Abstract
The chromosome 11q13.5 locus is frequently amplified in several types of human cancer. We have previously shown that 11q13.5 amplification was associated with significantly shorter overall survival in ovarian cancer patients but the molecular mechanisms of how amplification of this locus contributes to disease aggressiveness remain unclear. As ovarian cancer mortality is primarily related to resistance of chemotherapeutic agents, we screened the top six candidate genes within this amplicon for their contribution to drug resistance. Rsf-1 (also known as HBXAP) was found to be the only gene in which gene knockdown sensitized tumor cells to paclitaxel. Rsf-1 has been known to interact with hSNF2H to form an ISWI chromatin remodeling complex. We found that Rsf-1 was upregulated in paclitaxel resistant ovarian cancer cell lines and Rsf-1 immunoreactivity in primary ovarian carcinoma tissues correlated with in vitro paclitaxel resistance. Ectopic expression of Rsf-1 significantly enhanced paclitaxel resistance in ovarian cancer cells. Downregulation of hSNF2H or disruption of hSNF2H and Rsf-1 interaction enhanced paclitaxel sensitivity in tumor cells with Rsf-1 upregulation. Rsf-1 expression altered expression in several genes and activated certain signaling pathways that may contribute to drug resistance. In conclusion, our results suggest that Rsf-1 is the major gene within the 11q13.5 amplicon that contributes to paclitaxel resistance and the formation of the Rsf-1/hSNF2H complex is required for inducing this phenotype.
Keywords: Ovarian, Cancer, Genomics, Genetics, Pathogenesis
Introduction
Gene amplification is a common mechanism underlying oncogenic activation in human cancer (1, 2). Amplification of MYC family genes, EGFR, ERBB2 (Her2/neu), CCND1 (cyclin D1), CCNE (cyclin E), and FGFR has been reported in a variety of human neoplastic diseases. Amplifications in several of the oncogenes are associated with worse clinical outcome and are amenable to specific targeted therapy (3). In an effort to identify amplified chromosomal regions that may harbor novel cancer-associated genes, we have applied both digital karyotyping (4) and single nucleotide polymorphism (SNP) arrays (5) to analyze DNA copy number alterations in purified high-grade ovarian serous carcinoma, one of the most aggressive type of neoplastic diseases in women. Based on previous studies (6, 7), we found that the most common amplicons in high-grade ovarian serous carcinomas were those harboring CCNE (cyclin E1), Notch3, AKT2 and a discrete chromosomal region at 11q13.5 that contained several cancer-associated genes including p21/cdc42/Rad-activated kinase (PAK1) (8, 9), Gab2 (10) and Rsf-1 (HBXAP) (6). Fluorescence in situ hybridization analysis demonstrated 11q13.5 amplification in 13%-15% of high-grade ovarian serous carcinomas (6, 7). 11q13.5 amplification was significantly associated with worse clinical outcome in patients with high-grade serous carcinomas in independent retrospective cohorts (6, 11). Besides ovarian cancer, this amplicon is frequently detected in several human malignancies including breast, oral, esophageal, and head and neck carcinomas (12). These findings suggest that gene(s) within this amplicon may contribute to clinical aggressiveness in the neoplasms with 11q13.5 amplification.
The clinical outcome in ovarian cancer patients depends on several factors and the development of drug resistance is one of the main causes that account for the poor prognosis in patients who suffer from this disease. In order to identify the potential gene(s) within the 11q13.5 amplicon which may play a mechanistic role in developing drug resistance in high-grade ovarian serous carcinomas, we first selected the top six genes within this amplicon that demonstrated the most significant correlation between DNA and RNA copy number from a total of 14 genes within the amplicon. After knockdown of each gene using RNA interference, we determined their sensitivity to paclitaxel and carboplatin which are routinely used in treating ovarian cancer patients after cytoreduction surgery. We found that Rsf-1 knockdown significantly decreased the IC50 of paclitaxel in ovarian cancer cells. In this study, we further characterized the molecular mechanism of how Rsf-1 expression contributed to the development of drug resistance.
Materials and Methods
Cell lines and culture conditions
Ovarian cancer cells including OVCAR3, SKOV3, and A2780 cells were purchased from ATCC (Rockville, MD) and MPSC1 was previously established by us (13). Paclitaxel and carboplatin resistant cell lines were generated by selecting the viable MPSC1, A2780, OVCAR3, and SKOV3 cells three months after exposure to paclitaxel (0.25-0.5 μM) or carboplatin (0.5-2.0 μg/ml). The resistant cell clones from each cell line were maintained in culture medium containing paclitaxel (0.25 μM for SKOV3 and 0.5 μM for A2780, OVCAR3, and MPSC-1) or carboplatin (2.0 μg/ml for SKOV3 and MPSC1, 1.0 μg/ml for A2780, and 0.5 μg/ml for OVCAR3). All the cell lines used in this study were cultured in RPMI1640 medium containing 5% fetal bovine serum.
Selection of genes within the 11q13.5 amplicon for functional studies
As reported in our previous study (6), 10 high-grade ovarian serous carcinomas with 11q13.5 amplification and 53 control specimens without such amplification were analyzed to assess mRNA levels in all the genes within the minimal amplicon using quantitative real-time PCR. The control group included one freshly brushed ovarian surface epithelium (kind gift from Dr. M. Birrer), 6 benign cystadenomas, 10 serous borderline tumors and 36 high-grade serous carcinomas that did not harbor 11q13.5 amplification. In our previous report (6), we performed quantitative real-time PCR for all the genes except NARS2 because this gene has only recently been updated to the genome assembly database. Thus, the NARS2 expression level was analyzed in this study using the same method as used for other genes (6). The Mann-Whitney (rank sum) test was used to determine the significance in individual gene expression between 11q13.5 amplified and non-amplified specimens.
Rsf-1 inducible cell clones
The full-length Rsf-1 gene tagged with a V5 epitope tag at the C-terminal was cloned into the Tet-off expression vector, pTRE-hygro (Clontech, Mountain View, CA). Parental SKOV3 cells were first transfected with a tetracycline-controlled transactivator (tTA) expression vector and the tTA stable clones were selected by G418 (300 μg/ml). The inducible Rsf-1 vector was then introduced into the SKOV3-tTA cells, and the stable transfectants were selected by 150 μg/ml hygromycin and maintained in the culture medium containing 300 μg/ml G418, 150 μg/ml hygromycin and 2 μg/ml doxycycline (Dox). In this study, we selected two clones of Rsf-1 inducible cells and the pooled control cells with the tTA-vector only construct. To determine the efficiency of Rsf-1 induction in the Tet-off system, we performed Western blots to analyze Rsf-1 protein expression at different time points after the gene was turned-on using Dox-free media.
Immunohistochemistry
A mouse monoclonal anti-Rsf-1 antibody (clone 5H2/E4) was purchased from Upstate (Lake Placid, NY) and was used in immunohistochemistry at a dilution of 1:2,000. The specificity of the antibody has been validated in previous reports (6, 14). An EnVision+System peroxidase kit (DAKO, Carpinteria, CA) was used for staining following the manufacturer's instruction. Immunohistochemical staining was carried out on tissue microarrays containing 99 high-grade ovarian serous carcinoma tissues for which their in vitro drug resistance status was available. Immunointensity for Rsf-1 was scored as low (≤2) or high (>2). This criteria was used because of a significant correlation between immunointensity (score >2) and Rsf-1 amplification (6).
Real-time RT-PCR
Relative gene expression was measured by quantitative real-time PCR using an iCycler (Bio-Rad, Hercules, CA), and threshold cycle numbers (Ct) were obtained using the iCycler Optical system interface software. PCR primers were designed using the Primer 3 program. The primers for NARS2 real-time PCR were 5'-GACTCTGAGGGAGCTGGAGAAC-3' (forward) and 5'-AAGGTCGGACCAAAGGTAAACA-3' (reverse). The primers for CYR61 were 5'-CTCCCTGTTTTTGGAATGGA-3' (forward) and 5'-TGGTCTTGCTGCATTTCTTG-3' (reverse). The primers for osteopontin were 5'-TGAAATTCATGGCTGTGGAA-3' (forward) and 5'-ATG GTGCATACAAGGCCATC-3' (reverse). The primers for CTGF were 5'-CCTGGTCCAGACCA CAGAGT-3' (forward) and 5'-TGGAGATTTTGGGAGTACGG-3' (reverse). The other primers including those for Rsf-1, hSNF2H and beta-amyloid precursor gene (APP) were shown in previous reports (6, 15). The mean Ct of the gene of interest was calculated from replicate measurements and normalized with the mean Ct of a control gene, APP, for which expression is relatively constant among the SAGE libraries analyzed (16).
Gene knockdown using siRNA and shRNA
For functional screening of the top six genes, we purchased two small hairpin RNAs (shRNAs) for each gene from Sigma. hSNF2H specific small interfering RNAs (siRNAs) (UUCAAAUCGAGUGCAAACA) and (UUAAUAUCCGAGUAUACCA) and control siRNA that targeted the Luciferase gene (GAUUAAAUCUUCUAGCGACUGCUUCGC) were synthesized by the Integrated DNA Technologies (Coralville, IA). Cells were transfected with shRNA or siRNA at a final concentration of 2 μg or 200 nM, respectively, using lipofectamine (Invitrogen, Carlsbad, CA). Six hours after transfection, the cells were washed and harvested at the next day for cell growth and drug resistance assays. To enhance the silencing effect of Rsf-1 transcripts in the follow-up experiments, we used lentivirus carrying the Rsf-1 shRNA sequence templates (CCGGCCAGTTCTGAACTTTGAAGATCTCGAGATCTTCAAAGTTCAGAACT) and (CCGGCTTCTGAGACAAAGGGTTCTACTCGAGTAGAACCCTTTGTCTCAGA) which were inserted into the lentiviral plasmid (pLKO.1-puro).
Cell growth and drug resistance assay
Cells were grown in 96-well plates at a density of 3,000 cells per well. Cell number was measured by the incorporation of SYBR green I nucleic acid gel stain (Molecular Probes, Eugene, OR) using a fluorescence microplate reader (Fluostar from BMG, Durham, NC). Data was determined from four replicates and was expressed as the fold increase to the control group. For drug resistance assay, cells were seeded in 384-well plates at a density of 600 cells per well. After overnight culture, the cells were treated with a series of concentrations of paclitaxel or carboplatin. Four days after transfection (i.e., three days after drug treatment), 8 μl of cell-titer blue (Promega) was added and the plates were incubated for five hours. The absorbance was determined using a fluorescence microplate reader. The signal produced by conversion of resazurin to resorufin is directly proportional to viable cell number. Data was determined from four replicates and was expressed as the fold increase of the control group. IC50 was defined as the concentration that results in a 50% decreased in the number of cells as compared to that of the control cultures in the absence of the drug.
GeneChip analysis for transcript expression
RNA was prepared using a Qiagen RNAeasy kit from Rsf-1 inducible SKOV3 cells (15) in different experimental conditions. Affymetrix U133 Plus 2 arrays were used to analyze gene expression from Rsf-1 induced SKOV3 cells (48 hrs after induction), mock induced SKOV3 cells (48 hrs after mock induction) and vector-only control SKOV3 cells. Probe labeling, hybridization and scanning for the arrays were performed using the standard protocols at the Johns Hopkins Microarray Core. The dCHIP program was used to analyze the array data to select the differentially expressed genes that were overexpressed in the Rsf-1 induced group as compared to the mean of control groups including mock induced and vector-only control SKOV3 cells. The top up-regulated and down-regulated genes with corresponding expression values that were >2.5 folds of increase or decrease as compared to the control were input into the Ingenuity Pathways Analysis System (www.ingenuity.com). These genes, called focus genes, were overlaid onto a global molecular network developed from information updated in the Ingenuity Pathways Knowledge Base. Networks of these focus genes were then algorithmically generated based on their connectivity.
Drug resistance clinical assay
The assay was performed in Oncotech, Inc (Tustin, CA) using published protocol1. Briefly, a small piece of fresh tumor tissue was cultured in agar-based cell culture system and radioactive thymidine incorporation was used for measuring the cell growth in the presence of different chemotherapeutic drugs. The level of drug resistance was quantified and was classified into three groups: 1) low if tumor cells were inhibited by the tested agent and demonstrated less than median growth; 2) intermediate if there was moderate tumor growth; and 3) extreme if tumor cell growth was virtually unaffected by the chemotherapeutic agent. As defined in previous reports, extreme drug resistance (EDR) was interpreted as drug resistant and the intermediate and low drug resistance was interpreted as drug sensitive (1, 11).
Results
Selection of genes within the 11q13.5 amplicon for functional characterization
Using digital karyotyping to detect genome-wide DNA copy number alterations in high-grade ovarian serous carcinomas, we identified a minimal amplicon in the 11q13.5 chromosomal region (6). According to the UCSC Human Genome Browser Gateway in March 2006 (http://genome.ucsc.edu/cgi-bin/hgGateway), there were a total of 14 genes in which their complete coding sequences were located within this minimal amplicon. To identify the candidate cancer-driving genes from the 14 genes, we applied an approach based on the rationale that a tumor-driving gene, if amplified, is almost always over-expressed while the co-amplified “passenger” genes that are not involved in tumor development may or may not be so (17). Based on the Mann-Whitney (rank sum) test, we selected six genes showing the highest correlation between DNA and transcript copy number (p< 0.001). They included Rsf-1 (HBXAP) (p< 0.0001), INTS4 (p= 0.0001), CLNS1A (p= 0.0001), ALG8 (p= 0.0001), GAB2 (p= 0.0001), and PAK1 (p= 0.0002). The p- values for NDUFC2, C11orf67 and USP35 were 0.0015, 0.012 and 0.022, respectively. There was no statistical significance for the remaining genes including GFPD4, AQP11, KCTD14, THRSP and NARS2. Therefore, we selected Rsf-1 (HBXAP), INTS4, CLNS1A, ALG8, GAB2, and PAK1 for functional characterization.
Biological effects of gene knockdown of the six candidate genes
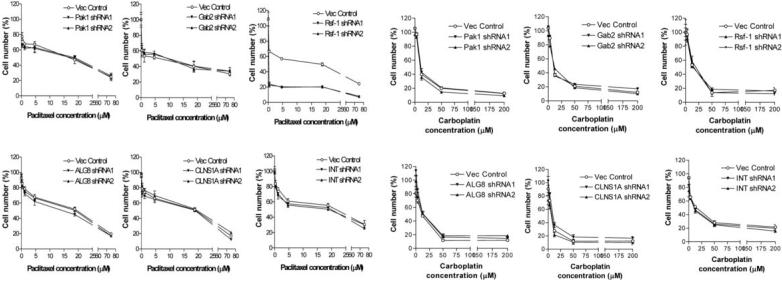
To determine the biological roles of those six genes in sustaining cell growth and in conferring drug resistance, we transfected OVCAR3 ovarian cancer cells, known to harbor a high level of 11q13.5 amplification, with shRNAs to knockdown each of the six genes. Two independent shRNAs with satisfied knockdown efficiency were used for each gene. Cell number of OVCAR3 cells was determined one day and four days after shRNA transfection. Supplementary Fig. 1 showed that cell number was most remarkably reduced in the cells transfected with Rsf-1 shRNAs as compared to PAK1, Gab2, ALG8, INTS4 and CLNS1A shRNAs. To determine if these six genes are potentially involved in drug resistance, we measured the IC50 of paclitaxel and carboplatin in OVCAR3 cells after shRNA transfection. Of note, the cell numbers in drug sensitivity assays were normalized to those in shRNA-treated SKOV3 cells without gene knockdown. For paclitaxel, we found that a significant decrease in the IC50 occurred only in Rsf-1 gene knockdown but not in other genes (Fig. 1A). For carboplatin, none of the shRNAs that target these six genes affected the IC50 (Fig. 1B). Therefore, we selected Rsf-1 to further study its role in enhancing paclitaxel resistance in ovarian cancer cells.
Figure 1. Effect of knockdown of the six genes within the ch11q13.5 on IC50 of paclitaxel (A) and carboplatin (B) in OVCAR3 cells.
Two shRNAs (shRNA1 and shRNA 2) to target each gene were used to knock down PAK-1, Gab2, Rsf-1, ALG8, CLNS1A and INTS4 in OVCAR3 cells with ch11q13.5 amplification. Vector-only was used as the negative control. Rsf-1 is the only gene in which shRNAs significantly reduce IC50 of paclitaxel.
Correlation of Rsf-1 protein expression and drug resistance in vitro and in vivo
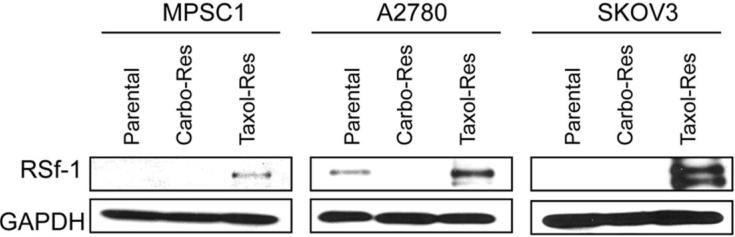
To determine whether Rsf-1 gene upregulation was correlated with paclitaxel resistance, we performed Western blot on the ovarian cancer cell lines including MPSC1, A2780, and SKOV3, and performed immunohistochemistry on clinical tumor specimens. First, we established three paclitaxel resistant ovarian cancer cell lines, SKOV3TR, A2780TR and MPSC1TR and three carboplatin resistant ovarian cancer cell lines, SKOV3CR, A2780CR and MPSC1CR by treating the parental cells with increasing concentrations of paclitaxel and carboplatin. We then compared Rsf-1 expression between drug resistant and parental cells. As shown in Fig. 2, Rsf-1 protein levels were significantly higher in paclitaxel resistant cells than their parental controls in all three cell lines. Like in OVCAR3 cells, the cell number was decreased in SKOV3TR cells after treatment with Rsf-1 shRNAs (p< 0.01) (supplementary Fig. 2). Next, we determined whether Rsf-1 expression correlated with paclitaxel resistance in ovarian cancer cells isolated from the primary tumors in patients with stage III or IV ovarian cancer. Based on immunohistochemistry, we were able to demonstrate that Rsf-1 immunointensity was significantly associated with increased paclitaxel resistance (Table 1). In contrast, there was no statistically significant correlation between Rsf-1 expression level and resistance to carboplatin. Rsf-1 immunoreactivity in representative specimens was shown in the supplementary Fig. 3.
Figure 2. Enhanced expression of Rsf-1 protein in paclitaxel-resistant ovarian cancer cells.
Western blot analysis shows that Rsf-1 protein levels are significantly higher in the paclitaxel-resistant ovarian cancer cell lines as compared to the parental and carboplatin-resistant cells. GAPDH was used as a loading control.
Table 1.
Correlation between in vitro drug resistance and Rsf-1 expression
| Staining Score ≤ 2 |
Staining Score > 2 |
Chi square test | |
|---|---|---|---|
|
Carboplatin-sensitive Carboplatin-resistant |
31 32 |
15 18 |
P=0.8305 |
|
Paclitaxel-sensitive Paclitaxel-resistant |
43 20 |
7 26 |
P<0.0001* |
Rsf-1 expression enhances paclitaxel resistance in ovarian cancer cells
To demonstrate the causal role of Rsf-1 in developing drug resistance, we used Tet-off Rsf-1 inducible SKOV3 ovarian cancer cells which expressed a very low level of Rsf-1 in the parental and non-induced cells (15). Western blot analysis showed that Rsf-1 protein was detected 6 hrs and 12 hrs after induction in two independent clones (Fig. 3A). Rsf-1 expression did not have a significant effect on cellular proliferation (supplementary Fig. 4); however, both clone 1 and clone 2 after Rsf-1 induction became more resistant to paclitaxel than the control clones without Rsf-1 induction (Fig. 3B). The IC50 of paclitaxel in Rsf-1 non-induced (doxycycline on) clone 1 and clone 2 cells were 0.15 μM and 0.11 μM, respectively, whereas the IC50 of paclitaxel in clone 1 and clone 2 after Rsf-1 induction (doxycycline off) was 2.8 μM and 7.2 μM, respectively. In contrast, the control cells had a similar IC50 in both doxycycline-on and -off conditions. Rsf-1 induction did not confer carboplatin resistance as reflected by similar IC50 curves between Rsf-1 induced and non-induced cells (Fig. 3C).
Figure 3. Rsf-1 induction in SKOV3 cells increases paclitaxel resistance.
A. Western blot analyses demonstrate that Rsf-1 protein expression is detectable as early as 6 hours after removal of doxycycline in two randomly selected clones (clone-1 and clone-2). B and C. Drug resistance assay shows that Rsf-1 expression confers resistance to paclitaxel (B) but not to carboplatin (C) in both Rsf-1 inducible SKOV3 clones.
The role of hSNF2H in Rsf-1 dependent paclitaxel resistance
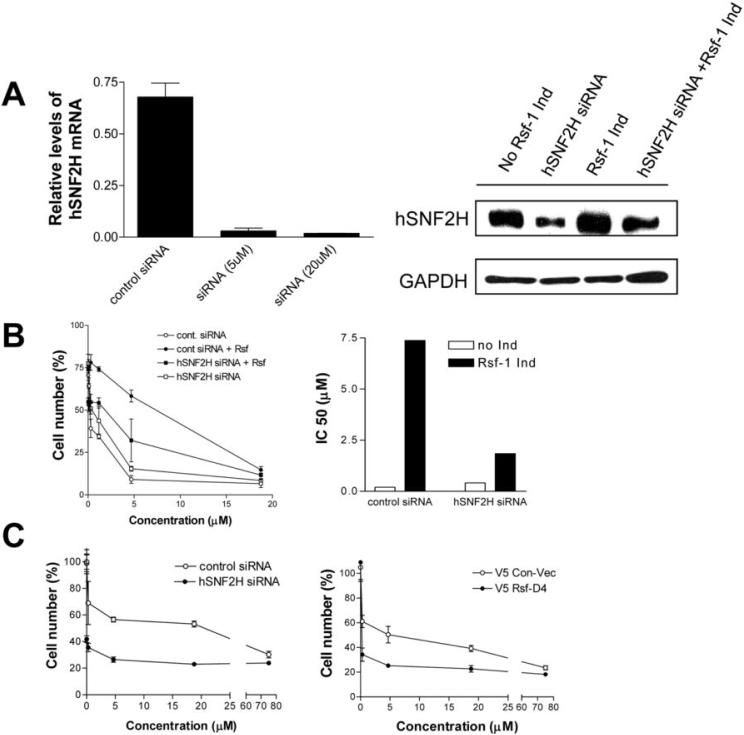
It has been known that Rsf-1 interacts with hSNF2H to form an ISWI chromatin remodeling complex in which Rsf-1 serves as a histone chaperone and hSNF2H as an ATPase (18-21). Here, we determined if hSNF2H was required for Rsf-1 to confer paclitaxel resistance. As shown in Fig. 4A, gene knockdown of hSNF2H reduced its expression at both mRNA and protein levels in SKOV3 cells. Reducing hSNF2H expression had only a minimal effect on cell proliferation in the control SKOV3 cell line (supplementary Fig. 5). Next, we measured the IC50 of paclitaxel in Rsf-1 inducible SKOV3 cells after transfection with hSNF2H or control siRNA in both Rsf-1 induced and non-induced conditions. We found that hSNF2H siRNA significantly decreased the IC50 of paclitaxel in Rsf-1 expressing SKOV3 cells. In contrast, hSNF2H siRNA had only a modest effect on the IC50 in cells without Rsf-1 induction (Fig. 4B). Next, we further tested if hSNF2H knockdown or expression of an Rsf-1 dominant negative protein, Rsf-D4 (15), could sensitize OVCAR3 cells with abundant endogenous Rsf-1 expression to paclitaxel. We found that either hSNF2H siRNA or Rsf-D4 reduced the cell proliferation to 81% and 70% of the controls, respectively. Interestingly, both approaches also enhanced the paclitaxel sensitivity in OVCAR3 cells (Fig. 4C). The above results indicated an important role of hSNF2H in Rsf-1 mediated paclitaxel resistance.
Figure 4. The role of hSNF2H in paclitaxel sensitivity.
A. Quantitative RT-PCR shows mRNA expression levels of hSNF2H are greatly reduced by hSNF2H siRNA in OVCAR3 cells with high level expression of endogenous hSNF2H (left panel). Similarly, Western blot analysis shows reduction of hSNF2H expression by hSNF2H siRNA 24 hours after siRNA transfection in the Rsf-1 inducible SKOV3 cells (right panel). siRNA against luciferase was used as the control. B. Rsf-1 inducible cells transfected with control or hSNF2H siRNAs were treated with various concentrations of paclitaxel under Rsf-1 turned-on or -off condition. Relative cell number in different treatment group was determined and presented as percentage of the control group at day 3 (left panel). IC50 of paclitaxel in Rsf-1 inducible SKOV3 cells transfected with control or hSNF2H siRNAs were determined under Rsf-1 turned-on or -off condition (right panel). hSNF2H siRNAs significantly decreases the IC50 of paclitaxel in Rsf-1 expressing SKOV3 cells. C. Effect of hSNF2H siRNA and Rsf-D4 expression on sensitivity of OVCAR3 cells to paclitaxel. In contrast to SKOV3 cells, OVCAR3 cells constitutively express Rsf-1 due to a high level of 11q13.5 amplification. Both hSNF2H siRNA (left panel) and Rsf-D4 (right panel) significantly sensitize OVCAR3 cells to paclitaxel treatment. The cell number at each concentration points was normalized to hSNF2H siRNA and Rsf-D4 treated cells without paclitaxel treatment.
Genes regulated by Rsf-1 expression
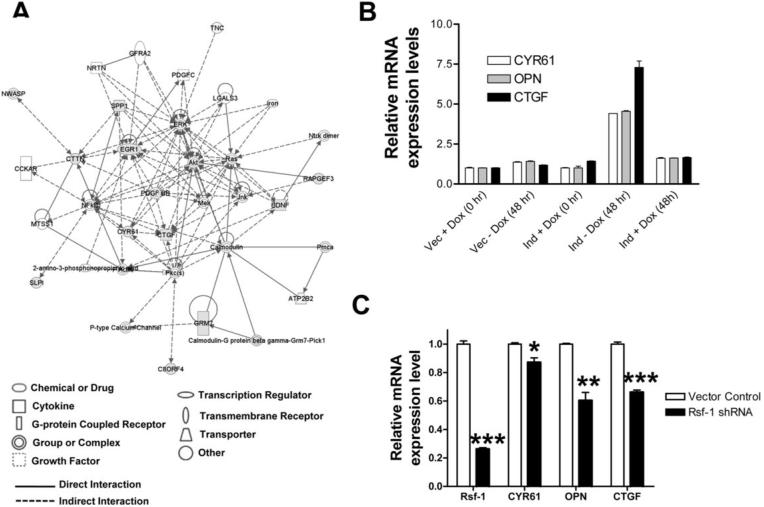
To further identify the Rsf-1 regulated genes and pathways that were potentially involved in drug resistance, we performed global transcript profiling to compare gene expression in Rsf-1 induced and control SKOV3 cells (mock induced and vector only controls) using the Affymetrix U133-Plus 2 microarrays. As compared to the Rsf-1 turn-off (mock induced) group and the vector-only control, Rsf-1 induced cells demonstrated upregulation and downregulation in 62 genes with at least 2.5 folds increase or decrease as compared to the controls (supplementary Table 1). To identify the hidden connections among the regulated genes, we performed a network analysis of the top 25 up-regulated and 37 downregulated genes using the Ingenuity Pathways Analysis Systems. Applying this Ingenuity Network filter, four functional networks could be constituted with significant Ingenuity scores. The network with the highest score was related to the Tissue Morphology, Cellular Assembly and Organization System (Fig. 5A). Several molecular hubs including NFkB, ERK, Akt and EGR1 to which many pathways converge were identified in the network. Among the candidate genes, three representative genes including osteopontin, cysteine rich protein 61 (CYR61) and connective tissue growth factor (CTGF) were selected for validation by quantitative real time PCR because these three genes have been implied in the pathogenesis of in human cancer (22-24). As shown in Fig. 5B and 5C, all three genes showed statistically significant upregulation in SKOV3 cells after Rsf-1 induction and demonstrated downregulation in OVCAR3 cells after Rsf-1 shRNA transfection, indicating that Rsf-1 proteins were required and sufficient to upregulate the expression of CYR61, CTGF, and osteopontin.
Figure 5. Candidate genes regulated by Rsf-1 and their interaction network.
SKOV3-Rsf-1-TRE (Rsf-1 inducible) cells and SKOV3-TRE (vector control) cells were incubated in culture medium with or without doxycycline and cells were harvested for mRNA extraction and Affymetrix array hybridization. A. The Ingenuity pathways analysis of Rsf-1 regulated genes listed in supplementary Table 1. A total of 25 up-regulated and 37 down-regulated genes with fold increase or decrease by at least 2.5 folds were input to the Ingenuity Pathways Analysis System. This system facilitates the identification of functional connections among Rsf-1-regulated genes with known relevance to the Tissue Morphology, Cellular Assembly and Organization System. Only the top network with a score of 34 is shown. Red shapes, up-regulated genes; green shapes, downregulated genes; white shapes, molecules added from Ingenuity's knowledge base. B. Quantitative real time PCR demonstrates the pattern of gene expression in three selected genes including osteopontin (OPN), CYR61 and connective tissue growth factor (CTGF) in Rsf-1 inducible (ind) and tTA-vector only (vec) SKOV3 cells in the presence (+Dox) or absence of Doxycyline (-Dox). C. The expression in OPN, CYR61 and CTGF and Rsf-1 decreases after transfection of Rsf-1 shRNAs in the OVCAR3 cells with a high level expression of endogenous Rsf-1. One asterisk: p = 0.018; two asterisks: p = 0.002; three asterisks: p < 0.001.
Discussion
Chromosome 11q13.5 amplification has been detected in several types of human cancer and the presence of the amplicon is associated with a worse clinical outcome. For example, our previous reports (6, 11) demonstrated that 11q13.5 amplification in ovarian serous carcinomas was significantly associated with shorter overall survival. However, the mechanisms accounting for the dismal clinical outcome are not clear. In the current study, we performed a functional genomic screening on genes located in the 11q13.5 amplicon for their roles in developing drug resistance to paclitaxel and carboplatin that are routinely used in treating patients at advanced stages of ovarian cancer. Our result showed that Rsf-1 (HBXAP) played an important role in conferring paclitaxel resistance in ovarian cancer cells.
Rsf-1/HBXAP encodes a nuclear protein containing several distinct structure motifs including a PHD domain (25). This protein was first identified as a novel cellular protein which bound to the pX nuclear protein of hepatitis B virus (26). HBXAP expression increases HBV transcription in a pX-dependent manner, suggesting its role in regulating the virus life cycle. Further studies demonstrated that the interaction between HBXAP and pX proteins regulated NF-κB activation (26). Soon after HBXAP was identified, an independent research group reported the full-length HBXAP (Rsf-1) protein as a subunit in a chromatin assembly factor, called ISWI-containing factor RSF (Remodeling and Spacing Factor) (18). The human RSF complex is composed of two subunits: the nucleosome-dependent ATPase, hSNF2H, and the histone chaperon, Rsf-1 (18, 20, 27). The Rsf-1/hSNF2H (or RSF) complex mediates nucleosome deposition and assembly, and participates in chromatin remodeling by mobilizing nucleosomes in response to a variety of growth modifying signals and environmental cues (18-21). Such nucleosome remodeling is essential for transcriptional regulation (25, 26, 28), DNA replication (29) and cell cycle progression (30). We have previously demonstrated that Rsf-1 and hSNF2H were co-upregulated in ovarian cancer tissues and that expression of Rsf-1 may likely prevent hSNF2H from protein degradation (15). Ectopic expression of Rsf-1 in SKOV3 ovarian cancer cells with a very low level of endogenous Rsf-1 expression increased hSNF2H protein levels and promoted SKOV3 tumor growth in a mouse tumor xenograft model (15).
Our findings from this study could have several biological and clinical implications. First, the current results provide a possible explanation for how 11q13.5 amplification in ovarian serous carcinomas contributes to shorter overall survival as compared to those without 11q13.5 amplification (6, 11). It is likely that tumors with increased DNA copy number of Rsf-1 overexpress Rsf-1 protein which renders the de novo paclitaxel resistance. The Rsf-1 expressing residual tumors after tumor cytoreduction surgery may survive better during chemotherapy and tend to recur earlier, and thus they are likely related to poor clinical outcome in ovarian cancer patients. Second, hSNF2H knockdown or expression of an Rsf-1 dominant negative protein containing the hSNF2H binding motif sensitized paclitaxel effects in cancer cells with Rsf-1 overexpression. These observations suggest that paclitaxel resistance in Rsf-1 upregulated cells is mediated by the chromatin remodeling activity from the Rsf-1/hSNF2H complex. This finding may have translational implications for new cancer therapy as enhancing the sensitivity of chemotherapy can be achieved by inactivating the Rsf-1/hSNF2H or by interrupting the complex formation in cancer cells. Third, our previous result showed that hSNF2H was required for cell proliferation and survival in Rsf-1 expressing OVCAR3 cells but not in SKOV3 cells with or without Rsf-1 induction. This finding is probably because OVCAR3 cells with Rsf-1 gene amplification and constitutive upregulation have become molecularly “addicted” to Rsf-1/hSNF2H expression, while SKOV3 cells which have a very low level of endogenous Rsf-1 expression (15) are less sensitive to hSNF2H knockdown even after an acute induction of Rsf-1. However, reducing hSNF2H expression sensitized both OVCAR3 cells and Rsf-1 expressing SKOV3 cells to paclitaxel, suggesting that different mechanisms are involved in promoting cell growth and drug resistance. Fourth, Rsf-1 expression in tumor specimens can be used as a surrogate marker alone or in combination with other markers to predict treatment response to paclitaxel. To this end, future multi-institutional cohort studies are required to validate the usefulness of Rsf-1 immunoreactivity as a potential diagnostic test for ovarian cancer or other cancer types with Rsf-1 amplification and overexpression.
There are at least two possible mechanisms that could explain how Rsf-1 overexpression contributes to tumor cell survival and growth in the presence of paclitaxel. First, an increased number of RSF complexes may concentrate on specific promoter and/or enhancer regions to modulate transcription activity of a set of genes which participate in drug resistance. In fact, based on comparison of gene profiles between Rsf-1 induced and non-induced SKOV3 ovarian cancer cells, we found that Rsf-1 expression was associated with changes in the expression of several genes. Among them, upregulation of CYR61 and osteopontin is of interest because both gene products were previously reported to be involved in developing drug resistance (22, 23). Furthermore, using interaction network analysis, it appears that several major molecular hubs were identified in this network and they included NFκB, ERK, Akt and EGR1. Among them, NFκB, ERK and Akt have been suggested to participate in developing chemoresistance in cancer cells (30-37). For example, the Akt pathway has been shown to play a causal role in the development of docetaxel and paclitaxel resistance in cancer cell lines including those derived from ovarian cancer (31-33). It is of great interest that Rsf-1 has been shown to interact with NF-κB (34). Further studies are required to demonstrate the detailed mechanisms how the Rsf-1/hSNF2H complex contributes to tumor development through their downstream mediators. Second, Rsf-1 overexpression may alter the dynamics of partnership between hSNF2H and hSNF2H binding proteins. Besides Rsf-1, hSNF2H is known to interact with several cellular proteins to form different hSNF2H-containing protein complexes that have diverse cellular functions. In our previous study, we demonstrated that excessive Rsf-1 molecules sequestered hSNF2H, leading to a loss or decrease in the abundance of other hSNF2H-binding complexes such as hSNF2H/BAZ1A and hSNF2H/BAZ1B (15). Because several members in the SNF family have been suggested as tumor suppressors and are found to be downregulated or inactivated in cancer tissues (35, 36), it is plausible that reduction of these hSNF2H complexes with tumor suppressor potential by excessive Rsf-1 contributes to the observed drug resistant phenotype in cancer cells. However, in this study, we found that expressing the Rsf-D4 deletion mutant which has the hSNF2H binding activity, enhanced growth inhibitory effect of paclitaxel in tumor cells. This observation suggests that sequestering hSNF2H from other protein complexes by the Rsf-D4 mutant alone is not able to confer paclitaxel resistance as seen in the full-length Rsf-1. Therefore, the paclitaxel resistant phenotype mediated by the full-length Rsf-1 is more likely due to an increase in RSF complex formation rather than a decrease in other hSNF2H-containing complexes.
Although the above represents our preferred views, alternative interpretations should be indicated. First, the shRNA screening approach used in this study suggests that Rsf-1 is the main gene within the 11q13.5 amplicon responsible for paclitaxel resistance. However, other gene(s) within the 11q13.5 amplicon might also be involved in the aggressive behavior of 11q13.5 amplified carcinomas. For example, PAK1, a member of serine/threonine kinase family, plays a critical role in controlling anchorage-independent growth and invasiveness in breast cancer cells (37), and in promoting mammalian tumorigenesis (38) and angiogenesis (39). Thus, PAK1 expression may lead to a more aggressive clinical course in ovarian cancer. It is thus plausible that Rsf-1 collaborates with PAK1 and other genes to propel tumor progression. Second, similar to other dominant negative approaches that are applied to modulate activity of endogenous proteins, the Rsf-D4 approach reported in this study may not be specific to interrupt the interaction between Rsf-1 and hSNF2H. It is possible that the Rsf-D4 mutant may complex with other protein(s) besides hSNF2H and that overexpression of Rsf-D4 can potentially interfere with the binding of these protein(s) to the endogenous full-length Rsf-1. Finally, Even though the correlation is statistically significant, seven tissues with high Rsf-1 didn't show a significant drug resistance. This finding suggests that other factors may also play a role in conferring taxol resistance phenotype that is independent of Rsf-1.
In summary, we demonstrated in this report that development of paclitaxel resistance in ovarian cancer cells was attributed at least in part by Rsf-1 which frequently amplifies in several types of human cancer. We further showed that the formation of Rsf-1 and hSNF2H chromatin remodeling complexes was essential for developing this resistance phenotype. Our results suggest that Rsf-1 can be used as a biomarker for paclitaxel resistance and can be exploited as a potential target for chemosensitization in carcinomas with Rsf-1 amplification or overexpression.
Supplementary Material
Acknowledgements
The authors appreciate the valuable comments from the laboratory members.
Financial support: This study was supported by grants from NCI/NIH (RO1 CA129080; RO1 CA103937 to IMS), American Cancer Society (RSG-08-174-01-GMC to TLW) and Department of Defense Research Council (OC0400600 to TLW).
Footnotes
References
- 1.Kinzler KW, Vogelstein B. The Genetic Basis of Human Cancer. 2nd edition McGraw-Hill; Toronto: 2002. [Google Scholar]
- 2.Meltzer PS, Kallioniemi A, Trent JM. Chromosome alterations in human solid tumors. In: Vogelstein B, Kinzler KW, editors. The genetic basis of human cancer. McGraw-Hill; New York: 2002. pp. 93–113. [Google Scholar]
- 3.Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358:502–11. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 4.Wang TL, Maierhofer C, Speicher MR, et al. Digital karyotyping. Proc Natl Acad Sci U S A. 2002;99:16156–61. doi: 10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bignell GR, Huang J, Greshock J, et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res. 2004;14:287–95. doi: 10.1101/gr.2012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shih Ie M, Sheu JJ, Santillan A, et al. Amplification of a chromatin remodeling gene, Rsf-1/HBXAP, in ovarian carcinoma. Proc Natl Acad Sci U S A. 2005;102:14004–9. doi: 10.1073/pnas.0504195102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakayama K, Nakayama N, Jinawath N, et al. Amplicon profiles in ovarian serous carcinomas. Int J Cancer. 2007;120:2613–7. doi: 10.1002/ijc.22609. [DOI] [PubMed] [Google Scholar]
- 8.Bekri S, Adelaide J, Merscher S, et al. Detailed map of a region commonly amplified at 11q13-->q14 in human breast carcinoma. Cytogenet Cell Genet. 1997;79:125–31. doi: 10.1159/000134699. [DOI] [PubMed] [Google Scholar]
- 9.Schraml P, Schwerdtfeger G, Burkhalter F, et al. Combined array comparative genomic hybridization and tissue microarray analysis suggest PAK1 at 11q13.5-q14 as a critical oncogene target in ovarian carcinoma. Am J Pathol. 2003;163:985–92. doi: 10.1016/S0002-9440(10)63458-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentires-Alj M, Gil SG, Chan R, et al. A role for the scaffolding adapter GAB2 in breast cancer. Nat Med. 2006;12:114–21. doi: 10.1038/nm1341. [DOI] [PubMed] [Google Scholar]
- 11.Brown LA, Kalloger SE, Miller MA, et al. Amplification of 11q13 in ovarian carcinoma. Genes Chromosomes Cancer. 2008;47:481–9. doi: 10.1002/gcc.20549. [DOI] [PubMed] [Google Scholar]
- 12.Schwab M. Amplification of oncogenes in human cancer cells. Bioessays. 1998;20:473–9. doi: 10.1002/(SICI)1521-1878(199806)20:6<473::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 13.Pohl G, Ho CL, Kurman RJ, Bristow R, Wang TL, Shih Ie M. Inactivation of the mitogen-activated protein kinase pathway as a potential target-based therapy in ovarian serous tumors with KRAS or BRAF mutations. Cancer Res. 2005;65:1994–2000. doi: 10.1158/0008-5472.CAN-04-3625. [DOI] [PubMed] [Google Scholar]
- 14.Mao TL, Hsu CY, Yen MJ, et al. Expression of Rsf-1, a chromatin-remodeling gene, in ovarian and breast carcinoma. Hum Pathol. 2006;37:1169–75. doi: 10.1016/j.humpath.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Sheu JJ, Choi JH, Yildiz I, et al. The Roles of Human Sucrose Nonfermenting Protein 2 Homologue in the Tumor-Promoting Functions of Rsf-1. Cancer Res. 2008;68:4050–7. doi: 10.1158/0008-5472.CAN-07-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buckhaults P, Zhang Z, Chen YC, et al. Identifying tumor origin using a gene expression-based classification map. Cancer Res. 2003;63:4144–9. [PubMed] [Google Scholar]
- 17.Hogarty MD, Brodeur GM. Gene amplification in human cancers: biological and clinical significance. In: Vogelstein B, Kinzler Kw, editors. The genetic basis of human cancer. 2 edition McGraw-Hill; New York: 2002. pp. 115–28. [Google Scholar]
- 18.Loyola A, Huang J-Y, LeRoy G, et al. Functional Analysis of the Subunits of the Chromatin Assembly Factor RSF. Mol. Cell. Biol. 2003;23:6759–68. doi: 10.1128/MCB.23.19.6759-6768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loyola A, LeRoy G, Wang YH, Reinberg D. Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes Dev. 2001;15:2837–51. doi: 10.1101/gad.937401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeRoy G, Loyola A, Lane WS, Reinberg D. Purification and characterization of a human factor that assembles and remodels chromatin. J Biol Chem. 2000;275:14787–90. doi: 10.1074/jbc.C000093200. [DOI] [PubMed] [Google Scholar]
- 21.LeRoy G, Orphanides G, Lane WS, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–4. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 22.Menendez JA, Vellon L, Mehmi I, Teng PK, Griggs DW, Lupu R. A novel CYR61-triggered `CYR61-alphavbeta3 integrin loop' regulates breast cancer cell survival and chemosensitivity through activation of ERK1/ERK2 MAPK signaling pathway. Oncogene. 2005;24:761–79. doi: 10.1038/sj.onc.1208238. [DOI] [PubMed] [Google Scholar]
- 23.Graessmann M, Berg B, Fuchs B, Klein A, Graessmann A. Chemotherapy resistance of mouse WAP-SVT/t breast cancer cells is mediated by osteopontin, inhibiting apoptosis downstream of caspase-3. Oncogene. 2007;26:2840–50. doi: 10.1038/sj.onc.1210096. [DOI] [PubMed] [Google Scholar]
- 24.Chu CY, Chang CC, Prakash E, Kuo ML. Connective tissue growth factor (CTGF) and cancer progression. J Biomed Sci. 2008 doi: 10.1007/s11373-008-9264-9. [DOI] [PubMed] [Google Scholar]
- 25.Shamay M, Barak O, Shaul Y. HBXAP, a novel PHD-finger protein, possesses transcription repression activity. Genomics. 2002;79:523–9. doi: 10.1006/geno.2002.6717. [DOI] [PubMed] [Google Scholar]
- 26.Shamay M, Barak O, Doitsh G, Ben-Dor I, Shaul Y. Hepatitis B virus pX interacts with HBXAP, a PHD finger protein to coactivate transcription. J Biol Chem. 2002;277:9982–8. doi: 10.1074/jbc.M111354200. [DOI] [PubMed] [Google Scholar]
- 27.Aihara T, Miyoshi Y, Koyama K, et al. Cloning and mapping of SMARCA5 encoding hSNF2H, a novel human homologue of Drosophila ISWI. Cytogenet Cell Genet. 1998;81:191–3. doi: 10.1159/000015027. [DOI] [PubMed] [Google Scholar]
- 28.Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flanagan JF, Peterson CL. A role for the yeast SWI/SNF complex in DNA replication. Nucleic Acids Res. 1999;27:2022–8. doi: 10.1093/nar/27.9.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 31.Riedel RF, Porrello A, Pontzer E, et al. A genomic approach to identify molecular pathways associated with chemotherapy resistance. Mol Cancer Ther. 2008;7:3141–9. doi: 10.1158/1535-7163.MCT-08-0642. [DOI] [PubMed] [Google Scholar]
- 32.Weng D, Song X, Xing H, et al. Implication of the Akt2/sruvivin pathway as a critical target in paclitaxel treatment in human ovarian cancer cells. Cancer Lett. 2008 doi: 10.1016/j.canlet.2008.08.027. in press. [DOI] [PubMed] [Google Scholar]
- 33.Kim SH, Juhnn YS, Song YS. Akt involvement in paclitaxel chemoresistance of human ovarian cancer cells. Ann N Y Acad Sci. 2007;1095:82–9. doi: 10.1196/annals.1397.012. [DOI] [PubMed] [Google Scholar]
- 34.Huang JY, Shen BJ, Tsai WH, Lee SC. Functional interaction between nuclear matrix-associated HBXAP and NF-kappaB. Exp Cell Res. 2004;298:133–43. doi: 10.1016/j.yexcr.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 35.Klochendler-Yeivin A, Muchardt C, Yaniv M. SWI/SNF chromatin remodeling and cancer. Curr Opin Genet Dev. 2002;12:73–9. doi: 10.1016/s0959-437x(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 36.Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–6. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vadlamudi RK, Adam L, Wang RA, et al. Regulatable expression of p21-activated kinase-1 promotes anchorage-independent growth and abnormal organization of mitotic spindles in human epithelial breast cancer cells. J Biol Chem. 2000;275:36238–44. doi: 10.1074/jbc.M002138200. [DOI] [PubMed] [Google Scholar]
- 38.Wang RA, Mazumdar A, Vadlamudi RK, Kumar R. P21-activated kinase-1 phosphorylates and transactivates estrogen receptor-alpha and promotes hyperplasia in mammary epithelium. Embo J. 2002;21:5437–47. doi: 10.1093/emboj/cdf543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bagheri-Yarmand R, Vadlamudi RK, Wang RA, Mendelsohn J, Kumar R. Vascular endothelial growth factor up-regulation via p21-activated kinase-1 signaling regulates heregulin-beta1-mediated angiogenesis. J Biol Chem. 2000;275:39451–7. doi: 10.1074/jbc.M006150200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.