel's laws, the central dogma of molecular biology, and the structure of DNA and its functions within cells and their anatomic, organismal, and environmental contexts. BI has emerged as the computational driving engine behind the analysis of massive data that support discovery science in genomics, proteomics, metabolomics, and the other biological “-omic” subfields. It is not surprising then, that BI is predominantly oriented to the support of research.
It is said often that the field of medicine is as much art as science.44 Although the field of medicine as a whole has seen dramatic advances in its scientific base during the last century (and last decades in particular), its focus on the management of individual patients means that it has yet to find a unified scientific framework for interpreting the great variety of evidence that such clinical practice entails. The large number of proposed paradigms for medicine—evidence-based medicine,45 molecular medicine,46 telemedicine,47 economic medicine,48 technology-driven care,49 and many others—illustrates the heterogeneity of scientific assumptions underpinning its different technologies, which advance unpredictably based on opportunities for improving the quality of individual patient care and, increasingly, containing their costs. The lack of unified theories in medicine may have also contributed to the extreme diversity of directions within MI, with very different subfields emphasizing medical practice, research, and educational applications. The area of informatics itself also covers a wide diversity of technologies and scientific approaches on how to model, represent, and manage information computationally, and there is as yet no unified underlying scientific theory for it any more than there is for medicine. MI's emphasis on understanding and developing practical medical information systems, thus, gets a double dose of breadth and diversity.
Data Quality and Analysis: Noise and Uncertainty
Medical data sets frequently are partial, noisy, and difficult to reproduce because of individual variability and the subjective nature of many clinical observations. Underlying medical knowledge often is uncertain, as shown in many MI studies of medical decision making.21,22,23,24,25 While biological data share, to a large measure, in all the above problems, they typically are gathered under more controlled conditions as the result of carefully designed experiments based on specific models of instrumentation, observation, and controllable replication.
In medicine, factors beyond the physiologic—e.g., psychological/cognitive, socioeconomic, ethical, geographical, environmental—introduce variables that are difficult to define and measure in clinical contexts. The very large range of variation in such factors, coupled with the fundamentally different reasoning problems18 involved in treating individual patients, may explain why many MI systems, such as decision support programs, frequently fail to generate similar or reproducible outcomes outside their institution of origin. Research in biology—and BI—has been laboratory centered with its own sources of variation, but these are more clearly differentiated according to whether they are “in vivo,” “in vitro,” or “in silico,” all recognized as generating different qualities of data, having differing levels of experimental control, and answering different types of questions with various degrees of evidential support.50
Integration of Data and Knowledge—Networks, Databases, Interoperability, and Information Retrieval
Medical informatics has made substantial contributions to data documentation, integration, and networking through the development of standards, medical terminologies, and coding systems (e.g., UMLS, SNOMED, HL7, MeSH) that have laid the foundation for research on data models and facilitated the interoperability of data and programs across heterogeneous computing platforms and systems. However, most clinical databases usually are special-purpose, restricted in access or private, and frequently not interconnected with other databases or the Web. Patient data are rarely freely exchanged and must be protected (usually a legal requirement) by security and confidentiality technologies and protocols. Despite this, a number of pioneering medical informatics projects have analyzed large clinical databases and used them to study patterns of longitudinal manifestations, tests, and treatments,26,27 and these have contributed to our understanding of medical data structures and analytical methodologies. More recently, medical informaticians have been in the forefront of developing ontology-building tools such as PROTÉGÉ,51 computerizing the major text on genetic components of disease52 and developing a highly detailed computational model for medical knowledge needed for the interpretation of increasingly complex and informative imaging modalities—the Foundational Model of Anatomy.53
Bioinformatics, in contrast, has only recently begun to work on developing ontologies, e.g., gene ontology (GO),54 and related initiatives. The collaborative efforts carried out within the HGP enhanced the exchange of genomic information as a means for accelerating research. Bioinformaticians have built databases for a variety of sequence, structure, and functional data,31 facilitating research efforts in genomics, proteomics, and all the other developing “-omic” technologies.
Given the shared concerns of genetics and medicine in understanding the mechanisms of organismal health and disease and their respective informatics methods and systems in BI and MI, one might expect that many standards, coding systems, and ontologies will have to be shared and updated jointly in the future. A case in point is the project in Iceland that is gathering genomic data from a largely genetically circumscribed population. Genomic data from such a controlled group of people should provide significant insight into relationships between genomic and phenotypic information. Researchers expect that this kind of analysis will yield new knowledge about the causes of various diseases and alternative therapeutic approaches.55,56
While this kind of work raises expectations, it also presents a serious challenge for molecular epidemiology, given the different types of data gathered under uncontrolled conditions and merged retrospectively in the database.
Tools to Support Medical Practice
Medical informatics professionals have engaged in many long-term efforts to develop systems for patient care, the management of hospital and other medical information,14,15,23 and its communication through telemedicine. Major concerns have been with practical issues of developing and understanding these systems and helping improve quality of care. They also contributed to develop the field of knowledge engineering, in which MI researchers did not intend to simulate human problem solving in a formal way57 but were able to capture expert reasoning heuristics with systems such as MYCIN,22 DIALOG24 (later INTERNIST-I), CASNET,21 and PIP,25 which introduced novel approaches to manage medical uncertainty using rule-based causal, hierarchical, and frame-based knowledge extracted from expert clinicians in their respective fields. AI approaches to scientific discovery in genomics seem to show other differences between MI and BI.58 While medical informaticians have had scientific goals of understanding how to model expert knowledge about individual decision-making practice and represent it in computer systems, they generally built practical applications that empirically supported their design hypotheses about both medical reasoning and related software engineering.
Work on digital medical libraries and the development of many reference texts and databases of biomedical information online following the pioneering example of Medline have revolutionized the routine way in which people can access and directly make use of the biomedical literature. Availability of OMIM online, the database for genetic causes of disease, provides one example of a bridge to research. Yet, connection to individual patient data is still almost entirely human-mediated. Search tools over the Web are increasingly sophisticated but still limited in their retrieval performance from text and even more so from image content.
BI has only recently become concerned with medical practice problems, as microarray gene expression data become increasingly available for incorporation into the medical record. Concerns about potential misuse of genetic data emphasize the importance of developing new combined BMI methods for ensuring their confidentiality without losing the opportunities for learning about genetic components of diseases through the aggregation of anonymized data.
Tools for Research and Practice: Signal, Image Processing, Visualization, Computational Modeling
Signal and image processing and analysis techniques have been used in MI and BI for many decades. From the perspective of methods and software techniques, no great differences exist between the disciplines, although BI has been closer to biophysical/chemical models of signal processing and analysis, while MI has been closer to the applied, biomedical engineering approaches to analysis and instrument/systems design. These involve devices such as electrocardiography, electroencephalography, computed tomography, and magnetic resonance imaging together with the informatics tools for signal analysis and decision support that are in routine use in most advanced clinical settings.
In BI, graphics modeling has been used widely for analyzing protein structures and building three-dimensional models of molecules. The objective of this research is focused on scientific inquiry, to understand better the connection of macromolecular structure and function. Once the structure of molecules or proteins were reconstructed in three dimensions, researchers can anticipate more precisely the functions of a molecule, which also is essential for drug discovery and design. In MI, different types of goals have predominated: two-dimensional and three-dimensional image analysis and reconstruction of images for diagnostic and treatment planning, and three-dimensional modeling (and virtual reality methods) for medical training and surgical simulation. Computational modeling of disease processes has been carried out in the context of consultation and decision-support systems and for qualitative description and explanation.59,60
Bioinformatics approaches may contribute new insights for MI research in computational modeling and analysis, while MI may contribute to advance research in medicine by building three-dimensional anatomic models that can be linked to specific functions and physiologic processes.
Professional and Patient Continuing Education
In contrast to MI professionals, those in BI have less occasion for teaching physicians or patients their informatics craft. While BI professionals may become involved in developing future health prevention and personalized therapy plans, these are rarely ready for deployment beyond the laboratory today.
One of the fundamental challenges for genomics to succeed in achieving significant results in medicine is to educate health practitioners, e.g., physicians and nurses, on its foundations and methods. Physicians will not easily change their reasoning approaches to think in terms of genetic and genomic information, and BI and MI should collaborate to develop methods to enhance this shift in clinical practice. Informatics tools may help by embedding the knowledge representation and problem-solving methods needed to better manage and combine genomic and clinical information. MI and BI developers can facilitate a shift from current practice to genomic medicine by creating user-friendly interfaces and advice systems that facilitate clinicians' work.
However, the fundamental challenge is to discover the essential new knowledge about natural and treated courses of illnesses for such technologies to be widely adopted. Such knowledge is centered around the elucidation of the connections between genotypes and phenotypes and the modeling of diseases by mathematical, logical, and semantic relationships that will provide both the ensemble predictive and explanatory power needed for scientific discovery. It also will provide some new type of prediction needed to make wise individual choices in a complex world of relationships between individuals at the cellular, organ, and whole organism levels and their multifaceted group and role memberships. Past experience suggests this is more likely to come from new observational/data sources, combined with powerful visualization and representation methods that make clinical significance direct and utility obvious and necessary. Informaticians who are also strong biomedical scientists—or biomedical scientists who are also strong informaticians—will be needed. Hybrids of both sorts are few, given the vast amount of knowledge that needs to be mastered, and this is the challenge that faces those involved in education in this new interdisciplinarity, as we discuss next.
BI and MI Education and Training
Given the professional differences that have evolved over time and the relatively small overlap of the MI and BI research communities, it is doubtful that a merging of the disciplines will happen easily or in the short term. Yet, a productive area of mixing and interchange may likely emerge even as they preserve their independence. Programs in biomedical informatics such as those at Stanford and Columbia involve common components with concentrations in either MI or BI. Because MI and BI often involve significant differences in background, specialization, and cognitive reasoning style in those who gravitate to and become successful in the two fields, only time will tell what types of combined programs will prove effective in providing good training for future leaders in MI and BI separately or the newly merged BMI discipline.
Cognitive studies of clinical reasoning have advanced over the past decade, but general models for describing it adequately have yet to emerge,61 and MI decision support systems are mainly empirically justified. In contrast, modern (postdescriptive) biologists appear to reason in a much more focused way based on scientifically grounded principles of molecular biology, genetics, and evolutionary theory, although they must also bring to bear considerable numbers of heuristics for each domain and type of problem.
In general, biologists have been exacting customers for computer applications, being usually directly involved in the efforts to develop the informatics tools that serve their needs of scientific discovery62 and, as such, more conscious of the need for specifications, data models, and software evaluation criteria. In contrast, physicians, on the whole, still tend to avoid analytical and computational methods and experience, which suggests that independent MI expertise will continue to be needed for developing informatics tools for future genomic-based medicine. MI and BI professionals need to be aware of these differences to systematize the development of informatics systems for medicine and biology.
Developing a Joint Biomedical Informatics Strategy: Toward “-omic” Medicine Based on Systems Biology and Informatics
Medical informatics and bioinformatics professionals have both made (separately) significant research contributions in areas such as knowledge-based systems, database design, data mining, sequence and structure analysis, and image processing.
There are similarities between the fields that bear more exploration. Through our analysis we have seen the potential that both disciplines pose for interaction. Not only do they share many interests, methods, and tools, but each also presents complementary needs and opportunities to the other. The following is a brief analysis exploring the possibilities of synergistic interactions between MI and BI based on their contributions and strengths in differing subfields of research and practice.
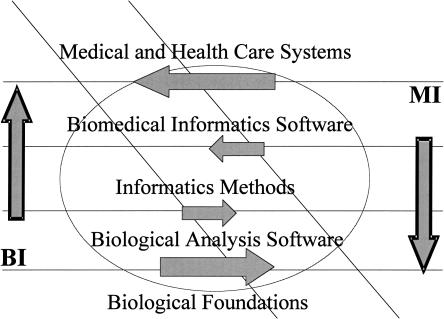
▶ illustrates a graphic metaphor of these potential interactions. It represents the relative strengths of BI and MI in dealing with biological foundations to expertise in building practical medical and more general (consumer Web-based, telemedicine) health care applications. It reflects how MI (on the right side), has historically emphasized the medical/health care system's design and implementation shown in the uppermost stratum of the diagram, leading to considerable development of biomedical informatics software over the last 30 to 40 years. The large arrow pointing from the MI right side to the BI left side of the diagram indicates how this experience might provide a net flow of useful experience, techniques, and methods to BI researchers as they move toward medical applications of gene expression and other array data. In contrast, BI, which is closer to the biological foundations (especially at the molecular and cellular levels) than MI, is represented with a larger area at the bottom-most stratum. Here, the large arrow going from left to right shows how MI might benefit from a net flow of experience and methods from BI at these foundational and biological analysis levels.
Figure 1.
Graphic metaphor of potential synergies and competition between medical informatics (MI) and bioinformatics (BI) indicating directions from current emphases toward increased interactions and the sharing of methods and tools within biomedical informatics. The diagonal zone represents the area of largest expected complementarities and synergies, while the lightly outlined central oval represents a zone of possible competitive challenges in subtopics of greatest overlap between MI and BI.
At the central stratum of informatics methods we have indicated how the exchange of methods, experience, and approaches is more balanced, with arrows of smaller size pointing in each direction toward the zone of potential synergies, which goes down the central diagonal of the diagram. We have deliberately placed the arrows at the boundaries between strata to illustrate some of the nuances of the situation. BI tends to use more mathematical and informatics models currently, while MI tends to be centered on clinical informatics with a predominance of systems, software, and knowledge engineering approaches. The lightly outlined oval superimposed on the central part of the figure represents the potential for competition, focused on the central stratum of informatics methods, which currently tend to be different in both goals and techniques between MI and BI.
▶ conveys some current opportunities for synergy along the diagonal zone where BI and MI interact. These can lead to fusion within the joint discipline of biomedical informatics (BMI) if the complementarities and opportunities for synergy win out, or continued separation of the disciplines if the contrasts and competitive pressures predominate, as we discuss next.
Conclusions
The compare-and-contrast summary of disciplinary features of MI and BI given in ▶ has led us to an analysis suggesting that differences between the disciplines are based on differing historical development, scientific content, and informatics goals. We have argued that the features of both disciplines are directly linked to the characteristics of their underlying fields of application: medicine and biology.11 Emphasis in MI has been more technological or engineering like and focused on medical practice, although many efforts (largely academic) have also been devoted to biomedical research activities. Meanwhile, BI has focused on building applications for supporting scientific research. From these historical and scientific viewpoints, both disciplines have grown separately.
In contraposition, there also are many areas in which similar objectives and complementarities raise opportunities for interaction and exchange between the disciplines. For instance, many efforts in database integration, information retrieval, standards, ontologies, decision support, multimedia tools, medical records, data mining, user interfaces, or image processing in both MI and BI could be shared and exchanged between the two disciplines. The most important collaboration will be to transfer some of the lessons learned by the professionals of both disciplines in decades of work in many applications and projects to novel BI–MI approaches to genomic-based medicine. In this way, MI can benefit from BI in its experience with problems of scientific inquiry, while MI can benefit BI with its expertise in developing informatics methods and tools for solving medical problems.
For a significant shift to occur in medicine, based on genomics, dramatic changes will have to take place in the methodologic and cognitive approaches to clinical care. As mentioned above, it is difficult to foresee a rapid and smooth shift taking place in medicine without the contribution of radically new perspectives. Informatics methods and tools, coming from both MI and BI, will contribute to and make possible this new vision and practice of biomedicine.
The authors thank Dr. Fernando Martin Sanchez and Dr. Ilias Iakovidis for many useful discussions on parts of the material presented in this article and the constructive suggestions from the JAMIA reviewers on the manuscript. Part of this work was supported by funding from the EC BIOINFOMED study and the INBIOMED project, Ministry of Health, Spain.
References
- 1.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 2.Altman R. Bioinformatics in support of molecular medicine. In: Chute CG. (ed.). Proc AMIA Symp 1998:53–61. [PMC free article] [PubMed]
- 3.Sander C. Genomic medicine and the future of health care. Science. 2000; 287:1977–8. [DOI] [PubMed] [Google Scholar]
- 4.Kohane I. Bioinformatics and clinical informatics: the imperative to collaborate. J Am Med Inform Assoc. 2000; 7:439–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulikowski C. The micro-macro spectrum of medical informatics. Challenges: from molecular medicine to transforming health care in a globalizing society. Methods Inf Med. 2001; 41:20–4. [PubMed] [Google Scholar]
- 6.Miller P. Opportunities at the intersection of bioinformatics and health informatics. J Am Med Inform Assoc. 2000; 7:431–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins FS, Guttmacher AE. Genetics moves into the medical mainstream. JAMA. 2001; 286:2322–4. [DOI] [PubMed] [Google Scholar]
- 8.McDonald CJ. Hickam 2000. The maturation of, and linkages between, medical informatics and bioinformatics. J Lab Clin Med. 2001; 138:359–66. [DOI] [PubMed] [Google Scholar]
- 9.Musen M, van Bemmel J (eds). Challenges in Medical Informatics. Successes and Failures. Shortliffe, EH: IMIA Satellite Working Conference, 29–31March2001.
- 10.Stead W. The challenge of bridging between disciplines [editorial]. J Am Med Inform Assoc. 2001; 8:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maojo V, Martin-Sanchez F, Crespo J, Iakovodis I, Kulikowsli C. Medical informatics and bioinformatics: European efforts to facilitate synergy. Brief report. J Biomed Inform. 2001; 34:423–7. [DOI] [PubMed] [Google Scholar]
- 12.Martin-Sanchez F (project coordinator). BIOINFOMED. Prospective Analysis on the Relationships and Synergy between Medical Informatics and Bioinformatics. Brussels: European Commission, 2002.
- 13.Barnett GO. The application of computer-based medical record systems in ambulatory practice. N Engl J Med. 1984; 310:1643–50. [DOI] [PubMed] [Google Scholar]
- 14.McDonald CJ, Overhage JM, Tierney WM, et al. The Regenstrief medical record system: a quarter century experience. Int J Med Inform. 1999; 54:225–53. [DOI] [PubMed] [Google Scholar]
- 15.Kuperman GJ, Gardner RM, Pryor TA. HELP: A Dynamic Hospital Information System. New York: Springer-Verlag, 1991.
- 16.Shortiffe EH, Perreault L, Fagan L, Wiederhold G (eds). Medical Informatics. Computer Applications in Health Care (ed 2). New York: Springer Verlag, 2001.
- 17.Weed LL. Medical records that guide and teach. N Engl J Med. 1968; 278:593–9. [DOI] [PubMed] [Google Scholar]
- 18.Ledley RS, Lusted L. Reasoning foundations of medical diagnosis. Science. 1959; 130:9–21. [DOI] [PubMed] [Google Scholar]
- 19.Warner HR, Olmsted CM, Rutherford BD. HELP—A program for medical decision-making. Comp Biomed Res. 1972; 5:65–74. [DOI] [PubMed] [Google Scholar]
- 20.De Dombal FT, Leaper DJ, Staniland JR, McCann AP, Horrocks JC. Computer-aided diagnosis of acute abdominal pain. Br Med J. 1972; 2:9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulikowski C, Weiss S. Computer-based Models of Glaucoma. Report no 3. Department of Computer Science, Computers in Biomedicine. New Brunswick, NJ: Rutgers University, 1971.
- 22.Shortliffe EH. Computer-Based Medical Consultations: MYCIN. New York: Elsevier, 1976.
- 23.Gorry GA. A System for Computer-Aided Diagnosis. Report no. MAC-44. Project MAC. Cambridge, MA: Massachusetts Institute of Technology, 1968.
- 24.Pople H, Myers J, Miller R. DIALOG: A Model of Diagnostic Logic for Internal Medicine. In: Proceedings of the Fourth International Joint Conference on Artificial Intelligence. Cambridge, MA: Massachusetts Institute of Technology, AI Laboratory, 1975, pp 845–55.
- 25.Szolovits P, Pauker SG. Categorical and probabilistic reasoning in medical diagnosis. Artif Intell. 1978; 11:115–44. [Google Scholar]
- 26.Rosati RA, Wallace GG, Stead EA. The way of the future. Arch Intern Med. 1973; 131:285–8. [PubMed] [Google Scholar]
- 27.Fries J. Time oriented patient records and a computer data bank. JAMA. 1972; 222:1436–542. [PubMed] [Google Scholar]
- 28.Lindberg DA, Siegel ER, Rapp BA, Wallingford KT, Wilson SR. Use of MEDLINE by physicians for clinical problem solving. JAMA. 1993; 269:3124–9. [PubMed] [Google Scholar]
- 29.Lindberg C. The Unified Medical Language System (UMLS) of the National Library of Medicine. J Am Med Rec Assoc. 1990; 61(5):40–2. [PubMed] [Google Scholar]
- 30.Cimino JJ, Patel VL, Kushniruk AW. Studying the human–computer–terminology interface. J Am Med Inform Assoc. 2001; 8:163–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ouzounis C. Two or three myths about bioinformatics [editorial]. Bioinformatics. 2000; 16:187–9. [DOI] [PubMed] [Google Scholar]
- 32.Mount D. Bioinformatics: Sequence and Genome Analysis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 2001.
- 33.Baxevanis AD, Ouellette BF (eds). Bioinformatics: A Practical Guide to the Analysis of Genes and Proteins. New York: Wiley and Sons, 1998.
- 34.Pearson W. Training for bioinformatics and computational biology. Bioinformatics. 2001; 17:761–2. [DOI] [PubMed] [Google Scholar]
- 35.Altman R. The interactions between clinical informatics and bioinformatics. J Am Med Inform Assoc. 2000; 7:439–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kay L. Who Wrote the Book of Life: A History of the Genetic Code. Stanford: Stanford University Press, 2000.
- 37.Crick FHC. The Biological Replication of Macromolecules. Symp Soc Exp Biol. 1958; XII:138. [Google Scholar]
- 38.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952; 117:500–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohler RR, Bruni C, Gandolfi A. A System Approach to Immunology. Proc IEEE. 1978; 68:964–90. [Google Scholar]
- 40.Garland LH, Miller ER, Swerling HB, et al. Studies on value of serial films in estimating progress of pulmonary disease. Radiology. 1952; 58:161–77. [DOI] [PubMed] [Google Scholar]
- 41.Blum B, Duncan K (eds). A History of Medical Informatics. Reading: Addison Wesley, 1990.
- 42.Collen M. A History of Medical Informatics in the United States: 1950 to 1990. American Medical Informatics Association. Bethesda: Hartman Publishing, 1995.
- 43.Lander ES, Waterman MS (eds). Calculating the Secrets of Life: Contributions of the Mathematical Sciences to Molecular Biology. Washington, DC: National Academy Press, 1995.
- 44.Weatherall D. Science and the Quiet Art. Medical Research & Patient Care. Oxford: Oxford University Press, 1995.
- 45.Sackett D. Evidence-Based Medicine: How to Practice & Teach Ebm. New York: Harcourt Brace, 1997.
- 46.Ross D. Introduction to Molecular Medicine. New York: Springer Verlag, 2002.
- 47.Bauer M, Ringel A. Telemedicine and the Reinvention of Healthcare (Healthcare Informatics Executive Management Series). New York: McGraw-Hill Professional, 1999.
- 48.Edwards RT. Paradigms and research programmes: is it time to move from health care economics to health economics?. Health Econ. 2001; 10:635–49. [DOI] [PubMed] [Google Scholar]
- 49.Greenes R. Future of medical knowledge management and decision support. Stud Health Technol Inform. 2002; 80:29–44. [PubMed] [Google Scholar]
- 50.Martín-Sanchez F, Maojo V, López-Campos G. Integrating genomics into health information systems. Methods Inf Med. 2002; 41:25–30. [PubMed] [Google Scholar]
- 51.Musen M. Domain ontologies in software engineering: Use of Protege with the EON architecture. Methods Inf Med. 1998; 37:540–550. [PubMed] [Google Scholar]
- 52.Hamosh A, Scott AF, Amberger J, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM). Hum Mutat. 2000; 15(1):57–61. [DOI] [PubMed] [Google Scholar]
- 53.Rosse C, Shapiro LG, Brinkley JF. The digital anatomist foundational model: principles for defining and structuring its concept domain. Proc AMIA Symp. 1998; 15:820–4. [PMC free article] [PubMed] [Google Scholar]
- 54.The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet. 2000; 25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Annas GJ. Rules for research on human genetic variation—lessons from Iceland. N Engl J Med. 2000; 342:1830–3. [DOI] [PubMed] [Google Scholar]
- 56.de Groen PC. In: Renee Berg. A healthy database; IBM creating a system for millions of Mayo Clinic patient files. Post-Bulletin, Rochester, MN, 25March2002.
- 57.Buchanan B, Shortliffe EH. Rule-Based Expert Systems: The Mycin Experiments of the Stanford Heuristic Programming Project. New York: Addison-Wesley, 1984.
- 58.Juvan P, Zupan B, Demsar J, et al. Web-Enabled Knowledge-Based Analysis of Genetic Data. Crespo J, Maojo V, Martin-Sanchez F (eds). Medical Data Analysis. Lecture Notes in Computer Science 2199. New York: Springer Verlag, 2001.
- 59.Kuipers BJ. Qualitative simulation as causal explanation. IEEE Trans Syst Man Cybernet. 1987; 17:432. [Google Scholar]
- 60.Lucas PIF. The representation of medical reasoning models in resolution-based theorem provers. Artif Intell Med. 1993; 5:395. [DOI] [PubMed] [Google Scholar]
- 61.Kassirer J, Kopelman R. Learning Clinical Reasoning. Philadelphia: Lippincott, Williams & Wilkins, 1991.
- 62.Hagen J. The origins of bioinformatics. Nat Rev Genet. 2000; 1:231–6. [DOI] [PubMed] [Google Scholar]