Abstract
Objective: Despite the advantages of structured data entry, much of the patient record is still stored as unstructured or semistructured narrative text. The issue of representing clinical document content remains problematic. The authors' prior work using an automated UMLS document indexing system has been encouraging but has been affected by the generally low indexing precision of such systems. In an effort to improve precision, the authors have developed a context-sensitive document indexing model to calculate the optimal subset of UMLS source vocabularies used to index each document section. This pilot study was performed to evaluate the utility of this indexing approach on a set of clinical radiology reports.
Design: A set of clinical radiology reports that had been indexed manually using UMLS concept descriptors was indexed automatically by the SAPHIRE indexing engine. Using the data generated by this process the authors developed a system that simulated indexing, at the document section level, of the same document set using many permutations of a subset of the UMLS constituent vocabularies.
Measurements: The precision and recall scores generated by simulated indexing for each permutation of two or three UMLS constituent vocabularies were determined.
Results: While there was considerable variation in precision and recall values across the different subtypes of radiology reports, the overall effect of this indexing strategy using the best combination of two or three UMLS constituent vocabularies was an improvement in precision without significant impact of recall.
Conclusion: In this pilot study a contextual indexing strategy improved overall precision in a set of clinical radiology reports.
Despite the increased emphasis on structured data entry that many electronic medical record systems (EMRS) mandate and the positive impact of such an approach on data accuracy,1 the majority of clinical data generated electronically as part of the patient care process is still in the form of unstructured or semistructured narrative text.2 It seems likely that this situation will continue into the foreseeable future, given the inherent difficulties and limitations of capturing many complex clinical observations as structured data. One could argue that the future EMRS will use a document-centric model that leverages a set of representational standards that preserve our ability to use narrative text as the primary mechanism for data capture. This document-centric approach currently is best represented by the Health Level Seven (HL7) Clinical Document Architecture3 (CDA) model.
The CDA addresses three major challenges inherent in a document-centric model of the clinical record: (1) representing the document itself and its relationships to the entire patient record; (2) representing internal document structure and, perhaps most importantly; (3) representing the important concepts contained within the text of the document. The first two issues are addressed by specifying that clinical documents be presented using eXtensible Markup Language (XML). The use of XML in the CDA guarantees that all clinical documents inherit a common representational model and that each document within a specific document class (e.g., radiology reports) share a common set of XML tags. The problem of concept representation within documents has been addressed using two alternative, but potentially complementary approaches: natural language processing4,5 and document indexing.6 In document indexing one generally is interested in an automated process of noun phrase identification. Because of the significant synonymy that exists in the medical vocabulary, there is often the additional desire to automatically map identified noun phrases to their canonical representation in one or more standard biomedical terminologies. The goal of this approach is to standardize the semantic representational model across a heterogeneous document set.
We have been interested in this problem for some time, in part, because of our work with Multimedia Electronic Medical Record Systems7 (MEMRS) and the related need to automatically identify important biomedical concepts found in a variety of clinical imaging reports. We have published work previously8,9 describing our experience using SAPHIRE,10 an automated indexing system, to represent biomedical concepts within radiology reports using the National Library of Medicine's (NLM) Unified Medical Language System (UMLS) Metathesaurus11,12 as a semantic model. While this work has confirmed that the UMLS can provide canonical terms for many important concepts found in clinical imaging reports, the performance of the automated indexing process itself has been hampered by generally low precision.
In analyzing the reasons for this poor precision, we were interested in examining whether the problem could be ameliorated by restricting, during the automated indexing process, the set of constituent UMLS terminologies used to represent concepts within specific sections of radiology reports. Our hope was that by narrowly restricting the set of UMLS constituent vocabularies chosen to represent each document section and by varying the members of this set on a document section-by-section basis, we could improve overall precision. We consider this approach to be an example of contextual indexing, by which we mean the application of a variety of strategies that attempt to improve automated concept recognition in clinical documents by identification of how semantic context varies between document types and within individual document categories.
In the course of this work we have decided to use the CDA as our basic document model. XML-based clinical documents, such as radiology reports, benefit from this approach in that we can leverage on an emerging representational standard to codify the document structure and associated representational tags required to support a uniform approach to contextual indexing of clinical documents. This approach not only allows us to represent the original clinical documents using a standards-based model, but also allows us to integrate the indexing rules required by our approach and the output of the automated indexing process itself back into the XML-based document.
Research Hypotheses and Methods
This study was designed to test two related hypotheses: (1) the precision of automatic concept assignment can be improved, without significant overall negative impact on recall, by indexing document sections using only a selected subset of the source vocabularies contained in U.S. National Library of Medicine's Unified Medical Language System (UMLS) Metathesaurus; and (2) the set of optimal UMLS source vocabularies that produce improved indexing precision will vary by document section and document class.
Indexing precision was defined as the percentage of correctly assigned concepts of all concepts assigned by the indexing engine. Recall was defined as the percentage of correctly assigned concepts of all concepts in the gold standard (as defined below). This definition has been used by other researchers.13,14 We also used eleven-point average precision, which is a common measurement of the combination of precision and recall. Eleven-point average precision is computed by averaging the precision (possible interpolated) of the indexing engine for a certain report at the recall levels 0.0, 0.1, 0.2,…, 0.9 and 1.0.
To test these hypotheses, we developed an indexing simulation model that allowed us to measure how using each of a large number of possible combinations of UMLS source vocabularies affected indexing precision and recall on a test set of radiology reports. The goal was to automatically identify combinations of source vocabularies that resulted in improved precision with minimal or no impact on recall.
The test document set for this pilot study consisted of 50 radiology imaging reports, chosen to represent the most common imaging modalities (10 conventional chest x-ray, 10 head computed tomography (CT) scans, 10 chest CT scans, 10 abdomen-pelvis CT scans, 5 head magnetic resonance images (MRI) and 5 whole-body radionuclide bone scans). These reports had been previously manually deidentified and then indexed using the National Library of Medicine's UMLS Knowledge Source Server (KSS)15 by two domain experts knowledgeable about UMLS and clinical content. The results of this manual UMLS indexing served as the gold standard in this experiment. This process has been described elsewhere.8 Throughout this report, we use a sample report as shown in ▶ (with human indexing results attached at the end) to illustrate how this methodology transforms the structure and content of the report. Because of space limitations, for illustration purposes, we have chosen to use a relatively simple radiology report format rather than the full, verbose, XML document format. The Appendix, the full XML representation of the indexed document used as the example in this paper, is available as an online data supplement at <www.jamia.org>.
-
Each radiology report was marked up with the following sections: Procedure, History, Technique, Findings, and Impression, using a script. The markups were inspected by the authors and corrected manually as required. The sample report after step 1 is shown in ▶, with the insertions of section heading such as PROCEDURE or FINDINGS.
At the end of the report text, is a list of indexing elements. Each element consists of the following components in order: the phrase in the report that generated the indexing term (e.g., Chest), the preferred UMLS concept name (e.g., Thorax), the UMLS Concept Unique Identifier (e.g., C0039992), and the UMLS Semantic Type (e.g., Body Location or Region).
This document set then was indexed automatically with UMLS terms (UMLS Metathesaurus 2000) using the SAPHIRE indexing engine.16 ▶ shows our sample radiology report indexed by SAPHIRE. Because SAPHIRE generates many indexing terms per document (for this report it generated over 100 possible terms), we have included only a sample of the indexing terms generated for illustration purposes. This indexing engine produced a number of UMLS concept attributes for each term that it generated: (1) the original phrase in the document that triggered the match (e.g., left lateral chest), (2) the preferred UMLS concept name (e.g., Lateral to the left), (3) the UMLS Concept Unique Identifier (e.g., C0450414), and (4) the list of UMLS source vocabularies containing the UMLS concept (e.g., RCD99). SAPHIRE also assigned a numeric score to each UMLS concept generated (not shown), which allowed us to calculate 11-point average precision as well as standard precision and recall.
-
Next, based on the markups, each report was partitioned into sections labeled as Procedure, History, Technique, Findings, and Impression. Taking the results of the manual UMLS indexing and SAPHIRE indexing for the whole report, the script then selected both the manual and SAPHIRE-generated UMLS concept descriptors for each section and appended them at the end of the report section, as explained below. Corresponding to the three sections of the sample report, there are three output files concatenated together shown in ▶.
From ▶, we can see that each record generated by manual indexing had four fields: a phrase in the document, the matched UMLS concept name, its UMLS CUI, and semantic type. A script took the first field (phrase in the document) and searched for that text in the content of all sections of that document. That manual indexing record then was assigned to each section containing the phrase (possibly more than one, as each UMLS concept was assigned to a document only once by human indexers even if it appeared in different sections repeatedly.)
As described earlier, each record of SAPHIRE indexing contained the following elements: (1) the original phrase in the document that triggered the match; (2) the preferred UMLS concept name; (3) the list of UMLS source vocabularies; and (4) the UMLS Concept Unique Identifier (CUI). We discovered that the text in the original phrase field was not always exactly the same as the original text in the document, because SAPHIRE performed some normalization on this field, such as case folding, removing stop words, and punctuations, and did not necessarily maintain the original phrase. To successfully locate each result record in the original document, the script applied the same lexical changes to the content of each document section and searched for the phrase normalized by SAPHIRE. In this way, we were able to match the original phrase as generated by SAPHIRE with the actual original phrase in the document itself.
SAPHIRE assigned concepts differently than did our human indexers. It followed the order of document content and assigned the same UMLS concepts repeatedly as long as phrases in different parts of the document matched the same concept. This is desirable since SAPHIRE allows partial matching, and slightly different phrases can trigger the match of the same concept with different scores. To calculate the 11-point average precision for the whole document, only the highest score for each concept was used.
Each document section was then output to disk as a separate file together with the concepts assigned to that section by both the human indexers and by SAPHIRE. Each section was therefore equivalent to a separate document for which we generated measures of precision, recall, and 11-point average precision.
-
The next step was to simulate SAPHIRE indexing using selected UMLS source vocabularies. We identified 19 major UMLS source vocabularies (▶) that we felt would be most likely used in indexing the content of radiology reports. Our simulated indexing experiments used only these 19 source vocabularies.
The performance of the simulated automatic indexing engine was measured using each combination of up to three of the selected 19 source vocabularies. Different vocabularies in the combination were used in the following way, for example, the combination of source A and B meant the indexing engine could assign concepts in source A or B to the document. We also measured the performance of indexing by all 60 English source vocabularies in the UMLS for comparison purposes. Thus, the total of 1160
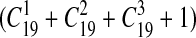 measurements were performed on each section of all reports. In each measurement, a combination of source vocabularies was chosen. A script checked the field of the source vocabulary list in each concept record assigned by SAPHIRE. The concept was selected only when there was at least one chosen vocabulary in the list. The script then measured the precision, recall, and 11-point precision, treating selected concepts as the output of SAPHIRE. A concept was considered a true-positive if its CUI matched one of the CUIs assigned to the same section by human indexers.
measurements were performed on each section of all reports. In each measurement, a combination of source vocabularies was chosen. A script checked the field of the source vocabulary list in each concept record assigned by SAPHIRE. The concept was selected only when there was at least one chosen vocabulary in the list. The script then measured the precision, recall, and 11-point precision, treating selected concepts as the output of SAPHIRE. A concept was considered a true-positive if its CUI matched one of the CUIs assigned to the same section by human indexers. The same measurements were performed for each section of all reports. The data then were collected, and comparisons were made across different types of sections, different types of reports, and different combinations of source vocabularies. An optimal combination of source vocabularies was determined (as explained in the following Results section) for each section of each report type. ▶ shows the output of this step. It shows all of the “correct” indexing terms (underlined in the figure) and a sample of the “incorrect” ones generated by SAPHIRE when using contextual indexing. One significant positive effect of this step is the dramatic reduction in the number of indexing terms generated (144 unique terms in the version shown in ▶, 41 unique terms in ▶, a 72% reduction).
Figure 1.
Sample radiology report with manually-derived UMLS descriptors.
Figure 2.
Radiology report shows partial output of SAPHIRE indexing.
Figure 3.
Partial SAPHIRE indexing results on the sample chest x-ray report, using all UMLS source vocabularies. The results from both manual and SAPHIRE indexing were split into report sections.
Table 1.
Selected 19 Major Source Vocabularies in Radiology Report Indexing
| UMLS SAB | Full Source Vocabulary Name |
|---|---|
| AIR93 | AI/RHEUM, 1993 |
| CCPSS99 | Canonical Clinical Problem Statement System (CCPSS), Version 1.0 |
| COS95 | Computer-Stored Ambulatory Records (COSTAR), 1995 |
| CPT2000 | Physicians' Current Procedural Terminology (CPT), 2000 |
| DXP94 | DXplain (an expert diagnosis program) |
| ICD10 | International Statistical Classification of Diseases and Related Health Problems (ICD-10), 10th rev. |
| MMSL99 | Multum MediSource Lexicon, 1999 |
| MSH2000 | Medical Subject Headings (MeSH), 2000 |
| NCI2000 | NCI Thesaurus, Version 2.0 |
| NDDF99 | National Drug Data File, 1999 |
| NEU99 | Neuronames Brain Hierarchy, 1999 |
| RAM99 | Randolph A. Miller Clinically Related Concepts, 1999 |
| RCD99 | Clinical Terms Version 3 (Read Codes) (Q199) |
| SNMI98 | SNOMED International, version 3.5 |
| SNM2 | Systematized Nomenclature of Medicine, 2nd edition |
| SPN99 | Standard Product Nomenclature (SPN), 1999 |
| UMD2000 | Universal Medical Device Nomenclature System: Product Category Thesaurus, 2000 |
| UWDA99 | University of Washington Digital Anatomist (UWDA), 1999 |
| WHO97 | WHO Adverse Drug Reaction Terminology (WHOART), 1997 |
Figure 4.
SAPHIRE indexing results (partial) on the sample chest x-ray report, using contextual strategy, with different selected source vocabularies in this case: Procedure section was indexed using ICD10 only; Findings section was indexed using MSH2000, SNMI98, and UMD2000; Impression section was indexed using MSH2000 and SNMI98. All of the correct indexing terms generated are shown. A representative sample of lower precision terms are also shown.
Results
The results of the experiment were aggregated and analyzed based on types of labeled document sections and the overall types of radiology reports. The optimal UMLS source vocabulary combinations were determined according to the total number of true-positive terms yielded by that combination, using the results of manual indexing as the gold standard. This approach allowed us to identify major contributing source vocabularies, and the results were also generally consistent with the results obtained by ranking the source vocabulary combinations using 11-point average precision. ▶ through ▶▶▶▶ show the optimal source vocabulary combination for each document section type and report type, together with 11-point average precision (11PP), changes in 11PP (delta11PP), changes in standard precision (deltaPrec), and changes in retrieval recall (deltaRec). For example, the second line of ▶ reads: for the Procedure section of Bone Scan reports, the best source vocabulary combination was found to be of MeSH 2000 and SNOMED International 98. Using these vocabularies for simulated indexing achieved an 11-point average precision of 27%, with improvements of 22% in 11-point average precision, 185% in precision, and 0% in recall compared with the results of indexing using all UMLS source vocabularies.
Table 2.
Optimal Source Vocabulary Combination for Procedure Section
| Report Type | Source Vocabulary Combination | 11PP | delta-11PP | delta-Prec | delta-Rec |
|---|---|---|---|---|---|
| Bone scan | MSH2000 + SNMI98 | 27% | 22% | 185% | 0% |
| Chest x-ray | MSH2000, ICD10 (tie) | 100% | 246% | 246% | 0% |
| CT abdomen | UWDA99 | 55% | 98% | 850% | 0% |
| CT chest | RCD99 | 65% | 80% | 263% | 0% |
| Head CT | MSH2000 + RCD99 | 79% | 1% | 305% | 0% |
| Head MRI | MSH2000 + RCD99 | 32% | 15% | 53% | 0% |
Table 3.
Optimal Source Vocabulary Combination for History Section
| Report Type | Source Vocabulary Combination | 11PP | delta-11PP | delta-Prec | delta-Rec |
|---|---|---|---|---|---|
| Bone scan | MSH2000 | 21% | 97% | 269% | −20% |
| Chest x-ray | MSH2000 | 17% | −50% | −3% | −75% |
| CT abdomen | MSH2000 + RCD99 | 42% | −7% | 67% | −26% |
| CT chest | CCPSS99 + MSH2000 + SNM2 | 30% | −21% | 21% | −30% |
| Head CT | MSH2000 + SNMI98 + SNM2 | 39% | 3% | 93% | −5% |
| Head MRI | MSH2000 + SNMI98 | 30% | 14% | 10% | 0% |
Table 4.
Optimal Source Vocabulary Combination for Technique Section
| Report Type | Source Vocabulary Combination | 11PP | delta-11PP | delta-Prec | delta-Rec |
|---|---|---|---|---|---|
| Chest x-ray | SNM2, UWDA99, ICD10 (tie) | 100% | 567% | 676% | 0% |
| CT abdomen | MSH2000 + SNMI98 | 32% | 13% | 103% | 0% |
| CT chest | AIR93 + MSH2000 + SNM2 | 50% | 49% | 276% | 0% |
| Head CT | MSH2000 + RCD99 + SNMI98 | 55% | 7% | 44% | 0% |
| Head MRI | MSH2000 + SNMI98 | 4% | −89% | −43% | −75% |
Table 5.
Optimal Source Vocabulary Combination for Findings Section
| Report Type | Source Vocabulary Combination | 11PP | delta-11PP | delta-Prec | delta-Rec |
|---|---|---|---|---|---|
| Bone scan | MSH2000 + RCD99 + SNMI98 | 11% | 17% | 24% | −18% |
| Chest x-ray | MSH2000 + SNMI98 + UMD2000 | 16% | 10% | 87% | −13% |
| CT abdomen | MSH2000 + RCD99 + SNMI98 | 13% | 7% | 38% | 0% |
| CT chest | CCPSS99 + MSH2000 + SNMI98 | 9% | 5% | 51% | −13% |
| Head CT | MSH2000 + RCD99 + SNMI98 | 20% | 8% | 21% | −8% |
| Head MRI | MSH2000 + RCD99 + SNMI98 | 28% | −3% | 14% | −16% |
Table 6.
Optimal Source Vocabulary Combination for Impression Section
| Report Type | Source Vocabulary Combination | 11PP | delta-11PP | delta-Prec | delta-Rec |
|---|---|---|---|---|---|
| Bone scan | MSH2000 + SNMI98 | 16% | 144% | 113% | −23% |
| Chest x-ray | MSH2000 + SNMI98 | 16% | 33% | 89% | −21% |
| CT abdomen | MSH2000 + RCD99 + SNMI98 | 34% | 6% | 40% | 0% |
| CT chest | CCPSS99 + MSH2000 + RCD99 | 16% | 16% | 60% | −23% |
| Head CT | MSH2000 + RCD99 + SNMI98 | 27% | 16% | 47% | −2% |
| Head MRI | MSH2000 + RCD99 + SNMI98 | 33% | 2% | 23% | 0% |
The Procedure section benefited most from this contextual indexing strategy. With no impact on recall, precision was improved by more than 100% in the Procedure sections of most radiology reports, the exception being only 50% improvements in head MRI reports. MeSH 2000, RCD 99, and SNOMED International 98 either alone or in various combinations were clearly superior in five of the six report types, the exception being CT abdomen reports in which UWDA99 was superior.
The contextual indexing strategy performed very differently on the History sections of the documents. For head CT and bone scan reports, this approach improved precision significantly. However, it produced only minor improvements on the History sections of head MRI reports (with no negative impact on recall) and resulted in lower precision on reports of chest x-ray, CT abdomen, and CT chest. MeSH 2000 was the most useful source vocabulary in the history section, sometimes complemented by SNOMED or RCD 99.
There were no Technique sections in bone scan reports. Significant improvement on precision was shown in the Technique sections of chest x-ray, CT abdomen, CT chest, and head CT reports without negative impact on recall. However, limited combination of source vocabularies produced negative impact on recall, precision, and 11-point average precision on the Technique section of head MRI reports.
In the Findings section, the positive impact on precision was balanced by a decrease in recall in most cases. For reports of chest x-ray, CT abdomen, and head CT, the trade-off was clearly worthwhile based on improvements in 11-point precision and precision over the negative impact in recall. The advantage was less marked for reports of bone scan and CT chest and not worthwhile for reports of head MRI. MeSH 2000 and SNOMED International 98 appeared in all top performing source vocabulary combinations for the Findings sections of all types of reports. The combination of MeSH 2000, RCD 99, and SNOMED International 98 was the best in four of six types of reports.
The Impression sections in most types of reports clearly benefited from the contextual indexing strategy with minor impact (less than 2%) on recall in reports of CT abdomen, head CT, and head MRI, but greater impact (about 20%) in the remaining three types. The precision–recall trade-off was considered acceptable, based on the improvements in 11-point average precision and precision over the negative impact on recall for most reports except for those of CT chest. The combination of MeSH 2000, RCD 99, and SNOMED International 98 excelled in the Impression sections of three types of reports, while MeSH 2000 and SNOMED International 98 achieved largest improvements on the other two types.
Discussion
As previous studies have found,8 the UMLS Metathesaurus provided adequate coverage for the majority of important biomedical concepts in radiology reports. UMLS indexing engines like SAPHIRE can achieve reasonable recall performance but often suffer from poor precision. As we have shown in this experiment, significantly limiting the number of UMLS source vocabularies used to index specific document sections can improve precision. We hypothesize that one factor related to the demonstrated poor precision performance of UMLS-based automated indexing engines is their “indiscriminate” use of a large set of overlapping terminologies, irrespective of the conceptual focus that many biomedical document sections contain. These terminologies introduce noise as well as signals. In the case of radiology reports, most matched concepts are contributed by different combinations of a few source vocabularies. By using only those source vocabularies, the indexing precision can be improved significantly without much negative impact on recall. Our simulated indexing model allowed us to identify the optimal source vocabularies for each document section.
Another interesting point is that the optimal combination depends on the section type and report type of the specific document type. Indexing radiology reports by sections enables the indexing engine to use different source vocabulary combinations and possibly other strategies based on the section type and report type, thus, achieving better indexing performance. We consider such an adaptive indexing strategy an example of “contextual indexing,” because it takes into account the semantic context of various document types and sections when selecting the appropriate vocabularies to use for representation. There usually are different types of documents in any biomedical document collection. The idea of contextual indexing is to use a training set to determine the best strategy for each document type instead of using only one strategy for the whole collection. The mappings between document types and optimal indexing strategies can be stored in a knowledge base. For each document in the test set, the indexing engine can index it using the best strategy that has been predetermined based on its document type and other information stored in the knowledge base. Thus, in the cases in which a few selected source vocabularies do not perform very well, the indexing engine still can use all UMLS source vocabularies.
We further hypothesized that within certain document classes (e.g., radiology reports), there are subclasses (e.g., chest CT scan) of documents that, while sharing the same document structure, require different combinations of UMLS source vocabularies to produce optimal indexing results. The results of this pilot study support this hypothesis and suggest that the simulated indexing approach that we have adopted to automatically determine optimal source vocabulary combinations needs to focus on these document subclasses rather than the document class itself.
In this experiment, the History section and the Technique section sometimes were very short or even omitted altogether. Thus, the results for these two types of sections are volatile and less convincing. For the results associated with other types of sections, the performance impact (i.e., impact on precision and recall) of using combinations of selected source vocabularies is relatively consistent across individual reports of the same report type and section type.
The size of the document set is an important factor in an experiment of this type. This pilot experiment was carried out on a relatively small document set, and this is the primary limitation of our study. However, deriving a gold standard for UMLS concept indexing is very time-consuming and fraught with potential confounding issues. Also, it is well known that using the results of human indexing as the gold standard is susceptible to inconsistency between different human indexers.17 Related research has shown that using more raters to cover the same content could largely improve the reliability of the result.18
Conclusion
This pilot study suggests a promising approach to improving precision by choosing small optimal subsets of the UMLS source vocabularies, based on the results of simulated indexing, and using only these vocabularies to index specific sections of clinical documents. Our results confirm that, at least for radiology reports, the optimal source vocabulary combinations vary by document section. We hope to confirm these findings using a larger set of radiology reports. We are also interested in extending this approach to other clinical documents. A challenge that remains is how to create the gold standard indexing that this methodology requires. We are currently examining a number of approaches to this problem.
Supplementary Material
Supported by contract N01-LM-9-3522 from the U.S. National Library of Medicine (NLM) and by the Office of Information Resources and Technology at Stanford University School of Medicine.
References
- 1.Cheung NT, Fung V, Chow YY, Tung Y. Structured data entry of clinical information for documentation and data collection. Medinfo. 2001;10(pt 1):609–13. [PubMed] [Google Scholar]
- 2.Tange HJ, Schouten HC, Kester AD, Hasman A. The granularity of medical narratives and its effect on the speed and completeness of information retrieval. J Am Med Inform Assoc. 1998;5:571–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dolin RH, Alschuler L, Beebe C, et al. The HL7 clinical document architecture. J Am Med Inform Assoc. 2001;8:552–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baud R, Ruch P. The future of natural language processing for biomedical applications. Int J Med Inf. 2002;67(1–3):1–5. [DOI] [PubMed] [Google Scholar]
- 5.Hripcsak G, Austin JH, Alderson PO, Friedman C. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology. 2002;224:157–63. [DOI] [PubMed] [Google Scholar]
- 6.Berrios DC, Cucina RJ, Fagan LM. Methods for semi-automated indexing for high precision information retrieval. J Am Med Inform Assoc. 2002;9:637–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe HJ. Multimedia electronic medical record systems. Acad Med. 1999;74:146–52. [DOI] [PubMed] [Google Scholar]
- 8.Lowe HJ, Antipov I, Hersh W, Smith CA, Mailhot M. Automated semantic indexing of imaging reports to support retrieval of medical images in the multimedia electronic medical record. Methods Inf Med. 1999;38:303–7. [PubMed] [Google Scholar]
- 9.Hersh WR, Mailhot M, Arnott-Smith C, Lowe HJ. Selective automated indexing of findings and diagnoses in radiology reports. J Biomed Inform. 2001;34:262–73. [DOI] [PubMed] [Google Scholar]
- 10.Hersh W, Hickam D. Information retrieval in medicine: the SAPHIRE experience. Medinfo. 1995;8 pt 2:1433–7. [PubMed] [Google Scholar]
- 11.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993;32:281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Humphreys B, Lindberg DA, Schoolman HM, Barnett GO. The Unified Medical Language Systems: an informatics research collaboration. J Am Med Inform Assoc. 1998;5:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sager N, Lyman M, Bucknall C, Nhan N, Tick LJ. Natural language processing and the representation of clinical data. J Am Med Inform Assoc. 1994;1:142–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman C, Alderson P, Austin J, Cimino J, Johnson S. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1:161–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCray AT, Razi AM, Bangalore AK, Browne AC, Stavri PC. The UMLS Knowledge Source Server—a versatile Internet-based research tool. Proc AMIA Fall Symp. 1996:164–8. [PMC free article] [PubMed]
- 16.Hersh WR, Leone TJ. The SAPHIRE server. Proc Annu Symp Comput Appl Med Care. 1995:858–62. [PMC free article] [PubMed]
- 17.Funk M, Reid C. Indexing consistency in MEDLINE. Bull Med Library Assoc. 1983;71:176–83. [PMC free article] [PubMed] [Google Scholar]
- 18.Hripcsak G, Kuperman GJ, Friedman C, Heitjan DF. A reliability study for evaluating information extraction from radiology reports. J Am Med Inform Assoc. 1999;6:143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






