Abstract
DNA polymerase X (pol X) from the African swine fever virus is a 174-amino acid repair polymerase that likely participates in a viral base excision repair mechanism, characterized by low fidelity. Surprisingly, pol X’s insertion rate of the G•G mispair is comparable to that of the four Watson-Crick base pairs. This behavior is in contrast with another X-family polymerase, pol β, which inserts G•G, mismatch poorly and has higher DNA repair fidelity. Using molecular dynamics simulations, we previously provided support for an induced fit-mechanism for pol X in the presence of the correct incoming nucleotide. Here, we perform molecular dynamics simulations of pol X/DNA complexes with different incoming incorrect nucleotides in various orientations (C•C, A•G and G•G (anti), A•G and G•G (syn)) and compare the results to available kinetic data and prior modeling. Intriguingly, the simulations reveal that the G•G mispair with the incoming nucleotide in the syn configuration undergoes large-scale conformational changes similar to that observed in the presence of correct base pair (G•C). The base pairing in the G•G mispair is achieved via Hoogsteen hydrogen bonding with an overall geometry that is well poised for catalysis. Simulations for other mismatched base pairs show that an intermediate closed state is achieved for the A•G and G•G mispair with the incoming dGTP in anti conformation, while the protein remains near the open conformation for the C•C and the A•G syn mismatches. In addition, catalytic site geometry and base paring at the nascent template-incoming nucleotide interaction reveal distortions and misalignments that range from moderate for A•G anti to worst for the C•C complex. These results agree well with kinetic data for pol X and provide a structural/dynamic basis to explain, at atomic level, the fidelity of this polymerase compared with other members of the X family. In particular, the more open and pliant active site of pol X, compared to pol β, allows pol X to accommodate bulkier mismatches such as guanine opposite guanine, while the more structured and organized pol β active site imposes higher discrimination which results in higher fidelity. The possibility of syn conformers resonates with other low fidelity enzymes like Dpo4 (from the Y family), which readily accommodate oxidative lesions.
Keywords: ASFV polymerase X, Pol X, molecular dynamics simulations, protein/DNA complex, induced-fit mechanism, mismatch base pair, base excision repair
Introduction
The African swine fever virus (ASFV) is an encapsulated deoxyvirus with icosahedral morphology, known to induce a lethal infection in domestic pigs1. Its natural hosts are warthogs, and bush pigs, in which ASFV causes unapparent persistent infections, and argasid ticks of the genus Ornithodoros that live on the suids2; 3. The disease is mostly confined to the area of sub-Saharan Africa, but the virus has been found also in the Iberian Peninsula and the Caribbean4; 5. The viral genome is a double stranded DNA molecule that encodes a total of 151 proteins6, including a minimal DNA repair system, composed of a apurinic/apyrimidinic (AP) endonuclease7; 8, an ATP-dependent DNA ligase6; 9, and a repair polymerase, pol X, which catalyses a single-nucleotide filling in gapped DNA10. Pol X, with 174 amino acids, is the smallest naturally occurring DNA-directed DNA polymerase described so far. It belongs to the X family of polymerases and shares sequence and structure similarity with the well studied human DNA polymerase β (pol β)11, an enzyme involved in Base Excision Repair (BER)12; 13; 14. Since the correct replication of DNA affects the genome integrity15, DNA polymerases have been the subject of numerous experimental16 and theoretical studies17 directed toward understanding the fidelity mechanism and enzymatic cycle.
The molecular structure of most polymerases resembles a hand (left handed in the X-family), with three distinct subdomains: palm, thumb, and fingers18; 19; 20. The palm and the fingers are instrumental in binding to the gapped DNA and positioning it into the right conformation for extension. A conserved catalytic triad, composed of three carboxylate residues, is located in the palm domain and, together with two metal ions, helps catalyze the template directed nucleotidyl transfer reaction to the primer strand 21; 22; 23; 24; 25. Overall, the reaction proceeds with an “induced-fit” mechanism 26; 27; 28; 29; 30, where only the correct incoming nucleotide induces the conformational change to a “closed” state that has the right positioning of the catalytic groups for proper synthesis; the incorrect nucleotide, on the other hand, causes a misalignment of the key residues so that repair is hampered.
NMR solutions structures of pol X showed that its three-dimensional structure resembles a simplified hand, missing one of the subdomain present in other X-family polymerases, either the fingers or the thumb, depending on the nomenclature used, important for DNA binding 11; 31. Experimental data show that pol X binds DNA tightly even though it is missing the putative DNA-binding domain. Chemical shift perturbation and fluorescence data that analyzed the binding of pol X to single-stranded-DNA revealed that two lysine-rich areas, helix C on the palm domain and helix E on the thumb domain, could be implicated in DNA binding 31; 32. However, recent studies on pol X double stranded-DNA complexes that employed quantitative fluorescent titrations and energy transfer techniques indicate that the proper DNA-binding subsite is the one located on the thumb subdomain 33. These findings further validate our initial model and the dynamics studies performed on DNA/pol X complexes that suggested cooperative interactions between helix E and the DNA strand 34. Recent ultracentrifugation and small-angle X-ray scattering studies on pol X/DNA complexes confirmed that pol X forms a 1:1 complex with DNA and further identified several positive residues on helix E and adjacent strand as important for protein/DNA interactions 35.
Despite structural similarities, pol X and pol β exhibit different fidelity profiles. While pol β is a moderate fidelity enzyme, pol X is much more error tolerant, with fidelities ranging from 7700 to 1.9 (see Table 1 for review of the kinetic data). It has been hypothesized that the error-proneness of this enzyme is important for the virus since it can contribute to genetic and antigenic diversity 36; 37.
Table 1.
Summary of kinetic data for pol X and correlation with molecular dynamics results.
| Base Pair | Kpol/Kd(M−1s−1) | Kd(mM) | Fidelity | Helix E movement (Å) (from dynamics simulations) |
|---|---|---|---|---|
| C•G | 2300a | 230a | n.a. | 6.89 |
| G•C | 200b | 270b | n.a. | 5.34c |
| G•G | 130b | 21b | 3.8b | 5.34 (syn);
3.07 (anti) |
| A•G | 30a | 20a | 30a | 2.76 (anti)
2.87 (syn) |
| C•C | 0.30a | 860a | 7700a | 1.96 |
Showalter & Tsai, 2001
Lamarche et al., 2006
Sampoli Benitez et al., 2006
Kinetic data suggest that pol X incorporates G versus G (G•G mismatch) with a catalytic efficiency comparable to that of the correct Watson and Crick base pair. For the A•G mismatch, while the enzyme binds well to the incoming nucleotide dGTP, the catalytic rate constant is low, giving rise to a considerably lower catalytic efficiency compared to the correct base pairing (see Table 1 for kinetics data). On the other hand, the C•C mispair is the least efficient, catalyzed with fidelity of 770038. The lack of structural data makes molecular modeling and dynamics simulations a welcome approach by which to investigate the molecular details of the interaction between pol X, DNA and incoming nucleotide for the mismatch base pairs. Earlier, our in silico studies of pol X with correct base pairing suggested an induced-fit mechanism consistent with experimental studies 34; 39. Here we investigate using molecular dynamics simulations the molecular details of several mismatched base pairings to help interpret the puzzling kinetic data at atomic level to understand the error-proneness of pol X. Our comparison of dynamics behavior and active site geometry for C•G and G•G versus A•G and C•C leads to systematic trends correlated with kinetics data that explain why the G•G is easily extended.
Results and discussion
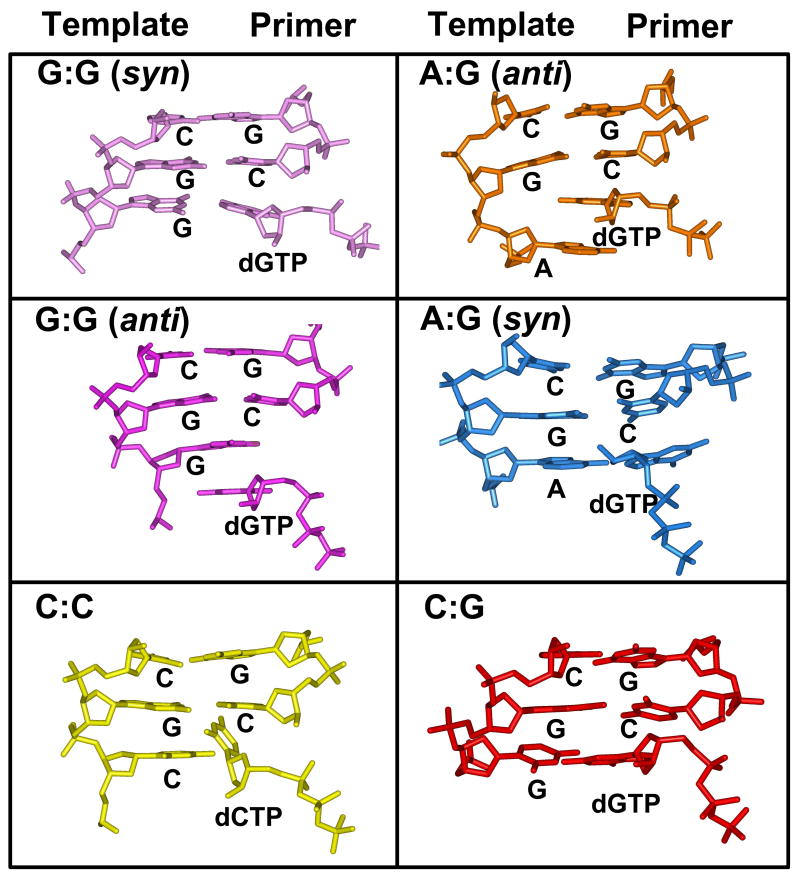
As described under Methodology, six ternary complexes of pol X (Figure 1) were set as starting configurations (Table 2), with G•G in both anti and syn orientations for the incoming nucleotide (Figure 2), A•G in both anti and syn orientations and C•G, and C•C in anti conformations. A reference pol b system with a G•G syn mismatch was also simulated for comparison.
Figure 1.
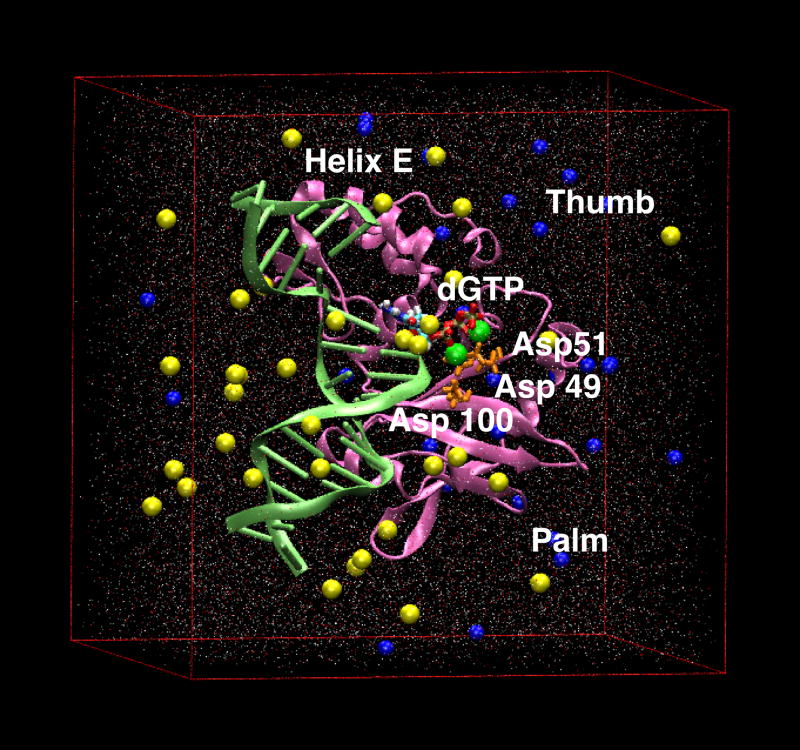
Starting pol X model for dynamics simulations. Pol X is shown in pink with key residues and areas highlighted. The catalytic triad (Asp 49, 51 and 100) is shown in orange. The incoming nucleotide, dGTP is colored by atom, the two divalent Mg ions are green and the gapped DNA, in light green is cartooned. The box of water molecules, colored by atom, is also shown as well as counter ions (Na+ in blue and Cl− in yellow) placed to achieve physiological ionic strength.
Table 2.
Summary of molecular dynamics simulations performed.
| System | Total Number of Atoms | Length of Simulation (ns) |
|---|---|---|
| C•G | 39441 | 10.5 |
| G•G (syn) | 39444 | 10.5 |
| G•G (anti) | 39444 | 10.5 |
| A•G (syn) | 39443 | 10.5 |
| A•G (anti) | 39443 | 10.5 |
| C•C | 39438 | 10.5 |
| Pol β: G•G (syn) | 39998 | 11.1 |
Figure 2.
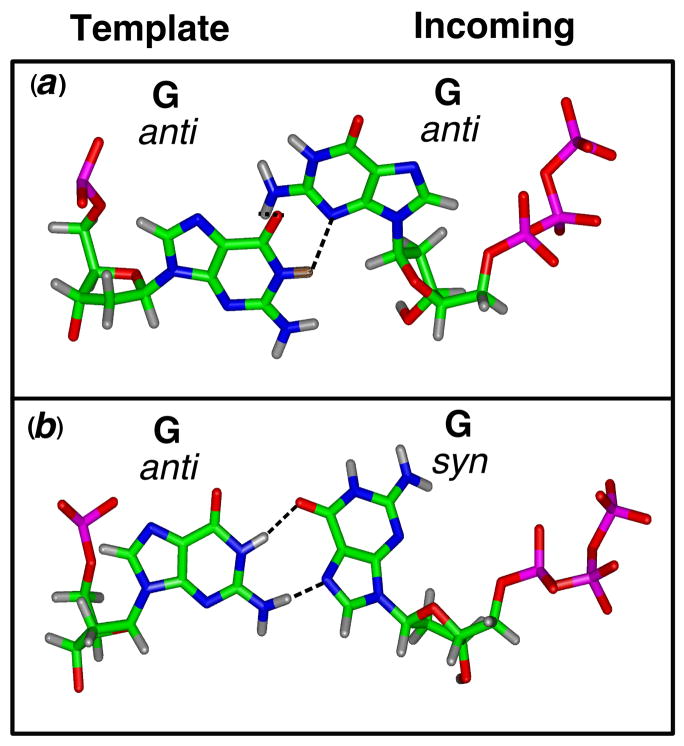
Different conformations for the G•G mispair. (a) Both template and incoming nucleotide are in anti conformation. (b) The template is in anti, while the incoming nucleotide has a syn conformation. In this latter arrangement, Hoogsteen hydrogen bonds can be formed (shown in black).
Pol X can accommodate G•G mismatch in a syn conformation
Previous simulations suggested that pol X undergoes a conformational change only in the presence of the correct incoming nucleotide. This conformational change occurs in the thumb subdomain and can be exemplified by the movement of helix E. Interestingly, when only the gapped DNA is present, no conformational change was observed 34.
Here, simulations of the G•G mispair show that a conformational change consistent with a transition from open to closed state is achieved with the incoming nucleotide (dGTP) in the syn conformation (Figures 3 and 4), but not from the anti conformation. The analysis of the Root Mean Square deviations (RMSD) (Figure 4, Table 1) clearly shows that the conformational change occurs not only in helix E but also in the entire thumb subdomain. The total helix movement, measured as RMSD between the starting conformation and the final simulated structure, obtained by averaging the RMSD values over the last 1 ns of the dynamics simulation, is reported in Table 1. The helix movement for the G•G mispair with the incoming nucleotide in syn conformation is comparable to that observed for the correct G•C structure and only slightly lower than that for the correct C•G structure. This result is in agreement with kinetic data that show a general preference for the incorporation of purines vs pyrimidines38; 40.
Figure 3.
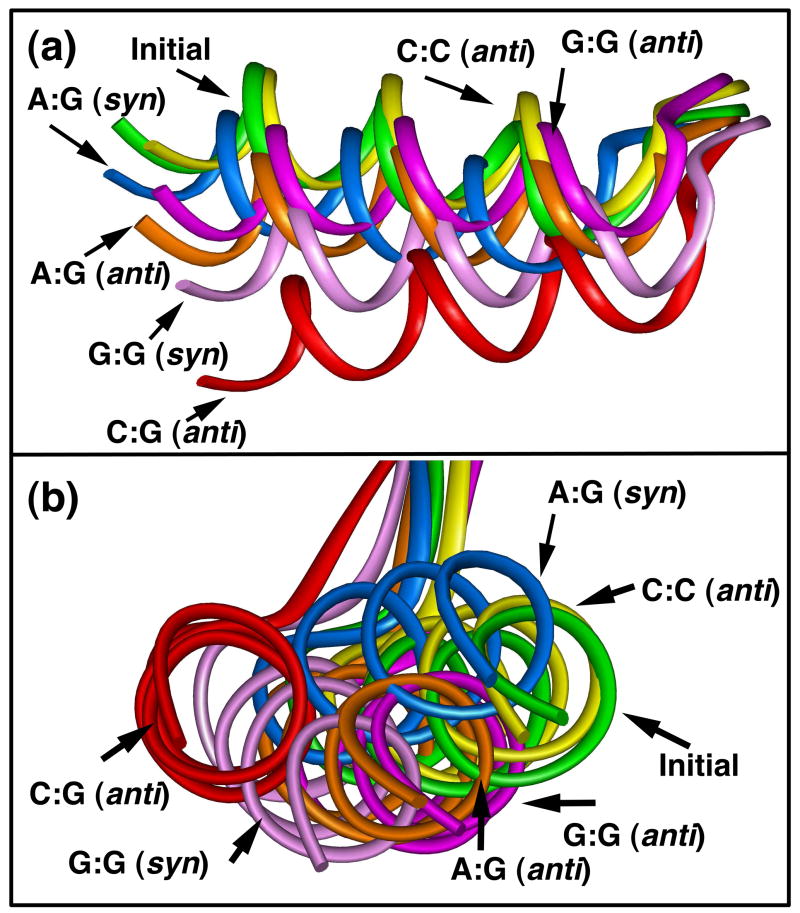
(a) Lateral and (b) front views of helix E movement for the correct base pairing and mismatched structures after 10.5 ns of dynamics simulation. The final structures were obtained by averaging over the last nanosecond of dynamics production. All of the structures are superimposed on the Cα atoms of the palm subdomain (1–105) of their initial structures. The colored ribbons identify the simulated initial structure (green), correct base pair C•G (red), and G•G syn (light purple), A•G anti (orange), A•G syn (light blue), G•G anti (pink), and C•C (yellow).
Figure 4.
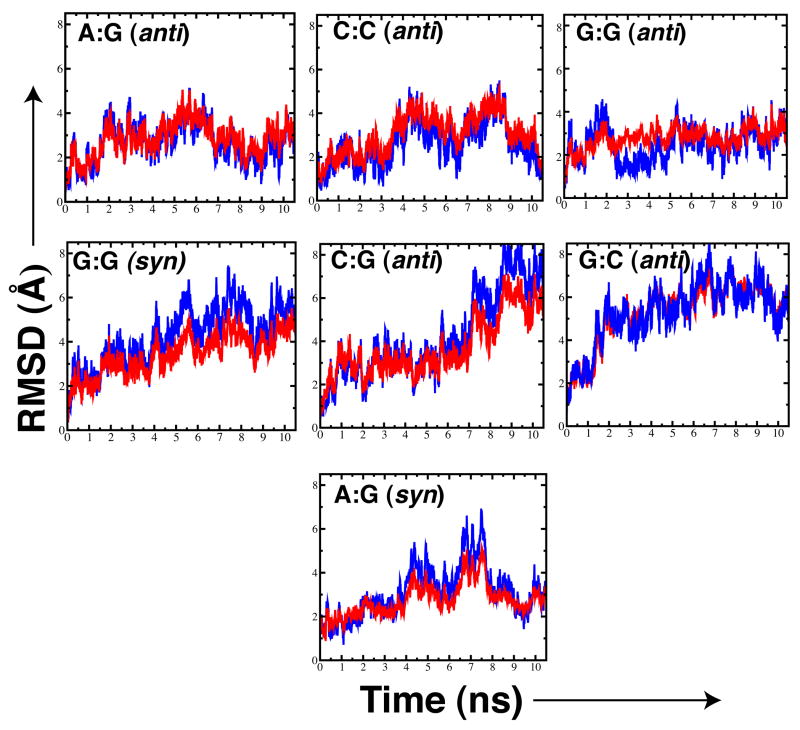
Time evolution plots of the Root Mean Square Deviation (RMSD) values for the α-helix E Cα atoms (blue) and the thumb subdomain (red) of the pol X mismatched structures and correct base pair matched structures. RMSD values were obtained by superimposing the palm subdomain (residues 1–105) with respect to their initial structures. RMSD plots of the mismatched structures (upper panels and lower left panel); RMSD plots of the correct base pair matched structures (two lower right panels). RMSD averages values for the α-helix E over the last nanosecond of the simulations are reported in Table 1.
With the incoming G nucleotide in anti conformation, the conformational change is about half that value (Table 1). The idea of considering an alternative syn geometry for the G•G and A•G mispairs came from a report of Hoogsteen base pairing in the G•G mismatch for BF polymerase 41. Since then, other studies have suggested that in mismatched purine-purine pairs the incoming nucleotide adopts a syn conformation, including in a pol b crystal structure with a G•A mismatch 42 and a kinetic study for E. coli DNA polymerase I with a G•G mispair 43. In this latter study, the A•G mispair appears to have an anti•anti geometry, but the authors could not exclude an anti•syn conformation.
Several crystallographic and molecular dynamics studies have also reported that the damaged templating base 8-oxoG preferentially assumes the syn when pairing with purine nucleotides 44; 45; 46.
A recent study on BF that employed molecular dynamics simulations and free energy calculations showed that the adoption of this less conventional syn conformation was also favored by the incoming nucleotide for adenine incorporation opposite the oxidatively damaged 8-oxoG in anti conformation 47.
Our results suggest that the G•G mismatch in pol X may assume an anti•syn conformation and that this geometry can be extended further, since a conformational change from open to closed state is observed with no major template strand distortions.
For the A•G mismatched pair, an intermediate state between the open and closed form is achieved when the incoming nucleotide is in anti conformation. However, helix E does not undergo a significant conformational change when the incoming nucleotide is in syn conformation and thus the system remains closely to the open form. The nonnegligible value of helix E RMSD over the last ns of simulation (2.87 Å, Table 1) is due to a more a translational movement rather than an open to closed transition, so that the protein remains in an open conformation. These results suggest that the incoming nucleotide will likely be in anti conformation rather than syn when the A•G mismatch is incorporated, as larger helix E movement and better active site geometry (see below) are observed in the simulated A•G anti structure.
Finally, for the C•C mispair, no large conformational change in helix E is observed (Figure 3).
Overall, simulation results are in agreement with the available kinetic data (Table 1). These data show that even though the A•G mismatch complex forms (low dissociation constant Kd for the complex), the catalysis does not occur readily (low catalytic efficiency, calculated as kpol/Kd). On the other hand, the C•C mismatch has both high Kd and low kpol/Kd and subsequently the highest fidelity with respect to this misincorporation. Although the movement of helix E and more broadly of the thumb subdomain is not a direct measurement of reaction catalysis, our data support an induced-fit mechanism, in which the right geometry of the active site is achieved only with correct pairing and G•G mismatch pairing, provided that the incoming base is in syn conformation.
Base pairing at the incorporation site reveals a distorted geometry for certain mismatches
As can be seen from Figure 5, the incorrect incoming nucleotide for A•G anti (orange), G•G anti (magenta), C•C (yellow) and the corresponding template residue assume distorted conformations that deviate from Watson-Crick arrangements. Instead, in these mismatches, the base pairs partially stack against one another; this distortion is most severe for C•C. The staggered conformation of base pairs is consistent with previously reported mismatched structures 41; 48. the other hand, we see that the G•G syn mismatch has an undistorted active site with the incoming base aligning well with the templating base, through a Hoogsteen hydrogen bond pattern (Figures 2 and 5). According to the geometric selection model, the size and shape complementarity of the nascent base pairs are more important than the ability to form Watson-Crick hydrogen bonds. It has been shown that a number of DNA polymerases can incorporate nonpolar nucleoside isosteres that cannot form Watson-Crick interaction 49; 50 and that this ability correlates with the fidelity of DNA synthesis 51. Therefore, it is not surprising that low fidelity pol X can incorporate mismatches with non Watson-Crick interactions. However, as can be inferred from the kinetic data, not all mismatches are equally incorporated. The distorted geometry of the nascent base pair correlates well with the misincorporation rate, with the more distorted being incorporated less frequently. In addition, the displacement of the incoming nucleotide, which results in a longer Pα-O3′ distance, further discourages incorrect nucleotide insertion 48, as will be discussed in the next paragraph.
Figure 5.
Geometry of template-primer DNA base pairs with the incoming nucleotide in the simulated structure (average structure over the last nanosecond of dynamics trajectory) The template-primer DNA base pairs with the incoming nucleotide in the simulated G•G syn (light purple), A•G anti (orange), A•G syn (light blue), G•G anti (pink), C•C (yellow) mismatches.
The A•G syn mispair is an interesting exception, because it does not follow the pattern described above. In this mismatched structure, good Hoogsteen hydrogen bonding is achieved and therefore the base pairing geometry is optimal. However, the incoming nucleotide is positioned away from the primer strand and, as a result, the active site is severely distorted, (see below), underscoring our hypothesis that the syn orientation is not likely to occur.
Active site analysis echoes above trends
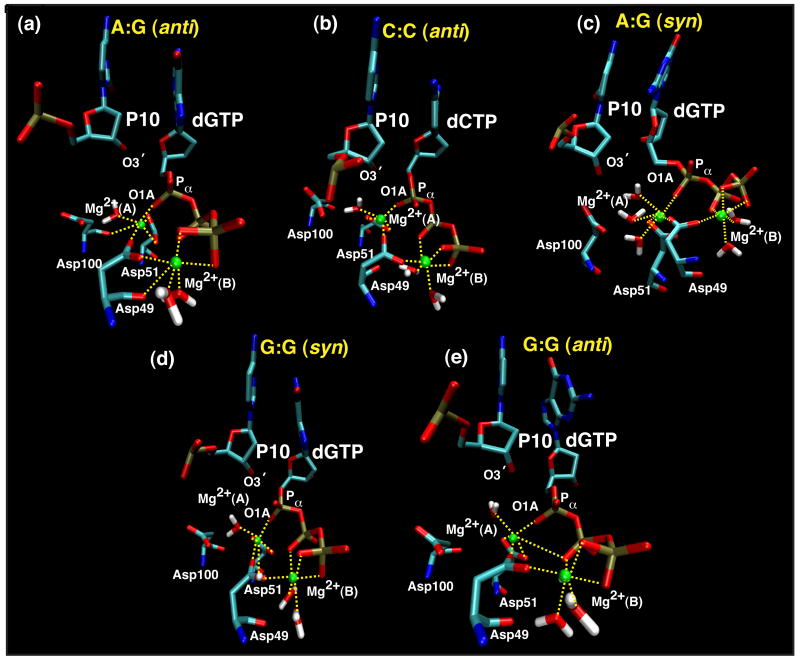
During the nucleotide incorporation, the basic chemical reaction is the formation of a covalent phosphodiester bond between the incoming dNTP and the terminal primer base. It has been shown that this nucleotidyl transfer reaction is favored when the distance between the Pα of the incoming nucleotide and the 3′-hydroxy group of the primer is 3 Å 30. Our Pα-O3′ distances, calculated by averaging the distances over the last ns of each simulation, are larger for all mismatched structures, but the closest to the favored distance is for the G•G syn mismatched structure (3.63 Å), while the furthest from the favored one is seen in the C•C mismatch (9.51 Å) (Table 3). Note that an ideal distance is not observed even when the incoming nucleotide is the correct one in any of the simulation structures. As discussed in our previous molecular dynamics studies, the active site arrangement is still not perfectly poised for the chemical reaction, as shown by the coordination of the catalytic magnesium ion. A similar situation was observed for pol b and this has led to our pre-chemistry avenue hypothesis17; 48. For the chemical reaction to occur, this catalytic ion has to coordinate with all three aspartate residues of the catalytic triad. In fact, only two aspartates (Asp 49 and Asp 51) are coordinated with the magnesium ion and the third, Asp 100, is father away, displaced in the Mg2+ coordination sphere by a water molecule. A similar situation is observed for all the mismatched pairs, in which a water molecule completes the hexa-coordination of the catalytic magnesium ion instead of Asp 100 (Figure 6).
Table 3.
Average active-site interatomic distances (final 1ns of production dynamics).
| Distance (Å)
|
||||||
|---|---|---|---|---|---|---|
| C•G | G•G syn | G•G anti | A•G anti | A•G syn | C•C | |
| Nucleotidyl transfer distance | ||||||
| dNTP (Pa) – P10 (O3′) | 3.55 | 3.63 | 6.74 | 3.65 | 7.85 | 9.51 |
|
| ||||||
| Catalytic magnesium ion coordination | ||||||
| Mg2+ (A) – P10 (O3′) | 4.81 | 4.37 | 7.53 | 4.46 | 6.93 | 9.34 |
| Mg2+ (A) – dNTP (O1A) | 1.80 | 1.84 | 1.86 | 3.90 | 4.03 | 1.86 |
|
| ||||||
| Nucleotide-binding magnesium ion coordination | ||||||
| Mg2+ (B) – dNTP (O1A) | 4.32 | 4.37 | 5.66 | 3.95 | 4.00 | 4.41 |
| Mg2+ (B) – dNTP (O2γ) | 1.92 | 1.91 | 1.84 | 3.94 | 1.91 | 1.94 |
dNTP, 2′-deoxynucleside triphosphate; P10, primer nucleotide.
Figure 6.
Coordination spheres of the catalytic Mg2+ (A) and the nucleotide binding Mg2+ (B) ions at the end of the dynamics trajectory for all pol X/DNA complex mispairs. The structures shown here are the last frame of the dynamics production.
That the critical distance dNTP Pα-O3′ in the G•G syn mispair system is comparable to the structure obtained with the correct incoming nucleotide reconfirms that this system has an overall geometry approaching the optimal one for nucleotide incorporation. Other distances that have been shown to be important for the chemical reaction are also reported in Table 3. All distances were obtained by averaging the values over the last ns of each simulation. In every case, distances in the G•G syn mispair system compare well with the correct C•G system. In contrast, all other mismatched pairs have one or two distances that are significantly larger.
Since the two magnesium ions are also required for the nucleotidyl transfer reaction according to the two-metal ion catalytic mechanism 22, monitoring the respective coordination spheres can help assess how ready the active site is for the chemical reaction (Table 3, Figure 6). Again, the G•G syn structure has distances that compare well with the C•G structure. On the other hand, the other mismatched structures reveal more significant distortion around the metal coordination spheres. For example, the A•G anti mismatch, which has a reasonable nucleotidyl transfer distance, displays distortions around the nucleotide-binding magnesium. Figure 6 shows that this ion is surrounded by three water molecules and is only loosely coordinated with the incoming nucleotide, dGTP (see distances in Table 3). The sixth coordination site is occupied by the side-chain of Asp 49. The other magnesium ion is instead coordinated with two water molecules, the side chains of Asp 51 and Asp 49 and O1A oxygen atom from the substrate Pa, more similar to structure with the correct incoming nucleotide. On the other hand, both the A•G syn and the C•C mispair structures show large nucleotidyl transfer reaction distances and overall bad geometries around the two metal ions, suggesting that these structures are not well poised for chemistry.
As noted above, the magnesium ions are hexa-coordinated in all simulated structures (Figure 6). However, the aspartate side chains of the catalytic triad are not always present in their coordination spheres, as they should for the proper alignment of the active site residues required for catalysis. As noted for the A•G anti mismatch structure, water molecules occupy the vacant positions giving rise to an overall active site cavity that is more exposed compared with the correct incoming nucleotide.
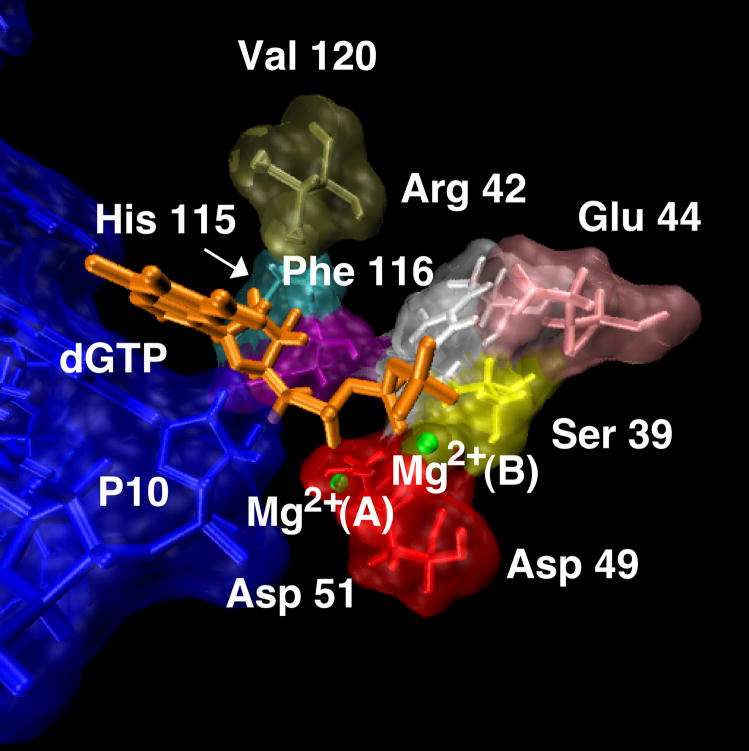
Pol X’s active site cavity is relatively open and only few protein residues contribute to stabilize the incoming nucleotide. As shown in Figure 7, which depicts the protein residues in the G•G syn mismatch structure within 4 Å of the incoming nucleotide, the residues that interact with the incoming dGTP are few (only 8) and mostly on one side of the nucleotide. Considering the paucity of residues defining this active site cavity, it is not surprising that pol X can accommodate mismatches more easily than higher fidelity polymerases. Interestingly, some of these residues, and in particular Val 120 and Glu 44, appear severely distorted in C•C and G•G anti mismatch structure, in agreement with the fact that these mispairs cannot be incorporated efficiently with the given geometry.
Figure 7.
Protein residues with 4 Å of the incoming nucleotide dGTP for the G•G syn mismatch system in the average structure over the last nanosecond of dynamics. The DNA is shown in blue. The two required magnesium ions are also displayed in green: (A) catalytic Mg2+ ion, and (B) nucleotide binding ion.
Comparison with pol β
DNA polymerase β is a well studied family X member involved in human base excision repair. The generally accepted mechanism for this enzyme involves cycling between an open (inactive) and a closed (active) conformation to initiate catalysis 26. As mentioned above, despite the structural similarity, pol β exhibits a quite different behavior when incorporating mismatched pairs, yielding fidelities which are substantially higher that the ones observed for pol X. Kinetic data show that the rate of incorporation of a wrong nucleotide is substantially lower than the corresponding rate for the right one 52. In particular, catalytic efficiency, calculated as kpol/Kd, for incorporation of G opposite G is 0.3 M−1s−1, compared to a value 7.5 × 105 M−1s−1 for the correct incorporation of C opposite G, thus making this mismatched pair one of the least favorable to be incorporated 53; 54. Molecular dynamics simulations for mismatched pairs in the pre-chemistry state reveal that the closing motion of helix N required for the catalytic cycling is disrupted in the presence of incorrect incoming nucleotides 48. In addition, active site distortions whose magnitude correlates well with the kinetic data are also observed. Our pol b studies long suggested that for mismatched pairs chemistry proceeds from partially open states or suboptimal activated complexes 48; 55. In a recent analysis of the relationship between the conformational landscape and fidelity using the empirical valence bond mapping (EVB), it was also concluded that the transition state for an incoming incorrect nucleotide such as G•G likely involves a different conformation of the protein, more similar to the open than to the closed one 56. In addition, studies on the G•G mismatch employing mixed quantum mechanics/molecular mechanics (QM/MM) techniques showed that the activation energy in this system is about 5 kcal/mol higher than in the correct system, due mainly to structural distortion of the active site 57.
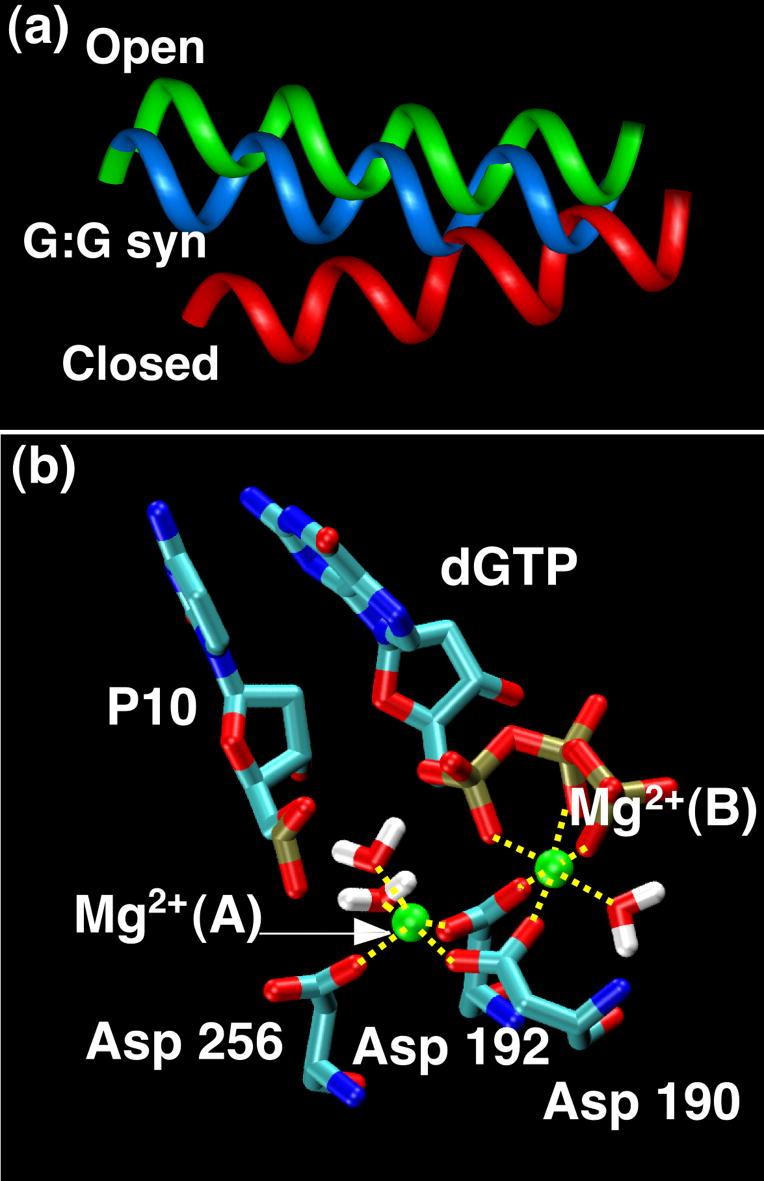
Thus, the body of evidence that suggests that pol b’s active site is more discriminating than pol X, explaining the difference in fidelity for the G opposite G misincorporation. However, all previous simulations on the G•G mismatch pair were performed with the incoming nucleotide in anti conformation. Here, based on the geometric observations for pol X, our simulations of the G•G mispair with the incoming nucleotide in syn conformation help illuminate the catalytic geometries of pol X and pol b in the presence of G•G mismatch. As Figure 8 shows, helix N of the G•G system for pol b does not move to a closed conformation, but rather stays in an intermediate state that is closer to open, as also found for the G•G anti mispair. Analysis of the active site geometry reveals that while the syn conformation allows a better geometry than the anti, severe distortions remain. In particular, the critical distance for catalysis Pα-O3′ is to 4.34 Å for G•G syn versus 4.97 Å for the G•G anti system. In addition, Hoogsteen base pairing affords a better base stacking for the nascent base instead of the staggered interactions seen in the anti system. However, the overall coordination of the two magnesium ions still departs from the geometry required for nucleotidyl transfer and important protein residues also appear distorted. A water molecule is positioned between the catalytic magnesium ion and the incoming nucleotide, so that direct coordination of the ion with the oxygen O1A of the dGTP is lost.
Figure 8.
Results from the G•G mismatch simulation of DNA pol b with the incoming nucleotide in syn conformation. In the upper panel (a) helix N of the simulated structure (average structure over the last nanosecond of production dynamics) is displayed (light blue) compared with the crystallographic open (green) and closed (red) conformation of the protein. In the lower panel, (b), the arrangement of the active site is shown. The coordination of the two magnesium ions clearly departs from that in the ideal geometry.
Conclusions and Future Directions
Our study of several mismatched base pairs systems for pol X reveals that the G•G mismatch structure with the incoming nucleotide in a syn conformation undergoes a conformational change and achieves an overall active site geometry similar to that observed with the correct incoming nucleotide G•C. These results suggest that pol X incorporates easily the G•G mismatch, in agreement with kinetic data. Other mismatches may not be incorporated as efficiently, because they give rise to more distorted base pairing and/or distorted active sites. These simulations are the only structural data available for pol X/DNA complexes in the presence of mismatches. Of course, experimental validation is critical to confirm our hypotheses.
It has been suggested that although the error proneness exhibited by pol X may be important for creating genetic diversity, it may not be the primary criterion of evolutionary selection. Rather, pol X and other polymerases might have been selected for translesion synthesis capabilities 58; 59; 60; 61. In particular, a recent study showed that pol X can bypass the 8-oxo-G lesion and is unique in incorporating 8-oxo-G•A slightly better than 8-oxo-G•C 62. This oxidative damage is particularly interesting for pol X since oxidative stress by reactive oxygen species (ROS) is hypothesized to be prevalent in the cellular environment where ASFV operates 8; 63. Intriguingly, the authors of ref. 60 suggested that this preference might be due to the fact that the incoming nucleotide, dATP, can utilize both the Watson and Crick face and the Hoogsteen face to base pair with 8-oxo-G, while dCTP uses only the Watson-Crick face. This possibility will be intriguing to explore with computational approaches.
Computational Methodology
Initial models
Cartesian coordinates for five ternary complexes (Table 2 excluding A•G syn), comprised of pol X, a 16-mer double stranded gapped DNA, and incoming nucleotide (correct and incorrect), were built using the Insight II Biopolymer module (Accelrys, CA). (See separate section for the A•G syn system, below). The starting structure was the ternary complex of pol X/DNA/dCTP previously used for dynamics simulations 34. In this structure, the template base at the abasic site is guanine, while the correct incoming nucleotide is cytosine. This system also contains the two necessary magnesium ions, and monovalent counter ions for electrical neutrality at ionic strength of 150 mM (35 Cl− and 28 Na+), in addition to a box of 11,783 molecules of water (Figure 1). The G•G mismatched model was built by substituting the base of the incoming nucleotide of the correct G•C ternary structure with guanine. In this model, both the incoming nucleotide and the template base are in the anti conformation (indicated as G•G anti structure). A model was also constructed in which the guanine base of the incoming mismatch nucleotide is rotated 180° with respect to the sugar moiety into a syn conformation (called G•G syn) (Figure 2). Usually, DNA base pairs form Watson-Crick hydrogen bonding when they are in anti conformation. However, an unconventional syn geometry is sometimes found in DNA polymerases’ purine-purine mismatched structures 41; 42; 43 or when oxidatively damaged bases (such as the 8-oxo-7,8-dihydro-2′-deoxyguanosine, or 8oxoG) are present 45; 47; 64. Other studies that investigated the steric effects involved in base pairing in DNA polymerase I, in the presence of benzimidazole and other nucleobase analogs, revealed that this alternative syn minimized steric clashes and afforded the best steric and hydrogen bonding complementarity opposite to guanine 65; 66.
The C•C mismatched structure was built from the original ternary model by substituting the base on the templating strand from G to C. Finally, the A•G mismatch was constructed using as a starting point the G•G anti model and then substituting the guanine on the templating strand with an adenine.
A simulation of a pol b/DNA complex with incorrect incoming nucleotide G•G (template•incoming dGTP) was started from the intermediate (“half-closed”) conformation before the nucleotidyl transfer reaction to capture pol b’s closing within 10 ns. For the correct base pair (G•C) 30 the intermediate model was constructed as an average of the crystallographic open, binary gapped complex (1BPX) and closed, ternary complex (1BPY) from the PBD/RCSB resource. Specifically, the model’s thumb subdomain is partially closed, with the correct base pair (G•C) in the active site. On the basis of this model, a complex of pol b with the G•G mispair was built by replacing the correct incoming nucleotide (dCTP) with incorrect nucleotides dGTP. The incoming nucleotide is in syn and template is in anti conformation.
Minimization, Equilibration and Molecular Dynamics
Energy minimizations, equilibrations, and dynamics simulations for the five systems above systems were performed using the program CHARMM (Chemistry Department, Harvard University, Cambridge, MA)67 with the all-atom forcefield 26a268; 69. All starting models were minimized with 10,000 steps of steepest descent keeping all the protein heavy atoms fixed, followed by further minimization with 20,000 steps of adapted basis Newton-Raphson minimization 67; 70, until the gradient of RMSD was 10−6 kcal/mol•Å. The minimization was then repeated, allowing all the atoms to move. The systems were then equilibrated for 30 ps at 300 K, using the Langevin multiple time-step LN integrator 71, minimized again and re-equilibrated for other 30 ps before starting the dynamics production. The parameters used during equilibrations and dynamics runs were 1 fs for the short time-step, Dt (used to update the bond, angle and dihedral energy terms): 2 fs for the medium time-step (used to update the non bonded interaction within a 7 Å distance); and 150 fs for the long time-step (used to update all the other non bonded interaction up to the global non-bonded interaction cutoff, here 14 Å). The SHAKE algorithm was employed in all runs to constrain the bond lengths involving hydrogen atoms 72. Electrostatic and van der Waals interactions were smoothed to zero at 12 Å with a shift function and a switch function respectively. A Langevin damping constant of g = 10 ps−1 was chosen. Dynamics simulations for all systems with the CHARMM program were run for 10.5 ns and required approximately 14.1 days per nanosecond of CPU time on four parallel processors of an Origin 3000, 120-MHz processor Silicon Graphic machine at New York University.
Average structures for the final systems were calculated in CHARMM using the last 1 ns of dynamics production.
System Preparation and Dynamics Simulations of Pol X A•G syn mismatch
The A•G syn mismatched model was built by substituting the base of the incoming nucleotide of the correct G•C ternary structure with guanine and template residue with adenine. The guanine base of the incoming mismatch nucleotide was rotated 180° with respect to the sugar moiety into a syn conformation (called A•G syn).
Energy minimizations and molecular dynamics simulations were performed using the molecular dynamics program NAMD73 with version C26a2 of the CHARMM force field69. First, the system was energy minimized for 10000 steps using the Powell algorithm. The system was then equilibrated for 20 ps at 300 K. Constant temperature was maintained at 300 K using weakly coupled Langevin dynamics of non-hydrogen atoms with a damping coefficient g of 10 ps−1; pressure was maintained at 1 atm using a Langevin piston Nosé-Hoover barostat with an oscillation period of 200 fs and a decay time of 100 fs. Bonds to all hydrogen atoms were kept rigid using SHAKE72, permitting a time step of 2 fs. The system was simulated in periodic boundary conditions, with full electrostatics computed using the Particle-Mesh-Ewald method74; 75 with grid spacing on the order of 1 Å or less. Short-range non-bonded terms were evaluated at every step using a 12 Å cutoff for van der Waals interactions and a smooth switching function. The total simulation length is 10.5 ns.
Acknowledgments
B.S.B. would like to thank Dr. Schlick for the generous use of her computational facilities. We also thank the reviewers for the suggestion to perform the A:G syn simulation and for helpful comments. Molecular images were generated using VMD 76 and the INSIGHT II package (Accelrys, San Diego, CA). This work was supported by NSF grant MCB-0316771, NIH grants R01 GM55164 and R01 ES012692, and the donors of the American Chemical Society Petroleum Research Fund to T.S. Partial support by Philip Morris USA and Philip Morris International to T.S. are also gratefully acknowledged. Finally, L.B. acknowledges the Rose Badgeley Trust Foundation for support.
Abbreviations
- ASFV
African Swine Fever Virus
- pol X
DNA Polymerase X
- BER
Base Excision Repair
- pol β
DNA polymerase β
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Salas ML. African swine fever virus (Asfaviridae) In: Granof RWaA., editor. Encyclopedia of Virology. 2. Academic Press; London: 1999. pp. 30–38. [Google Scholar]
- 2.Wilkinson PJ. The persistence of African swine fever virus in Africa and the Mediterranean. Prev Vet Med. 1984;2:71–82. [Google Scholar]
- 3.Kleiboeker SB. Swine fever: classical swine fever and African swine fever. Vet Clin North Am Food Anim Pract. 2002;18:431–51. doi: 10.1016/s0749-0720(02)00028-2. [DOI] [PubMed] [Google Scholar]
- 4.Vinuela E. African swine fever virus. Curr Top Microbiol Immunol. 1985;116:151–70. doi: 10.1007/978-3-642-70280-8_8. [DOI] [PubMed] [Google Scholar]
- 5.Dixon LK, Abrams CC, Bowick G, Goatley LC, Kay-Jackson PC, Chapman D, Liverani E, Nix R, Silk R, Zhang F. African swine fever virus proteins involved in evading host defence systems. Vet Immunol Immunopathol. 2004;100:117–34. doi: 10.1016/j.vetimm.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Yanez RJ, Rodriguez JM, Nogal ML, Yuste L, Enriquez C, Rodriguez JF, Vinuela E. Analysis of the complete nucleotide sequence of African swine fever virus. Virology. 1995;208:249–78. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]
- 7.Lamarche BJ, Tsai MD. Contributions of an endonuclease IV homologue to DNA repair in the African swine fever virus. Biochemistry. 2006;45:2790–803. doi: 10.1021/bi051772g. [DOI] [PubMed] [Google Scholar]
- 8.Redrejo-Rodriguez M, Garcia-Escudero R, Yanez-Munoz RJ, Salas ML, Salas J. African swine fever virus protein pE296R is a DNA repair apurinic/apyrimidinic endonuclease required for virus growth in swine macrophages. J Virol. 2006;80:4847–57. doi: 10.1128/JVI.80.10.4847-4857.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamarche BJ, Showalter AK, Tsai MD. An error-prone viral DNA ligase. Biochemistry. 2005;44:8408–17. doi: 10.1021/bi047706g. [DOI] [PubMed] [Google Scholar]
- 10.Oliveros M, Yanez RJ, Salas ML, Salas J, Vinuela E, Blanco L. Characterization of an African swine fever virus 20-kDa DNA polymerase involved in DNA repair. J Biol Chem. 1997;272:30899–910. doi: 10.1074/jbc.272.49.30899. [DOI] [PubMed] [Google Scholar]
- 11.Showalter AK, Byeon IJ, Su MI, Tsai MD. Solution structure of a viral DNA polymerase X and evidence for a mutagenic function. Nat Struct Biol. 2001;8:942–6. doi: 10.1038/nsb1101-942. [DOI] [PubMed] [Google Scholar]
- 12.Wilson SH. Mammalian base excision repair and DNA polymerase beta. Mutat Res. 1998;407:203–15. doi: 10.1016/s0921-8777(98)00002-0. [DOI] [PubMed] [Google Scholar]
- 13.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–6. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 14.Lindahl T. Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat Res. 2000;462:129–35. doi: 10.1016/s1383-5742(00)00024-7. [DOI] [PubMed] [Google Scholar]
- 15.Mol CD, Parikh SS, Putnam CD, Lo TP, Tainer JA. DNA repair mechanisms for the recognition and removal of damaged DNA bases. Annu Rev Biophys Biomol Struct. 1999;28:101–28. doi: 10.1146/annurev.biophys.28.1.101. [DOI] [PubMed] [Google Scholar]
- 16.Beard WA, Wilson SH. Structural insights into the origins of DNA polymerase fidelity. Structure. 2003;11:489–96. doi: 10.1016/s0969-2126(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 17.Radhakrishnan R, Arora K, Wang Y, Beard W, SH W, T S. Regulation of DNA repair fidelity by molecular checkpoints: “gates” in DNA polymerase beta’s substrate selection. Biochemistry. 2006;45:15142–56. doi: 10.1021/bi061353z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beard WA, Wilson SH. Structural design of a eukaryotic DNA repair polymerase: DNA polymerase beta. Mutat Res. 2000;460:231–44. doi: 10.1016/s0921-8777(00)00029-x. [DOI] [PubMed] [Google Scholar]
- 19.Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313:762–6. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 20.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–8. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 21.Delarue M, Poch O, Tordo N, Moras D, Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990;3:461–7. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 22.Beese LS, Steitz TA. Structural basis for the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. Embo J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steitz TA, Smerdon SJ, Jager J, Joyce CM. A unified polymerase mechanism for nonhomologous DNA and RNA polymerases. Science. 1994;266:2022–5. doi: 10.1126/science.7528445. [DOI] [PubMed] [Google Scholar]
- 24.Bolton EC, Mildvan AS, Boeke JD. Inhibition of reverse transcription in vivo by elevated manganese ion concentration. Mol Cell. 2002;9:879–89. doi: 10.1016/s1097-2765(02)00495-1. [DOI] [PubMed] [Google Scholar]
- 25.Yang L, Arora K, Beard WA, Wilson SH, Schlick T. Critical role of magnesium ions in DNA polymerase beta’s closing and active site assembly. J Am Chem Soc. 2004;126:8441–53. doi: 10.1021/ja049412o. [DOI] [PubMed] [Google Scholar]
- 26.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–15. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 27.Doublié S, Ellenberger T. The mechanism of action of T7 DNA polymerase. Curr Opin Struct Biol. 1998;8:704–712. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. Embo J. 1998;17:7514–25. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koshland DE. The key-lock theory and the induced-fit theory. Angew Chem Int Ed Engl. 1994;33:2375–2378. [Google Scholar]
- 30.Arora K, Schlick T. In silico evidence for DNA polymerase-beta’s substrate-induced conformational change. Biophys J. 2004;87:3088–99. doi: 10.1529/biophysj.104.040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maciejewski MW, Shin R, Pan B, Marintchev A, Denninger A, Mullen MA, Chen K, Gryk MR, Mullen GP. Solution structure of a viral DNA repair polymerase. Nat Struct Biol. 2001;8:936–41. doi: 10.1038/nsb1101-936. [DOI] [PubMed] [Google Scholar]
- 32.Jezewska MJ, Marcinowicz A, Lucius AL, Bujalowski W. DNA polymerase X from African swine fever virus: quantitative analysis of the enzyme-ssDNA interactions and the functional structure of the complex. J Mol Biol. 2006;356:121–41. doi: 10.1016/j.jmb.2005.10.061. [DOI] [PubMed] [Google Scholar]
- 33.Jezewska MJ, Bujalowski PJ, Bujalowski W. Interactions of the DNA Polymerase X of African Swine Fever Virus with Double-stranded DNA. Functional Structure of the Complex. J Mol Biol. 2007;373:75–95. doi: 10.1016/j.jmb.2007.06.054. [DOI] [PubMed] [Google Scholar]
- 34.Sampoli Benitez BA, Arora K, Schlick T. In silico studies of the African swine fever virus DNA polymerase X support an induced-fit mechanism. Biophys J. 2006;90:42–56. doi: 10.1529/biophysj.105.071944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang K, Niebuhr M, Aulabaugh A, Tsai M. Solution structures of 2: 1 and 1: 1 DNA polymerase-DNA complexes probed by ultracentrifugation and small-angle X-ray scattering. Nucleic Acids Res. 2008;36:849–60. doi: 10.1093/nar/gkm1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon L, Wilkinson P. Genetic diversity of African swine fever virus isolates from soft ticks (Ornithodoros moubata) inhabiting warthog burrows in Zambia. J Gen Virol. 1988;69:2981–93. doi: 10.1099/0022-1317-69-12-2981. [DOI] [PubMed] [Google Scholar]
- 37.García-Barreno B, Sanz A, Nogal M, Viñuela E, Enjuanes L. Monoclonal antibodies of African swine fever virus: antigenic differences among field virus isolates and viruses passaged in cell culture. J Virol. 1986;58:385–92. doi: 10.1128/jvi.58.2.385-392.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Showalter AK, Tsai MD. A DNA polymerase with specificity for five base pairs. J Am Chem Soc. 2001;123:1776–7. doi: 10.1021/ja005758x. [DOI] [PubMed] [Google Scholar]
- 39.Bakhtina M, Roettger MP, Kumar S, Tsai MD. A unified kinetic mechanism applicable to multiple DNA polymerases. Biochemistry. 2007;46:5463–5472. doi: 10.1021/bi700084w. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Escudero R, Garcia-Diaz M, Salas ML, Blanco L, Salas J. DNA polymerase X of African swine fever virus: insertion fidelity on gapped DNA substrates and AP lyase activity support a role in base excision repair of viral DNA. J Mol Biol. 2003;326:1403–12. doi: 10.1016/s0022-2836(03)00019-6. [DOI] [PubMed] [Google Scholar]
- 41.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–16. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 42.Batra V, Beard W, Shock D, Pedersen L, Wilson S. Nucleotide-induced DNA polymerase active site motions accommodating a mutagenic DNA intermediate. Structure. 2005;13:1225–33. doi: 10.1016/j.str.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 43.Kretulskie A, Spratt T. Structure of purine-purine mispairs during misincorporation and extension by Escherichia coli DNA polymerase I. Biochemistry. 2006;45:3740–6. doi: 10.1021/bi052306u. [DOI] [PubMed] [Google Scholar]
- 44.Zang H, Irimia A, Cho iJ, Angel K, Loukachevitch L, Egli M, Guengerich F. Efficient and high fidelity incorporation of dCTP opposite 7, 8-dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J Biol Chem. 2006;281:2358–72. doi: 10.1074/jbc.M510889200. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Reddy S, Beard W, Wilson S, Schlick T. Differing conformational pathways before and after chemistry for insertion of dATP versus dCTP opposite 8-oxoG in DNA polymerase beta. Biophys J. 2007;92:3063–70. doi: 10.1529/biophysj.106.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brieba L, Eichman B, Kokoska R, Doublié S, Kunkel T, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–61. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venkatramani R, Radhakrishnan R. Effect of oxidatively damaged DNA on the active site preorganization during nucleotide incorporation in a high fidelity polymerase from Bacillus stearothermophilus. Proteins. 2008;71:1360–72. doi: 10.1002/prot.21824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arora K, Beard WA, Wilson SH, Schlick T. Mismatch-induced conformational distortions in polymerase beta support an induced-fit mechanism for fidelity. Biochemistry. 2005;44:13328–41. doi: 10.1021/bi0507682. [DOI] [PubMed] [Google Scholar]
- 49.Moran S, Ren RXF, Kool ET. A thymidine triphosphate shape analog lacking Watson-Crick paring ability is replicated with high sequence selectivity. Proc Natl Acad Sci USA. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kool ET. Hydrogen bonding, base stacking and steric effects in DNA replication. Annu Rev Biophys Struct. 2001;30:1–22. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- 51.Morales JC, Kool ET. Functional hydrogen bonding map of the minor groove binding tracks of six DNA polymerases. Biochemistry. 2000;39:12979–12988. doi: 10.1021/bi001578o. [DOI] [PubMed] [Google Scholar]
- 52.Beard WA, Shock DD, Vande Berg BJ, Wilson SH. Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J Biol Chem. 2002;277:47393–8. doi: 10.1074/jbc.M210036200. [DOI] [PubMed] [Google Scholar]
- 53.Vande Berg BJ, Beard WA, Wilson SH. DNA structure and aspartate 276 influence nucleotide binding to human DNA polymerase beta. Implication for the identity of the rate-limiting conformational change. J Biol Chem. 2001;276:3408–16. doi: 10.1074/jbc.M002884200. [DOI] [PubMed] [Google Scholar]
- 54.Beard WA, Shock DD, Wilson SH. Influence of DNA structure on DNA polymerase beta active site function: extension of mutagenic DNA intermediates. J Biol Chem. 2004;279:31921–9. doi: 10.1074/jbc.M404016200. [DOI] [PubMed] [Google Scholar]
- 55.Radhakrishnan R, Schlick T. Fidelity discrimination in DNA polymerase beta: differing closing profiles for a mismatched (G:A) versus matched (G:C) base pair. J Am Chem Soc. 2005;127:13245–52. doi: 10.1021/ja052623o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiang Y, Goodman M, Beard W, Wilson S, Warshel A. Exploring the role of large conformational changes in the fidelity of DNA polymerase beta. Proteins. 2008;70:231–47. doi: 10.1002/prot.21668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alberts I, Wang Y, Schlick T. DNA polymerase beta catalysis: are different mechanisms possible? J Am Chem Soc. 2007;129:11100–10. doi: 10.1021/ja071533b. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Wu X, Guo D, Rechkoblit O, Taylor JS, Geacintov NEZW. Lesion bypass activities of human DNA polymerase mu. J Biol Chem. 2002;277:44582–7. doi: 10.1074/jbc.M207297200. [DOI] [PubMed] [Google Scholar]
- 59.Haracska L, Prakash LSP. Role of human DNA polymerase kappa as an extender in translesion synthesis. Proc Natl Acad Sci U S A. 2002;99:16000–5. doi: 10.1073/pnas.252524999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson R, Washington M, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–9. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 61.Nair D, Johnson RE, Prakash S, Prakash LAKA. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;309:377–80. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 62.Kumar S, Lamarche BJ, Tsai MD. Use of damaged DNA and dNTP substrates by error-prone DNA polymerase X from African Swine Fever Virus. Biochemistry. 2007;46:3814–25. doi: 10.1021/bi061501l. [DOI] [PubMed] [Google Scholar]
- 63.Beckman KB, Ames BN. Oxidative decay of DNA. J Biol Chem. 1997;272:19633–6. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, Schlick T. Distinct energetics and closing pathways for DNA polymerase beta with 8-oxoG template and different incoming nucleotides. BMC Struct Biol. 2007;7 doi: 10.1186/1472-6807-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kincaid K, Beckman J, Zivkovic A, Halcomb R, Engels J, Kuchta R. Exploration of factors driving incorporation of unnatural dNTPS into DNA by Klenow fragment (DNA polymerase I) and DNA polymerase alpha. Nucleic Acids Res. 2005;33:2620–8. doi: 10.1093/nar/gki563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sintim H, Kool E. Remarkable sensitivity to DNA base shape in the DNA polymerase active site. Angew Chem Int Ed Engl. 2006;45:1974–9. doi: 10.1002/anie.200504296. [DOI] [PubMed] [Google Scholar]
- 67.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM a program for macromolecular energy, minimization and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 68.MacKerell ADJ, Brooks BR, Brooks CL, III, Nilsson L, Roux B, Won Y, Karplus M. CHARMM: The energy function and its parameterization. In: Schleyer PvR., editor. Encyclopedia of Computational Chemistry. John Wiley and Sons, Ltd; New York, NY: 1998. pp. 271–277. [Google Scholar]
- 69.MacKerell ADJ, Banavali NK. All-atom empirical force-field for nucleic acids. II Applications to molecular dynamics simulations of DNA and RNA in solution. J Comp Chem. 2000;21:105–120. [Google Scholar]
- 70.Schlick T. Optimization methods in computational chemistry. In: Boyd KBLaDB., editor. Reviews in Computational Chemistry. Vol. 3. VCH Publishers; New York, NY: 1992. pp. 1–71. [Google Scholar]
- 71.Yang L, Beard W, Wilson S, Roux B, Broyde S, Schlick T. Local deformations revealed by dynamics simulations of DNA polymerase Beta with DNA mismatches at the primer terminus. J Mol Biol. 2002;321:459–78. doi: 10.1016/s0022-2836(02)00617-4. [DOI] [PubMed] [Google Scholar]
- 72.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 73.Phillips J, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel R, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Darden T, York DM, Pedersen LG. Particle mesh Ewald: An N•logN method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 75.Norberg J, Nilsson L. On the truncation of long-range electrostatic interactions in DNA. Biophys J. 2000;79:1537–53. doi: 10.1016/S0006-3495(00)76405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]