Abstract
As part of the innate immune defense against HIV infection, OTK18, a zinc finger protein, is upregulated in human macrophages and reduces viral replication through suppression of viral long-terminal repeat promoter activity. Although we know that the processing products of OTK18 accumulate in the cytoplasm of brain perivascular macrophages in advanced HIV encephalitis cases, the molecular mechanisms behind its post-translational processing are still poorly understood. To characterize OTK18 processing, we assessed a panel of protease inhibitors to identify the candidates involved in the OTK18 processing using human monocyte-derived macrophages (MDM) over-expressing OTK18 by recombinant adenoviral gene transfer. Viral infection of MDM strongly increased the processing of OTK18 into its N-terminal fragment. Treatment of OTK18-expressing MDM with calpain- and proteasome- inhibitors significantly accumulated either full-length or processed OTK18 fragments in time and dose-dependent manners. A series of OTK18 truncation mutants and synthetic peptides were tested to locate the calpain cleavage sites after arginine 359. Finally, we developed an enhanced cyan and yellow fluorescent protein (ECFP and EYFP)-based intramolecular fluorescent resonance energy transfer (intramolecular FRET) system to monitor the OTK18 endoproteolysis in human microglia cell line. Inhibition of proteasome activity significantly increased the intramolecular FRET signal in the nucleus. These data suggest that calpain and proteasome are involved in OTK18 endoproteolysis and degradation. Additionally, intramolecular FRET has proven to be a useful tool for monitoring the processing in live cells.
Introduction
Our previous studies demonstrated that OTK18/ZNF175, a 711 amino acid Kruppel associated box (KRAB) C2H2 type zinc finger protein (ZNF), is specifically expressed in brain perivascular macrophages but not in microglia of HIV encephalitis brain (for review, please refer to (Buescher et al., 2007)). Expressed OTK18 potently suppresses HIV-1 replication partly due to its direct inhibition of HIV-1 Tat mediated HIV-1 long terminal repeat (LTR) activation (Carlson et al., 2004a). Using LTR mutant screening and transcription-DNA duplex binding assays, we recently showed that OTK18 suppresses LTR via direct binding to two distinct regulatory regions: negative regulatory element and Ets element (Horiba et al., 2007).
When full-length OTK18 is expressed in certain mammalian or insect cells, we observed endoproteolysis of OTK18, generating 35kD N-terminal fragments (OTK18N), which is enhanced by viral infection. The processing site is unknown; however, considering the size of the processed fragment and the existence of a putative nuclear localization signal (NLS)--RKKP-- at position 359 between zinc fingers 1 and 2, it is likely that OTK18 was processed prior to the NLS. Thus, OTK18N may be exported to the cytoplasm for subsequent degradation. In HIV encephalitis brain, OTK18 immunoreactivity was specifically detected in the cytoplasm of the perivascular macrophages (Carlson et al., 2004b), suggesting that viral infection and brain inflammation synergistically enhance OTK18 processing and accumulation. We hypothesize that viral infection enhances OTK18 processing by activating specific proteases, which regulates its nuclear localization and transcriptional activity.
Through the screening of protease inhibitors we focused on the calpain and proteasomal degradation of OTK18. Calpains constitute a cysteine protease family and are activated by calcium at neutral pH (Huang and Wang, 2001). Calpain 1 (μ-calpain) and calpain 2 (m-calpain) are particularly abundant in brains and differ in their calcium sensitivity for activation (1-20μM for μ-calpain and 0.25-0.75 mM for m-calpain) (Stracher, 1999). Calpain 1 is involved in synaptic function, memory formation, calcium-mediated neurotoxicity, N-methyl-D-aspartate (NMDA) receptor signaling, cleavage of p35 and activation of cycline-dependent kinase 5 (CDK5), cleavage and activation caspase cascade, and is implicated neurodegenerative disorders, such as Alzhiemer's, Parkinson's, and Huntington's disease (for review, see (Liu et al., 2008)).
Recently, the relationship between the ubiquitin-proteasome system and critical steps of HIV budding has been under intense investigation. Ubiquitination of viral particles is critically important in the last steps of viral replication, and proteasome inhibitors can block viral budding through Endosomal Sorting Complex Required for Transport (ESCORT), which mediates the budding of many enveloped viruses (for review, see (Martin-Serrano, 2007)). Proteasome also mediates the interleukin-10-induced degradation of cyclin T1, which is critical for viral replication in macrophages (Wang and Rice, 2006).
This study suggests that HIV-1 infection strongly enhances OTK18 processing in human macrophages, and that calpain is involved in the cleavage at amino acids 275-300. As predicted, expressed OTK18N is mainly localized in cytoplasm, while full-length OTK18 is mostly in the nucleus. Both full-length and processed OTK18 were degraded by the proteasomal degradation pathway, which regulates its nuclear retention as determined by enhanced cyan and yellow fluorescent protein (ECFP and EYFP)-based intramolecular fluorescent resonance energy transfer (intramolecular FRET) system in live cells. These data suggest calpain and proteasomal regulation of KRAB-ZNF family.
Materials and Methods
Cell Line and Tissue Culture
Monocytes were obtained from leukophoresis of HIV-1, -2, and hepatitis B seronegative donors and purified by counter current centrifugal elutriation (Gendelman et al., 1988). Cell suspensions were documented >98% monocytes using the cell morphology criteria in Wright-stained cytosmears and CD68 immunolabeling. Monocytes were cultured as adherent cells at 5 × 105 cells per well in 24-well plates for virus titration and 2 × 106 cells per well in 6-well plates. Monocytes were cultured in Media A: Dulbecco's modified eagle medium (DMEM, Invitrogen, Carlsbad, CA) with 10% heat-inactivated pooled human serum, 1 mM glutamine, 50 μg/ml gentamicin, 10 μg/ml ciprofloxacin (all from Invitrogen), and 1000 U/ml highly purified recombinant human macrophage colony stimulating factor (MCSF, a generous gift from Genetics Institute, Cambridge, MA). MCSF was supplemented in the culture medium only for the initial 7 days of cultivation in order to promote differentiation to macrophages (MDM) and then withdrawn (Media B). Culture medium was half-exchanged every 3 days. All tissue culture reagents were screened before use and found negative for endotoxin (<10 pg/ml; Associates of Cape Cod, Inc., Woods Hole, MA) and mycoplasma contamination (Gen-probe II; Gen-probe Inc., San Diego, CA).
The human microglia cell line (a gift from M. Tardieu) (Janabi et al., 1995) was cultured at 37°C in a humidified 5% (v/v) CO2-air environment in DMEM supplemented with 10% fetal bovine serum (FBS), 10U/ml penicillin, and 10μg/ml streptomycin (all reagents from Invitrogen). The cells were seeded in 6-well plates (2 × 106 cells per well) for DNA transfection and biochemical studies, or seeded in poly-D-lysine-coated 35 mm MatTek dishes for live imaging as described below.
Recombinant Adenovirus Generation and Viral Infection
Green fluorescent protein (GFP) and OTK18-expressing adenovirus were constructed and produced as previously reported (Carlson et al., 2004a). After 7 days differentiation, adenovirus titration was performed on MDM for each new donor. Virus (1 × 105, 2 × 105, 3 × 105, 4 × 105, 5 × 105, 6 × 105, 7 × 105, and 8 × 105 viral particles/cell) plus 500 μl Media B were added to each well and incubated for 1 hour at 37°C. Virus media was replaced with 500 μl fresh Media B, and cells were incubated overnight at 37°C. Sixteen hours later, the optimal viral dose was determined by GFP expression. Viral doses used for each experiment are listed in figure legends. Once a viral dose was determined, experimental cells were infected by adenovirus plus 2 ml Media B to each well and incubated for 1 hour at 37°C. Virus media was replaced with 2 ml fresh Media B, and cells were incubated for 24 hr at 37°C.
For HIV infection, the MDM were infected with HIV-1ADA at 0.1 mutiplicity of infection (MOI) 24 hrs after the Ad-GFP or Ad-OTK18 infection and subjected to immunoblotting at 3 and 10 days post-infection (DPI) as described (Carlson et al., 2004a). For protease inhibitor study, virus-infected MDM were treated with 10μM Calpain Inhibitor I, 100nM staurosporine, 10μM MG132, or 0.1%v DMSO at 7 day post-infection for 24hrs and the cell lysates were subjected to immunoblotting.
Chemicals
Cells were incubated as described in figure legends with various inhibitors: chloroquine (lysosomal inhibitor, Sigma); Calpain Inhibitor I, MG132 (proteasome inhibitor); Z-VAD-FMK (pan-caspase inhibitor); Staurosporine (apoptosis inducer and non-specific protein kinase inhibitor); Calpeptin (calpain inhibitor); Cathepsin L Inhibitor IV; or CA-074 Me (cathepsin B Inhibitor IV, all from Calbiochem/EMD Chemicals, Inc., Gibbstown, NJ).
Gene Construction
OTK18 deletion mutants
GAL4 binding domain (GBD)-OTK18 fusion proteins of various lengths (GBD 90-115, GBD90-241, GBD90-300) were constructed by restriction digestion and blunt-end ligation of the original GBD-full-length OTK18 (1-711) as described previously (Carlson et al., 2004a). Construction of OTK18-GFP was described previously (Carlson et al., 2004a), and OTK18 (1-300)-GFP was constructed by subcloning the proof-reading PCR amplified DNA fragment containing the OTK18 1-300 amino acid coding sequence into the BglII and BamHI sites of the pEGFP-N1 vector (Invitrogen) as in-frame ligation.
FRET Constructs
Flanked full-length OTK18 cDNA was subcloned into pECFP-C1 vector (Clontech/Invitrogen) at SalI and BamHI sites by Pfu proofreading PCR-based technique to generate ECFP-OTK18 fusion protein expression vector (pECFP-OTK18). pECFP-OTK18-EYFP and pECFP-EYFP (positive control of intramolecular FRET) vectors were generated by proof-reading PCR-based subcloning of EYFP gene into the pECFP-OTK18 and pECFP-C1 vectors at BamH I and Xba I sites as in-frame ligation.
All the PCR-subcloned regions were DNA sequenced, and expression of fusion proteins was confirmed by transient expression in mammalian cells, followed by fluorescent imaging and immunoblotting using anti-CFP/GFP/YFP and anti-OTK18 antibodies.
Transfection and Preparation of Cell Lysates
The cells were plated on poly-D-lysine-coated 6-well plates at 2 × 105 cells/well. Cells were transfected using 2 μg DNA and 6 μl FuGENE 6 (Roche Diagnostics, Indianapolis, IN) according to kit instructions. Twenty-four hours after transfection, inhibitors were added and treated as indicated in the figure legends. Cells were lysed in 200 μl of lysis buffer (20 mM Tris-HCl pH 7.6, 0.3 M KCl, 2 mM EDTA, 1% Triton, 0.5 mM Na3VO4, and 2 mM NaF, plus protease inhibitor cocktail, Roche), and total protein concentration was determined by BCA Protein Assay Kit (Pierce, Rockford, IL). Lysates were added to equal volumes of 2X Laemmli Buffer and 10% β-Mercaptoethanol (Sigma).
For the subcellular fractionation of MDM at days 3, 7, and 10 after Ad-OTK18 infection, the cell pellet was resuspended in hypotonic buffer, followed by homogenization using Dounce Homogenizer as described (Rosati et al., 2001). The homogenate was centrifuged at 1,850 × g for 10 min to collect the cytoplasmic fraction (S100). The nuclei pellet was incubated in one volume of 0.3M KCl buffer at 4°C for 30 min to release DNA-bound molecules, and centrifuged at 25,000 × g for 30 min to collect the nuclear extract (NE).
Immunoblotting
Ten μg total protein was loaded per well. Samples were run on a 10% polyacrylamide or 4-15% gradient gel (Bio-Rad) and transferred to a PVDF membrane (Millipore, Billerica, CA). Membranes were blocked for a minimum of 1 hour at room temperature in TBST + 5% milk. Membranes were incubated with 1:1000 anti-OTK18 monoclonal antibody (mAb) (Carlson et al., 2004a) or 1:500 anti-Gal4 DNA binding domain (GBD) mAb (Santa Cruz Biotechnology, Santa Cruz, CA) in 5% milk overnight at 4°C. Membranes were washed 3 × 10 minutes with TBST and incubated with 1:4000 donkey anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 hour at room temperature. Then, membranes were again washed as previously described and developed with ECL Plus (Amersham/GE Healthcare, Piscataway, NJ).
Membranes were stripped with restore Western Blot Stripping Buffer (Pierce), blocked for a minimum of 1 hour at room temperature and TBST + 5% milk and incubated with 1:5000 anti-GFP monoclonal antibody (Clontech/Invitrogen) or 1:100,000 anti-β-actin mAb (Sigma) in 5% milk overnight at 4°C. Membranes were washed as previously described and incubated with 1:10,000 donkey anti-mouse secondary antibody (Jackson ImmunoResearch) for 1 hour at room temperature. Membranes were washed as previously described and developed with ECL Plus (GE Healthcare).
Calpain-1 cleavage of OTK18 peptides
Two purified synthetic peptides in the region of OTK18 301-378, 20 μg each, all from Alpha Diagnostic Inc, San Antonio, TX) were incubated with calpain-1 (400ng, Calbiochem) in a reaction buffer (20mM Hepes, pH 7.9, 60mM KCl, 1mM Ca2+, 0.2mM EDTA, 20% glycerol, and 0.5mM phenylmethylsulfonylfluoride) at 30°C for 30min, and the reaction was stopped by addition of 3mM EDTA (final concentration).
MALDI Method
One microliter of sample was placed on a stainless steel 384 spot MALDI plate and allowed to air dry. Then 0.5μL of α-cyano-4-hydroxy-cinnamic acid (CHCA) matrix (saturated solution in 60% ACN + 0.2% TFA) was added, allowed to air dry, then repeated once. A 4800 MALDITOF/TOFTM analyzer (Applied Biosystems, Framingham, MA), in reflector positive mode was used for all mass analyses. Each mass spectrum was generated by averaging 400 laser shots over a mass range m/z 800-5000. Tandem MS was obtained with a precursor ion of 1054.595 m/z using 1kV positive mode collision-induced dissociation (CID) and high gas pressure with air as the collision gas. Six hundred and twenty-five laser shots were averaged for each MS/MS spectrum and the resulting sequence interpretation was manually performed.
Live Imaging and Sensitized Emission-Fluorescence Resonance Energy Transfer (SE-FRET) analysis
The cells were plated on poly-D-lysine-coated 35 mm MatTek dishes (MatTek Corp, Ashland, MA) at 2 × 105 cells/dish and transfected as described. Twenty-four hours post-transfection, 10 μM MG132 in CO2-independent media (Invitrogen) was added just before imaging. Live CFP, CFP/YFP FRET, and YFP images were taken by a Dual-View Micro-Imager (Optical Insights, Santa Fe, NM) positioned between the Nikon TE-2000U inverted fluorescent microscope (Nikon Instruments, Melville, NY) output port and cooled charge-coupled device camera (Coolsnap HQ, Roper Scientific, Duluth, GA) as described with minor modifications (Shyu et al., 2008). Excitation was conducted using a 436/20 exciter and OI-04-EM emission filter set (Chroma Technology, Rockingham, VT). Images were taken every 5 minutes for 2 hours. Simultaneous CFPex/CFPem (donor excitation/donor emission: CFP), CFPex/YFPem (donor excitation/acceptor emission: CFP/YFP FRET), and YFPex/YFPem (acceptor excitation/acceptor emission: YFP) imaging was taken in the time-lapse experiments.
For the experiment with translational inhibitor (puromycin) treatment, cells were plated and transfected as described. Twenty-four hours post-transfection, CO2-independent media (Invitrogen) + 20 μg/ml puromycin (Sigma) was added just before imaging, and cells were incubated for 30 minutes. Next, 10 μM MG132 (Calbiochem) + 20 μg/ml puromycin was added and cells were imaged every 5 minutes for 2 hours.
For the sensitized emission-FRET (SE-FRET) analysis, all images were acquired using the multiple wavelength acquisition feature of MetaMorph (Molecular Devices, Sunnyvale, CA) in combination with a three-filter setup, for consecutive acquisition of YFP, CFP, and FRET, all at same exposure times. Corrected FRET images and calculated correction coefficients A and B were obtained and applied, respectively, using the FRET dialogue box of MetaMorph to eliminate bleed-through. Note that corrected FRET images are commonly known as net FRET (nF). The nF was calculated as: nF = (Intensity of the image acquired from the ECFP/EYFP complex using FRET filter setup) - [CoB x (Intensity of the image acquired from the ECFP/EYFP complex by using donor filter setup] - [CoA x (Intensity of the image acquired from the ECFP/EYFP complex using donor filter setup)], where CoB and CoA are the correction coefficients for CFP and YFP, respectively (Gordon et al., 1998). Specifically, CoB = (Average intensity of the image acquired from the donor-only sample using FRET filter setup)/(Average intensity of the image acquired from the donor-only sample by using donor filter setup). CoA = (Average intensity of the image acquired from the acceptor-only sample using FRET filter setup)/(Average intensity of the image acquired from the acceptor-only sample using acceptor filter setup). At least 6 spots per nucleus were measured for the average nF intensity calculation (n=3 cells per group per time point). The data were presented as fold increase of average nF at t=0 and 120 min time points over t=0 time point with or without puromycin. After intensity measurements for quantitative analysis, FRET images were background-subtracted using Metamorph, pseudocolored using Photoshop CS3 (Adobe Systems Inc., San Jose, CA), and plotted on 3-D Surface Plots using freeware ImageJ Software (NIH) for qualitative analysis.
Statistics
All experiments were repeated at least three times with three different donors for human MDM. All the data were normally distributed. In case of multiple mean comparisons, data were analyzed by analysis of variance (ANOVA), followed by Newman-Keuls multiple comparison tests using statistics software (Prism 4.0, Graphpad Software, La Jolla, CA). In case of single mean comparison, data were analyzed by Student's t test. In case of time-course study on FRET imaging, data were analyzed by two-way repeated measure ANOVA, followed by Bonferroni posttest. P values less than 0.05 were regarded as significant.
Results
HIV-1 infection enhances OTK18 endoproteolysis and gene expression
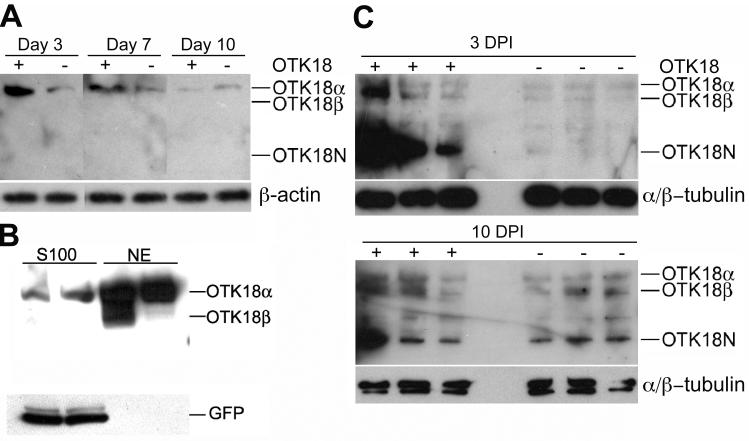
We first examined OTK18 processing in human MDM with or without HIV-1 infection. For this purpose, full-length OTK18 was transiently expressed using recombinant adenovirus (Ad-OTK18/GFP) expressing OTK18 and GFP as separate molecules under two different CMV promoters as described (Carlson et al., 2004a). The cells were subjected to immunoblotting for OTK18 at 3, 7, and 10 days post infection using mAb against amino acids 1-180 of OTK18 protein. This antibody detects recombinant OTK18 at 75kD (OTK18α), 65kD (OTK18β), and 35 kD (OTK18N). OTK18α is full-length OTK18 1-711, whereas OTK18β presumably encodes 56-711, starting from the alternative translational initiation site, and OTK18N is the endoproteolysed N-terminal fragment. OTK18a was the main band in the Ad-OTK18/GFP infected (+ lanes) and Ad-GFP infected (- lanes) groups when expressed in human MDM from day 3 to 10 (Fig. 1A). Recombinant OTK18 expression peaked around day 3 (100%), and the band intensity was diminished to 60% on day 7 and 1% on day 10 post-infection. We examined the localization of the full-length OTK18 by subcellular fractionation of MDM at 48 hrs after infection of Ad-OTK18/GFP; as expected, the majority of OTK18a and b was localized in nuclear extract fraction (NE) (Fig. 1B). In comparison, GFP was exclusively localized in the cytoplasmic fraction (S100), confirming the nuclear localization of OTK18. Next, we challenged the Ad-OTK18/GFP or Ad-GFP infected MDM with viral infection (HIV-1ADA, 0.1 MOI) 24 hrs after the adenoviral infection, and cells were collected for immunoblotting at 3 and 10 days post HIV-1 infection (DPI). As shown in Fig. 1C, significant accumulation of OTK18N was observed in Ad-OTK18/GFP-infected group (+ lanes, 3 DPI upper panels). At 10 DPI, we observed upregulation of OTK18a, b, and OTK18N in both Ad-OTK18/GFP and Ad-GFP infected groups (+ and - lanes, 10 DPI lower panels), and OTK18N was the major fragment in Ad-OTK18/GFP infected group (+ lanes). These data suggest enhanced endoproteolysis of OTK18 and upregulation of endogenous OTK18 by HIV-1 infection, consistent with our previous studies (Carlson et al., 2004a; Carlson et al., 2004b).
Figure 1. Processing of OTK18 upon HIV-1 infection of MDM.
A, Immunoblotting of OTK18 (upper panel) and β-actin (lower panel) 3, 7, and 10 days after Ad-OTK18/GFP (+) or Ad-GFP (-) adenovirus infection using anti-OTK18 mAb specific to OTK18 1-180 amino acids. Only OTK18α (75 kD), but neither OTK18β (65 kD) nor OTK18N (35 kD), was detected. B, Immunoblotting of cytoplasmic (S100) and nuclear extract (NE) of OTK18-expressing MDM after subcellular fractionation. 40 μg of each fraction was loaded onto 4-15% SDS-PAGE and subjected to immunoblotting using anti-OTK18 (upper panel) or anti-GFP (lower panel) monoclonals. OTK18N was undetected in these fractions. C, Immunoblotting of OTK18 (upper panel) and α/β-tubulin (lower panel) at 3 and 10 days post HIV infection (DPI). Thirty million MDM were infected with adenovirus (Ad-OTK18/GFP (+) or Ad-GFP adenovirus (-)), followed by infection with HIV-1ADA (one day later). OTK18α (75 kD), OTK18β (65 kD) and OTK18N (35 kD) were detected in both Ad-OTK18/GFP and Ad-GFP-infected MDM at Day 10.
Proteasome and calpain are involved in OTK18 metabolism
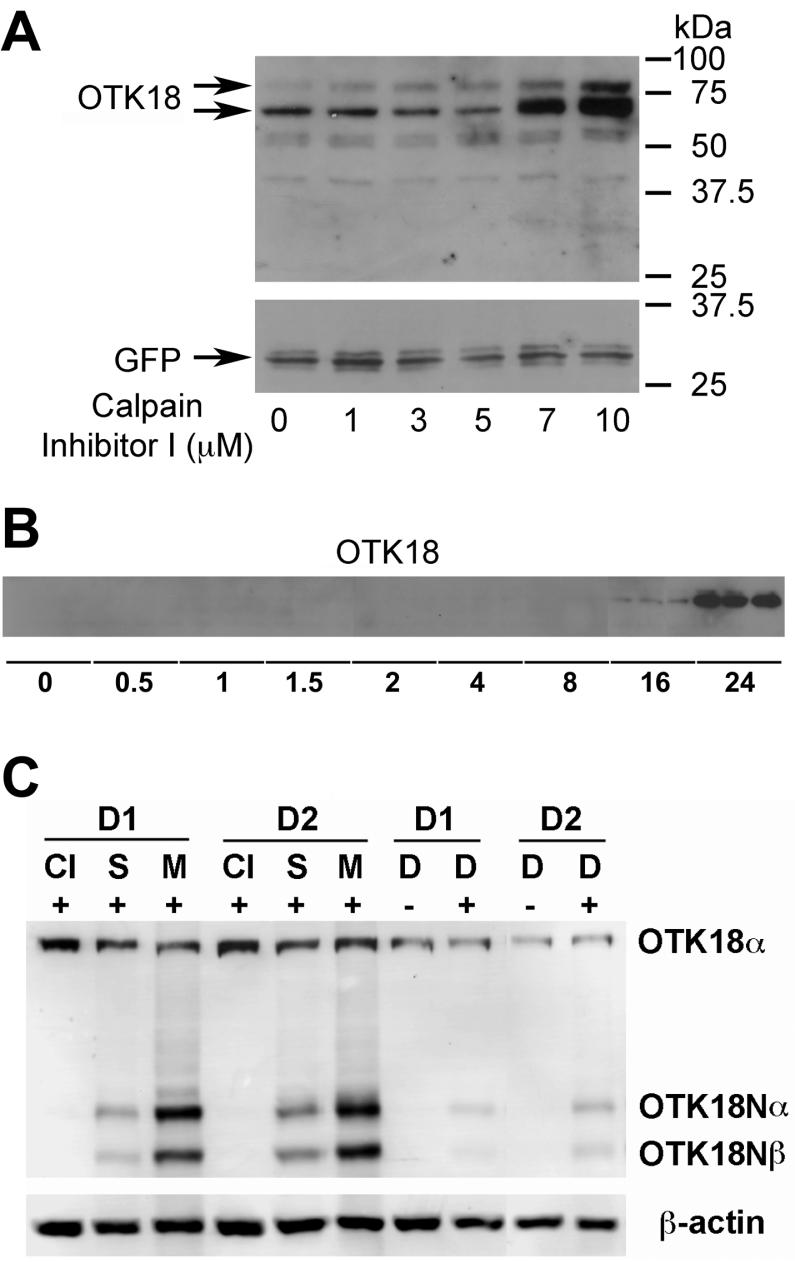
A number of intracellular molecules are functionally regulated by degradation or endoproteolysis, via lysosomal or proteosomal degradation for the former, or site-specific cleavage by defined endopeptidases (e.g. serine/cysteine/aspartate proteases and metalloendopeptidases) for the latter. To characterize the molecules involved in the OTK18 endoproteolysis and degradation, we tested a series of protease inhibitors on Ad-OTK18/GFP infected MDM. There is strong accumulation of both full-length and processed OTK18 by treatment with MG132, a proteasome inhibitor, whereas calpain inhibitor I specifically accumulated OTK18α/β (Fig. 2A). In contrast, chloroquine, a lysosomal inhibitor, had no effect on OTK18 metabolism. This suggests that calpain (EC 3.4.22.17), a cysteine protease, mediates the endoproteolysis of OTK18, and both OTK18α/β and OTK18N undergo proteasomal degradation in MDM. We also found that staurosporine accumulated over-expressed OTK18α/β in MDM, while Z-VAD FMK, a caspase inhibitor, had no effect by itself (Fig. 2B). Interestingly combination of calpain ihibitor I with staurosporine and or Z-VAD FMK showed synergistic enhancement of OTK18 accumulation, suggesting that caspase activity is also involved in the OTK18 endoproteolysis but secondary to the calpain-mediated cleavage. Other cathepsin inhibitors (cathepsin L inhibitor IV and CA074 Me) had limited effect on OTK18 accumulation (data not shown). Calpain inhibitor I accumulated OTK18α/β in dose- and time-dependent manners (Fig. 3A-B), and the accumulation peaked between 16-24 hr post treatment period (Fig. 3B).
Figure 2. Calpain-dependent OTK18 endoproteolysis in MDM and human microglia cell line.
A, MDM were infected with adenovirus expressing GFP (Ad-GFP) or Ad-OTK18, and treated with 10 μM lysosomal inhibitor (chloroquine), 10 μM proteosomal inhibitor (MG132), or 10 μM Calpain Inhibitor I for 16 hrs. B, Human microglia cell line was infected with Ad-OTK18 and treated with varying combinations of 100 nM staurosporine, 10 μM or 20 μM Caspase Inhibitor (Z-VAD-FMK), and 10 μM Calpain Inhibitor I for 16 hrs.
Figure 3. Accumulation of OTK18 in HIV-1-infected MDM by calpain inhibitor.
A, MDM were infected with Ad-OTK18 and treated with increasing doses of Calpain Inhibitor I for 16 hrs. B,Immunoblotting of OTK18. MDM were infected with Ad-OTK18 and treated with 10 μM Calpain Inhibitor I for the times indicated (in hours). C, Accumulation of OTK18 in HIV-1-infected MDM. MDM from donor 1 and 2 (D1 and D2, respectively) were infected with HIV-1ADA at 0.1 MOI (+ lanes), and treated with 10μM Calpain Inhibitor I (CI), 100nM staurosporine (S), 10μM MG132 (M), or 0.1%v DMSO (D) at 7 day post-infection for 24hrs. The cell lysates were subjected to immunoblotting for OTK18 (upper panel) or β-actin (lower panel).
Calpain cleaves OTK18 protein, which regulates cytoplasmic translocation
We also examined out finding in a human microglia cell line (Janabi et al., 1995; Janabi et al., 1998). However, this cell line expresses CD68, a macrophage maker (Holness and Simmons, 1993) but CD45-CD163-CD14low as determined by FACS (see accompanying manuscript by Buescher JL et al, JNIP #157). In contrast, primary cultured human MDM and human microglia are CD45+CD163+CD14+ and CD45+CD163-CD14+, respectively, suggesting that the original microglia-like phenotype of the cell line is lost. This cell line expresses endogenous OTK18 in low levels (Fig. 4A, middle panel), and can be easily transfected with DNA plasmids and suitable for transient expression studies. We employed a series of Gal4 Binding domain (GBD)-fusion protein OTK18 deletion mutants (90-115, 90-240, 90-300), or non-tagged full-length OTK18 (1-711) to identify the region responsible for the calpain-mediated endoproteolysis. After transient expression of GBD-OTK18 fusion mutants in human brain macrophage cell line, cells were treated with or without calpain inhibitor I, and subjected to immunoblotting using anti-GBD, anti-OTK18, and anti-β-actin mAbs (Fig. 4A-B). Anti-OTK18 mAb recognizes the N-terminal region of OTK18 and could not detect GBD-OTK18 fusion deletion mutants (Fig. 4A, upper panel). We detected accumulation of fusion protein containing OTK18 90-300 (300) and 1-711 (FL), but not of 90-115 (115) or 90-240 (240) as determined by immunoblotting (Fig. 4A upper and lower panels). The band intensity measurement confirmed the significant differences in protein accumulation of OTK18 90-300 (300) and 1-711 (FL) by calpain inhibitor I treatment, compared to non-treated (Fig. 4B). Since the nuclear localization signal is located at position 359, this prompted us to investigate if this cleavage translocates OTK18 protein from nucleus to cytoplasm. For that purpose, OTK18-GFP or OTK18(1-300)-GFP fusion protein, expressing either full-length OTK18 1-711 or 1-300 fused to GFP, was transiently transfected into human embryonic kidney 293 cells, and localization was confirmed by intrinsic fluorescent signal. OTK18-GFP was specifically localized in the nucleus as determined by the Hoechst 33342 staining, a nuclear marker, whereas OTK18(1-300)-GFP distributed mainly in the cytoplasm as either intact fusion protein or cleaved GFP (Fig. 4C), suggesting that OTK18 is cleaved after amino acid 300.
Figure 4. Effect of Calpain Inhibitor I on OTK18 deletion mutants.
A, The cells were transfected with pFA-CMV, pGBD-OTK18 deletion mutants corresponding to GBD-OTK18 90-115 (115), 90-240 (240), 90-300 (300), or pcDNA3.1-OTK18 expressing non-GBD-tagged full length OTK18 (FL). Twenty-four hours after transfection, cells were treated with 0.05% DMSO (- CI) or 10 mM Calpain Inhibitor I (+ CI) for 16 hrs. Total cell extracts were subjected to immunoblotting using anti-GBD (upper panel), anti-OTK18 (middle panel), or anti-b-actin mAbs (lower panel). B, Quantification of relative band intensities (*denotes P<0.001). C, Fluorescent images of human embryonic kidney 293 cells transfected with OTK18-GFP (left panel) or OTK18 (1-300)-GFP (right panel). Phase: phase contrast image, GFP: intrinsic GFP signal of OTK18 fusion protein, Hoeschst 33342: nuclear staining. Original magnification: 200 X.
To directly demonstrate OTK18 cleavage by calpain, we have synthesized two peptides corresponding to amino acid region 301-378, and confirmed that one of the peptides was digested by purified calpain-I after arginine 359 as determined by MALDI and MALDI-TOF/TOF (Figure 5 A-D). Since the amino acid 359-378 region of the peptide is completely degraded and show no other peak (Fig. 5B) and this region contains KPY+CXDCGK+F (+ as charged residue and X as random residue) sequence which are found in 10 C2H2 rings in the C-terminal of OTK18, it is possible that the C-terminal region after position 359 were completely degraded.
Figure 5. Mass spectrometric identification of a fragment derived by caplain from OTK18 peptide.
A, Mass spectrum of OTK peptide (GKVFI-PHTRKKPYKCHDCGKAFFQMLSLLLYECS) analyzed by MALDI 4800 TOF. B, A predominant 1054.59 m/z ion (MALDI-TOF) was detected after digestion of OTK18 peptide with calpain. C, MS/MS (MALDI-TOF/TOF) spectra of generated by collision induced dissociation of a precursor ion 1054.59 m/z showing corresponding y- and b- ions. D, Amino acid sequence derived from MS/MS spectra of precursor ion 1054.59 m/z.
Visualization of OTK18 endoproteolysis by Intramolecular FRET in live cells
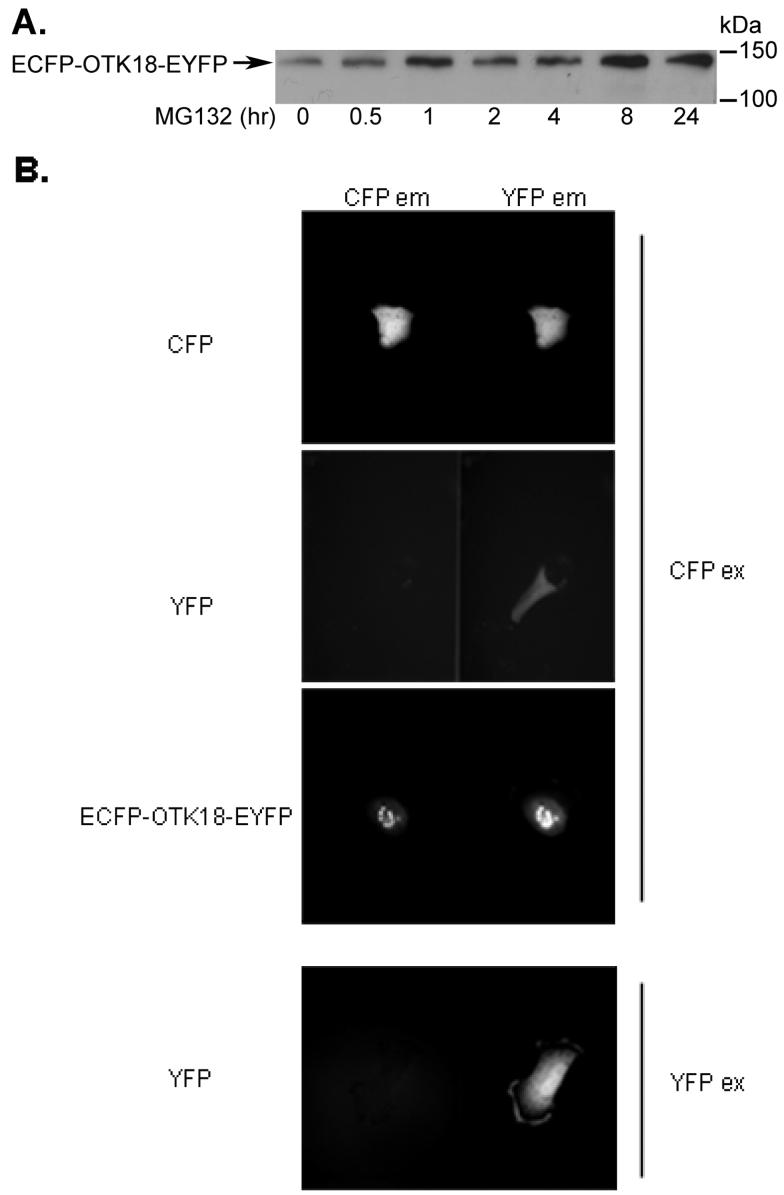
Finally, we developed a tool to monitor the endoproteolysis and degradation of OTK18 in live cells by constructing ECFP-OTK18-EYFP fusion gene. Excitation of N-terminal ECFP using CFP filter emits fluorescence from CFP, which overlaps the excitation wavelength of C-terminal EYFP located within FRET distance. Emission from the EYFP completes the energy transfer from ECFP to EYFP within the ECFP-OTK18-EYFP fusion protein. Endoproteolysis or degradation of OTK18 results in separation of ECFP and EYFP, and loss of intramolecular FRET signal. Thus, calibrated FRET intensity corresponds to the amount of intact ECFP-OTK18-EYFP molecule in the nucleus. The expressed ECFP-OTK18-EYFP in human brain macrophage cell line was very stable with high basal expression level, showing accumulation due to MG132 treatment in a time-dependent manner (Fig. 6A). However, ECFP-OTK18-EYFP was insensitive to calpain inhibitor I up to 24 hr of incubation, presumably due to its conformational changes by ligation of two fluorescent molecules on both terminals (data not shown). Thus, we focused on the effect of MG132 on ECFP-OTK18-EYFP metabolism. Net FRET (nF) intensity was calculated based on the images of ECFP-OTK18-EYFP in CFP, YFP, and FRET filter set-ups, and images of ECFP (donor) and EYFP (acceptor) alone in CFP, YFP, and FRET filter set-ups (see Materials and Methods). This calculation corrects the nF intensity of ECFP-OTK18-EYFP FRET image which can be affected by bleed-through signals of ECFP itself (CFPex/YFPem image) or EFYP itself (CFPex/YFPem image, Fig. 6B). Expressed ECFPOTK18-EYFP in the cell line was exclusively localized in nucleus (Fig. 7A), and its SE-FRET intensity was not enhanced by MG132 in the absence of puromycin (Fig. 7A, upper panels and Fig. 6B, left panel). To avoid the potential complication of monitoring the turnover of SE-FRET signal due to the new protein synthesis, we treated cells with puromycin, a protein translation inhibitor, with or without MG132 (Fig. 7A, lower panels), and observed significant increase in SE-FRET peaks as shown qualitatively by the 3D histogram (Fig. 7A, lower right panels) and quantitatively by average nF intensity by MG132 treatment (Fig. 7B right panel) at a 120 min time point. This demonstrates that proteasomal inhibition accumulates intact ECFP-OTK18-EYFP molecule in the nucleus.
Figure 6. Expression and FRET analysis of ECFP-OTK18-EYFP.
A, The cells were transfected with ECFP-OTK18-EYFP and treated with 10 mM MG132 for the times indicated. Total cell extract was subjected to immunoblotting using anti-OTK18 mAb. B, The cells were transfected with ECFP, EYFP, or ECFP-OTK18-EYFP 24 hours prior to imaging, and excited with CFP filter (CFPex for ECFP, EYFP, or ECFP-OTK18-EYFP) or YFP filter (EYFP only), and images were taken by CFP filter (CFPem, left panel) and YFP filter (YFPem, right panel) using Dual-View imaging system. Original magnification: 400X.
Figure 7. Detection of OTK18 accumulation by SE-FRET.
A, The cells were transfected with ECFP-OTK18-EYFP 24 hours prior to imaging, and treated with or without 20 µg/ml puromycin at t = -30 min, then with or without 10µM MG132 t = 0 min (the start time of imaging). Images were taken in 5-minute increments up to 120 minutes. Cells were excited with CFP, and both CFP and YFP emissions are shown as well as the calculated SE-FRET image. 3D surface plots were constructed using ImageJ. B, Average nF intensity was calculated from time-lapse images using MetaMorph software and plotted as fold increase over original average nF at t=0 in the absence of puromycin (left panel) or presence of 20 mg/ml puromycin (right panel). Solid triangle (black) or square dots (green) represent MG132 untreated or treated groups, respectively. Linear regression and repeated measurement ANOVA (RMANOVA) were performed by Prism (p > 0.05 for no puromycin-treated experiment, and p < 0.0001 for puromycin-treated experiment).
Discussion
Role of zinc finger proteins on viral replication
Regulation of the viral life cycle in human mononuclear phagocytes (brain perivascular macrophages and microglia) is mediated by multiple mechanisms (Buescher et al., 2007), including gene induction of interferons (Meylan et al., 1993; Baca-Regen et al., 1994), interaction with RNA-activated kinase (Roy et al., 1990), and induction of apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) (Peng et al., 2006). Our previous studies have shown that expression of zinc finger proteins, containing KRAB (Krüppel-associated box), which associate with tandem arrays of Krüppel-type (C2H2) zinc finger motifs, represents yet another antiviral mechanism in human macrophages. Previously, functional genes and pseudogenes in the human chromosome19p12-13.1 KRAB ZNF cluster were identified from the human KRAB-ZNF gene catalog (Huntley et al., 2006). This large primate-specific gene cluster contains over 70 ZNF loci, including 39 intact genes as well as pseudogenes (Hamilton et al., 2006). OTK18 is not a unique molecule in that aspect, since a number of other zinc finger proteins are located in this region and were up-regulated by HIV-1 infection of human MDM as determined by human high-density cDNA microarray study, including ZNF14, ZNF43, ZNF45, ZNF85, ZNF91, ZNF134, ZNF136, ZNF137, ZNF146, ZNF211, ZNF223, ZNF255, ZNF264, ZNF274, ZNF304, and ZNF361 (unpublished observations). The principles of our findings on post-translational regulation of OTK18 in human macrophages may be applied to the other ZNF family members upregulated by viral infection.
Calpain and proteasomal regulation of viral replication and neuropathogenesis
Calpain is a cysteine protease significantly involved in multiple steps of the viral life cycle. Calpain degrades IκB and enhances NFκB binding to the HIV long-terminal repeat, which is critically important for the promoter activation (Teranishi et al., 2003). Calpain activation is also involved in the LFA-1 and ICAM-1 mediated viral adhesion and fusion into CD4+ T cells (Stewart et al., 1998; Tardif and Tremblay, 2005). Our study revealed another function of calpain on the viral life cycle, by processing the KRAB-ZNF transcriptional suppressor. Since OTK18N is significantly upregulated in perivascular macrophages of advanced HIVE, calpain activity may be highly upregulated in these cells and serve as a focal point of viral production in the brain. Thus, this study suggests the importance of HIV+ perivascular macrophages for the progression of HIV-associated neurocognitive disorders. Calpain is also involved in NMDA receptor-mediated neurotoxicity by HIV-infected macrophages (O'Donnell et al., 2006; Wang et al., 2007). Activation of the NMDA receptor by glutamate or gp120 results in Ca2+ influx, which activate calpain and subsequent cleavage of p35 cyclin-dependent kinase 5 (CDK5) activator to p25, which induces neuronal cell death (Wang et al., 2007). The calpain inhibitor is also neuroprotective (O'Donnell et al., 2006). The role of calpain in HIV neuropathogenesis, however, is poorly studied and can be an important subject in the field. Taken together, these studies suggest calpain inhibition as a novel target of anti-viral therapy for suppressing viral entry, replication, and neurotoxicity.
Viral infection of macrophages induces caspase activation, which leads to activation of calpain. Thus, we tested if staurosporine, a potent caspase activator, enhances OTK18 endoproteolysis. However, staurosporine unexpectedly enhanced accumulation of OTK18 molecules in both MDM and microglia cell line. Since Z-VAD-FMK, a pan caspase inhibior, did not accumulate OTK18 but rather enhanced calpain inhibitor I or staurosporine-mediated OTK18 accumulation, caspase may not directly cleave OTK18 but rather enhances the calpain activity. There is no report of staurosporine-mediated protein accumulation in the literature to understand this mechanism. However, one potential explanation of this finding is that since calpain is normally inactivated by forming a complex with calpastatin, some staurosporine-sensitive protein kinase may mediate dissociation of the complex for calpain activation. However, this mechanism is beyond the scope of this study.
Proteasome also plays an important role in the viral life cycle, especially budding and cyclin T1-mediated gene expression. It directly regulates NF-κB activation through degradation of IκBα and also mediates suppression of viral replication through degradation of cyclin T1, a cellular cofactor essential for viral Tat function (Liou et al., 2004; Liou et al., 2006; Wang and Rice, 2006). Increased levels of ubiquitinated proteins are correlated with the reduction of synaptic protein levels in aged HIV brains (Gelman and Schuenke, 2004). Proteasome also mediates HIV gp120-indcued degradation of tight junction molecules in human brain microvascular endothelial cells, suggesting the role of proteasome activity in virus-mediated blood brain barrier degradation and HIV neuropathogenesis (Nakamuta et al., 2008). However, the role of ubiquitin or proteasome system on HIV neurotoxicity is poorly studied and could be a future subject or investigation.
Our data suggest the potential mechanism of cytoplasmic accumulation of OTK18N in the perivascular macrophages in the HIV encephalitis brain (Carlson et al., 2004b). In the encephalitic brain, activation of virus-infected cells with pro-inflammatory cytokines, such as interferon-γ, leads to upregulation of immunoproteasome and activation of viral replication. This may also lead to enhanced proteolysis of OTK18, disruption of its own negative gene regulation system, and accumulation of processed OTK18 (OTK18N) in the cytoplasm of macrophages. Besides the post-translational processing mechanism identified in this paper, we have also studied how viral infection activates the promoter of OTK18 gene (Buescher et al, 2008). Indeed, we found that primary cultured microglia have a different gene expression profile of transcriptional factors (YY1, FoxD3, and c-Ets-1) upon viral infection, which differentially regulates OTK18 promoter activation. These dynamic regulation mechanisms—both pre- and post-translational—of a zinc finger molecule might be applicable to understanding the function of other KRAB-ZNF molecules clustered in the same chromosome region in the context of their viral regulation.
Acknowledgment
We would like to thank M. Tardieu for human microglia cell line, Y. Tot for supporting the quantification of SE-FRET images, J. Buescher for assisting with tissue culture and purification of DNA plasmids, and Meg Marquardt for manuscript editing. This work is supported in part by NIH Grants R01 MH072539 (T.I.) and NCRR P20RR15635 (T.I.).
References
- Baca-Regen L, Heinzinger N, Stevenson M, Gendelman HE. Alpha interferon-induced antiretroviral activities: restriction of viral nucleic acid synthesis and progeny virion production in human immunodeficiency virus type 1-infected monocytes. J Virol. 1994;68:7559–7565. doi: 10.1128/jvi.68.11.7559-7565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher J, Gross S, Gendelman H, Ikezu T. Neuropathogenesis of HIV. In: Berger JR, Portegies P, editors. Handbook of Clinical Neurology. Elsevier B. V.; Amsterdam, Netherlands: 2007. pp. 45–68. [DOI] [PubMed] [Google Scholar]
- Carlson K, Leisman G, Limoges J, Pohlman G, Horiba M, Buescher J, Gendelman H, Ikezu T. Molecular Characterization of a Putative Anti-Retroviral Transcriptional Factor, OTK18. J Immunol. 2004a;172:381–391. doi: 10.4049/jimmunol.172.1.381. [DOI] [PubMed] [Google Scholar]
- Carlson KA, Limoges J, Pohlman GD, Poluektova LY, Langford D, Masliah E, Ikezu T, Gendelman HE. OTK18 expression in brain mononuclear phagocytes parallels the severity of HIV-1 encephalitis. J Neuroimmunol. 2004b;150:186–198. doi: 10.1016/j.jneuroim.2004.01.021. [DOI] [PubMed] [Google Scholar]
- Buescher JL, Martinez LB, Sato S, Okuyama S, Ikezu T. YY1 and FoxD3 regulate antiretroviral zinc finger protein OTK18 promoter activation induced by HIV-1 infection. J Neuroimmune Pharmacol. 2008 doi: 10.1007/s11481-008-9139-x. in press doi: 10.1007/s11481-008-9139-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman BB, Schuenke K. Brain aging in acquired immunodeficiency syndrome: increased ubiquitin-protein conjugate is correlated with decreased synaptic protein but not amyloid plaque accumulation. J Neurovirol. 2004;10:98–108. doi: 10.1080/13550280490279816. [DOI] [PubMed] [Google Scholar]
- Gendelman HE, Orenstein JM, Martin MA, Ferrua C, Mitra R, Phipps T, Wahl LA, Lane HC, Fauci AS, Burke DS, et al. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J Exp Med. 1988;167:1428–1441. doi: 10.1084/jem.167.4.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon GW, Berry G, Liang XH, Levine B, Herman B. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys J. 1998;74:2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AT, Huntley S, Tran-Gyamfi M, Baggott DM, Gordon L, Stubbs L. Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome Res. 2006;16:584–594. doi: 10.1101/gr.4843906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holness CL, Simmons DL. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood. 1993;81:1607–1613. [PubMed] [Google Scholar]
- Horiba M, Martinez L, Buescher JL, Sato S, Limoges J, Jiang Y, Jones C, Ikezu T. OTK18, a zinc finger protein, regulates human immunodeficiency virus type I long terminal repeat through two distinct regulatory regions. J Gen Virol. 2007;88:236–241. doi: 10.1099/vir.0.82066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Wang KK. The calpain family and human disease. Trends Mol Med. 2001;7:355–362. doi: 10.1016/s1471-4914(01)02049-4. [DOI] [PubMed] [Google Scholar]
- Huntley S, Baggott DM, Hamilton AT, Tran-Gyamfi M, Yang S, Kim J, Gordon L, Branscomb E, Stubbs L. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–677. doi: 10.1101/gr.4842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janabi N, Peudenier S, Heron B, Ng KH, Tardieu M. Establishment of human microglial cell lines after transfection of primary cultures of embryonic microglial cells with the SV40 large T antigen. Neurosci Lett. 1995;195:105–108. doi: 10.1016/0304-3940(94)11792-h. [DOI] [PubMed] [Google Scholar]
- Janabi N, Di Stefano M, Wallon C, Hery C, Chiodi F, Tardieu M. Induction of human immunodeficiency virus type 1 replication in human glial cells after proinflammatory cytokines stimulation: effect of IFNgamma, IL1beta, and TNFalpha on differentiation and chemokine production in glial cells. Glia. 1998;23:304–315. [PubMed] [Google Scholar]
- Liou LY, Herrmann CH, Rice AP. Human immunodeficiency virus type 1 infection induces cyclin T1 expression in macrophages. J Virol. 2004;78:8114–8119. doi: 10.1128/JVI.78.15.8114-8119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou LY, Haaland RE, Herrmann CH, Rice AP. Cyclin T1 but not cyclin T2a is induced by a post-transcriptional mechanism in PAMP-activated monocyte-derived macrophages. J Leukoc Biol. 2006;79:388–396. doi: 10.1189/jlb.0805429. [DOI] [PubMed] [Google Scholar]
- Liu J, Liu MC, Wang KK. Calpain in the CNS: from synaptic function to neurotoxicity. Sci Signal. 2008;1:re1. doi: 10.1126/stke.114re1. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J. The role of ubiquitin in retroviral egress. Traffic. 2007;8:1297–1303. doi: 10.1111/j.1600-0854.2007.00609.x. [DOI] [PubMed] [Google Scholar]
- Meylan PR, Guatelli JC, Munis JR, Richman DD, Kornbluth RS. Mechanisms for the inhibition of HIV replication by interferons-alpha, -beta, and -gamma in primary human macrophages. Virology. 1993;193:138–148. doi: 10.1006/viro.1993.1110. [DOI] [PubMed] [Google Scholar]
- Nakamuta S, Endo H, Higashi Y, Kousaka A, Yamada H, Yano M, Kido H. Human immunodeficiency virus type 1 gp120-mediated disruption of tight junction proteins by induction of proteasome-mediated degradation of zonula occludens-1 and -2 in human brain microvascular endothelial cells. J Neurovirol. 2008;14:186–195. doi: 10.1080/13550280801993630. [DOI] [PubMed] [Google Scholar]
- O'Donnell LA, Agrawal A, Jordan-Sciutto KL, Dichter MA, Lynch DR, Kolson DL. Human immunodeficiency virus (HIV)-induced neurotoxicity: roles for the NMDA receptor subtypes. J Neurosci. 2006;26:981–990. doi: 10.1523/JNEUROSCI.4617-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, Lei KJ, Jin W, Greenwell-Wild T, Wahl SM. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J Exp Med. 2006;203:41–46. doi: 10.1084/jem.20051512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati M, Valentin A, Patenaude DJ, Pavlakis GN. CCAAT-enhancer-binding protein beta (C/EBP beta) activates CCR5 promoter: increased C/EBP beta and CCR5 in T lymphocytes from HIV-1-infected individuals. J Immunol. 2001;167:1654–1662. doi: 10.4049/jimmunol.167.3.1654. [DOI] [PubMed] [Google Scholar]
- Roy S, Katze MG, Parkin NT, Edery I, Hovanessian AG, Sonenberg N. Control of the interferon-induced 68-kilodalton protein kinase by the HIV-1 tat gene product. Science. 1990;247:1216–1219. doi: 10.1126/science.2180064. [DOI] [PubMed] [Google Scholar]
- Shyu YJ, Suarez CD, Hu CD. Visualization of AP-1 NF-kappaB ternary complexes in living cells by using a BiFC-based FRET. Proc Natl Acad Sci U S A. 2008;105:151–156. doi: 10.1073/pnas.0705181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MP, McDowall A, Hogg N. LFA-1-mediated adhesion is regulated by cytoskeletal restraint and by a Ca2+-dependent protease, calpain. J Cell Biol. 1998;140:699–707. doi: 10.1083/jcb.140.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracher A. Calpain inhibitors as therapeutic agents in nerve and muscle degeneration. Ann N Y Acad Sci. 1999;884:52–59. doi: 10.1111/j.1749-6632.1999.tb08635.x. [DOI] [PubMed] [Google Scholar]
- Tardif MR, Tremblay MJ. Regulation of LFA-1 activity through cytoskeleton remodeling and signaling components modulates the efficiency of HIV type-1 entry in activated CD4+ T lymphocytes. J Immunol. 2005;175:926–935. doi: 10.4049/jimmunol.175.2.926. [DOI] [PubMed] [Google Scholar]
- Teranishi F, Liu ZQ, Kunimatsu M, Imai K, Takeyama H, Manabe T, Sasaki M, Okamoto T. Calpain is involved in the HIV replication from the latently infected OM10.1 cells. Biochem Biophys Res Commun. 2003;303:940–946. doi: 10.1016/s0006-291x(03)00447-9. [DOI] [PubMed] [Google Scholar]
- Wang Y, Rice AP. Interleukin-10 inhibits HIV-1 LTR-directed gene expression in human macrophages through the induction of cyclin T1 proteolysis. Virology. 2006;352:485–492. doi: 10.1016/j.virol.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Wang Y, White MG, Akay C, Chodroff RA, Robinson J, Lindl KA, Dichter MA, Qian Y, Mao Z, Kolson DL, Jordan-Sciutto KL. Activation of cyclin-dependent kinase 5 by calpains contributes to human immunodeficiency virus-induced neurotoxicity. J Neurochem. 2007;103:439–455. doi: 10.1111/j.1471-4159.2007.04746.x. [DOI] [PubMed] [Google Scholar]