SUMMARY
HIV-1 Nef plays important roles in HIV-1 replication and pathogenesis. It is translated from completely spliced HIV-1 RNA, its expression is inherently regulated at the levels of viral DNA transcription and RNA splicing. Here we show that Sam68 cytoplasmic mutants potently suppress Nef expression. The suppression requires Sam68 domain aa269-321 and is correlated with its ability to induce stress granules. In addition, the suppression is specific to Nef, and direct binding to nef mRNA 3′UTR confers the suppression specificity. Furthermore, nef mRNA is targeted to and enriched in these induced stress granules. Importantly, Nef suppression occurs in the context of HIV-1 infection of CD4+ T lymphocytes with little MHC I and CD4 down-regulation. Taken together, these results demonstrate that stress granule induction and nef mRNA sequestration account for this translational suppression of Nef expression and offers a new strategy for development of anti-HIV therapeutics to buttress our fight against HIV/AIDS.
INTRODUCTION
Gene expression from integrated HIV-1 provirus involves transcription, pre-mRNA splicing, nuclear export, and translation, it is regulated by the myriads of interactions between viral elements and cellular proteins. The basal transcription is initiated by interactions of cellular transcriptional factors with the specific DNA sequences within the HIV-1 long terminal repeat (LTR) (Garcia et al., 1987). HIV-1 Tat interacts with the TAR RNA and the CDK9/Cyclin T complex and transactivates the basal level of transcription to synthesize full-length HIV-1 pre-mRNA transcripts (Wei et al., 1998). Some transcripts undergo alternative pre-mRNA splicing to generate a variety of smaller RNA species, while others remain unspliced (Stoltzfus and Madsen, 2006). The pre-mRNA splicing is regulated by cis-acting elements within the viral RNA and their interaction with cellular proteins such as those in hnRNP A/B family and SR family (Stoltzfus and Madsen, 2006). Completely spliced RNA that encodes HIV-1 Tat, Rev and Nef is exported into the cytoplasm via the constitutive mRNA export pathway. In turn, HIV-1 Rev and its interaction with cellular proteins such as CRM-1 facilitate nuclear export of unspliced and partially spliced HIV RNA into the cytoplasm (Fischer et al., 1995), where they serve as genomic RNA for progeny viruses or as mRNA for expression of all other HIV-1 proteins. A great deal of efforts has been directed to understand HIV-1 transcription, pre-mRNA splicing, and nuclear export and their control. In contrast, little is known about the translational control of HIV-1 RNA in the cytoplasm (Yilmaz et al., 2006).
Unlike Tat and Rev, Nef does not directly participate in regulation of HIV-1 gene transcription, although they are all translated from completely spliced HIV-1 RNA. Nef is, if not more, equally important for HIV pathogenesis and AIDS disease progression. The nef gene is relatively conserved in all primate lentiviruses HIV-1, HIV-2 and SIV, and its open reading frame is partially overlapped with HIV-1 3′LTR (Trono, 1995). Infection of HIV-1 strains defective in the nef gene is linked to delayed disease progression (Deacon et al., 1995). An intact nef gene is required for development of the pathology in SIV-infected rhesus macaques (Kestler et al., 1991). The roles of Nef in HIV/AIDS pathogenesis have been attributed to its ability to down-regulate several immunologically important cell surface molecules such as CD4 (Garcia and Miller, 1991), MHC I molecules (Schwartz et al., 1996) and CD3 (Schindler et al., 2006) and to activate intracellular signaling pathways (Arora et al., 2000). Anti-HIV therapeutics has so far mainly been targeted at virally encoded polymerases including reverse transcriptase and protease (Piacenti, 2006). It is highly conceivable that intervention of Nef expression likely offers new strategies to complement the current anti-HIV therapy for better treatment outcomes.
A number of the host cellular proteins are involved in HIV-1 infection, replication and pathogenesis. Among them is Src-associated protein in mitosis of 68 kDa (Sam68). Sam68 belongs to the protein family of the signal transduction and activation of RNA (STAR) (Lukong and Richard, 2003). It was first demonstrated to be capable of substituting for and synergizing with HIV-1 Rev function (Reddy et al., 1999). Sam68 has been shown to be responsible for the Rev functional defect in astrocytes (Li et al., 2002b). A consensus has begun to emerge that Sam68 is an indispensable cellular factor for HIV-1 Rev nuclear export function and HIV replication (Li et al., 2002a; Li et al., 2002b; Modem et al., 2005). Sam68 cytoplasmic mutant Δ410 lacking a nuclear signal (NLS) inhibits HIV-1 replication in a dominant negative fashion (Reddy et al., 1999; Soros et al., 2001; Zhang et al., 2005). Interestingly, this mutant and other NLS deletion mutants do not prevent constitutive Sam68 from entering the nucleus (Zhang et al., 2005). These findings suggest that Sam68 may serve additional function in the cytoplasmic processes of HIV-1 life cycle. In this study, we demonstrate that NLS-deleted Sam68 mutants induce stress granule (SG) formation and specifically interacts with nef mRNA and as a result, leads to sequestration of nef mRNA in these granules and eventual suppression of Nef expression.
RESULTS
Suppression of HIV-1 Nef expression by Sam68 mutant Δ410
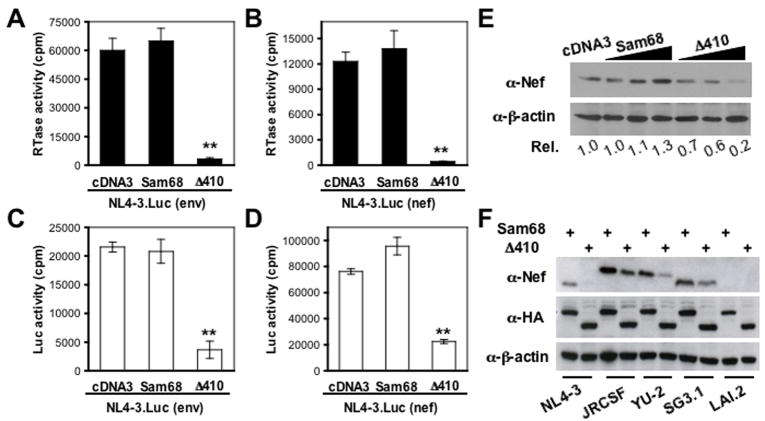
To determine possible function of Sam68 in cytoplasmic processes of HIV replication, we constructed two luciferase reporter viruses NL4-3.Luc(env) in which the firefly luciferase (Luc) reporter gene was inserted in frame into the env gene, and NL4-3.Luc(nef) in which the Luc gene was inserted in frame into the nef gene (Fig. S1). Both insertions were designed to disrupt env and nef gene expression. Thus, Luc expression in these two reporter viruses represents Rev-dependent Env expression and Rev-independent Nef expression, respectively. We transfected 293T cells with NL4-3.Luc(env) or NL4-3.Luc(nef) with Sam68 or Δ410 and determined HIV-1 production in the culture supernatants and Luc expression in the cells. In agreement with published findings (Reddy et al., 1999; Zhang et al., 2005), Δ410 completely abrogated HIV-1 production in both NL4-3.Luc(env) and NL4-3.Luc(nef) transfected cells, while Sam68 had little effects (Fig 1A & B). In addition, Δ410 significantly inhibited Luc expression in NL4-3.Luc(env) transfected cells (Fig. 1C), as it inhibits Rev-mediated nuclear export of HIV-1 intron-containing singly spliced env RNAs (Reddy et al., 1999; Soros et al., 2001; Zhang et al., 2005). To our surprise, Δ410 also inhibited Luc activity in NL4-3.Luc(nef) transfected cells by about ~70% (Fig. 1D), even though the Luc in NL4-3.Luc(nef) is expressed from completely spliced HIV-1 RNA and is Rev-independent.
Figure 1. Δ410 effects on HIV-1 replication and Nef expression.
A–D. 293T cells were transfected with NL4-3.Luc(env) (A & C) or NL4-3.Luc(nef) (B & D), and Sam68 or Δ410, cell culture supernatants were collected for the RT activity assay 42 hr after transfection (A & B), and the cells were harvested for the luciferase activity assay (C & D). The data were mean ± SEM. E. 293T cells were transfected with NL4-3, and an increasing amount of HA-tagged Sam68 or Δ410. The cells were harvested for Western blot analysis using anti-Nef, anti-HA or anti-β-actin antibodies 42 hr after transfection. F. 293T cells were transfected with NL4-3, JRCSF, YU2, SG3.1, or LAI.2 and Sam68 or Δ410 and similar Western blot analysis was performed.
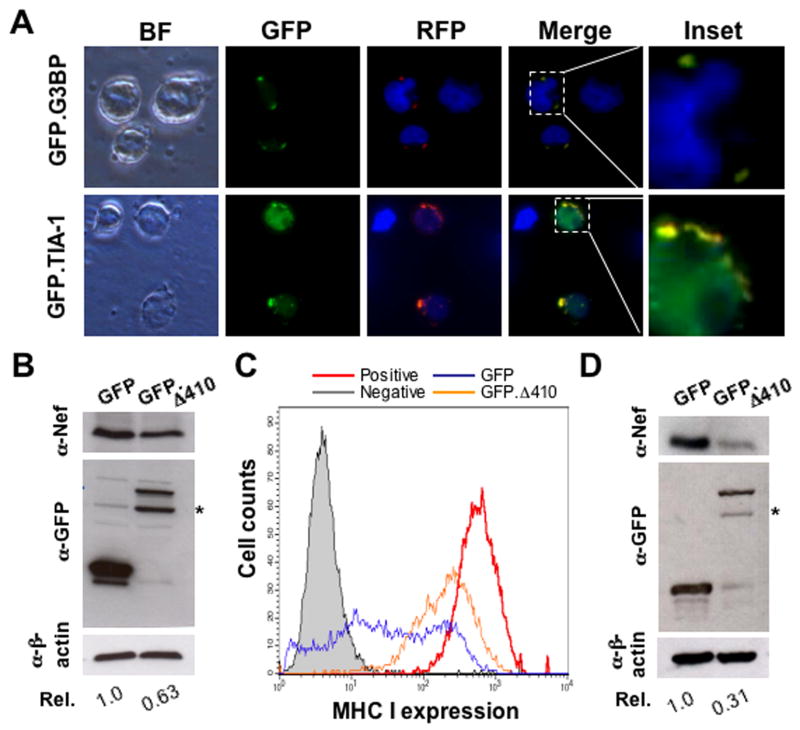
To ascertain that this observation was not an artifact resulting from use of these Luc reporter viruses, we transfected 293T cells with NL4-3 and Sam68 or Δ410 and determined Nef expression by Western blot analysis. Similarly, Δ410 expression led to a decrease of Nef expression by 80%, while Sam68 expression did not decrease Nef expression (Fig. 1E). To determine whether this only happened to NL4-3 Nef, we performed similar experiments with 7 additional HIV-1 isolates, some of which were primary HIV isolates. Δ410 expression exhibited similar suppression of Nef expression from JRCSF, YU-2 and SG-3.1 (Fig. 1F).
The anti-Nef serum that was used in these experiments was not able to recognize LAI.2 Nef (Fig. 1F), as well as Nef proteins from HIV isolates p90CF402.1.8, p94UG114.1.6, and pMJ4 (data not shown). These results show that Δ410 suppresses Rev-independent Nef expression from completely spliced HIV-1 RNA.
Requirement of Sam68 domain between aa269 and aa321 for Nef suppression
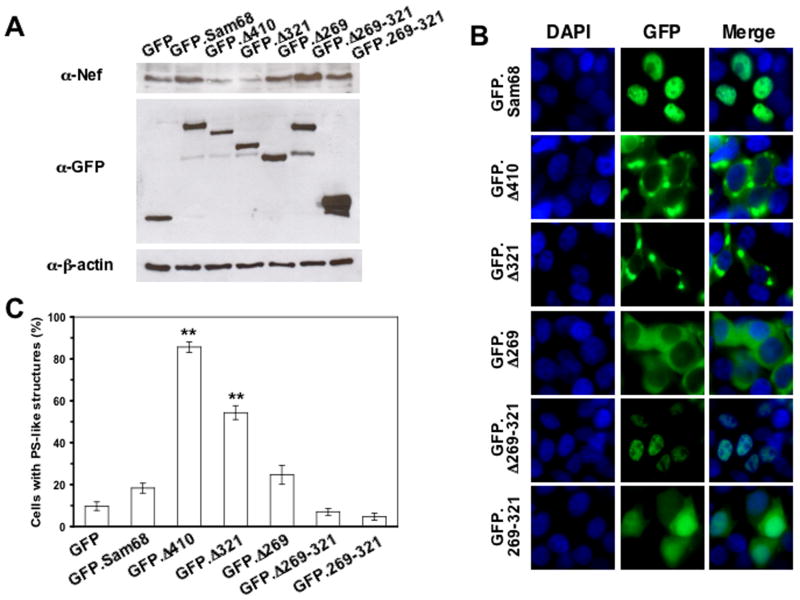
Structurally, Sam68 contains the heteronuclear ribonucleoprotein particle K homology (KH) domain, an RG-rich region, six proline-rich domains (P0 to P5), a tyrosine-rich domain, and a nuclear localization signal (NLS) (Fig. S2). Our early studies have shown that a domain located between aa269 and aa321 of Sam68 is directly involved in Δ410-induced inhibition of HIV-1 replication (Zhang et al., 2005). To determine the roles of this domain on Δ410-induced Nef suppression, we co-transfected 293T cells with NL4-3 and a panel of GFP-tagged Sam68 and its mutants (Fig. S2), and determined Nef expression. Similarly to Δ410, Δ321 that was deleted of P4, P5 and NLS also suppressed Nef expression, whereas Δ269 containing additional deletion of P3 and the RG rich region had no effects on Nef expression (Fig. 2A). Moreover, Δ269-321 that contains the NLS did not affect Nef expression, nor did the 269–321 domain alone. Similar results on Nef expression and subcellular localization were obtained with a panel of HA-tagged Sam68 and mutants (data not shown). These results suggest that both cytoplasmic localization and the aa269-321 domain that contains the P3/RG-rich region are required for Nef suppression.
Figure 2. Effects of Sam68 deletion mutation on HIV-1 Nef expression and their subcellular localization.
A. 293T cells were transfected with NL4-3 and GFP-tagged Sam68 or each of the mutants, followed by Western blot analysis for anti-Nef, anti-GFP or anti-β-actin. B & C. COS-7 cells were transfected with each of the GFP-tagged plasmids in triplicates. Cells were stained in 100 ng/ml DAPI for nuclei. The images were representative of each GFP fusion protein (B). Random 10 fields were counted for the total number of transfected cells and the cells exhibiting granule-like structures and expressed as the percentage of the granule-containing cells (C). The data were mean ± SEM.
We then attempted to determine the mechanisms of this domain-mediated Nef suppression. We began by careful examination of the intracellular localization of these mutants using the same panel of GFP fusion constructs. As expected, full-length Sam68 that contains the intact NLS was detected in the nucleus, and Δ410, Δ321, and Δ269 that all lack the NLS were localized in the cytoplasm (Fig. 2B). Moreover, Δ269-321 was expressed in the nucleus, while the 269–321 domain was detected throughout the cells. Of note was that Δ410 and Δ321 appeared in a unique granule-like structure in the cytoplasm, whereas a similar mutant Δ269 showed an evenly cytoplasmic distribution, a typical pattern of cytoplasmic protein. The apparent association between formation of the granule-like structure and Nef suppression prompted us to quantitate the percentage of the granule-like structure-containing cells in each transfection. More than 85.67 ±2.49% Δ410-transfected cells exhibited this unique structure, while the structure was detected in 54.33 ±3.30% Δ321-transfected cells (Fig. 2C). Both were significantly higher than GFP alone (9.67 ±2.05%), Sam68 (18.33 ±2.36%), Δ269 (24.67 ± 4.50%), Δ269-321 (7.00 ± 1.63%) and 269–321 (4.67 ± 1.70%). There were no significant differences among GFP alone, Sam68, Δ269, Δ269-321 and 269–321. These results reveal the first link between Nef suppression and formation of the granule-like structure.
Δ410 localization into stress granules
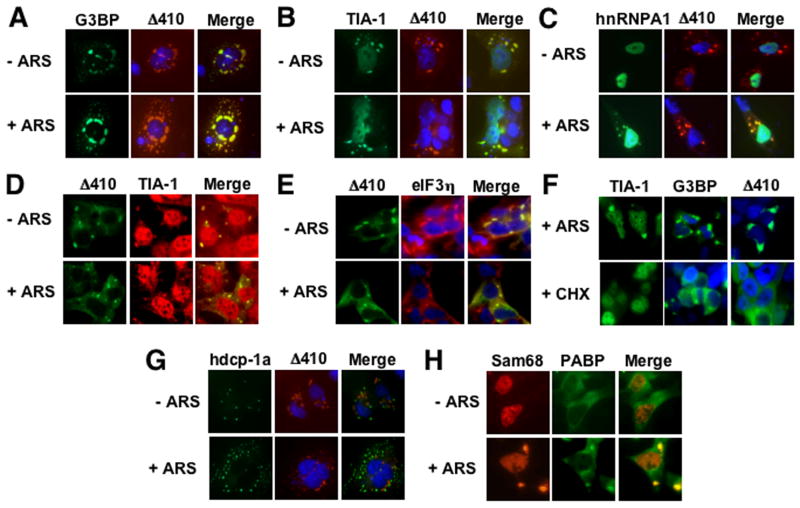
Stress granules (SG) is one of the mechanisms to regulate gene expression at the translational level in response to a variety of external stimuli (Anderson and Kedersha, 2002). Our findings that Δ410 expression resulted in Nef suppression and formation of a granule-like structure prompted us to determine whether Δ410 expression directly induced stress granules. We transfected COS-7 cells with HA-tagged Δ410, and GFP-tagged SG markers GFP-G3BP, GFP-TIA-1, or GFP-hnRNP A1. G3BP and TIA-1 induce SG assembly when over-expressed in the absence of external stress stimuli (Anderson and Kedersha, 2008), while hnRNP A1 is localized into the SG only under stress conditions (Guil et al., 2006). Thus, we then briefly exposed the transfected cells to sodium arsenite (ARS), a widely used oxidative stressor. We then performed immunofluorescence staining for Δ410 localization using an anti-HA epitope antibody. As expected, G3BP and TIA-1 were localized in the SG irregardless of ARS treatment (Fig. 3A & B), whereas hnRNP A1 was expressed in the nucleus of cells in the absence of ARS treatment and was re-localized into the SG when the cells were exposed to ARS (Fig. 3C). In contrast, Δ410 was found to be localized in the granule-like structure in all these three co-transfections, and ARS treatment did not affect this localization. Moreover, co-localization analysis revealed that Δ410 was co-localized with G3BP and TIA-1 in both ARS-treated and untreated cells and only with hnRNP A1 in ARS-treated cells, confirming that the granule-like structure induced by Δ410 expression is SG. To exclude the possibility that Δ410-induced SG formation results from over-expression of these SG markers, we transfected 293T cells with GFP.Δ410 and performed immunofluorescence staining for endogenous TIA-1 using an anti-human TIA-1 antibody. Similarly, Δ410 was co-localized with endogenous TIA-1 in the SG, even though a majority of the TIA-1 protein was detected in the nucleus (Fig. 3D). Localization of Δ410 in stress granules was further confirmed using another endogenous SG marker eIF3η(Fig. 3E). In addition, we also transfected cells with GFP.Δ410 and treated these cells with cyclohexamide (CHX) that is shown to dissolve SG. Similarly to GFP.TIA-1 and GFP.G3BP-expressing cells, CHX treatment led to SG disappearance in Δ410-induced granules (Fig. 3F). Furthermore, we determined whether Δ410 was also localized in the processing bodies (P bodies), as the P bodies share and exchange many structural components with SG and regulate protein translation and/or mRNA degradation in the cytoplasm (Kedersha et al., 2005). We transfected COS-7 cells with a GFP-tagged P body marker hdcp-1α(Fenger-Gron et al., 2005) and HA-tagged Δ410 and performed similar immunofluorescence staining for Δ410. We also treated cells with ARS as a control, as ARS is known to increase the size and number of P bodies. There was no co-localization between hdcp-1α and Δ410 in both ARS-treated and untreated cells (Fig. 3G). Lastly, we determined effects of oxidative stress on intracellular localization of the wild-type Sam68 protein that is predominantly localized in the nucleus. We transfected cells with RFP.Sam68 and treated the cells with ARS. We also included GFP-tagged poly (A)-binding protein (PABP) as a SG marker in these experiments. ARS treatment changed Sam68 from nuclear localization to both nuclear and cytoplasmic SG-like localization, while PABP formed SG in the cytoplasm under ARS treatment (Fig. 3H). Taken together, these results demonstrate that Δ410 expression induces SG formation and suggest that SG formation likely contributes to Δ410-mediated Nef suppression.
Figure 3. Δ410 co-localization with stress granule markers G3BP, TIA-1, and hnRNP A1.
COS-7 cells were transfected with HA-tagged Δ410 and GFP.G3BP (A), GFP.TIA-1 (B), or GFP.hnRNP A1 (C). Prior to fixation, the cells were treated with or without 0.5 mM sodium arsenite (ARS) for 1 hr. The cells were then stained with an anti-HA antibody followed by a phycoerythrin (PE)-conjugated secondary antibody. D & E. 293T cells were transfected with GFP.Δ410 only and treated with ARS as described above. The cells were then stained using an anti-TIA-1 antibody (D) or an anti-eIF3η antibody (E) and PE-conjugated secondary antibody. F. 293T cells were transfected with GFP.Δ 410, GFP.TIA-1 or GFP.G3BP and treated with ARS as above, or with 100 μg/ml cyclohexamine for 90 min. G. COS-7 cells were transfected with HA-tagged Δ410 and GFP.hdcp-1α. H. 293T cells were transfected with RFP.Sam68 or GFP.PABP and treated with ARS as above. In all transfections, cells were stained in DAPI for nuclei and the images were representative of each transfection. The co-localization composite was shown as “Merge”.
Specificity of Δ410-induced Nef suppression
We then wished to determine the underlying mechanisms of Δ410-induced HIV-1 Nef suppression. Sam68 regulates CD44 expression through its effects on pre-mRNA alternative splicing (Cheng and Sharp, 2006). HIV-1 gene expression involves complex alternate splicing of HIV-1 RNA. Meanwhile, STAR family proteins and KH domain-containing proteins are known to regulate mRNA stability (Gherzi et al., 2004; Jan et al., 1999; Larocque et al., 2005; Schumacher et al., 2005). Thus, we first compared HIV-1 RNA splicing, stability, and translation of a reporter gene containing only the 289 nt. leader sequence shared by all HIV-1 transcripts, and found that Δ410 suppression of Nef expression was not through inhibition of HIV-1 RNA splicing to completely spliced RNAs or through changes in the stability and translation of HIV-1 LTR-derived RNA (Fig. S3 A & B). We also utilized the siRNA strategy to knock down Sam68 and determined the relationship between Sam68 and Nef expression. We showed that Δ410-mediated Nef suppression was not likely a loss of function of this mutant in the cytoplasm (Fig. S4).
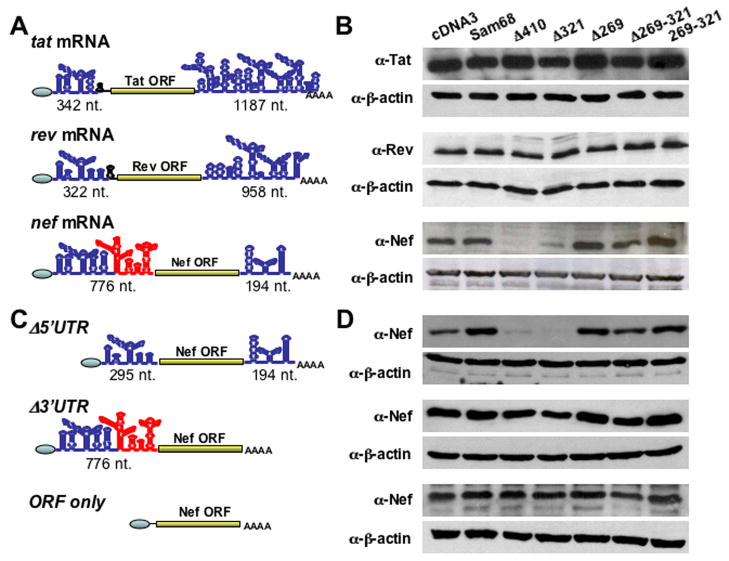
We then switched to understand the molecular mechanisms of Δ410-induced HIV-1 Nef suppression by focusing on the specificity of this suppression. To address the specificity issue, we constructed tat, rev and nef minigenes (Fig. 4A & S5) and compared Δ410 effects on their expression. We transfected each of these minigenes with Sam68 or its mutants and determined the minigene expression using Western blot analysis. In agreement with our earlier findings (Fig. 2A), both Δ410 and Δ321 suppressed Nef expression (Fig. 4B). However, Δ410 and Δ321 had little effects on Tat and Rev expression. As the primary sequences of tat, rev and nef mRNAs extensively overlap, we then turned our attention to the 5′ and 3′ untranslated regions (5′UTR and 3′UTR) of these mRNAs. When we performed the secondary structure analysis of these mRNAs using the RNAfold program (Hofacker, 2003), we found that both 5′UTR and 3′UTR regions of these mRNAs significantly differed from each other in their secondary structure and length (Fig. 4A). Of particular note is part of 5′UTR of nef mRNA that is absent in tat and rev mRNAs (marked in red, Fig. 4A). Thus, we deleted this unique 5′UTR region from nef minigene (Fig. 4C). Meanwhile, we also constructed a nef minigene mutant that was deleted of the entire 3′UTR of nef mRNA, which is also significantly different from those of tat and rev mRNAs (Fig. 4A & C). We then compared Nef expression from these nef minigene deletion mutants using the similar co-transfection and Western blot analysis approaches. The 5′UTR deletion showed little effects on Δ410- and Δ321-induced Nef suppression, while the 3′UTR deletion completely abolished Nef suppression by these two Sam68 mutants (Fig. 4D). Further corroborated with these findings was that Δ410 or Δ321 did not suppress Nef expression from an expressing cassette that only contains the nef ORF (Fig. 4D). These results show that 3′UTR of nef mRNA is the molecular determinant for the specificity of Δ410- and Δ321-induced Nef suppression.
Figure 4. The specificity of Δ410 suppression to Nef expression over Tat and Rev.
A & C. The secondary structure of 5′UTR and 3′UTR of all minigenes were predicted by the RNAfold program. The length in nucleotides for each 5′UTR and 3′UTR was given beneath each minigene and mutants. B & D. 293T cells were transfected with each of the minigenes, Sam68 or each of its mutants. Cells were harvested to determine Tat, Rev, or Nef expression by Western blot analysis.
Δ410 interaction with nef mRNA 3′UTR
The above results from the minigene experiments (Fig. 4) prompted us to further determine whether Δ410 bound to nef mRNA 3′UTR and whether the binding was correlated with its suppressive effects. We took advantage of a RNA-protein interaction-based yeast three hybrid reporter gene assay (SenGupta et al., 1996), which allows in vivo detection of RNA-protein binding. This assay system consists of three components: one hybrid protein of bacteriophage MS2 coat protein fused to LexA DNA-binding domain, the other hybrid protein containing the protein to be tested fused to the Gal4 DNA-activation domain, and one hybrid RNA that has the RNA target for the MS2 coat protein as well as the RNA to be tested. When these three components are introduced into and expressed in the appropriate yeast strain, direct binding of the test protein and the test RNA will activate the LacZ reporter gene expression in the yeast. We constructed Δ410 fusion expression plasmid and the MS2 RNA-nef mRNA 3′UTR hybrid RNA plasmid (MS2-nef 3′UTR). We also constructed MS2-nef 3′RTU hybrid RNA plasmid in which the nef mRNA 3′UTR is in a reverse orientation and used it as a cognate control. In addition, we also constructed a fusion expression plasmid of Δ269 which only slightly induced slight stress granule formation and did not suppress Nef expression. We then transfected respective plasmids into the yeast strain L40 and determined β-Gal activity of the transformants. We also included iron regulatory protein (IRP) and its iron responsive element (IRE) RNA-binding target as positive controls in these experiments. As expected, co-transfection of IRP fusion plasmid with MS2-IRE hybrid RNA activated LacZ gene expression and β-Gal activity (Table 1). Co-transfection of Δ410 fusion plasmid and MS2-nef 3′UTR RNA plasmid activated LacZ gene expression and β-Gal activity, albeit to much less extent. Importantly, there was no detectable β-Gal activity in co-transfection of Δ410 fusion plasmid and MS2-nef 3′RTU and in all other co-transfections expressing the Δ269 fusion plasmid. Taken together, these studies show that Δ410 suppresses Nef expression through its specific binding to the 3′UTR of nef mRNA and suggest Δ410-induced Nef suppression likely result from nef mRNA localization and sequestration into stress granules through binding to Δ410 and subsequent being unavailable for protein translation.
Table 1.
Δ410 binding to 3′UTR of nef mRNA
| Hybrid protein 1 | Hybrid RNA | Hybrid protein 2 | Relative β-Gal activity |
|---|---|---|---|
| MS2-coat | MS2 | IRP | 0 |
| MS2-IRE | IRP | 6.763 ± 0.429 | |
| MS2 | Δ410 | ND | |
| MS2-nef 3′UTR | Δ410 | 0.122 ± 0.025 | |
| MS2-nef 3′RTU | Δ410 | ND | |
| MS2 | Δ269 | ND | |
| MS2-nef 3′UTR | Δ269 | ND | |
| MS2-nef 3′RTU | Δ269 | ND |
The yeast strain L40 was transfected with hybrid protein 2 and hybrid RNA expression plasmids as indicated. The transformants were grown on an SD plate lacking tryptophan, leucine, uracil, and histidine for 5 days. Fifteen colonies from each transformation were inoculated into the same SD liquid medium, triplicates of each inoculation were determined for the β-Gal gene expression using the liquid culture β-Gal assay. Data are mean ± SEM. ND: no detectable β-Gal activity or no colony growth.
Localization of HIV-1 nef RNA transcripts in Δ410-induced SG
Over-expression of several RNA binding proteins induce SG formation, e.g., G3BP, TIA-1, TTP, Caprin-1, FAST, FRMP, LINE-1 ORF, and smaug (Anderson and Kedersha, 2008). Thus, it is very important to further ascertain whether expression of these SG inducing proteins also leads to Nef suppression. We tested two well-characterized SG-inducing proteins that are also often used as SG markers, TIA-1 and G3BP. We transfected 293T cells with NL4-3 with GFP-TIA-1 or GFP-G3BP and determined Nef expression by Western blot analysis. As expected, over-expression of TIA-1 and G3BP induced SG formation (Fig. S6A). But, compared to the GFP control, expression of either TIA-1 or G3BP did not alter Nef expression (Fig. S6B). These results show that not every SG formation results in Nef suppression and provide additional evidence to support the specificity of Δ410 toward to Nef.
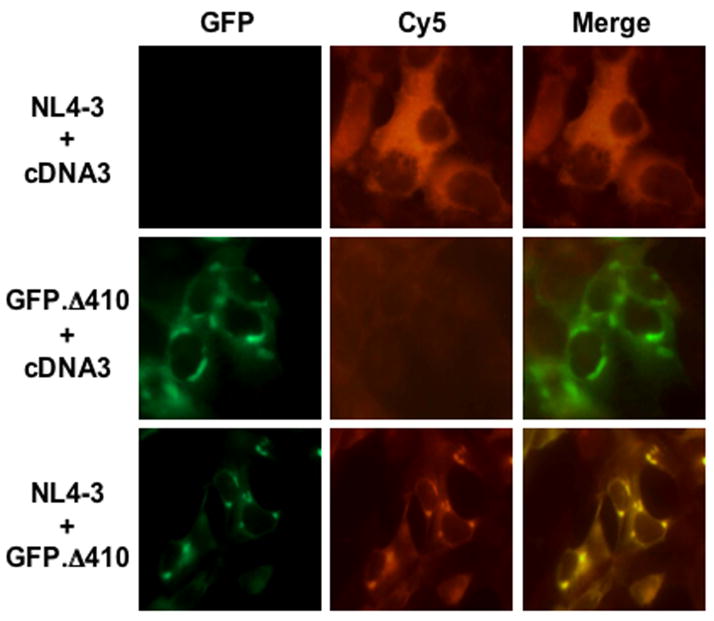
Translational suppression often occurs as a result of SG formation and selective targeting of mRNA (Kawai et al., 2006). Based on the findings (Fig. 4, & Table 1), we next determined whether this was indeed the case for Δ410-mediated Nef suppression. We examined the intracellular distribution of HIV-1 nef RNA transcripts in Δ410-expressing cells. We transfected 293T cells with HIV-1 NL4-3, GFP.Δ 410, or both. NL4-3 was used as the source for nef mRNA transcripts. We performed the fluorescence in situ hybridization using a nef-specific Cy-5-labelled oligonucleotide probe. This probe was designed to ensure a maximal specificity and accessibility to nef mRNA transcripts using an algorithm-based program for mRNA secondary structure (Hofacker, 2003). As expected, nef-Cy5 was exclusively detected throughout the cytoplasm of cells that were only transfected with NL4-3, and only a background level of hybridization signals was detected in GFP.Δ 410-transfected cells (Fig. 5). Co-expression of NL4-3 and GFP.Δ410 led to marked changes of nef-Cy5 localization to a more distinct cytoplasmic pattern and its co-localization with Δ410. These results validate the notion that Δ410-mediated Nef suppression results from Δ410-induced SG formation and nef mRNA targeting and sequestration in SG.
Figure 5. Effects of Δ410-induced SG formation on localization of nef RNA transcripts.
293T cells were transfected with GFP.Δ 410, NL4-3, or both. The cells were then processed for the fluorescence in situ hybridization as described in Experimental Procedures using a Cy5-labelled nef oligonucleotide. The images were representative of each co-transfection. Co-localization of Cy5-nef and Δ410 was shown in the column marked as “Merge”.
Δ410 localization to SG and inhibition of Nef expression and MHC I down-regulation in HIV-infected T lymphocytes
CD4+ T lymphocytes are natural target cells for HIV-1 infection. Thus, we next determined whether Δ410 expression induced SG formation in Jurkats, commonly used CD4+ human T lymphocytes for HIV infection. We transfected Jurkats with RFP-tagged Δ410, in combination with GFP-tagged SG markers such as GFP-G3BP or GFP-TIA-1. We then performed the fluorescence microscopic imaging for RFP- and GFP-tagged proteins. In agreement with our previous findings (Fig. 3), G3BP and TIA-1 co-localized with Δ410 in cytoplasmic SG in the absence of ARS treatment (panels “merge” and “inset”, Fig. 6A). Similar results were obtained in cells treated with ARS (data not shown). We next determined effects of Δ410 expression on Nef expression in HIV-infected cells. To distinguish Δ410 effects on HIV-1 replication from those on Nef expression, we performed a single round infection assay that has previously been established to detect Nef expression in HIV-infected cells (Wei et al., 2005). We transfected Jurkats with either GFP or GFP.Δ410 and incubated the cells for 24 hr, at which time point the transfection efficiency was estimated to be about 70% with the cell viability of over 85% and no differences were noted between these GFP and GFP.Δ410 transfections (Fig. S7). We then infected the transfected cells with replication defective VSV-G pseudotyped NL4-3.Luc(env) virus and determined Nef expression by Western blot analysis 48 hr post infection. As shown in 293T cells (Fig. 1 & 2), Δ410 expression led to a decrease of Nef expression by about 40% in Jurkats (Fig. 6B). Meanwhile, we also performed cell surface staining and flow cytometry analysis for MHC I expression. Δ410 expression in HIV-infected cells showed a significant reduction of MHC I down-regulation, when compared to the cells expressing GFP only (Fig. 6C). Similar results were also obtained with CD4 expression (data not shown).
Figure 6. Δ410-induced SG formation and its effect on HIV-1 Nef expression in Jurkats and PBMC.
A. Jurkats were transfected with GFP.G3BP or GFP.TIA-1 with RFP.Δ410 and processed for microscopic imaging 42 hr post transfection. Co-localization of GFP and RFP Δ410 was shown in the columns marked as “Merge” along with high mag “insets”. B & C. Jurkats were transfected and then infected with 50,000 cpm RT count of VSV-G pseudotyped NL4-3.Luc(env) viruses. The cells were harvested for Western blot analysis (B) and for cell surface staining for MHC I followed by flow cytometry analysis (C) 48 hr post infection. Negative and positive MHC I staining were performed in mock transfected cells. Only GFP-positive cells that were infected with VSV-G pseudotyped NL4-3.Luc(env) viruses were gated for MHC I expression. D. Human PBMC was cultured in the presence of PHA (3 μg/ml) and IL-2 (50 units/ml) for 4 days, transfected with GFP or GFP.Δ410 expression plasmid, infected with VSV-G pseudotyped NL4-3.Luc(env) viruses for 6 hr, and harvested for Western blot analysis. B & D. *, degraded GFP.Δ 410. All data were representative of three independent experiments.
To extend the physiological significance from the established CD4+ Jurkats to primary CD4+ lymphocytes, we performed similar experiments in human PBMC. The transfection efficiency was estimated to be about 55%, while the cell viability was about 70%. Compared to the GFP control, GFP.Δ410 suppressed Nef suppression by about 70% in these primary cells (Fig. 6D). The difference of Δ410-induced Nef suppression between Jurkats and human PBMC is likely due to the different susceptibility of these two cells to HIV infection. Furthermore, we also determined whether Δ410 had any deleterious effects on cells. We expressed Δ410 in a stable fashion and analyzed cell survival and cell cycle using the propidium iodide staining and flow cytometry. There was little cell death in Δ410-expressing cells (Fig. S8). Despite modest accumulation of cells at the G2 phase in Δ410-expressing cells, the ratio of G0G1 to (G2+S) was very similar between Δ410 cells and the control cells, which were 1.71 and 1.75, respectively (Fig. S8). Similar results were obtained in transiently transfected cells (data not shown). These results further validate our findings that Δ410 suppresses HIV-1 Nef expression through SG formation and support possible translation of these findings into anti-HIV therapeutic development.
DISCUSSION
Using a pair of NL4-3-based luciferase reporter viruses, we demonstrated that expression of NLS-deleted Δ410 mutant led to a significant decrease in Luc gene expression in place of HIV-1 nef gene (Fig. 1D) and HIV-1 Nef expression (Fig. 1E & F). Removal of the domain between aa269 and 321 abrogated the suppressive effect (Fig. 2A) and formation of a granule-like structure in the cytoplasm (Fig. 2B & C). The granule-like structure induced by this mutant was verified to be SG (Fig. 3). A unique minigene assay demonstrated that Δ410-induced suppression was specific to HIV-1 Nef through a uniquely structured 3′UTR of nef mRNA (Fig. 4). This was further corroborated by the findings that Δ410 bound to nef mRNA 3′UTR in vivo in a direct manner (Table 1). Moreover, HIV-1 nef mRNA transcripts were co-localized with Δ410-induced SG (Fig. 5). Importantly, SG induction and Nef suppression by Δ410 also occurred in the context of HIV-1 infection of CD4+ T lymphocytes (Fig. 6). Thus, we concluded that Δ410 suppressed HIV-1 Nef expression through induction of stress granule formation and sequestration of HIV-1 nef mRNA transcripts from being translated to Nef protein.
Sam68 possesses a non-conventional NLS at its C terminus, and deletion of this NLS results in Sam68 cytoplasmic localization (Ishidate et al., 1997). Sam68 is absolutely required for HIV-1 gene expression, replication and Rev-dependent gene expression (Li et al., 2002a; Li et al., 2002b; Modem et al., 2005). Therefore, it is not surprising that inhibition of HIV-1 replication by NLS-deleted Sam68 cytoplasmic mutant Δ410 has been shown to be in a dominant negative fashion, resulting in the retention of the wild-type Sam68 in the cytoplasm and preventing it from functioning in Rev-mediated nuclear export of HIV RNAs (Reddy et al., 1999; Zhang et al., 2005). Moreover, Δ410 is also shown to sequester incompletely spliced HIV-1 RNAs from the translation machinery in perinuclear bundles (Soros et al., 2001). Furthermore, wild-type Sam68 is found to be capable of localizing into the nucleus when it is co-expressed with NLS-deleted mutants (Zhang et al., 2005). The current study demonstrates that Δ410 suppresses HIV-1 Nef translation. Taken together, these studies suggest that Δ410 negatively regulates HIV replication at two distinct steps of the virus life cycle: Rev-mediated nuclear export of incompletely spliced HIV-1 RNA and translation of HIV-1 nef mRNA in the cytoplasm.
SG is one type of RNA granules and is formed in response to a variety of external stimuli including over expression of multiple RBP and has recently been recognized as an effective way to regulate gene expression by storing translationally arrested mRNAs (Anderson and Kedersha, 2008; Kawai et al., 2006). Sam68 has recently been shown to be in RNA granules in hippocampal neurons and during early embryogenesis (Grange et al., 2008; Paronetto et al., 2008). In addition, indirect evidence indicates Sam68 localization in cytoplasmic granules during stressful conditions such as that in poliovirus infection and ischemia (Daoud et al., 2002; DeGracia et al., 2007; Mazroui et al., 2006; McBride et al., 1996). Interestingly, hnRNP A1 and FAST, two previously characterized SG components, have recently been shown to directly interact with Sam68 (Paronetto et al., 2007; Simarro et al., 2007), suggesting that Sam68 could be recruited into SG under stress conditions through binding to other SG components. Our results show for the first time that over-expression of Δ410 and Δ321 mutants was capable of inducing SG assembly (Fig. 2 & 3), while Sam68 was re-localized to SG upon oxidative stress (Fig. 3H and data not shown). Our results also show that the RG/P3-rich region (aa269-aa321) was required for the SG formation (Fig. 2B & C). Interestingly, RG-rich regions have been shown to be essential for SG assembly/recruitment of different RBP (Tourriere et al., 2003). Therefore, the fact that deletion of NLS from Sam68 such as Δ410 and Δ321mutants give rise to spontaneous SG induction in the cytoplasm even in the absence of any other stresses strongly supports the notion that Sam68 possesses the inherent ability to induce stress granules. Nevertheless, the exact molecular mechanisms whereby Sam68 is recruited to SG or its mutants induce SG remain to be elucidated.
In general, newly transcribed and spliced mRNA is translationally repressed to prevent ectopic expression while en route into the cytoplasm from the nucleus (Kwon et al., 1999). Viral mRNA including HIV-1 RNA shall be no exception. The movement of the viral mRNA in the cytoplasm involves interaction with multiple host proteins, which often give rise to RNA granules (Cochrane et al., 2006; Mouland et al., 2001). RNA granules including SG all contain translationally repressed mRNA and various RBP (Anderson and Kedersha, 2006). RNA targeting into these granules is understandably determined by the specificity of RNA-protein interaction, and the RNA secondary structure is considered to be more important than its primary sequence in determining the specificity of this interaction. The sequences of tat, rev and nef mRNA share a significant homology. However, the length and secondary structure of their 5′UTR and 3′UTR considerably differ from each other (Fig. 4A), which predispose them to different mRNA scanning mechanisms or offer the differential specificity to translational silencer proteins or miRNA. Using a newly designed minigene strategy, we unequivocally showed that Δ410 only suppressed Nef not Tat or Rev expression (Fig. 4A & B) and that the 3′UTR of nef mRNA was directly involved (Fig. 4C & D) through specific binding (Table 1). Further corroborating these findings was that HIV-1 nef mRNA transcripts were detected in these induced SG (Fig. 5). These results provide the first evidence to show that completely spliced HIV-1 RNA transcripts are subjected to translational regulation as a result of their unique UTR secondary structure.
Nef expression in lymphoid and monocytic cells controls two key aspects of HIV-1 infection and AIDS pathogenesis: evasion of the humoral and cellular immune response, and sustained hyperactivation of the immune system (Schindler et al., 2006). Although, clinically successful antiretroviral therapy can reduce viral replication to undetectable levels in plasma, Nef is continuously being expressed in latently infected PBMC (Fischer et al., 2004). These findings qualify Nef as an ideal target for development of antiretroviral therapeutics. We showed that Δ410 also induced SG formation in CD4+ T lymphocytes (Fig. 6A) and suppressed Nef expression in these cells (Fig. 6B) and in human PBMC (Fig. 6D). In parallel, Δ410 expression considerably restored surface expression of CD4 and MHC I in HIV-1 infected cells (Fig. 6C). These results show that Nef suppression by Δ410 occurs in the natural target cells for HIV-1. Meanwhile, Δ410 expression did not show any apparent adverse effects on the host cells (Fig. S8). Therefore, it is conceivable that targeting Δ410 or Δ410-based therapeutics to HIV-1-infected cells could provide additional protection against HIV/AIDS through suppression of Nef expression and function.
EXPERIMENTAL PROCEDURES
Plasmids, cell culture and transfections and antibodies
The details on all plasmids were provided in the supplement. 293T, Jurkats and COS-1 cells were purchased from the American Tissue Culture Collection (Vanassas, VA) and transfected by the standard calcium phosphate precipitation method (293T), Lipofectamine 2000 (COS-7, Invitrogen, Carlsbad, CA), or a Nucleofactor kit (Jurkats and PBMC, Gaithersburg, MD). Annealed oligonucleotide duplex control and Sam68 siRNAs were purchased from Dharmacon (Lafayette, CO) and transfected as described previously (Cheng and Sharp, 2006). Wherever appropriate, pcDNA3 was included to equalize the amount of plasmid DNA, while pCMVβ-Gal was included to normalize the transfection variations. Anti-JR-CSF Nef (1:750, donated by Dr. K. Krohn and Dr. V. Ovod), anti-Tat (1:500, donated by Dr. J. Karn) and anti-Rev antibodies (1:500, donated by Dr. A. Szilvay) were obtained from the NIH AIDS Research and Reference Reagent Program. Anti-HA (1:1000), anti-β-actin (1:3500), goat polyclonal anti-eIF3η (N20, 1:75) and anti-Sam68 (1:1000) were from (Santa Cruz Biotechologies, Santa Cruz, CA). Anti-GFP (1:250) was from Clontech.
Immunofluorescence staining and imaging
Cells were fixed in 4% paraformaldehyde at room temperature (RT) for 25 min and then permeabilized in 0.3% Triton X-100 for 5 min. The cells were then blocked in 1% fetal bovine serum at 37°C for 15 min, followed by incubation with mouse monoclonal anti-HA (1:50, Santa Cruz), goat polyclonal anti-TIA-1 (1:100, Santa Cruz), or goat polyclonal anti-eIF3η at 37°C for 16 hr. Then, the cells were incubated with phycoerythrin (PE)-conjugated rabbit anti-mouse IgG (1:50, Santa Cruz), or PE-conjugated donkey anti-goat at room temperature for 1 hr. To stain the nuclei, the cells were incubated in 100 ng/ml 4′,6′-diamidino-2-phenylindole (DAPI) at 37 °C for 25 min. Fluorescence images were captured using a digital video imaging microscope system consisting of an Axiovert 200 M microscope with a 100 × 1.4 UVF objective and three different excitation/emission filters (D360/40-D460/50, D470/40-D535/40, D546/10-D590), and an Axiovision video camera (Carl Zeiss, Thornwood, NY). For quantification, cells were plated in triplicate wells, 100 cells of each well were counted for perinuclear granules containing GFP+ cells.
Fluorescence in situ hybridization (FISH)
Nef-Cy3 oligonucleotide probe 5′-TGG GAG CAG TAT CTC GAG ACC TAG A-3′ (nt. 8875-8900 of NL4-3) was synthesized, fluorescently labeled and PAGE purified (Operon Biotechnologies, Huntsville, AL). FISH was performed in 293T cells 42 hr post-transfection. Briefly, transfected cells were fixed with 4% paraformaldehyde at RT for 30 min and then permeabilized with 70% ethanol at 4°C for 24 hr. Then, the cells were rehydrated in 2X SCC (300 mM NaCl, 30 mM sodium citrate, pH 7.0) and 50% formamide at RT for 10 min and hybridized with 100 ng Nef-Cy3 probe at 37°C overnight in a vol. of 40 μl containing 40 μg of E. coli tRNA, 10% dextran sulfate, 50% formamide, 2X SCC, and 20 U RNasin. The Nef-Cy3 probe was denatured at 80°C for 75 sec before it was added to the cells. After sequential washes with 0.5X SCC and 0.1X SCC, each for 7 min at RT, fluorescent images were captured as above.
Data analysis
All values expressed as mean ± SEM. Comparisons among groups were made using two-tailed Student’s t-test. A p value of < 0.05 was considered statistically significant (*), and p < 0.01 highly significant (**).
Acknowledgments
We would like to thank Dr. Janice Blum and Dr. Ann Roman for advices. This work was supported by the grants R01NS039804 and R01MH065158 (to JJH) from the National Institutes of Health. We would also like to apologize for not being able to cite all literature due to the limited space.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson P, Kedersha N. Stressful initiations. J Cell Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Arora VK, Molina RP, Foster JL, Blakemore JL, Chernoff J, Fredericksen BL, Garcia JV. Lentivirus Nef specifically activates Pak2. J Virol. 2000;74:11081–11087. doi: 10.1128/jvi.74.23.11081-11087.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Sharp PA. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol Cell Biol. 2006;26:362–370. doi: 10.1128/MCB.26.1.362-370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane AW, McNally MT, Mouland AJ. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology. 2006;3:18. doi: 10.1186/1742-4690-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daoud R, Mies G, Smialowska A, Olah L, Hossmann KA, Stamm S. Ischemia induces a translocation of the splicing factor tra2-beta 1 and changes alternative splicing patterns in the brain. J Neurosci. 2002;22:5889–5899. doi: 10.1523/JNEUROSCI.22-14-05889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon NJ, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker DJ, McPhee DA, Greenway AL, Ellett A, Chatfield C, et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- DeGracia DJ, Rudolph J, Roberts GG, Rafols JA, Wang J. Convergence of stress granules and protein aggregates in hippocampal cornu ammonis 1 at later reperfusion following global brain ischemia. Neuroscience. 2007;146:562–572. doi: 10.1016/j.neuroscience.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Fischer M, Joos B, Hirschel B, Bleiber G, Weber R, Gunthard HF. Cellular viral rebound after cessation of potent antiretroviral therapy predicted by levels of multiply spliced HIV-1 RNA encoding nef. J Infect Dis. 2004;190:1979–1988. doi: 10.1086/425983. [DOI] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Foster JL, Garcia JV. Role of Nef in HIV-1 replication and pathogenesis. Adv Pharmacol. 2007;55:389–409. doi: 10.1016/S1054-3589(07)55011-8. [DOI] [PubMed] [Google Scholar]
- Garcia JA, Wu FK, Mitsuyasu R, Gaynor RB. Interactions of cellular proteins involved in the transcriptional regulation of the human immunodeficiency virus. EMBO J. 1987;6:3761–3770. doi: 10.1002/j.1460-2075.1987.tb02711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JV, Miller AD. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature. 1991;350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Grange J, Belly A, Dupas S, Trembleau A, Sadoul R, Goldberg Y. Specific interaction between Sam68 and neuronal mRNAs: Implication for the activity-dependent biosynthesis of elongation factor eEF1A. J Neurosci Res. 2008 doi: 10.1002/jnr.21824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Long JC, Caceres JF. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol Cell Biol. 2006;26:5744–5758. doi: 10.1128/MCB.00224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishidate T, Yoshihara S, Kawasaki Y, Roy BC, Toyoshima K, Akiyama T. Identification of a novel nuclear localization signal in Sam68. FEBS Lett. 1997;409:237–241. doi: 10.1016/s0014-5793(97)00455-9. [DOI] [PubMed] [Google Scholar]
- Jan E, Motzny CK, Graves LE, Goodwin EB. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 1999;18:258–269. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol Cell Biol. 2006;26:3295–3307. doi: 10.1128/MCB.26.8.3295-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kestler HW, 3rd, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, Desrosiers RC. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- Kwon S, Barbarese E, Carson JH. The cis-acting RNA trafficking signal from myelin basic protein mRNA and its cognate trans-acting ligand hnRNP A2 enhance cap-dependent translation. J Cell Biol. 1999;147:247–256. doi: 10.1083/jcb.147.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocque D, Galarneau A, Liu HN, Scott M, Almazan G, Richard S. Protection of p27(Kip1) mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nat Neurosci. 2005;8:27–33. doi: 10.1038/nn1359. [DOI] [PubMed] [Google Scholar]
- Li J, Liu Y, Kim BO, He JJ. Direct participation of Sam68, the 68-kilodalton Src-associated protein in mitosis, in the CRM1-mediated Rev nuclear export pathway. J Virol. 2002a;76:8374–8382. doi: 10.1128/JVI.76.16.8374-8382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Y, Park IW, He JJ. Expression of exogenous Sam68, the 68-kilodalton SRC-associated protein in mitosis, is able to alleviate impaired Rev function in astrocytes. J Virol. 2002b;76:4526–4535. doi: 10.1128/JVI.76.9.4526-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong KE, Richard S. Sam68, the KH domain-containing superSTAR. Biochim Biophys Acta. 2003;1653:73–86. doi: 10.1016/j.bbcan.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Mazroui R, Sukarieh R, Bordeleau ME, Kaufman RJ, Northcote P, Tanaka J, Gallouzi I, Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol Biol Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride AE, Schlegel A, Kirkegaard K. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc Natl Acad Sci U S A. 1996;93:2296–2301. doi: 10.1073/pnas.93.6.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modem S, Badri KR, Holland TC, Reddy TR. Sam68 is absolutely required for Rev function and HIV-1 production. Nucleic Acids Res. 2005;33:873–879. doi: 10.1093/nar/gki231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouland AJ, Xu H, Cui H, Krueger W, Munro TP, Prasol M, Mercier J, Rekosh D, Smith R, Barbarese E, et al. RNA trafficking signals in human immunodeficiency virus type 1. Mol Cell Biol. 2001;21:2133–2143. doi: 10.1128/MCB.21.6.2133-2143.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Bianchi E, Geremia R, Sette C. Dynamic expression of the RNA-binding protein Sam68 during mouse pre-implantation development. Gene Expr Patterns. 2008;8:311–322. doi: 10.1016/j.gep.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Piacenti FJ. An update and review of antiretroviral therapy. Pharmacotherapy. 2006;26:1111–1133. doi: 10.1592/phco.26.8.1111. [DOI] [PubMed] [Google Scholar]
- Reddy TR, Xu W, Mau JK, Goodwin CD, Suhasini M, Tang H, Frimpong K, Rose DW, Wong-Staal F. Inhibition of HIV replication by dominant negative mutants of Sam68, a functional homolog of HIV-1 Rev. Nat Med. 1999;5:635–642. doi: 10.1038/9479. [DOI] [PubMed] [Google Scholar]
- Schindler M, Munch J, Kutsch O, Li H, Santiago ML, Bibollet-Ruche F, Muller-Trutwin MC, Novembre FJ, Peeters M, Courgnaud V, et al. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell. 2006;125:1055–1067. doi: 10.1016/j.cell.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Hanazawa M, Lee MH, Nayak S, Volkmann K, Hofmann ER, Hengartner M, Schedl T, Gartner A. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120:357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard JM. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- SenGupta DJ, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci U S A. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simarro M, Mauger D, Rhee K, Pujana MA, Kedersha NL, Yamasaki S, Cusick ME, Vidal M, Garcia-Blanco MA, Anderson P. Fas-activated serine/threonine phosphoprotein (FAST) is a regulator of alternative splicing. Proc Natl Acad Sci U S A. 2007;104:11370–11375. doi: 10.1073/pnas.0704964104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soros VB, Carvajal HV, Richard S, Cochrane AW. Inhibition of human immunodeficiency virus type 1 Rev function by a dominant-negative mutant of Sam68 through sequestration of unspliced RNA at perinuclear bundles. J Virol. 2001;75:8203–8215. doi: 10.1128/JVI.75.17.8203-8215.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus CM, Madsen JM. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr HIV Res. 2006;4:43–55. doi: 10.2174/157016206775197655. [DOI] [PubMed] [Google Scholar]
- Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Trono D. HIV accessory proteins: leading roles for the supporting cast. Cell. 1995;82:189–192. doi: 10.1016/0092-8674(95)90306-2. [DOI] [PubMed] [Google Scholar]
- Wei BL, Arora VK, Raney A, Kuo LS, Xiao GH, O’Neill E, Testa JR, Foster JL, Garcia JV. Activation of p21-activated kinase 2 by human immunodeficiency virus type 1 Nef induces merlin phosphorylation. J Virol. 2005;79:14976–14980. doi: 10.1128/JVI.79.23.14976-14980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- Yilmaz A, Bolinger C, Boris-Lawrie K. Retrovirus translation initiation: Issues and hypotheses derived from study of HIV-1. Curr HIV Res. 2006;4:131–139. doi: 10.2174/157016206776055039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu Y, Henao J, Rugeles MT, Li J, Chen T, He JJ. Requirement of an additional Sam68 domain for inhibition of human immunodeficiency virus type 1 replication by Sam68 dominant negative mutants lacking the nuclear localization signal. Gene. 2005;363:67–76. doi: 10.1016/j.gene.2005.06.043. [DOI] [PubMed] [Google Scholar]