Abstract
DNA methylation is an indispensible epigenetic modification of mammalian genomes. Consequently there is great interest in strategies for genome-wide/whole-genome DNA methylation analysis, and immunoprecipitation-based methods have proven to be a powerful option. Such methods are rapidly shifting the bottleneck from data generation to data analysis, necessitating the development of better analytical tools. Until now, a major analytical difficulty associated with immunoprecipitation-based DNA methylation profiling has been the inability to estimate absolute methylation levels. Here we report the development of a novel cross-platform algorithm – Bayesian Tool for Methylation Analysis (Batman) – for analyzing Methylated DNA Immunoprecipitation (MeDIP) profiles generated using arrays (MeDIP-chip) or next-generation sequencing (MeDIP-seq). The latter is an approach we have developed to elucidate the first high-resolution whole-genome DNA methylation profile (DNA methylome) of any mammalian genome. MeDIP-seq/MeDIP-chip combined with Batman represent robust, quantitative, and cost-effective functional genomic strategies for elucidating the function of DNA methylation.
Introduction
Modulation of the epigenome is one of the key mechanisms by which cells generate functional diversity from an essentially static genome1. The epigenome – the combination of DNA- and chromatin-associated epigenetic modifications that exist within a cell – is a dynamic entity, influenced by pre-determined genetic programs, or external environmental cues. Given the diversity of cell types within complex organisms such as mammals, it is staggering to think of how many epigenomes exist, or are possible. Unraveling this complexity remains one of biology's most important challenges.
DNA methylation is the only known epigenetic system that modifies the DNA molecule itself. In mammals, it occurs predominantly at CpG dinucleotides and is involved in diverse processes such as development, genomic integrity, X-inactivation, and imprinting2. Furthermore, perturbed DNA methyation is a hallmark of several human diseases, including cancer. Consequently, there is great interest in experimental and analytical tools for genome-wide/whole-genome DNA methylation profiling. In the last few years, a variety of experimental approaches have emerged for genome-wide, and very recently whole-genome, DNA methylation profiling (reviewed in Ref. 3). These can be classified into 3 main categories: (i) Restriction enzyme-based methods use one or more enzymes that will restrict DNA only if it is unmethylated (e.g. HpaII or NotI), or methylated (e.g. McrBC). These methods, coupled with either microarrays4-10 or capillary sequencing11, have been applied to genome-wide DNA methylation profiling of several organisms, but are limited to the analysis of CpG sites located within the enzyme recognition site(s). (ii) The second group of techniques is based on the reaction between genomic DNA and sodium bisulfite, which results in the conversion of unmethylated cytosines to uracil (and eventually thymine following amplification), whereas methylated cytosines remain unconverted12. Bisulfite-conversion based approaches offer single CpG resolution, and have been applied to microarrays13-16, high-throughput PCR sequencing17,18, and more recently to next-generation sequencing (BS-seq)19, resulting in an almost complete DNA methylation profile (DNA methylome) for the ~120 Mb genome of Arabidopsis thaliana. However, the reduction of sequence complexity following bisulfite conversion means that it is difficult to design enough unique probes to analyze bisulfite-converted DNA comprehensively on a genome-wide scale on microarrays, whereas the BS-seq approach is currently prohibitively expensive for the routine analysis of large genomes such as human. (iii) Immunoprecipitation-based methods use either 5-methylcytosine-specific antibodies (Methylated DNA Immunoprecipitation20, MeDIP or mDIP21), or methyl-binding domain proteins22-24, to enrich for the methylated (or unmethylated25) fraction of the genome. MeDIP/mDIP, combined with microarrays (MeDIP-chip), was used to delineate the first high-resolution whole-genome DNA methylation profile of any genome (Arabidopsis22,26) and the first high-resolution DNA methylation profile of human promoters27. However, until now, it has not been possible to estimate absolute methylation levels from MeDIP, and analysis of regions with low CpG density has been assumed to be problematic27.
The development of these various methods means that the bottleneck is rapidly shifting from data generation to data analysis, necessitating the development of more powerful analytical tools. Although no single experimental method offers the ‘perfect solution’, MeDIP-chip has quickly become a widely used20-22,26-31, and cost-effective approach for genome-wide/whole-genome DNA methylation analysis. Here we report the development of a novel cross-platform algorithm – Bayesian Tool for Methylation Analysis (Batman) – that can estimate absolute DNA methylation levels, across a wide range of CpG densities, from MeDIP-based experiments. We first demonstrate Batman's performance on MeDIP-chip, and then show it can also be used to analyze MeDIP profiles generated from next generation sequencing – a new technique we have developed, called MeDIP-seq. Our MeDIP-seq data represent the first high-resolution whole-genome DNA methylation profile of any mammalian genome. Batman is therefore a powerful cross-platform analytical tool for data generated from microarrays or next-generation sequencing, and will aid future studies aiming to understand the role of DNA methylation in the wider context of the epigenome.
Results
Generation of Human Genome-wide MeDIP-chip Data
MeDIP was performed on 3 biological replicates of mature spermatozoa from normal human donors (Supplementary Table 1 online), using a modified version of the original MeDIP protocol20 (Methods and Supplementary Figs 1 and 2 online). Human spermatozoa are relatively homogenous, easily obtained, and of interest from the point of view of understanding the role of DNA methylation during gametogenesis, fertilization, and early embryogenesis. Following MeDIP, samples were hybridized to custom high-density oligonucleotide microarrays (Nimblegen Systems, Inc.) that contained 42,144 regions of interest (ROIs), each typically 500 – 1000 bp in length, containing 5 – 10 unique 50mer probes. The ROIs overlapped 82% of all known transcriptional start sites (TSSs), 72% of non-promoter CpG islands, and a number of exonic, intronic and intergenic regions in the human genome (Ensembl genome browser32, Homo sapiens release 45.36g, NCBI36) (see Methods for further information regarding the array design). The correlation coeffecients (Pearson's) ranged from 0.54 to 0.72 among the 3 biological replicates, and 0.82 between a pair of technical replicates (dye-swaps), suggesting our MeDIP-chip experiments were reproducible.
Bayesian Tool for Methylation Analysis (Batman)
The efficiency of immunoprecipitation in MeDIP depends on the density of methylated CpG sites, which vary greatly within any given mammalian genome, making it difficult to distinguish variations in enrichment from confounding CpG density effects27. Consequently, until now, it has been impossible to estimate absolute methylation levels from MeDIP experiments, and the analysis of CpG-poor regions, in particular, has been assumed to be difficult27. Therefore, to analyze our MeDIP-chip data, we developed a new algorithm that models the effect of varying densities of methylated CpGs on MeDIP enrichment. This results in the transformation of normalized MeDIP-chip log2-ratios into a quantitative measure of DNA methylation across a wide range of CpG densities. We call this algorithm Bayesian Tool for Methylation Analysis (Batman), which is implemented as a suite of Java scripts (freely available from http://td-blade.gurdon.cam.ac.uk/software/batman/ under the GNU LPGL license).
Batman relies on the fact that we know the DNA sequences for many mammalian genomes, and that almost all DNA methylation in mammals occurs at CpG dinucleotides. Furthermore, we also know the range of the DNA fragment sizes generated during the initial shearing step in MeDIP (typically 400 – 700 bp). We define the coupling factor, Ccp, between probe p and CpG dinucleotide c as the fraction of DNA molecules hybridizing to probe p that contain the CpG c. Since we know the approximate range of DNA fragment sizes used in the MeDIP experiment, and assume that there are no fragment-length biases, then this is simply a function of the distance between the probe's genomic location and the CpG dinucleotide. This can be estimated empirically by sampling from the fragment length distribution and randomly placing each fragment such that it overlaps the probe: the resulting distribution is shown in Figure 1a. For a given probe, the sum of coupling factors, which we call Ctot, gives a measure of local CpG density. Plotting this parameter against the normalized log2-ratios from a typical MeDIP-chip experiment shows a fairly complex relationship (Fig. 1b). However, if we note that most CpG-poor regions of the genome are generally methylated, whereas the CpG-richest regions (CpG islands) are generally unmethylated, we can focus on the low-CpG portion of this plot and observe that there is an approximately linear relationship between the MeDIP-chip output and the density of methylated CpGs as measured by Ctot. Based on this observation, and assuming that it is only methylated CpGs which contribute to the observed signal, we developed a model whereby the signal observed at each array probe should depend on the methylation states of all nearby CpGs, weighted by the coupling factors between those CpGs and the probe. If we let mc indicate the methylation state at position c, and assume that the errors on the microarray are normally distributed with precision, then we can write a probability distribution for a complete set of array observations, A, given a set of methylation states, m, as:
Where G (x|μ, σ2) is a Gaussian probability density function. We can now use any standard Bayesian inference approach to find f(m|A), the posterior distribution of the methylation state parameters given the array (MeDIP-chip) data, and thus generate quantitative methylation profile information.
Figure 1.

Calibration of the Batman model against MeDIP-chip data (a) Estimated CpG coupling factors for a MeDIP-chip experiment as a function of the distance between a CpG dinucleotide and a microarray probe. (b) Plot of array signal against total CpG coupling factor, showing a linear regression fit to the low-CpG portion, as used in the Batman calibration step. This plot shows all data from one array on chromosome 6.
In order to reduce the computational cost of analyzing regions with very high CpG density, we took advantage of the fact that CpG methylation state is generally very highly correlated over a scale of hundreds of bases18. Instead of modeling every CpG individually, we grouped together all CpGs in 50 or 100bp windows and assumed that they would have the same methylation state. Inferring the methylation status at each CpG is now a deconvolution problem somewhat analogous to that considered when analyzing chromatin immunoprecipitation data33. Standard Bayesian techniques can be used to infer f(m|A), i.e. the distribution of likely methylation states given one or more sets of MeDIP-chip outputs. Our implementation of the Batman model uses Nested Sampling (http://www.inference.phy.cam.ac.uk/bayesys/), a highly robust Monte Carlo technique, to solve this inference problem. For each tiled region of the genome, we used a Nested Sampler-based approach to generate 100 independent samples from f(m|A). We then summarized the most likely methylation state in 100 bp windows by fitting Beta distributions to these samples. The modes of the most likely Beta distributions were used as our final methylation calls.
We assessed Batman's quantitative performance by comparing the Batman-analyzed MeDIP-chip data with bisulfite-PCR sequencing, a technique that allows DNA methylation measurements at individual CpG sites. We considered 667 bisulifte-PCR amplicons (spanning a wide range of CpG densities) from the Human Epigenome Project (HEP)18 that overlapped 1,481 50mer probes our microarray. The HEP bisulifte-PCR amplicons were generated from sperm samples different to those used in our MeDIP-chip experiments. Figure 2a shows a different version of the Batman calibration plot in which the MeDIP-chip log2-ratios have been colored according to HEP methylation levels, confirming that the calibration system we use provides a very good fit to the methylated section of the data. Figure 2b shows how Batman transforms LOESS-normalized log2-ratios into more quantitative results (R2 = 0.82, Pearson's) by increasing the dynamic range in low-CpG regions (although some noise still remains in this region). This is a significant improvement over using (i) LOESS-normalized log2-ratios (in a 100bp window centered around a 50mer probe that overlaps a HEP amplicon, (R2 = 0.46, Pearson's), or (ii) simple averaging of the LOESS-normalized log2-ratios for all probes within a 500bp window (R2 = 0.55, Pearson's), or (iii) averaging of the LOESS-normalized log2-ratios for all probes within a 500bp window and then dividing by the number of CpG sites within that window (R2 = 0.50, Pearson's). There are two likely explanations for the poor performance of the last method: it isn't a Bayesian method so there's no propagation of uncertainty (consequently noise in low-CpG regions is amplified), and the CpG influence isn't necessarily the same for all probes in a 500bp window. Batman addresses both of these issues. It is important to note that, in addition to estimating methylation levels in CpG-poor regions, Batman also effectively estimates methylation levels in CpG-dense methylated regions. Of the 667 bisulfite-PCR amplicons mentioned above, 15 are classified as CpG islands in the Ensembl genome browser, and display >80% methylation in the HEP. Batman identified all 15 as being heavily methylated (81 – 100% methylation, Supplementary Table 2 online). We further validated the Batman analysis by bisulfite-PCR sequencing of the same sperm samples used for MeDIP-chip. We selected 29 ROIs spanning a range of CpG densities, and again a very good correlation was observed (R2 = 0.85, Supplementary Fig. 3 and Supplementary Table 3 online).
Figure 2.

Comparison of Batman-analyzed MeDIP-chip data with bisulfite-PCR sequencing data from the Human Epigenome Project (a) Plot of MeDIP-chip data against CpG coupling factor, with points colored by methylation values from the HEP bisulfite-sequencing data. All probes that did not overlap at least one CpG annotated in HEP were excluded. (b) Comparisons of MeDIP-chip data with HEP using a range of processing strategies: LOESS-normalized log2-ratios in a 100bp window centered around a 50mer probe that overlaps a HEP amplicon (top left), simple averaging of the LOESS-normalized log2-ratios for all probes within a 500bp window (top right), averaging of the LOESS-normalized log2-ratios for all probes within a 500bp window and then dividing by the observed/expected CpG density (bottom left), Batman analyzed (bottom right). This analysis was derived from 1481 MeDIP-chip probes that overlapped 667 bisulfite-PCR amplicons from the HEP. HEP methylation values for all CpGs that overlapped any given 100bp MeDIP-chip window were averaged. Furthermore, to reduce noise in the HEP dataset, all 100 bp windows were required to have at least 2 HEP scores (i.e. data from the top and bottom bisulfite-PCR strands for windows containing a single CpG site, or from at least 2 different CpG sites) that differed by <50%. The purple – yellow (0 – 30) color bar on the right of each figure shows the total CpG coupling factor for each probe (c) Comparison of Batman-quantified MeDIP data with bisulfite data from HEP. Points show the mean Batman output for regions with a given HEP methylation level. Error bars show 95% bootstrap credible intervals.
We also tested Batman's performance on an independently generated MeDIP-chip dataset27. Weber et al. (2007) analyzed MeDIP profiles of ~16,000 promoters in human WI38 primary lung fibroblasts using high-density oligonucleotide arrays. We applied Batman to their MeDIP-chip data and analyzed promoters for which they also generated bisulfite-sequencing data (Supplementary Fig. 4 online). Batman was able to estimate absolute methylation levels over a wide range of CpG densities including low CpG density promoters (CpGo/e ~0.2, defined as LCPs27).
There is still a degree of noise in the Batman results, so we also show the mean Batman score for all regions with a given bisulfite methylation state (Fig. 2c), demonstrating Batman's output correlates almost linearly with the bisulfite results. It should be noted that Batman rarely outputs very extreme values (close to 0% or 100%) from MeDIP-chip data. This is a consequence of the Bayesian approach taken by Batman: each methylation call is associated with some degree of uncertainty, as represented by a credible interval. Since methylation levels below 0% or over 100% are meaningless, the entire credible interval must fit within a 0-100% scale. This means that the most credible estimates of methylation state are displaced away from the extremes. In principle, it would be possible to correct for this “compression” artifact by reading values off a curve such as that shown in Figure 2c. However, this transformation would complicate any consideration of the uncertainties attached to each methylation estimate. Since we do not find the compression to be a major problem when working with MeDIP-chip data, we report the output of the Bayesian model directly.
A human methylome generated using MeDIP-seq
Recently, next-generation sequencing technologies have emerged as powerful tools for whole-genome profiling of epigenetic modifications. They have been combined with chromatin immunoprecipitation (ChIP-Seq)34,35 for the analysis of histone modifications in human and mouse, and with bisulfite sequencing (BS-seq)19 to elucidate the DNA methylation profile of the 120Mb Arabidopsis genome. Inspired by these approaches, we combined MeDIP with next-generation sequencing – an approach we term MeDIP-seq – to generate the first high-resolution whole-genome DNA methylation profile (DNA methylome) of any mammalian genome, and show that Batman can also be used to estimate absolute DNA methylation levels from MeDIP-seq DNA methylome data.
We performed a second MeDIP on one of the sperm samples used in our MeDIP-chip experiments (sample SP3, Supplementary Table 1 online). The immunoprecipitated fraction was then subjected to next-generation sequencing using an Illumina Genome Analyzer (refer to Methods for detailed protocol). We obtained ~34.2 million single- and ~12 million paired-end reads that were mapped to the human genome using the Maq software [http://maq.sf.net/ and Li et al., manuscript submitted]. Only high quality read placements (Maq quality >= 10) were used, resulting in a total of ~26.5 million reads meeting this criterion. To maximize coverage, given the relatively short reads generated by the Illumina Genome Analyzer, we performed a smoothing step on the data by extending each paired-end read to a constant length of 500bp, and representing each singleton read as a 500bp block centred around the single read's mapping position. We do not expect this step to be necessary if longer fragments are selected.
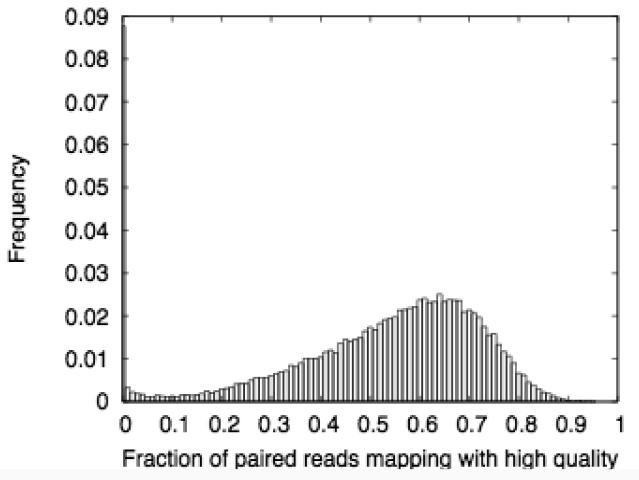
Assessment of the mapping quality revealed a degree of non-uniformity. Note in Figure 3a that there is a secondary peak of windows with extremely low mapping quality (<10% of reads map with q>=10). Many of these windows occur in large (megabase-scale) blocks. Investigation of representative examples suggests that they correspond with known duplications/structural variations in the human genome32 (data not shown). We chose to mask out these regions, representing approximately 75Mb of the genome. These regions are not included in the MeDIP-seq web display or any of our subsequent analyses. Such regions are also likely to be difficult to handle using other DNA methylation profiling strategies.
Figure 3.
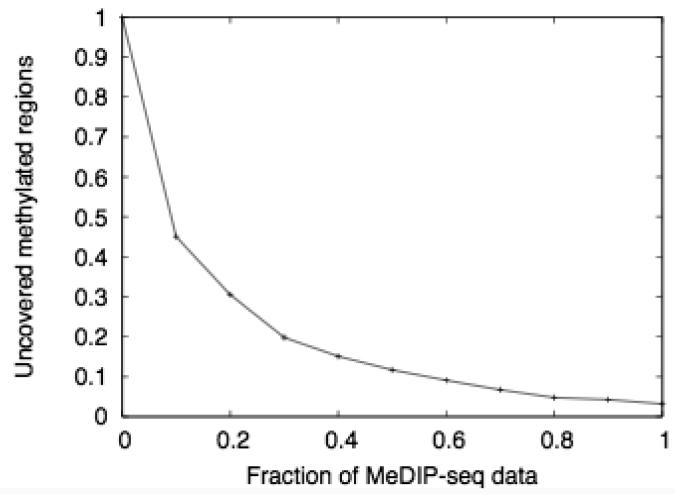
Mapping quality and genomic coverage of the MeDIP-seq data (a) Histogram showing the fractions of high-quality paired-end read mappings in 50kb windows across the genome. (b) Fraction of methylated regions (>60% methylation) which are not covered by reads in our MeDIP-seq dataset. As with all the MeDIP-seq analyses, the reads are extended to a length of 500bp.
We then assessed whether sufficient read-depth had been obtained, as an insufficient number of reads would result in some parts of the genome being incorrectly called as unmethylated. We therefore considered regions of the genome that were called methylated by the MeDIP-chip, and calculated the fraction of these regions that were covered by either the complete set of MeDIP-seq reads, or by randomly-chosen subsets of various sizes (Fig. 3b). This shows that our MeDIP-seq dataset covers >97% of methylated regions. We consider this coverage to be good, supported by subsequent comparisons of our MeDIP-seq results with bisulfite-PCR sequencing data from the HEP (described below). A further increase in read depth would possibly yield slightly more accurate results, but any such improvement would be subject to rapidly diminishing returns.
Batman Analysis of MeDIP-seq
Two slight modifications to the MeDIP-chip version of Batman were required when handling MeDIP-seq data. Firstly, since the read-out we use for MeDIP-seq is an absolute read density (which we sampled at arbitrary 50bp intervals along the genome) rather than a log2-ratio, a different model was required. Based on visual inspection of the MeDIP-seq data (Fig. 4a), we used polynomial model of order 2 instead of the linear model used for MeDIP-chip (Figs 1b and 2a). Secondly, the Gaussian error model was no longer appropriate (since the read density can never fall below zero), but a rectified Gaussian model could be used in a closely analogous manner. Following these two modifications, inference as described performed as above. We selected an output resolution of 100bp as a good compromise between fast computation and high resolution. This resolution is likely to be sufficient for many applications, since the methylation status of CpG sites within <1000 bp is significantly correlated (e.g. ~75% for ≤100 bp). Initially, this process gave results covering the entire human genome, except for assembly gaps and regions containing no CpGs. However, since the short reads from the Illumina genome Analyzer cannot be unambiguously mapped onto some repetitive parts of the genome, we expect Batman to under-call the methylation levels of the interior parts of large repeats. In recognition of this, we discarded all Batman results overlapping 500bp genomic tiles with >50% repeat coverage. Comparison of the Batman-analyzed MeDIP-seq results with sperm data from the HEP revealed a strong overall correlation (R2 = 0.85) (Fig. 4b and Supplementary Fig. 5 online).
Figure 4.

Comparison of Batman-analyzed MeDIP-seq data with bisulfite-PCR sequencing data from the Human Epigenome Project (a) MeDIP-seq read depth (i.e. the number of confidently placed reads overlapping a given point in the genome) for points overlapping HEP amplicons, plotted against total CpG coupling factor. Points are colored according to sperm DNA methylation (yellow – blue represents 0 – 100% methylation), as measured by in HEP16. (b) MeDIP-seq versus sperm bisulfite-PCR sequencing data from the Human Epigenome Project (HEP)16. 100 bp MeDIP-seq tiles are plotted against 1,322 overlapping HEP bisulfite-PCR amplicons. As in Figure 2b, HEP methylation values for all CpGs that overlapped any given 100bp MeDIP-seq tile were averaged, and all 100 bp windows were required to have at least 2 HEP scores (i.e. either data from the top and bottom strand for a single CpG site, or at least 2 CpG sites) that differed by <50%. The purple – yellow (0 -30) color bar on the right of each figure shows the total CpG coupling factor for each 100 bp tile. The same data stratified by CpG density is displayed in Supplementary Figure 4 online.
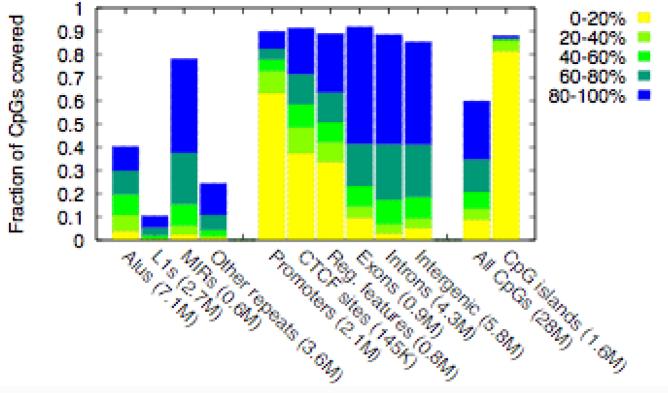
Figure 5a shows that our MeDIP-seq data provides high-resolution, quantitative coverage for ~90% of all CpG sites within CpG islands, promoters and other regulatory sequences, exons, and introns, and ~60% of all CpGs in the human genome. This represents a ~20X improvement in coverage over existing methods. The use of paired-end sequencing allowed us to measure DNA methylation levels of some small repeat-element families. Several recent studies on human36,37 and mouse38,39 have reported epigenetic variability at repeat-elements, and these could have phenotypic consequences. Furthermore, epigenetic silencing of repeat-elements is thought to be critical for genomic-integrity40. MeDIP-seq thus provides a means of analyzing such repeat-elements in future DNA methylome studies. Consistent with previous observations from genome-wide studies18,27, Batman analysis of the MeDIP-seq data reveals that promoters display an inverse correlation between CpG density and methylation (Supplementary Fig. 6 online), CpG islands are predominantly unmethylated, a significant proportion of CTCF (a DNA binding protein involved in insulator activity) sites are unmethylated41, and most other regions of the genome are methylated. Our Batman-analyzed MeDIP-seq data (and the MeDIP-chip data) are freely accessible via the Ensembl Genome Browser (Fig. 5b and www.ensembl.org), representing a useful resource for the scientific community.
Figure 5.
Genomic coverage and web display of the MeDIP-seq data (a) Genomic coverage of MeDIP-seq (measured as fraction of CpGs). Genomic features are from the Ensembl genome database (release 45). The first ten bars are mutually exclusive, i.e. repeats are not included when considering subsequent features. Numbers in parentheses indicate the total number of CpGs within the human genome in that category. Promoters are defined as 2kb regions centered on annotated transcriptional start sites, and Reg. features represent non-promoter Regulatory Features in Ensembl. The colors represent the range of DNA methylation levels (b) MeDIP-seq data integrated into Ensembl along with MeDIP-chip data of the same sperm DNA sample, and sperm bisulfite-PCR data from the Human Epigenome Project16. The yellow – green – blue color gradient represents 0 – 100% methylation.
Discussion
Unravelling the complexities of the epigenome is a very important objective, and recent years have seen the development of several strategies for genome-wide analysis of epigenetic marks, including DNA methylation. One of the principal challenges now is to develop more powerful analytical tools to interpret the vast amounts of data that are being/will be generated.
Here, we have reported the development and validation of Batman – a novel cross-platform algorithm for the quantitative analysis of MeDIP data generated using either arrays (MeDIP-chip) or next-generation sequencing technologies (MeDIP-seq, representing the first high-resolution DNA methylome of any mammalian genome). Batman, combined with MeDIP-chip or MeDIP-seq, provides estimation of absolute methylation levels over a wide range of CpG densities. This is a very useful property for DNA methylome analyses, as it will allow more effective genome-wide/whole-genome profiling, including CpG-poor regions that have traditionally been overlooked in most DNA methylome studies to date. Furthermore, estimation of absolute DNA methylation levels will facilitate cross-platform comparisons.
Although several strategies now exist for DNA methylation profiling, there are, to the best of our knowledge, two others that compare with MeDIP-chip/MeDIP-seq + Batman in terms of genomic coverage and quantitative performance. The first is Comprehensive High-throughput Arrays for Relative Methylation (CHARM), which was recently reported by Feinberg and colleagues42. CHARM combines a tiling-array design strategy with statistical procedures that average information from neighboring genomic locations. The authors applied CHARM to the McrBC assay in which the DNA is digested with McrBC, which restricts methylated DNA (recognition sequence RmC(N)55–103RmC). The enzyme is used on size-selected (1.5 – 4.0 kb) DNA to fractionate unmethylated DNA after digestion, which is co-hybridized on arrays with DNA similarly processed but not cut with the enzyme. The authors demonstrated that CHARM correlates well with bisulfite-conversion based data (R2 = 0.76). However, CHARM is not a ‘stand-alone’ algorithm but rather a strategy that requires the use of a particular array design and it is unclear whether it can be adapted to next-generation sequencing technologies. It does not estimate absolute DNA methylation levels and, as the authors note, CHARM suffers to some degree in the ability to discriminate highly methylated from highly unmethylated CpG islands. Interestingly, the authors also tested MeDIP-chip and concluded that it cannot be used to analyze CpG-poor regions42. Our results show that MeDIP + Batman can be used to provide absolute DNA methylation levels across a range of CpG densities (including CpG-poor regions) from arrays or next-generation sequencing.
Another recently reported approach is BS-seq, which Jacobsen and colleagues used to delineate a DNA methylome for the ~120 Mb Arabidopsis genome19. BS-seq has the ability to provide single-base pair resolution DNA methylation profiles, which is indeed a very useful property. However, at current sequencing costs, such an approach is still prohibitively expensive to analyze larger genomes such as the human which is ~25X bigger than the Arabidopsis genome. Based on our results, we estimate that ~40 million paired-end reads (less than a single run of an Illumina Genome Analyzer) are sufficient to generate a high-quality mammalian methylome, whereas approximately ~3.8 Gb of sequence (which would equate to > 40 million paired-end reads) was required to generate a single-base pair resolution (~20 X coverage) methylome for the ~120 Mb Arabidopsis genome using BS-seq19. Also, even though single-CpG resolution is desirable, the fact that the methylation status of CpG sites within <1000 bp is significantly correlated18 (e.g. ~75% for ≤100 bp), means that the ~100 bp resolution is suitable for many applications.
Although Batman in its present form performs well, we see opportunities for future development of MeDIP – post-processing platforms, especially with regard to the use of sequencing technologies. In particular, when analyzing paired-end MeDIP-seq data, it should be possible to take advantage of the exact mapping positions of each read, rather than summarizing the data as a set of read-depth samples, thereby improving the resolution. Also, it would be interesting to apply Batman to the analysis of Arabidopsis MeDIP data. Although both CpG and non-CpG methylation is found in Arabidopsis22,26, gene bodies contain predominantly the former, and therefore it should be possible to use Batman for the analysis of genic regions.
In the near future, the integration of (epi)genomic and functional approaches is going to be crucial for elucidating the biological role of DNA methylation. The need for such an integrated approach is also evident from the recently announced NIH Epigenome Roadmap Initiative calling for mapping of reference DNA methylation profiles on an unprecedented scale (http://nihroadmap.nih.gov/epigenomics/). We believe that the Batman algorithm combined with MeDIP-chip or MeDIP-seq will provide powerful and cost-effective strategies for quantitative, high-resolution DNA methylome analysis, and will contribute towards elucidating the role of the epigenome in health and disease.
Methods
Sperm Samples
Human mature spermatozoa were obtained as part of the MHC Haplotype Project (www.sanger.ac.uk/HGP/Chr6/MHC/) under Cambridge Local Research Ethics Committee approvals LREC-03/094 and LREC-04/Q0108/46).
Methylated DNA Immunoprecipitation
MeDIP was performed using a previously published protocol20, but we also included a ligation-mediated PCR (LM-PCR) step43 to amplify the material (the LM-PCR step was not performed for MeDIP-seq). Hybridizations of pre- and post-LM-PCR samples on custom tile-path arrays (2kb resolution) for the human Major Histocompatibility Complex (MHC) showed that the LM-PCR did not introduce significant bias (Supplementary Fig. 1 online). A detailed protocol is provided in the Supplementary Methods online.
Custom oligonucleotide array design and pre-Batman processing
Our microarray consists of 382,178 50-bp probes. Although we aimed to target all annotated TSSs and non-promoter CGIs, we were unable to design suitable unique probes for 18% of the TSSs and 28% of non-promoter CGIs. The array also contained 50-mer probes tiled at ~100 bp density across the entire human Major Histocompatibility Complex, and promoters and non-promoter CpG islands on the X- and Y-chromosomes. Analyses of these regions will be presented elsewhere. The array was originally designed using the NCBI build 35 version of the human genome assembly, but then mapped to NCBI build 36 using Exonerate44. To be mapped, probes were required to align full length and without gaps or mismatches. Probes that aligned more than once to the NCBI36 sequence were removed from the analysis. Tiled regions were defined by clustering uniquely mapped probes within 200bp of one another. Singleton probes were discarded. The tiled regions were then divided into 500bp ROIs. Following hybridization (performed by Nimblegen, Iceland using their standard conditions), arrays were LOESS-normalized using custom R-scripts prior to Batman anlaysis of the resulting log2 ratios.
Illumina Genome Analyzer sequencing
Based on the manufacturer's recommended protocol, we nebulised 10 μg sperm DNA (SP3 in Supplementary Table 1) with compressed nitrogen for 6 minutes at 32psi, giving fragments of <800bp. We then end repaired, phosphorylated and A-tailed the fragmented DNA and ligated Illumina paired end adapters to fragments. Of this we used ~1μg of adapter-ligated DNA for subsequent MeDIP enrichment (performed as described above). LM-PCR was not performed after MeDIP enrichment. Because the quantity of DNA obtained after MeDIP was low (~30ng) we deviated from the standard Illumina protocol and amplified the sample using Illumina paired-end PCR primers before gel electrophoresis and size selecting libraries. We excised bands from the gel to produce libraries with insert sizes of 85-160bp, and quantified these libraries using an Agilent Bioanalyzer 2100. We prepared paired-end flowcells with 3.2pM DNA (using 2 primer chemistry) using the manufacturer's recommended protocol and sequenced for 36 cycles on an Illumina Genome Analyzer fitted with a paired-end module. The reads were mapped onto the human genome reference sequence using the high-performance alignment software ‘maq’ (http://maq.sf.net/) prior to Batman analysis (described in the main text).
Statistics
All correlation coefficients were computed using Pearson's product-moment formula. All credible intervals were estimated by bootstrapping. All other statistical procedures related to Batman are described in the main text.
Supplementary Material
Acknowledgements
TAD, EMT, LB, KLH, DKJ, MMM, HK, TJPH, NPC, SB, DJT, HL, RD were supported by the Wellcome Trust. VKR was supported by the Barts and The London Charitable Trust, and a C.J. Martin Fellowship from the NHMRC, Australia. SG, NJ, and MH were supported by an EU grant (High-throughput Epigenetic Regulatory Organization in Chromatin (HEROIC), LSHG-CT-2005-018883) under the 6th Framework Program to SB (MH) and EB (SG, NL). NPT, JCM and ST were supported by grant C14303/A8646 from Cancer Research UK.
Footnotes
Author Contributions
T.A.D. co-conceived the study, wrote the Batman algorithm, co-analyzed data and co-wrote the paper; V.K.R. co-conceived the study, performed the bulk of the experimental work, co-analyzed data, co-wrote the paper, and provided overall project management; D.J.T. performed the Illumina Genome Analyzer sequencing; H.L. performed the maq analysis; P.F., E.K., S.G., N.J., J.H. designed the Ensembl web display for the data reported here; E.M.T., L.B., M.H. performed experimental work, K.L.H. and D.K.J. assisted with array design, N.P.T. and J.C.M. performed preliminary array analysis, M.M.M. supplied materials, E.B., T.J.P.H., R.D., S.T. provided intellectual input, S.B. co-conceived the study, co-wrote the paper, and provided overall project management. T.A.D. and V.K.R. contributed equally to this work.
References
- 1.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 3.Beck S, Rakyan VK. The methylome: approaches for global DNA methylation profiling. Trends Genet. 2008;24:231–237. doi: 10.1016/j.tig.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Tompa R, et al. Genome-wide profiling of DNA methylation reveals transposon targets of CHROMOMETHYLASE3. Curr Biol. 2002;12:65–68. doi: 10.1016/s0960-9822(01)00622-4. [DOI] [PubMed] [Google Scholar]
- 5.Lippman Z, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 6.Khulan B, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumacher A, et al. Microarray-based DNA methylation profiling: Technology and applications. Nucleic Acids Res. 2006;34:528–542. doi: 10.1093/nar/gkj461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ordway JM, et al. Comprehensive DNA methylation profiling in a human cancer genome identifies novel epigenetic targets. Carcinogenesis. 2006;27:2409–2423. doi: 10.1093/carcin/bgl161. [DOI] [PubMed] [Google Scholar]
- 9.Ching TT, et al. Epigenome analyses using BAC microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nat Genet. 2005;37:645–651. doi: 10.1038/ng1563. [DOI] [PubMed] [Google Scholar]
- 10.Shen L, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rollins RA, et al. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frommer M, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gitan RS, et al. Methylation-specific oligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome Res. 2002;12:158–164. doi: 10.1101/gr.202801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adorjan T, et al. Tumour class prediction and discovery by microarray-based DNA methylation analysis. Nucleic Acids Res. 2002;30:e21. doi: 10.1093/nar/30.5.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bibikova M, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinders J, et al. Genome-wide, high-resolution DNA methylation profiling using bisulfite-mediated cytosine conversion. Genome Res. 2008;18:469–476. doi: 10.1101/gr.7073008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rakyan VK, et al. DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project. PLoS Biol. 2004;2:e405. doi: 10.1371/journal.pbio.0020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 21.Keshet I, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Gebhard C, et al. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res. 2006;66:6118–6128. doi: 10.1158/0008-5472.CAN-06-0376. [DOI] [PubMed] [Google Scholar]
- 24.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 25.Illingworth R, et al. A Novel CpG Island Set Identifies Tissue-Specific Methylation at Developmental Gene Loci. PLoS Biology. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2006;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 27.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 28.Yasui DH, et al. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci U S A. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacinto FV, Ballestar E, Ropero S, Esteller M. Discovery of epigenetically silenced genes by methylated DNA immunoprecipitation in colon cancer cells. Cancer Res. 2007;67:11481–11486. doi: 10.1158/0008-5472.CAN-07-2687. [DOI] [PubMed] [Google Scholar]
- 30.Cheng AS, et al. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008;68:1786–1796. doi: 10.1158/0008-5472.CAN-07-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fouse SD, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flicek P, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi Y, et al. High-resolution computational models of genome binding events. Nat. Biotechnol. 2006;24:963–970. doi: 10.1038/nbt1233. [DOI] [PubMed] [Google Scholar]
- 34.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandovici I, et al. Interindividual variability and parent of origin DNA methylation differences at specific human Alu elements. Hum Mol Genet. 2005;14:2135–2143. doi: 10.1093/hmg/ddi218. [DOI] [PubMed] [Google Scholar]
- 37.Flanagan JM, et al. Intra- and interindividual epigenetic variation in human germ cells. Am J Hum Genet. 2006;79:67–84. doi: 10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 39.Rakyan VK, et al. Transgenerational inheritance of epigenetic states at the murine AxinFu allele occurs after maternal and paternal transmission. Proc Natl Acad Sci USA. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bestor TH. The host defence function of genomic methylation patterns. Novartis Found Symp. 1999;214:187–195. doi: 10.1002/9780470515501.ch11. [DOI] [PubMed] [Google Scholar]
- 41.Irizarry RA, et al. Comprehensive High-throughput Arrays for Relative Methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukhopadhyay R, et al. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res. 2004;14:594–602. doi: 10.1101/gr.2408304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oberley MJ, Farnham PJ. Probing chromatin immunoprecipitates with CpG-island microarrays to identify genomic sites occupied by DNA-binding proteins. Methods Enzymol. 2003;371:577–596. doi: 10.1016/S0076-6879(03)71043-X. [DOI] [PubMed] [Google Scholar]
- 44.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;15:31–34. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.