Abstract
Biologists rely on morphology, function and specific markers to define the differentiation status of cells. Transcript profiling has expanded the repertoire of these markers by providing the snapshot of cellular status that reflects the activity of all genes. However, such data have been used only to assess relative similarities and differences of these cells. Here we show that principal component analysis of global gene expression profiles map cells in multidimensional transcript profile space and the positions of differentiating cells progress in a stepwise manner along trajectories starting from undifferentiated embryonic stem (ES) cells located in the apex. We present three ‘cell lineage trajectories’, which represent the differentiation of ES cells into the first three lineages in mammalian development: primitive endoderm, trophoblast and primitive ectoderm/neural ectoderm. The positions of the cells along these trajectories seem to reflect the developmental potency of cells and can be used as a scale for the potential of cells. Indeed, we show that embryonic germ cells and induced pluripotent cells are mapped near the origin of the trajectories, whereas mouse embryo fibroblast and fibroblast cell lines are mapped near the far end of the trajectories. We suggest that this method can be used as the non-operational semi-quantitative definition of cell differentiation status and developmental potency. Furthermore, the global expression profiles of cell lineages provide a framework for the future study of in vitro and in vivo cell differentiation.
Keywords: embryonic stem, embryonic germ, induced pluripotent stem, mouse embryo fibroblast, embryonal carcinoma, retinoic acids, neural stem/progenitor, trophoblast stem, principal component analysis, leukemia inhibitory factor, epigenetic landscape, Waddington, developmental potency, cell lineage trajectory, gene expression profiling, DNA microarray analysis
1. Introduction
Developmental biologists have long held a view that development naturally progresses from totipotent fertilized eggs with unlimited differentiation potential to terminally differentiated cells, like a ball rolling from high to low points on a slope as depicted in Waddington's epigenetic landscape.1 The epigenetic landscape also points to another important aspect of development, which is the emergence of different cell lineages during the development; cells in specific developmental lineages are thought to take discrete paths (‘chreodes’) on the imaginary slope. Analogy of cell's developmental potency to the potential energy is thus widely accepted2,3 This analogy is also relevant to the fact that converting differentiated cells into pluripotent cells is difficult. In mammals, nuclear transplantation (cloning)4,5 had been the only way to achieve such a ‘up-hill battle’ reprogramming, until the successful production of induced pluripotent stem (iPS) cells by infecting MEFs with retroviruses carrying expression cassettes for four genes (Myc, Pou5f1, Sox2 and Klf4).6 Notwithsanding its importance, the potency has only been defined operationally by in vitro and in vivo cell differentiation assays as ‘the total of all fates of a cell or tissue region which can be achieved by any environmental manipulation'.7 Nuclear transplantation experiments, where the success rate gradually decreases according to developmental stages of donor cells, provide yet another operational definition of developmental potential.8–10 We previously showed a possibility to derive a scale of developmental potency from the global gene expression (transcript) profile data, but the data could not be that quantitative because of the use of a limited number of expressed sequence tags (ESTs) for the analysis.11 The work also did not address the issue of cell linege separations.
Mouse embryonic stem (ES) cells12,13 and embryonic germ (EG) cells14,15 are prototypical stem cells. These cells can be maintained as undifferentiated state in culture (self-renewal) and have the capacity to differentiate into essentially all the cell types (pluripotency). Therefore, these pluripotent stem cells provide tractable systems to study the developmental potency and cell lineage separation. It has been shown that the manipulation of cell culture condition or a single-gene expression level can differentiate ES cells into relatively homogenous cell population that are similar to the first three lineages in mammalian development:16 primitive ectoderm/neural ectoderm,17,18 trophoblast19,20 and primitive endoderm.21 In the first system, ES cells are cultured in monolayer in N2B27 medium, which drives undifferentiated ES cells into neural lineages.17 Previous DNA microarray analysis indicates that this in vitro ES cell differentiation process mimics in vivo cell differentiation to primitive ectoderm, neural ectoderm and subsequently neurons/glia cells.18 In the second system, ES cells are engineered to downregulate Pou5f1 (Oct3/4, Oct4) expression in a tetracycline-controllable manner (ZHBTc4 cell line19). It has been shown that repression of Pou5f1 induces the differentiation of ES cells into trophoblast lineage.19,20 In the third system, ES cells that are engineered to overexpress Gata6 in a dexamethasone-inducible manner differentiate into primitive endoderm (extraembryonic endoderm).21 Although the analyses of these ES cell differentiation systems have revealed the detailed changes of gene expression patterns, it remains to see whether the global comparison among these individual systems provide any further insights into developmental potency and cell lineage separation.
Here we show that principal component analysis (PCA), which can reduce the dimensionality of the gene expression profiles,22 maps cells in a multidimensional transcript profile space where the positions of differentiating cells progress in a stepwise manner along trajectories starting from undifferentiated ES cells located in the apex to the first three lineages in mammalian development: primitive endoderm, trophoblast and primitive ectoderm/neural ectoderm. Furthermore, EG cells and iPS cells are mapped near the origin of the trajectories, whereas mouse embryo fibroblast (MEF) and fibroblast cell lines are mapped near the far end of the trajectories.
2. Materials and methods
2.1. Cells and RNAs
For the majority of cells used in this study, we used a stock of Cy3-labeled cRNA samples that were used in our previous studies. The details of each cell types, their culture conditions, RNA extractions and Cy3-labeling can be found in the main text of this manuscript and in earlier publications.18,20,23–25 Cells cultured for this study and the culture conditions are as follows. G0–G5 cells: Production and characterization of 5G6GR ES cell clones that are engineered to overexpress Gata6 in a dexamethasone-inducible manner are described previously.21 ES cells were harvested every 24 h (Day 0–5) during differentiation in the presence of 100 mM dexamethasone (Sigma). F0–F5 cells: Undifferentiated F9 EC cells (ATCC number: CRL-1720) were treated with 100 nM all-trans-retinoic acid (RA, Sigma) and 1 mM dibutyryl cAMP (dbcAMP, Sigma) on adherent condition as reported.26 Photos of F9 EC cells during differentiation are available as Supplementary Fig. S1 of the published paper.27 F9 cells were harvested every 24 h (Day 0–5) during the differentiation. Both iPS-Fbxo156 and iPS-Nanog28 were cultured on the STO feeder cells as described previously. To remove the feeder cells, the iPS cells were passaged twice on the gelatin-coated culture dish and harvested for RNA extraction. NIH3T3, STO, MEF_BL6 and MEF_DR4 were cultured under the standard condition and harvested for RNA extraction. Total RNAs isolated for this study were labeled with Cy3-dye and used for the DNA microarray hybridization.
2.2. Microarray data analysis
DNA microarray analysis was carried out as described previously,18 except for the addition of ES cell total RNAs to the Universal Mouse Reference RNA (UMRR) and the use of a 4 × 44K microarray platform. In our previous DNA microarray studies,23 we used the Universal Mouse Reference RNA (UMRR: Stratagene), which is a mixture of total RNAs from 11 different mouse cell lines. However, to increase the representation of genes expressed in ES cells for the current study, we mixed the UMRR with total RNAs from ES cells (MC1, derived from 129S6/SvEvTac strain) cultured in the undifferentiated condition with LIF at 2:1 ratio. These UMRR plus ES RNAs were labeled with Cy5-dye, mixed with Cy3-labeled samples and used for DNA microarray hybridization. To maximize the uniformity of the microarray data, all the samples, including the ones analyzed by DNA microarray previously, were hybridized to the same platform (the NIA Mouse 44K Microarray v3.023 manufactured by Agilent Technologies #015 087). The intensity of each gene feature per array was extracted from scanned microarray images using Feature Extraction 9.5.1.1 software (Agilent Technologies) as described previously.23 Hierarchical clustering analysis and PCA (see Section 2.3) were carried out using an application developed in-house to perform ANOVA and other analyses (NIA Array Analysis software; http://lgsun.grc.nia.nih.gov/ANOVA/).22 Three-dimensional PCA figures were generated using the virtual reality modeling language (VRML) function of the NIA Array Analysis software22 and visualized by Cortona Vrml client (http://www.parallelgraphics.com/products/cortona/). To produce plots and lists of genes for Fig. 2, we used the R-statistical package29 and only non-redundant set of oligonucleotide probes (a total of 25 164 probes/genes). Of these, 21 890 genes were significant in ANOVA with FDR < 0.05 and used for further analyses. Genes correlated to each cell lineage trajectory were identified based on the correlation of >0.90. All the DNA microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number (GSE11 523) and the NIA Array Analysis software website (http://lgsun.grc.nia.nih.gov/ANOVA/).22
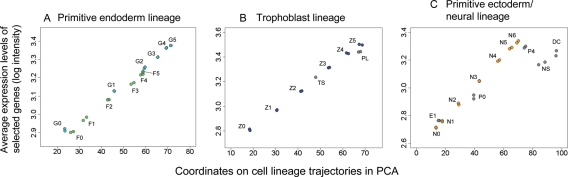
Figure 2.
Average expression levels of selected genes correlate well with the locations of each cell type in PCA coordinates. See Fig. 1A for the abbreviated names of the cells. Each point shows DNA microarray data for each sample; therefore, each cell type is represented by two points. (A) Primitive endoderm lineage, represented by the list of 1738 genes (Supplementary Table S1). (B) Trophoblast lineage, represented by the list of 2311 genes (Supplementary Table S2). (C) Primitive ectoderm/neural lineage, represented by the list of 2463 genes (Supplementary Table S3).
2.3. Principal component analysis
Principal component analysis is a statistical method to find major patterns in data variation, which is increasingly used for the analysis of gene expression microarray data.22,30–32 Each principal component (PC) is a linear combination of log-transformed expression values of all genes, and all components are orthogonal, i.e. mutually independent. The first few PCs, which explain most of the observed variance, are the most important; the remaining PCs often represent random fluctuations. Therefore, by plotting data against the first two or three PCs, one can reduce the dimensionality of the data without losing much of information. Contribution of genes to each PC (which may be positive or negative) is determined by singular value decomposition algorithm applied to the covariance matrix. Knowing these contributions, the entire gene expression profile of a given cell type can be represented by a single point in a two- or three-dimensional space. If two cell types are represented by closely located points in the PCA plot, global gene expression profiles of these cells are very similar. In contrast, if corresponding points in the PCA plot are far apart, their global expression profiles are very different. The detailed discussion of PCA plots and interpretation of DNA microarray data can be found in the previous publication.32
3. Results and discussion
To obtain accurate, comprehensive and comparable expression profiles, we carried out all the DNA microarray analyses within a few weeks using the same platform containing essentially all genes encoded on the mouse genome.23 Although two ES cell differentiation systems have been profiled previously using the earlier version of the array platform,18,20 we carried out the DNA microarray analysis again for the data consistency. In the first system, ES cells are cultured in monolayer in N2B27 medium for 6 days, which drives undifferentiated ES cells into neural lineages.17 RNAs were isolated and analyzed every day during the induction and RNAs (N0–N6: Fig. 1A). Previous DNA microarray analysis has been carried out using the array platform containing a limited number of genes, but indicates that this in vitro ES cell differentiation process mimics in vivo cell differentiation to primitive ectoderm, neural ectoderm and, subsequently, neurons/glia cells.18 In the second system, ES cells that are engineered to downregulate Pou5f1 (Oct3/4, Oct4) expression in a tetracycline-controllable manner (ZHBTc4 cell line differentiate into trophoblast lineage for 5 days).19,20 RNAs were isolated and analyzed every day during the induction (Z0–Z5 in Fig. 1A). To include the first three lineages of mammalian development,16 we added a third system: ES cells that are engineered to overexpress Gata6 in a dexamethasone-inducible manner differentiate into primitive endoderm (extra-embryonic endoderm) for 5 days21 (G0–G5 in Fig. 1A). We also carried out microarray analysis of a fourth system: F9 embryonal carcinoma (EC) cells differentiate into parietal endoderm—one of the extra-embryonic endoderm lineages for 5 days26,27 (F0–F5 in Fig. 1A). For comparison, we also analyzed P19 EC cells before (P0) and after 4 days of RA induction (P4); neural stem/progenitor (NS) cells derived from adult mouse brain and their differentiated cells21 (DC); and trophoblast stem (TS) cells26 and E12.5 placenta (PL).27 Altogether we obtained the DNA microarray data of 32 different cell types in duplicate.
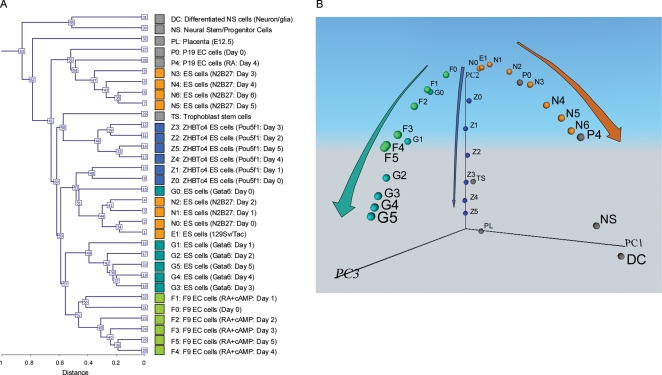
Figure 1.
Global gene expression profiles of 32 different cell types. Names of the cells and their abbreviated forms are shown. Color coding corresponds with the colors in all other figures. DNA microarray analysis was carried out in duplicate: for each cell type, two independent cell cultures were prepared and used for separate DNA microarray analysis. Reproducibility between two microarray data for each cell type was very high as shown by the Pearson correlation coefficient (mean 0.9927; SD 0.0043; range 0.9788–0.9966). The average of two samples is shown here. (A) Hierarchical clustering analysis. (B) Principal component analysis (PCA). Individual cell types are mapped in the 3D space according to the first three principal components (PC1, PC2 and PC3). A movie file is available as Supplementary Video S1.
We first carried out the hierarchical clustering analysis of these microarray data. Although the hierarchical clustering analysis grouped cells mostly into each differentiation system (Fig. 1A), it showed only the degree of similarity between different cell types and did not provide any insights into the relationship between the different cell lineages. To further analyze the data, we employed PCA, which can reduce the dimensionality of the gene expression profiles and map individual cells in a multidimensional space according to their global gene expression patterns (see Section 2.3 for the explanation of PCA). We first mapped the cells in the 3D transcript profile space (PC1, PC2 and PC3) (Fig. 1B). PCA of all these microarray data uncovered three major paths of differentiation from undifferentiated ES cells in 3D transcript profile space (Fig. 1B; Supplementary Video S1). Each path showed progressive changes in global expression patterns toward specific cell lineages: trophectoderm (Z0–Z5), extra-embryonic endoderm (G0–G5; F0–F5) and neural tissues (N0–N6). We call these paths ‘cell lineage trajectories,32’ because the cell differentiation process can be visualized as a trajectory in the multidimensional transcript profile space.
What do these trajectories represent? Because the transcript profile space is constructed from the expression patterns of genes, the changes in gene expression patterns are reflected in the changes in the position of a cell. To find gene expression changes that are correlated with these trajectories, we first drew a line for each cell lineage so that the squared distances from the cells of the lineage were minimized. The locations of other cell types, such as TS and NS, were projected onto the closest lineage trajectory (Fig. 1B). The x-axis of Fig. 2 shows the coordinates of these cell types on each trajectory (Fig. 2A–C). We then searched for sets of genes that were positively correlated with each trajectory line. We identified 1738 genes for the primitive endoderm lineage (Fig. 2A; Supplementary Table S1), 2311 genes for the trophoblast lineage (Fig. 2B; Supplementary Table S2) and 2463 genes for the primitive ectoderm/neural lineage (Fig. 2C; Supplementary Table S3) with a correlation of >0.90. The expression levels of these genes gradually increased as ES cells differentiated into each lineage (see Supplementary data for examples of gene expression changes). The average expression levels of these genes (y-axis in Fig. 2: cell lineage index32) can thus be used as surrogates for the coordinates of cell's positions on each trajectory (x-axis in Fig. 2). This lineage trajectory, therefore, not only defines a path to the fates of individual cell lineages, but also provides a scale for the extent of commitment/differentiation.
This notion was further supported by examining the locations of cells cultured and profiled independently in the transcript profile space: P19 EC cells before (P0) and after 4 days of RA induction (P4); NS cells derived from adult mouse brain and their differentiated cells18 (DC); and TS cells25,33 and E12.5 PL.25 These cells were mapped onto the transcript profile space based on their global gene expression profiles. As we expected, we found that P0, P4, NS and DC mapped near the primitive ectoderm/neural lineage trajectory (Figs 1B and 2C). Interestingly, the position of P0 was consistent with the notion that in contrast to ES cells, undifferentiated P19 EC cells are more committed and equivalent to primitive ectoderm cells.18 The position of P4 was also consistent with the notion that P19 EC cells differentiate into neuron-like cells after the induction with RA.34 Consistent with their known differentiation status, TS and PL mapped near the trophoblast lineage trajectory (Figs 1B and 2B). Two extra-embryonic endoderm lineages, which were represented by F9 EC cell differentiation26,35 (F0–F5) and ES cells driven by Gata621 (G0–G5), also showed lineage trajectories close to each other (Figs 1B and 2A). Taken together, these data support the notion that cell differentiation can be visualized as cell lineage trajectories in 3D transcript profile space.
We also noticed that these cell lineage trajectories represent the gradual loss of developmental potency or potential of cells (Fig. 1B). To examine this point further, we carried out microarray analysis of 12 additional cell types, including three undifferentiated ES cell lines (MC1, derived from 129S6/SvEvTac; MC2, derived from C57BL/6J; D3_GL; and EBRTcH3) and two undifferentiated EG cell lines [TGC8.5_5, derived from the primordial germ cell (PGC) of E8.5 C57BL/6J embryo; and EG1, derived from the PGC of E9 129 embryo].24 We also analyzed the expression profiles of MEF cells, stromal cells (STO) and a fibroblast cell line (NIH3T3). We then analyzed iPS cells, which have been established by infecting MEFs with retroviruses carrying expression cassettes for four genes (Myc, Pou5f1, Sox2 and Klf4).6,28 We used two different iPS lines: one was established by using Fbxo15 promoter-driven GFP as an indicator of pluripotency6 and the other was established by using Nanog promoter-GFP as an indicator of pluripotency.28
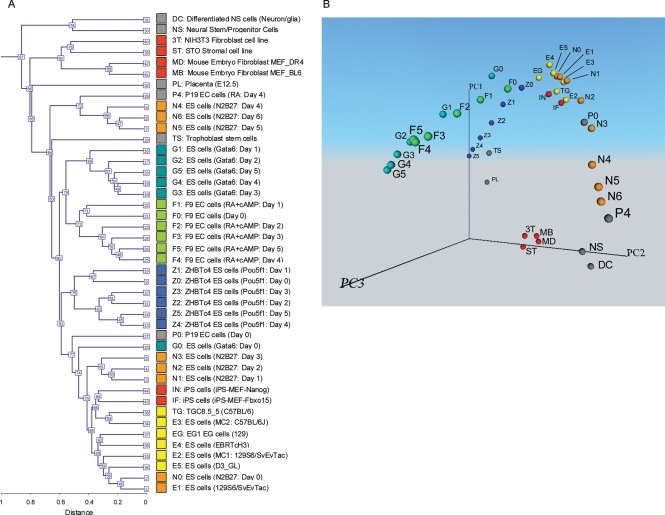
Hierarchical clustering analysis of the microarray data of all 44 cell types (original 32 cell types and 12 additional cell types; see Supplementary data for detail) showed that undifferentiated ES and EG cells were clustered together with iPS cells, whereas NIH3T3 fibroblast cells, STO stromal cells and MEF cells were clustered together (Fig. 3A). However, it was difficult to assess the overall relationship among all the cell types analyzed here. We, therefore, analyzed the microarray data of all 44 cell types by PCA (Fig. 3B; Supplementary Video S2). Interestingly, an overall structure of the transcript profile space remained essentially the same as that obtained by 32 cell types (Fig. 1B). The majority of the cells were aligned along the three major cell lineage trajectories, except for fibroblast and stromal cells (NIH3T3, STO and MEFs). The fact that these cells did not fall into any of the three cell lineage trajectories is consistent with their mesodermal origin. Their locations near the bottom of all three trajectory lines also support the general trend that these cell lineage trajectories represent the gradual loss of developmental potency of cells (Fig. 1B). Indeed, undifferentiated ES and EG cells mapped near the apex of the trajectory figures, as expected (Fig. 3B). Similarly, the iPS cells mapped near the apex of the trajectories. It has been demonstrated that these iPS cells have the complete phenotype of pluripotent cells, including germline competency in the case of iPS-Nanog.28 Considering that the iPS cells are derived from MEFs, changing the locations of cells from positions with low developmental potency (MEFs) to those with high developmental potency (iPSs) strongly supports our notion that PCA analysis can visualize and predict the developmental potency of cells.
Figure 3.
Global gene expression profiles of 44 different cell types. In addition to 32 cell types in the Fig. 1, 12 cell types are added for the analysis. Names of the cells and their abbreviated forms are shown. Color coding of cells are consistent throughout the paper. Average of two samples is shown. (A) Hierarchical clustering analysis. (B) Principal component analysis (PCA). Individual cell types are mapped in the 3D space according to the first three principal components (PC1, PC2 and PC3). A movie file is available Supplementary Video S2.
The work presented here provides a concept and a method to visualize and quantitate the differentiation status and developmental potency of cells. The PCA figures (Figs 1B and 3B) show remarkable similarity to the conceptual picture of Waddington's epigenetic landscape, where cell lineage trajectories represent creodes.1 Such a concept has been taken for granted by embryologists for over a century, but it now seems to be substantiated by the global gene expression patterns. The locations of cells in the multidimensional transcript profile space can be used to identify cells of unknown origin, determine the differentiation status of cells and to assess the developmental potency of cells, possibly without going through experimental manipulation and testing of the cells. The present work also provides a framework for the future study of ES cell differentiation by visualizing the direction to which cells are going to differentiate.
Supplementary Data
Supplementary data are available online at www.dnaresearch.oxfordjournals.org.
Funding
This work was entirely supported by the Intramural Research Program of National Institutes of Health, National Institute on Aging (Z01 AG000662).
Supplementary Material
Acknowledgements
We thank Drs. Tilo Kunath and Janet Rossant for providing RNAs of TS, and Dr. Angelo Vescovi for providing RNAs of NS and DC, Dr. Brigid Hogan for providing TG cells and Dr. Colin Stewart for providing EG1 cells. We also thank Dawood Dudekula and Yong Qian for assisting data analysis.
Footnotes
Edited by Osamu Ohara
References
- 1.Waddington C. H. The Strategy of the Genes. London: George Allen & Unwin Ltd; 1957. [Google Scholar]
- 2.Slack J. M. Conrad Hal Waddington: the last Renaissance biologist? Nat. Rev. Genet. 2002;3:889–895. doi: 10.1038/nrg933. [DOI] [PubMed] [Google Scholar]
- 3.Andrews P. W. From teratocarcinomas to embryonic stem cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002;357:405–417. doi: 10.1098/rstb.2002.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell K. H., McWhir J., Ritchie W. A., Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 5.Wakayama T., Perry A. C., Zuccotti M., Johnson K. R., Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Slack J. M. W. From Egg to Embryo: Determinative Events in Early Development. Cambridge University Press; 1991. Cambridge, UK. [Google Scholar]
- 8.Gurdon J. B., Byrne J. A. The first half-century of nuclear transplantation. Biosci. Rep. 2004;24:545–557. doi: 10.1007/s10540-005-2744-5. [DOI] [PubMed] [Google Scholar]
- 9.Hochedlinger K., Jaenisch R. Nuclear transplantation: lessons from frogs and mice. Curr. Opin. Cell Biol. 2002;14:741–748. doi: 10.1016/s0955-0674(02)00380-0. [DOI] [PubMed] [Google Scholar]
- 10.Yanagimachi R. Cloning: experience from the mouse and other animals. Mol. Cell. Endocrinol. 2002;187:241–248. doi: 10.1016/s0303-7207(01)00697-9. [DOI] [PubMed] [Google Scholar]
- 11.Sharov A. A., Piao Y., Matoba R., Dudekula D. B., Qian Y., VanBuren V., Falco G., Martin P. R., Stagg C. A., Bassey U. C., et al. Transcriptome analysis of mouse stem cells and early embryos. PLoS Biol. 2003;1:E74. doi: 10.1371/journal.pbio.0000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans M. J., Kaufman M. H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 13.Martin G. R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsui Y., Zsebo K., Hogan B. L. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70:841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- 15.Resnick J. L., Bixler L. S., Cheng L., Donovan P. J. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359:550–551. doi: 10.1038/359550a0. [DOI] [PubMed] [Google Scholar]
- 16.Pfister S., Steiner K. A., Tam P. P. Gene expression pattern and progression of embryogenesis in the immediate post-implantation period of mouse development. Gene Expr. Patterns. 2007;7:558–573. doi: 10.1016/j.modgep.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Ying Q. L., Stavridis M., Griffiths D., Li M., Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 18.Aiba K., Sharov A. A., Carter M. G., Foroni C., Vescovi A. L., Ko M. S. Defining a developmental path to neural fate by global expression profiling of mouse embryonic stem cells and adult neural stem/progenitor cells. Stem Cells. 2006;24:889–895. doi: 10.1634/stemcells.2005-0332. [DOI] [PubMed] [Google Scholar]
- 19.Niwa H., Miyazaki J., Smith A. G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 20.Matoba R., Niwa H., Masui S., Ohtsuka S., Carter M. G., Sharov A. A., Ko M. S. Dissecting Oct3/4-regulated gene networks in embryonic stem cells by expression profiling. PLoS One. 2006;1:e26. doi: 10.1371/journal.pone.0000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimosato D., Shiki M., Niwa H. Extra-embryonic endoderm cells derived from ES cells induced by GATA factors acquire the character of XEN cells. BMC Dev. Biol. 2007;7:80. doi: 10.1186/1471-213X-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharov A. A., Dudekula D. B., Ko M. S. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 2005;21:2548–2549. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- 23.Carter M. G., Sharov A. A., VanBuren V., Dudekula D. B., Carmack C. E., Nelson C., Ko M. S. Transcript copy number estimation using a mouse whole-genome oligonucleotide microarray. Genome Biol. 2005;6:R61. doi: 10.1186/gb-2005-6-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharova L. V., Sharov A. A., Piao Y., Shaik N., Sullivan T., Stewart C. L., Hogan B. L., Ko M. S. Global gene expression profiling reveals similarities and differences among mouse pluripotent stem cells of different origins and strains. Dev. Biol. 2007;307:446–459. doi: 10.1016/j.ydbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka T. S., Kunath T., Kimber W. L., Jaradat S. A., Stagg C. A., Usuda M., Yokota T., Niwa H., Rossant J., Ko M. S. Gene expression profiling of embryo-derived stem cells reveals candidate genes associated with pluripotency and lineage specificity. Genome Res. 2002;12:1921–1928. doi: 10.1101/gr.670002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strickland S., Smith K. K., Marotti K. R. Hormonal induction of differentiation in teratocarcinoma stem cells: generation of parietal endoderm by retinoic acid and dibutyryl cAMP. Cell. 1980;21:347–355. doi: 10.1016/0092-8674(80)90471-7. [DOI] [PubMed] [Google Scholar]
- 27.Yan Z., Wang Z., Sharova L., Sharov A. A., Ling C., Piao Y., Aiba K., Matoba R., Wang W., Ko M. S. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–1165. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 29.Team R. D. C. R Foundation for Statistical Computing. Vienna, Austria: 2007. [Google Scholar]
- 30.Seko Y., Azuma N., Takahashi Y., Makino H., Morito T., Muneta T., Matsumoto K., Saito H., Sekiya I., Umezawa A. Human sclera maintains common characteristics with cartilage throughout evolution. PLoS One. 2008;3:e3709. doi: 10.1371/journal.pone.0003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonnalagadda S., Srinivasan R. Principal components analysis based methodology to identify differentially expressed genes in time-course microarray data. BMC Bioinformatics. 2008;9:267. doi: 10.1186/1471-2105-9-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ko M. S. Expression profiling of the mouse early embryo: reflections and perspectives. Dev. Dyn. 2006;235:2437–2448. doi: 10.1002/dvdy.20859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka S., Kunath T., Hadjantonakis A. K., Nagy A., Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 34.Jones-Villeneuve E. M., McBurney M. W., Rogers K. A., Kalnins V. I. Retinoic acid induces embryonal carcinoma cells to differentiate into neurons and glial cells. J. Cell Biol. 1982;94:253–262. doi: 10.1083/jcb.94.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alonso A., Breuer B., Steuer B., Fischer J. The F9-EC cell line as a model for the analysis of differentiation. Int. J. Dev. Biol. 1991;35:389–397. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.