Abstract
Calcineurin (Cn) is a Ca2+/calmodulin-dependent serine/threonine phosphatase that regulates differentiation-specific gene expression in diverse tissues, including the control of fiber-type switching in skeletal muscle. Recent studies have implicated Cn signaling in diminishing skeletal muscle pathogenesis associated with muscle injury or disease-related muscle degeneration. For example, use of the Cn inhibitor cyclosporine A has been shown to delay muscle regeneration following toxin-induced injury and inhibit regeneration in the dystrophin-deficient mdx mouse model of Duchenne muscular dystrophy. In contrast, transgenic expression of an activated mutant of Cn in skeletal muscle was shown to increase utrophin expression and reduce overall disease pathology in mdx mice. Here we examine the effect of altered Cn activation in the context of the δ-sarcoglycan-null (scgd−/−) mouse model of limb-girdle muscular dystrophy. In contrast to results discussed in mdx mice, genetic deletion of a loxP-targeted calcineurin B1 (CnB1) gene using a skeletal muscle-specific cre allele in the scgd−/− background substantially reduced skeletal muscle degeneration and histopathology compared with the scgd−/− genotype alone. A similar regression in scgd-dependent disease manifestation was also observed in calcineurin Aβ (CnAβ) gene-targeted mice in both skeletal muscle and heart. Conversely, increased Cn expression using a muscle-specific transgene increased cardiac fibrosis, decreased cardiac ventricular shortening, and increased muscle fiber loss in the quadriceps. Our results suggest that inhibition of Cn may benefit select types of muscular dystrophy.
The Ca2+/calmodulin-activated serine/threonine protein phosphatase calcineurin (Cn) is expressed in many different cell types and is involved in a number of processes including lymphocyte development and proliferation, neuronal and muscle development, cardiac hypertrophy, and skeletal muscle fiber-type switching (1). Cn exists as a heterotrimer consisting of a 57−61 kDa catalytic A subunit (CnA) and two smaller 16−19 kDa EF-hand containing Ca2+-binding proteins referred to as calcineurin B (CnB) and calmodulin (2). In vertebrates, three unlinked loci encode the catalytic subunit (CnAα, CnAβ, and CnAγ), while two loci encode the regulatory subunit (CnB1 and CnB2) (3). The , CnAβ, and CnB1 genes are each expressed in a ubiquitous pattern throughout the body, while CnAγ and CnB2 expression are more restricted to the brain and testis (4-7). Once activated, Cn directly dephosphorylates a family of transcription factors referred to as nuclear factor of activated T cells (NFAT) within the cytoplasm, exposing a nuclear localization sequence and promoting translocation into the nucleus where they participate in transcriptional induction events (8). There are four Cn-regulated NFAT transcription factors, NFATc1-c4, each of which is expressed in skeletal muscle (9,10). Cn activity and the ability to activate NFAT are directly antagonized by the pharmacologic inhibitors FK506 and cyclosporine A (CsA) (8).
In skeletal muscle, augmented or sustained patterns of muscle contraction and fiber recruitment maintains or promotes the slow/oxidative fiber-type program through increased Ca2+ fluxing that facilitates Cn activation (11-21). However, Cn-NFAT signaling is not believed to regulate hypertrophic growth of skeletal muscle (18,20), in contrast to its well-characterized hypertrophic function in cardiac muscle (22). Cn-NFAT signaling has also been implicated in regulating the differentiation, developmental maturation, and regeneration of skeletal muscle (9,13,18,23,24). Consistent with these observations, Cn signaling can modify the progression and severity of skeletal muscle pathology associated with genetic mutations that cause Duchenne and other forms of muscular dystrophy (see below).
Duchenne and Becker muscular dystrophies are X-linked recessive disorders that arise due to mutations in the dystrophin gene, leading to destabilization in structural attachment between the underlying contractile units with the dystrophin-glycoprotein complex (DGC) and the basal lamina, hence altering the integrity of the cell membrane and promoting myofiber degeneration (25,26). The mdx mouse, which has a single point mutation within an exon of the dystrophin gene, is a widely employed model of muscular dystrophy. Disease in the mdx model shows a continuum of skeletal muscle disease severity with the diaphragm being the most affected. However, unlike human Duchenne patients, mdx mice live well into adulthood. Studies in mdx mice have suggested that Cn activation is protective and benefits the degree of muscle pathology. For example, transgenic mice expressing a constitutively active calcineurin protein (Cn*) in skeletal muscle were crossed with mdx mice, showing significant reductions in the extent of myotube central nucleation, fiber size variability and inflammation, as well as demonstrating an improvement in membrane integrity compared with mdx mice alone (27). As a potential mechanism underlying this protective effect, Cn* transgenic mice showed increased NFATc1 nuclear localization in skeletal muscle cells, and a 2-fold upregulation in utrophin protein levels that helped restore the integrity of the DGC in mdx mice (27). Studies of Cn inhibition found that treatment of young mdx mice with CsA led to a severe impairment in muscle regeneration, as assessed by fewer centrally-nucleated fibers, reduced muscle mass, decreased force output, increased collagen replacement, and increased inflammation relative to vehicle-treated littermates (28). In support of these findings, the muscle-sparing effects associated with the glucocortocoid deflazacort in mdx mice could be abrogated by the co-administration of CsA (29). Thus, Cn activity appears to be protective in the mdx mouse model.
To further characterize the role of Cn signaling in muscular dystrophy and to establish the relevance of Cn activity in non-dystrophin deficient models of muscular dystrophy, we employed δ-sarcoglycan-null (scgd−/−) mice. The δ-sarcoglycan gene encodes a member of the sarcoglycan complex of transmembrane proteins that is assembled into the DGC and is critical for maintaining membrane stability (25,26). The loss of scgd causes reductions or complete loss of the other members of the sarcoglycan complex, an effect that is not uniformly seen in the absence of dystrophin (30,31). Scgd−/− mice have cardiomyopathy and muscular dystrophy (corresponding to a human limb-girdle myopathy) with many hallmarks of progressive disease including cell death, muscle regeneration, inflammation, fibrosis, and reduced survival (30,31).
Contrary to the previously observed protective influence of Cn activation in mdx mice, here we describe an entirely opposite effect associated with Cn signaling in scgd−/− mice. Genetic disruption of Cn signaling, using two different genetic approaches, greatly benefited both skeletal and cardiac muscle disease in scgd−/− mice, as measured by a reduction or even absence (depending on the degree of Cn reduction) of fibrosis in both skeletal and cardiac muscle, decreased skeletal muscle degeneration and inflammation, and improved cardiac disease. Conversely, Cn* transgenic mice showed worsened skeletal muscle histopathology and heart disease in the scgd null background.
MATERIALS AND METHODS
Animal Models
Scgd−/− (30), CnB1fl/fl (18,32), myosin light chain1f-cre (MLC-cre) (18,33), CnAβ−/− (34), NFAT-luciferase (35) and muscle creatine kinase Cn*transgene (Tg) (MCK-Cn*Tg) (16) mice have been described previously. Animals had free access to food and water, and all experimentation was performed in the Cincinnati Children's Hospital Research Foundation animal care facility in accordance with the guidelines of the National Institutes of Health. Experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committee. Both male and female mice were used for all analyses. For decreased Cn protein expression studies, all analyses were performed on mice approximately 8−10 months of age. For increased Cn expression studies, mice 6-months of age were examined.
Western Analysis
Muscle protein extracts were prepared by homogenizing tissue in cell lysis buffer containing 20 mM Tris, pH 7.4, 137 mM sodium chloride, 25 mM β-glycerophosphate, 2 mM sodium pyrophosphate, 2 mM ethylenediaminetetraacetic acid, 1 mM sodium orthovanadate, 1% Triton X-100, 10% glycerol, 1 mM PMSF, 5 μg/ml leupeptin, 5 μg/ml aprotinin, and 2 mM benzamidine. Proteins (50 to 100 μg) were resolved on a sodium dodecyl sulfate-polyacrylamide gel, and transferred to a polyvinylidine difluoride membrane, and immunodetected by using an enhanced chemifluorescence kit as specified by the manufacturer (Amersham Bioscienes, Piscataway, NJ). The following antibodies were used: α-tubulin monoclonal antibody (1:2500, Santa Cruz Biotechnology, Santa Cruz, CA), CnB1 rabbit polyclonal antibody (1:500, Upstate, Waltham, MA), and calcineurin pan-A rabbit polyclonal antibody (1:1000, Chemicon International, Inc., Temecula, CA). Western blot reactivity was quantified on a Storm860 PhosphorImager (Molecular Dynamics, Piscataway, NJ) using Image Quant software.
Determination of Hydroxyproline Content
The method used is a modified version of that described by Woessner et al. (36). Briefly, a small piece of muscle (between 5−8 mg for diaphragm and 200−300 mg for gastrocnemius and quadriceps) was weighed and placed into a glass Pyrex tube containing 500 μl 6 N hydrochloric acid. The tubes were capped loosely and placed into a 100°C oven for 20 min. Tubes were then capped tightly and baked overnight to completely hydrolyze the tissue. The following day the tubes were removed, uncapped and placed in a dessicator containing sodium hydroxide pellets. The dessicator was placed into the oven at 50°C and connected to a vacuum going through a sodium hydroxide trap. The tissue was kept under vacuum until dry (24 h to 1 wk) and then resuspended in 1 ml 5 mM hydrochloric acid. An aliquot of sample (25 μl) was combined with 180 μl of milliQ H2O in 12×75 mm glass tubes. To this, 100 μl chloramine T solution was added (0.14 g chloramine-T, 2 ml H2O, and 8 ml hydroxyproline assay buffer). Hydroxyproline assay buffer was prepared by combining 11.4 g sodium acetate anhydrous, 7.5 g trisodium citrate dihydrate, 40 ml H2O (pH adjusted to 6.0) and 77 ml isopropanol and bringing to a final volume of 200 ml with H2O. Following addition of chloramine T solution, 1.25 ml Erlich's reagent was added (6.0 g p-dimethylaminobenzaldehyde, 18 ml 60% perchlorate, and 78 ml isopropanol). The samples were then vortexed for 15 seconds and incubated at 55°C for 20−25 min. Once cooled to room temperature, sample absorbance was read at 558 nm. A standard curve was run alongside the samples using trans-4-hydroxy-L-proline(Sigma, St Louis MO) standards (ranging from 0 to 4 μg) to determine hydroxyproline concentration. Readings were normalized to original tissue weight.
Luciferase Assay
Excised skeletal muscle (∼8 to 30 mg) was placed in 400 μl ice cold lysis buffer (1% Triton-X 100, 100 mM Tris-HCl pH 7.8, 2 mM ethylenediaminetetraacetic acid, 2 mM dithiothreitol, 2 μg/ml aprotinin, 2 μg/ml leupeptin, 1 μg/ml pepstatin, and 100 μg/ml phenylmethylsulphonylfluoride). Tissue was then finely minced on ice using scissors and sonicated. Homogenates were spun for 10 minutes at 14,000 rpm at 4°C. In a 96-well plate, 200 μl ice-cold reaction buffer (4 mM adenosine triphosphate [ATP], pH 7.0, 15 mM magnesium sulfate, 30 mM Tricine pH 7.8, and 10 mM dithiothreitol) was combined with 20 μl of tissue supernatant. To measure luciferase enzymatic activity in tissue extracts, 100 μl of 1 mM luciferin (Promega, Madison, WI) was injected into each well, and light intensity was measured with a luminometer (Microlumat LB 96B, Berthold, Wildbad, Germany) over 10 sec and expressed as relative light units (RLU) over 10 sec/μg proteins. Luciferase assays were performed on skeletal muscle from mice 5−7 weeks of age.
Evans Blue Dye Uptake
Evans blue dye (10 mg/ml in PBS) was injected intraperitoneally into 5−7-month old mice (0.1 ml/10 g body weight). The mice were sacrificed 6 h after injection. Muscles were excised and embedded in O.C.T. compound (Tissue-Tek, Torrence, CA), and snap-frozen in liquid nitrogen-cooled isopentane. Blocks were then sectioned into 8-μm thick slices, dried for 10 min, and washed briefly in PBS. To visualize muscle fibers, sections were then incubated with fluorescein isothiocyanate-labeled wheat germ agglutinin (50 μg/ml in 1X PBS, Sigma) in the dark for 1 h. Slides were then washed 3 times, 5 minutes each, and mounted with VectaShield (Vector Laboratories, Burlingame, CA) and coverslips.
Histology
Paraffin-embedded sections (8-μm thick) were cut at the mid-belly of the muscle. Masson's trichrome staining was carried out at the Molecular Pathology core facility at Cincinnati Children's Hospital Medical Center. Unoccupied area was determined using NIH ImageJ software. Briefly, the unstained area within a quadriceps section stained with Masson's trichrome was calculated along with the total area of the section The ratio of the unstained area / total area was calculated, with the resulting number used as an index of “unoccupied area”.
Echocardiography
Mice were anesthetized with 2% isoflurane, and hearts were visualized using a Hewlett Packard Sonos 5500 instrument and a 15-MHZ transducer. Cardiac ventricular dimensions were measured on M-mode three times for the number of animals indicated.
Analysis of Centrally-Located Nuclei and Fiber Area
Diaphragm muscles from mice were collected, fixed in 10% formalin containing PBS, and embedded in paraffin. Tissues were cut crosswise through the mid-belly of the muscle, and embedded cut side down. Muscle sections were cut at a thickness of 8-μm. Sections were deparaffinized in xylene and rehydrated in dilutions of ethanol. To visualize fibers, sections were incubated with tetramethyl rhodamine isothiocyanate-labeled wheat germ agglutinin (Sigma, St. Louis, MO) at 50 μg/ml in 1X PBS for 1 h, washed three times for 10 min each in 1X PBS, with the second wash containing 1 μg/ml bis-benzamide for nuclei visualization, if necessary. Sections were then mounted with VectaShield and coverslips. Fiber area analysis was performed using ImageJ analysis software (NIH), and approximately 250 fibers were measured per section. For central nuclei analysis, 2−3 areas at 10X magnification were examined and counted, averaging approximately 650 fibers per animal.
Real Time PCR
Total RNA was extracted from quadriceps muscles using TRIZOL® reagent (Invitrogen, Carlsbad, CA). One microgram of total RNA from each muscle was converted into double-stranded cDNA by using SuperScript First-Strand Synthesis System (Invitrogen) with an oligo-dT primer. Two microliters of the synthesized cDNA was then used for quantitative real-time PCR. The LightCycler® FastStart DNA MasterPLUS SYBR Green kit (Roche) was used for amplification of utrophin A, myocyte enhancer factor 2C (MEF2C), and 18S rRNA using the LightCycler 2.0 System. Primers used for amplification were: MEF2C 5′GGCCATGGTACACCGAGTACAACGAGC3′ and 5′GGGGATCCCTGTGTTACCTGCACTTGG3′; Utrophin A 5′GGCAGGAAGATTGCACAAGT3′ and 5′CTGCTAGCCAAGTCCCAGAG3′; 18S 5′AGTCCCTGCCCTTTGTACACA3′ and 5′CGATCCGAGGGCCTCACTA3′. The mRNA levels of 18S were measured as an internal control gene. Each cDNA sample was subjected to two individual PCR analyses using either the MEF2C or utrophin A primer pair. The target gene and the internal control gene were amplified in separate tubes. The increase in fluorescence was measured in real-time during the extension step. The threshold cycle (Cτ) was calculated, and the relative gene-expression was calculated using the 2-ΔΔCT method. For each sample, 18S CT values were subtracted from Utrophin A or MEF2C CT values to derive a delta CT value. The average value for wild-type (Wt) muscle was then subtracted from the values obtained from Cn*Tg, scgd−/−, or scgd−/− Cn*Tg muscles to derive a delta-delta CT value. The expression of these genes relative to Wt was then evaluated using the designation 2-ΔΔCT.
In situ ligation of hairpin 1
Frozen diaphragm sections (7 μm thick) were incubated with a biotin labeled blunt-end hairpin oligo (kit component) and T4 DNA ligase (kit component) as specified in the ApopTag® Peroxidase In Situ Oligo Ligation (ISOL) Apoptosis Detection Kit (Chemicon International). Ligation of the hairpin 1 oligo was detected using stepavidin-FITC conjugate (Alexa, 1:400 dilution). Total nuclei and membranes were stained with TO-PRO-3 iodide (Molecular Probes) and wheat germ agglutinin-TRITC (Sigma), respectively. Sections were analyzed using a confocal microscope, with pictures taken at 200X magnification. Six pictures per animal were counted for positive myonuclei. The analysis of ligation rates was performed in 4 Wt, 5 scgd−/−, 6 scgd−/− Cn*Tg, and 6 scgd−/− CnB1fl/flMLC-cre mice.
Statistical Analysis
Data are expressed as the means ± standard errors of the means. Statistical significance was determined by Student's t test at the P ≤ 0.05 level.
RESULTS
Cn Protein Expression and Activity in scgd−/− Skeletal Muscle
To begin to characterize the potential involvement of Cn signaling in altering muscular dystrophy disease progression in scgd−/− mice, the levels of CnA (catalytic) and CnB1 (regulatory) protein were examined. In young (6 wk.) scgd−/− skeletal muscle there was no detectable difference in CnB1 expression compared to Wt levels, although CnA (pan antibody) was subtly but significantly decreased (Fig. 1A). This reduction in CnA was observed in gastrocnemius, quadriceps, and diaphragm. However, CnA protein levels were increased in the hearts of scgd−/− mice (Fig. 1A), consistent with previous reports that diseased hearts can show increased Cn protein content. Since Cn protein expression is not necessarily predictive of activity, we also evaluated activity in Wt and scgd−/− skeletal and cardiac muscle. Wt and scgd−/− mice were crossed with transgenic mice containing an NFAT binding site-dependent luciferase reporter transgene (35). Surprisingly, NFAT luciferase activity significantly decreased by 57% in dystrophic skeletal muscle (soleus) and 60% in scgd−/− cardiac muscle (Fig 1B, see discussion). In control experiments, we determined that the presence of the NFAT-luciferase transgene itself did not directly alter CnA or CnB1 expression levels at baseline or in the presence of the scgd null alleles (data not shown).
Figure 1.
Analysis of Cn protein levels and NFAT reporter activity in scgd−/− muscle. (A) Western blots of total CnA (pan) and CnB1 expression in gastrocnemius, quadriceps, diaphragm, and heart in 6 week-old Wt ( n=4) and scgd−/− (n=4 or 5) mice. GAPDH or α-tubulin were included as a loading control and brain was used as a positive control (not shown). (B) NFAT-luciferase reporter activity in soleus and hearts of Wt and scgd−/− mice. The number of muscles assayed is shown for each group in the panel. *P<0.05 versus background (bkgrnd); †P<0.05 versus Wt.
Characterization of Cn Protein Expression and Activity in Cn-Deficient, scgd−/− Mice
We have previously shown that skeletal muscle from CnAβ−/− mice exhibits approximately a 40−50% reduction in total CnA and CnB protein expression (18). Similarly, the hearts of CnAβ−/− mice display a 50% loss of Cn protein expression and an 80% reduction in total Cn phosphatase activity (34). We also previously demonstrated that skeletal muscle from CnB1fl/fl mice crossed with a skeletal muscle-specific cre knock-in allele (in the myosin light chain 1f [MLC1f] locus) exhibit an even greater loss of CnA and CnB1 protein in skeletal muscle (at least an 80% reduction) (18). However, the MLC-cre knock-in allele is not expressed in slow-twitch skeletal muscle or the heart, so these tissues were only analyzed in globally deleted CnAβ−/− mice. Both CnB1fl/flMLC-cre and CnAβ−/− mice were crossed into the NFAT-luciferase transgene background to assess the effect on activity in the heart or skeletal muscle. Diaphragm muscle from both CnAβ−/− and CnB1fl/flMLC-cre mice showed a reduction of NFAT activity to background levels (Fig. 2A). A similar phenomenon was observed in heart, which has a higher basal level of NFAT reporter activity when compared to skeletal muscle (Fig. 2A). Loss of CnAβ in cardiac tissue reduced NFAT reporter activity by 93% (Fig. 2A), a value comparable to the 80% reduction observed with the traditional Cn enzymatic assay (34).
Figure 2.
NFAT reporter activity is reduced similarly in Cn-deficient and scgd−/− muscle. (A) NFAT-luciferase reporter activity in diaphragm and cardiac muscle in Wt, CnAβ−/−, and CnB1fl/flMLC-cre mice. The number of muscles assayed is shown for each group in the panel. *P<0.05 versus background (bkgrnd); †P<0.05 versus Wt. (B) Western blots for total CnB1 protein in scgd−/−, scgd−/− CnAβ−/−, and scgd−/− CnB1fl/flMLC-cre muscle. α-tubulin was included as a loading control and brain extract as a positive control. Quantitation of results from this and another independent western blot is shown below, with the number of total samples indicated in the bars. *P<0.05 versus scgd−/−. (C) Cardiac NFAT-luciferase reporter activity in Wt, scgd−/−, CnAβ−/−, and scgd−/− CnAβ−/− mice. The number of muscles assayed is shown for each group in the panel. *P<0.05 versus Wt; †P<0.05 versus scgd−/−.
We previously observed that loss of either the Cn catalytic or regulatory subunit led to the reciprocal destabilization and loss of the other protein subunit (18). To ensure a comparable loss of Cn expression in the scgd−/− background for subsequent phenotypic experiments, both CnAβ−/− and CnB1fl/flMLC-cre mice were evaluated for CnB1 protein expression in scgd−/− skeletal muscle. CnAβ−/− skeletal muscle showed a complete loss of CnAβ protein (data not shown) and a 50% reduction in CnB1 protein, while CnB1fl/flMLC-cre mice showed a greater than 80% loss in the scgd−/− background (Fig. 2B), which is similar to reductions observed by us previously (18). As a further control, loss of CnAβ from the heart significantly and comparably reduced NFAT-luciferase reporter activity in both Wt and scgd−/− mice (Figs. 2A and 2C).
Loss of CnAβ−/− Improves Histopathology in scgd−/− Mice
To assess the long-term effect of reduced Cn activity on dystrophic skeletal muscle histopathology, we analyzed diaphragm muscles of scgd−/− and scgd−/− CnAβ−/− mice at 36−40 weeks of age. Masson's trichrome staining (blue areas are fibrotic) of scgd−/− CnAβ−/− diaphragm sections demonstrated an obvious improvement in disease, such as maintained muscle fiber density and reduced collagen replacement, compared with scgd−/− mice (Fig. 3A). Histological sections from Wt controls showed no fibrotic staining whatsoever or alterations in fiber densities (data not shown). Dystrophic skeletal muscle is normally characterized by an overall reduction in fiber area due to increased regenerating fiber numbers. Quantification of fiber areas revealed the expected decrease in average fiber area in scgd−/− mice relative to Wt, although a trend was observed in scgd−/− CnAβ−/− diaphragm such that the average fiber area increased (Fig. 3B). However, measurement of collagen content by assessing hydroxyproline content in a biochemical assay did show a significant reduction in fibrotic tissue replacement in scgd−/− CnAβ−/− diaphragm compared to scgd−/− (Fig. 3C). Taken together, these results suggest a protective effect of Cn inhibition on dystrophic muscle pathology.
Figure 3.

CnAβ-deficient dystrophic muscle displays improved histopathology and decreased fibrosis. (A) Representative Masson's trichrome staining of paraffin sections from the diaphragm of age-matched scgd−/− and scgd−/− CnAβ−/− mice. (B) Analysis of average muscle fiber area in the diaphragm of Wt, scgd−/− and scgd−/− CnAβ−/− mice. The number of muscles assayed is shown for each group in the panel. *P<0.05 versus Wt. (C) Analysis of μg hydroxyproline / mg of diaphragm tissue in Wt, scgd−/−, CnAβ−/− and scgd−/− CnAβ−/− mice. The number of muscles assayed is shown for each group in the bars. *P<0.05 versus Wt; †P<0.05 versus scgd−/−.
Extensive and Muscle-Specific Loss of Cn Improves Skeletal Muscle Pathology in scgd−/− Mice
Examination of muscle from scgd−/− CnB1fl/flMLC-cre crossed mice at 36−40 weeks of age revealed a remarkable improvement in gross muscle pathology. Masson's trichrome staining of diaphragm sections from scgd−/− CnB1fl/flMLC-cre mice showed no evidence of fibrosis and muscle fibers appeared normal and healthy, while scgd−/− diaphragms showed extensive fibrotic tissue replacement (Fig. 4A). Histological examination of Wt sections showed no fibrosis (data not shown). Examination of fiber areas within the diaphragms of scgd−/− CnB1fl/flMLC-cre mice showed a dramatic increase in average fiber area compared to scgd−/− mice (Fig. 4B). Quantitation of centrally-nucleated fibers in the diaphragm, which indicates the extent of ongoing regeneration, demonstrated a significant reduction in scgd−/− CnB1fl/flMLC-cre mice compared to scgd−/− mice (Fig. 4C). Finally, measurement of hydroxyproline content showed essentially no increases in fibrosis in the diaphragm, quadriceps and gastrocnemius of CnB1fl/flMLC-cre mice, supporting the dramatic phenotype observed by Masson's trichrome staining of histological sections (Fig. 4D).
Figure 4.

Skeletal muscle specific deletion of CnB1 increases average fiber area, decreases centrally-nucleated fibers and ameliorates fibrosis in scgd−/− mice. (A) Representative Masson's trichrome staining of paraffin sections from diaphragm of age-matched scgd−/− and scgd−/− CnB1fl/flMLC-cre mice. (B) Analysis of average muscle fiber area in the diaphragm of Wt, scgd−/− and scgd−/− CnB1fl/flMLC-cre mice. The number of muscles assayed is shown for each group in the panel. *P<0.05 versus Wt; †P<0.05 versus scgd−/−. (C) Quantitation of fibers containing central nuclei in the diaphragm of Wt, scgd−/− and scgd−/− CnB1fl/flMLC-cre mice. The number of muscles assayed is shown for each group in the panel. *P<0.05 versus Wt; †P<0.05 versus scgd−/−. (D) Analysis of μg hydroxyproline / mg of tissue for the indicated muscle groups from Wt, CnB1fl/flMLC-cre, scgd−/−, and scgd−/− CnB1fl/flMLC-cre mice. The number of muscles assayed is shown for each group in the panel. *P<0.05 versus Wt; †P<0.05 versus scgd−/−.
Loss of Cn Does Not Stabilize the scgd−/− Muscle Membrane or Alter Apoptosis
Mechanistic studies were initiated to investigate why loss of Cn might benefit muscular dystrophy disease progression. The ongoing muscle degeneration/regeneration that occurs in muscular dystrophy is associated with contractile-induced damage of the sarcolemma. This damage permits passive Ca2+ influx or channel-mediated influx, which may in turn activate Ca2+-dependent signaling pathways that could trigger necrosis (25,26,37). Alternatively, increased intracellular Ca2+ could lead to the activation of degradative enzymes and the overload and dysfunction of Ca2+ cycling and storage systems (38). To examine whether loss of Cn protein could alter the characteristic increase in membrane permeability seen in muscular dystrophy, mice were injected with Evans blue dye, which under normal conditions is a membrane-impermeant molecule and cannot cross into skeletal muscle fibers. While muscle from Wt (not shown) and single CnB1fl/flMLC-cre mice showed no Evans blue dye uptake, both scgd−/− and scgd−/− CnB1fl/flMLC-cre mice had a similar increase in susceptibility to Evans blue dye uptake (Fig. 5A). This result suggests that the amelioration of muscle disease observed in scgd−/− CnB1fl/flMLC-cre mice is not associated with a stabilization of the plasma membrane.
Figure 5.

Assessment of plasma membrane integrity and apoptosis. (A) Confocal fluorescence of Evans blue dye uptake in the diaphragm of CnB1fl/flMLC-cre, scgd−/−, and scgd−/− CnB1fl/flMLC-cre mice. Histological sections from three separate mice are shown (1,2,3). Evan's blue dye uptake fluoresces red while the green fluorescence results from wheat germ agglutinin-fluorescein isothiocyanate staining to show membranes. (B) Percentage of nuclei with hairpin 1 oligonucleotide ligation in the indicated genotypes of mice from diaphragm histological sections (6 weeks of age). The number of mice analyzed in each group is present in the Materials and Methods. Approximately 1000−3000 nuclei were counted for each animal. There were no significant differences between the groups.
Degeneration of myofibers in muscular dystrophy has been associated with increased indexes of apoptosis, such as TUNEL and caspase 3 cleavage. Here we performed an analysis of hairpin 1 in situ ligation to measure DNA fragmentation consistent with apoptosis in histological sections from diaphragm (6 weeks old) that were also co-stained for nuclei and myofiber borders. Careful quantitation of total hairpin 1 ligation labeling showed approximately 0.36%−0.99% of myonuclei in Wt, scgd−/−, and in scgd−/− that also lacked CnB1 or that contained the Cn*TG (discussed later) (Fig. 5B). There were no significant differences in labeling rates between the 4 groups. These results suggest that while loss of CnB1 is protective to muscular dystrophy in scgd−/− mice, the mechanism appears to be independent of apoptosis at this age.
Loss of CnAβ in scgd−/− Mice Is Sufficient to Reduce Fibrosis and Improve Cardiac Disease
Mice lacking scgd also show substantial disease in cardiac muscle, a feature of most forms of human muscular dystrophy. To determine if loss of Cn in the heart could also impart a similar beneficial effect as seen in skeletal muscle, hearts from scgd−/− and scgd−/− CnAβ−/− mice were examined. Masson's trichrome staining of hearts from scgd−/− CnAβ−/− mice demonstrated a profound reduction in fibrosis (blue staining) compared to scgd−/− hearts, while sections from Wt hearts showed no appreciable staining whatsoever (Fig. 6A, and data not shown). This finding was confirmed by quantification of hydroxyproline content, showing the dramatic reduction of collagen present in scgd−/− CnAβ−/− hearts compared to scgd−/− hearts (Fig. 6B). Consistent with these results, the reduction in cardiac fractional shortening characteristically observed in scgd−/− mice was restored to near Wt levels by deletion of CnAβ (Fig. 6C). These results suggest that reduced Cn signaling is beneficial in both dystrophic skeletal and cardiac muscle.
Figure 6.

Loss of Cn reduces fibrosis and improves fractional shortening in hearts of dystrophic mice. (A) Representative Masson's trichrome staining of paraffin sections from separate hearts of age-matched scgd−/− and scgd−/− CnAβ−/− mice. Fibrosis is shown in blue. (B) Quantification of μg hydroxyproline / mg of tissue from heart for the indicated groups of mice. The number of hearts assayed is shown for each group in the bars. *P<0.05 versus Wt; †P<0.05 versus scgd−/−. (C) Echocardiographic assessment of fractional shortening in the hearts of the indicated groups of mice. The number of hearts measured is shown for each group in the bars. *P<0.05 versus Wt; †P<0.05 versus scgd−/−.
Characterization of Constitutively-Active Cn Transgenic Mice
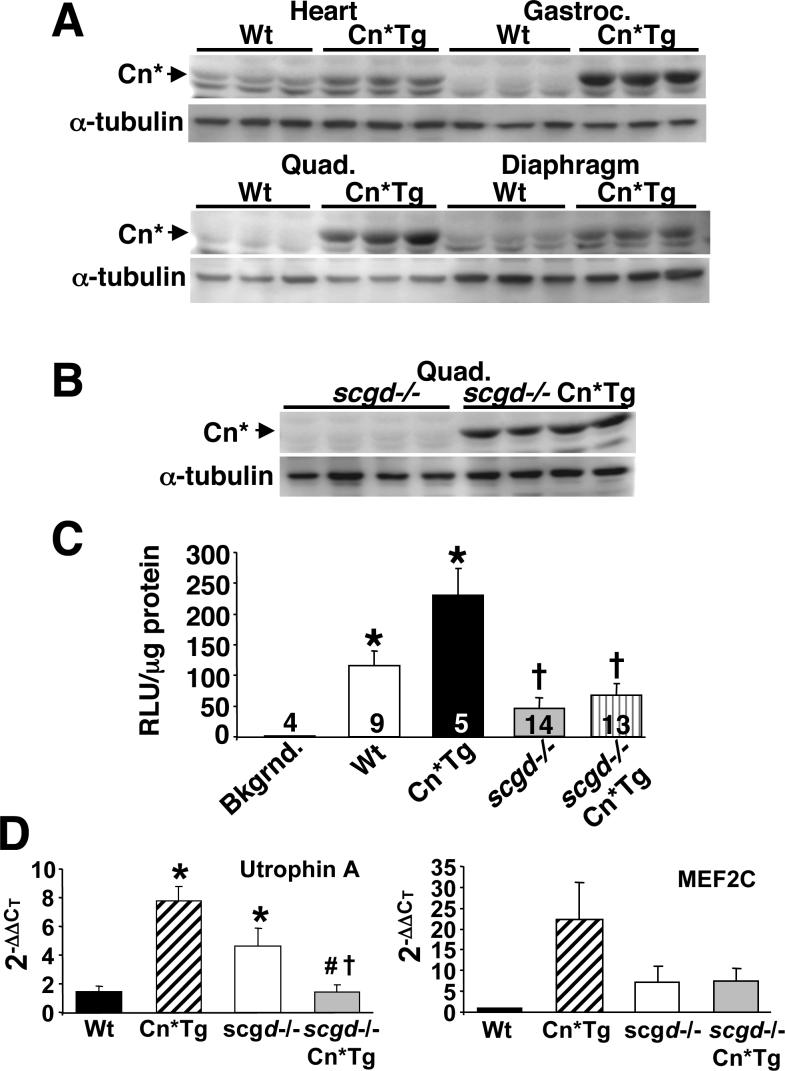
Having determined that loss of Cn expression is advantageous in dystrophic skeletal and cardiac muscle, we next wanted to assess the effects of increased Cn activity in the disease progression of scgd−/− mice. Mice containing a Cn*Tg (MCK promoter) were used, which have high levels of expression in skeletal muscle and slightly lower levels in the heart (16). Skeletal muscles of these mice show increases in slow muscle fibers and minor changes in muscle mass (16,39). Expression of the Cn*Tg protein was evident in a variety of normal skeletal muscles and heart (Fig. 7A). To avoid confusion, it should be noted that the Cn* protein co-migrates with background proteins in certain muscle types, such as heart, that non-specifically bind the pan CnA antibody and appear as a doublet (Fig. 7A). As a control, expression of the Cn*Tg was observed to be equally robust when expressed in the scgd−/− background (Fig. 7B).
Figure 7.
Assessment of Cn* transgene expression and activity. (A) Western blot analysis using a pan-CnA antibody to show expression of the Cn* protein in the indicated tissue of Wt and MCK-Cn*Tg mice. Each lane was generated from a separate muscle/animal. (B) Western blot analysis of Cn* protein expression in dystrophic muscle (from scgd−/− mice) shows robust expression of the transgene. Each lane was generated from a separate muscle. (C) Analysis of NFAT-luciferase reporter activity in hearts of mice from the indicated groups expressing the Cn*Tg. The number of hearts measured is shown for each group in the panel. *P<0.05 versus background (bkgrnd); †P<0.05 versus Wt or Cn*Tg. (D) Real time PCR for utrophin A and MEF2C mRNA from skeletal muscle of the indicated genotypes. *P<0.05 versus Wt; †P<0.05 versus Cn*Tg; #P<0.05 versus scgd−/−.
Because expression of Cn is a well-known inducer of cardiac hypertrophy (22), we examined the hearts of these mice for any increase in mass. Even though moderately expressed, the MCK-Cn* transgene was sufficient to induce a 41.3% increase in the heart-to-body weight ratio compared to Wt (data not shown). When crossed with the NFAT-luciferase transgenic mice, Cn*Tg mice showed a 100% increase in activity in the heart compared with Wt mice (Fig. 7C). However, as presented earlier, dystrophic heart muscle showed a significant reduction in NFAT-luciferase activity, even in the presence of the MCK-Cn* transgene (Fig. 7C) (see discussion).
To investigate downstream effects of Cn-NFAT signaling, we quantified the mRNA levels of utrophin A and MEF2C. As previously observed, utrophin A mRNA levels were increased by Cn activation in skeletal muscle, as was MEF2C. Utrophin A, but not MEF2C, was also upregulated by the scgd null mutation, although the combination of the Cn*Tg with the scgd null mutation produced a downregulation relative to either allele (Fig. 7D).
Expression of Cn* in scgd−/− Mice Does Not Increase Fibrosis Or Affect Fiber Area
In light of our data suggesting that loss of Cn is beneficial to scgd−/− muscle, we analyzed the histopathology of diaphragm muscle from Cn*Tg mice. Masson's trichrome staining of diaphragm sections from mice 24 weeks of age did not show obvious differences in fibrotic tissue replacement between scgd−/− Cn*Tg and scgd−/− mice (Fig. 8A). Once again, Wt histological sections were omitted because they showed no fibrotic staining (data not shown). However, areas of muscle fiber loss with suggestive fatty-tissue replacement were apparent throughout nearly every muscle section in scgd−/− Cn*Tg mice. Although fatty-tissue replacement of muscle is not an uncommon phenomenon in dystrophic muscle, its appearance was far more prevalent in the presence of the Cn*Tg (Fig. 8A). Analysis of average fiber area of muscle revealed the expected decrease in scgd−/− mice compared to Wt, which was similarly decreased in the presence of the Cn*Tg (Fig. 8B). Quantification of hydroxyproline content confirmed the Masson's trichrome sections, establishing that there is no significant difference in diaphragm collagen content between scgd−/− and scgd−/− Cn*Tg mice (Fig. 8C). This conclusion is supported by analysis of histopathology in the quadriceps muscle of scgd−/− and scgd−/− Cn*Tg mice. While fibrosis was similar, quadriceps showed a noticeable increase in areas of profound fiber loss and suggestive fatty tissue replacement in scgd−/− Cn*Tg mice compared with scgd−/− mice alone (Fig. 9A,B). Sections from Wt quadriceps showed no such events (data not shown). Thus, expression of Cn* in the scgd−/− background mildly enhances histopathology through a specific increase in fiber loss.
Figure 8.

Mild increase in diaphragm histopathology in scgd−/− Cn*Tg mice. (A) Representative Masson's trichrome staining of paraffin sections from separate diaphragms of age-matched scgd−/− and scgd−/− Cn*Tg mice. (B) Analysis of average fiber area in the diaphragm of Wt, scgd−/−, and scgd−/− Cn*Tg mice. The number of muscles measured is shown for each group in the bars. *P<0.05 versus Wt. (C) Quantification of μg hydroxyproline / mg of tissue from diaphragm for the indicated groups of mice. The number of muscles assayed is shown for each group in the bars. *P<0.05 versus Wt.
Figure 9.

Histological analysis of quadriceps muscles reveals increased fatty tissue replacement in scgd−/− Cn*Tg mice. (A) Representative Masson's trichrome staining of paraffin sections from quadriceps of age-matched scgd−/− and scgd−/− Cn*Tg mice. (B) Histological sections from quadriceps muscle were digitized and the area not occupied by muscle fiber was calculated for the indicated genotype. *P<0.05 versus Cn*Tg or scgd−/−. Number of samples analyzed was 12 for Wt, 6 for Cn*Tg, 10 for scgd−/−, and 6 for scgd−/− Cn*TG.
Increased Cardiac Fibrosis and Worsened Heart Disease in scgd−/− Cn*Tg Mice
In contrast to skeletal muscle, expression of the Cn*Tg in the heart is sufficient to induce hypertrophy and pathology by itself (22). Analysis of Masson's trichrome-stained sections showed significantly more fibrosis and myocyte disorganization in the hearts of scgd−/− Cn*Tg mice compared with scgd−/− mice alone, while Wt sections showed no visual fibrosis (Fig. 10A, and data not shown). Quantification of hydroxyproline content confirmed this observation, showing a 148.6% increase in collagen content in scgd−/− Cn*Tg mice over Wt hearts and an 80.4% increase over scgd−/− hearts (Fig. 10B). Furthermore, echocardiographic analysis of cardiac fractional shortening revealed that both scgd−/− hearts and Cn*Tg hearts have a comparable decrease, while hearts of scgd−/− Cn*Tg mice showed an even greater decrease (Fig. 10C). In conclusion, activation of Cn in cardiac muscle significantly worsens the pathology associated with scgd−/− dependent muscular dystrophy.
Figure 10.

Expression of Cn*Tg in hearts of dystrophic mice leads to increased fibrosis and decreased fractional shortening. (A) Representative Masson's trichrome staining of paraffin sections from hearts of age-matched scgd−/− and scgd−/− Cn*Tg mice. Each section is from a separate heart. (B) Quantification of μg hydroxyproline / mg of tissue from heart for the indicated groups of mice. The number of hearts assayed is shown for each group in the bars. *P<0.05 versus Wt; †P<0.05 versus scgd−/− or Cn*Tg. (C) Echocardiographic assessment of fractional shortening (%) in the hearts of the indicated groups of mice. The number of hearts measured is shown for each group in the bars. *P<0.05 versus Wt; †P<0.05 versus scgd−/− or Cn*Tg.
DISCUSSION
The most dramatic result of the current study is that genetic disruption of Cn reduced histopathology in both skeletal muscle and hearts of mice lacking scgd. Two genetic approaches were used to reduce Cn activity in vivo as a means of assessing its role in scgd-deficient muscular dystrophy. The first involved a total somatic deletion of the CnAβ gene, which comprises at least 50% or more of the total Cn activity in heart and skeletal muscle (18,19,34). The potential limitation of a total somatic deletion approach is that immune cells or other non-muscle cell-types (stem cells) could secondarily impact the inflammatory response and/or the regenerative response of skeletal muscle during muscular dystrophy disease progression. In addition, loss of CnAβ only partially reduces total Cn activity, since the CnAα gene is not affected, which might be sufficient to facilitate some/all Cn-dependent pathologic responses in heart and skeletal muscle. However, the CnAβ−/− model was critical because it permitted investigation of the cardiac phenotype associated with reduced Cn activity, and since it also affected skeletal muscle satellite cells and stem cells that might contribute to regenerating muscle. The second approach involved a skeletal muscle differentiation-specific deletion of the CnB1 gene using a cre-loxP approach. The CnB1 gene is absolutely required for Cn activity and for CnA subunit stability, so that its deletion in skeletal muscle using the MLC-cre knock-in allele (33) permits a greater than 80% deletion of Cn content (18). The strengths of this approach are that total Cn activity is reduced substantially more than with the CnAβ−/− gene-targeted model, and that Cn is only deleted from differentiated skeletal muscle, suggesting the cellular autonomy of any observed effect. A significant limitation of the CnB1fl/flMLC-cre model, however, is that deletion is specific to skeletal muscle and does not occur in the heart, so that any cardiac manifestations associated with muscular dystrophy could not be evaluated. Despite the intrinsic differences between the CnAβ−/− and CnB1fl/flMLC-cre models, both approaches showed a protective effect in skeletal muscle when crossed into the scgd−/− background (Fig. 3 versus Fig. 4). We believe that the greater profile of skeletal muscle protection observed in scgd−/− mice lacking CnB1 versus CnAβ is simply the result of a much greater total decrease in Cn activity associated with the CnB1fl/flMLC-cre strategy.
The overall conclusion that loss of Cn activity is protective in scgd−/− mice is further supported by the antithetic effect observed with the Cn*Tg. Here, the gain-of-function for Cn activity in skeletal muscle showed enhanced histopathology in the form of greater myofiber drop-out. However, Cn*Tg mice did not show increased tissue fibrosis, collectively suggesting that while increased Cn activity did not dramatically exacerbate dystrophy-related muscle pathology, it may specifically affect a regulatory circuit that promotes fiber replacement with non-muscle tissue, such as fat. We speculate that as the scgd−/− Cn*Tg mice age, they could eventually show greater disease compared with scgd−/− mice since the greater loss of muscle fibers and the accumulation of fatty tissue would hasten functional deterioration. Collectively, both the gain and loss-of-function models support the overall conclusion that Cn inhibition would benefit skeletal muscle disease in this model of limb-girdle muscular dystrophy. This contention is even more obvious in the heart, where Cn inhibition significantly benefited histopathology and improved fractional shortening, while expression of the Cn*Tg worsened cardiac disease and enhanced functional deterioration.
Another interesting observation is that NFAT transcriptional reporter activity was uniformly and significantly decreased in both the skeletal muscle and heart of scgd−/− mice, even in the presence of the Cn*Tg. Consistent with this observation, downstream targets of Cn-NFAT signaling, utrophin A and MEF2C, were also downregulated in scgd−/− mice in the presence of the Cn*Tg. The NFAT-dependent reporter consists of 9 multimerized NFAT-only sites upstream of a minimal promoter fused to the luciferase cDNA (35). This transgene is expressed ubiquitously and is exquisitely sensitive to Cn activity in vivo (35). However, NFAT activity towards this reporter or even the utrophin A promoter is negatively regulated by a handful of kinases that directly phosphorylate the N-terminal regulatory domain in NFATs, including glycogen synthase kinase 3β, protein kinase A, casein kinase I, c-Jun N-terminal kinase (JNK), and p38 mitogen-activated protein kinase (MAPK) (8). Alteration in the activity of these kinases can profoundly impact NFAT activity, such that inhibition of either JNK or p38 MAPK enhances endogenous NFAT in the heart and spontaneously induces hypertrophy (40). These observations are of particular interest given reports that JNK1 activity is significantly increased in both skeletal muscle and heart of the mdx mouse (41,42). Skeletal muscle from mdx mice also showed upregulation of p38 MAPK activity (43). Thus, Cn-NFAT signaling could be specifically antagonized in muscular dystrophy by increased JNK1 and p38 activity, leading to the observed down-regulation in NFAT-luciferase activity in both heart and skeletal muscle of scgd−/− mice. If Cn activity is indeed detrimental to muscular dystrophy disease progression, such a mechanism of downregulated Cn activity could represent a protective compensatory effect.
It is also interesting that calcineurin activity is reduced in skeletal muscle disease, while in many forms of heart disease activity and/or protein levels are significantly increased (indeed it is increased in the hearts of scgd−/− mice). However, this dichotomous observation between heart and skeletal muscle likely reflects the highly specialized role that calcineurin plays in each tissue. For example, in the heart calcineurin directly regulates myocyte growth, yet overexpression of the same activated calcineurin transgene in skeletal muscle had no effect on growth or fiber size (16,22). In contrast, calcineurin functions as a critical regulator of fiber-type switching in skeletal muscle, while the heart lacks such a biologic process (16). Given these dramatically different functional roles, it is not surprising that calcineurin's activity could be antithetically regulated during disease in cardiac versus skeletal muscle.
Our observation that Cn inhibition largely rescues skeletal muscle and heart disease manifestations in the scgd−/− mouse is in dramatic contrast to conclusions reached in the mdx mouse. For example, muscles from mdx mice expressing the same MCK-Cn* transgene that we used showed significant reductions in the extent of central nucleation, fiber size variability, inflammation, and an improvement in membrane integrity (27). These results suggested that Cn activation, not inhibition, is protective in dystrophic muscle. A potential reason underlying this difference likely relates to inherent differences in the mdx and scgd−/− mouse models themselves. Indeed, we have previously observed that myostatin inhibition or deletion has little effect or may be even detrimental to muscular dystrophy in scgd−/− mice (44), while disease in mdx mice is partially rescued by myostatin inhibition/deletion (45). Both mdx and scgd−/− mice have different disease characteristics that relate to a different phenotypic spectrum in human muscular dystrophy. For example, mdx mice are thought to have greater regenerative capacity in their skeletal muscle compared with dystrophic humans (46,47). Moreover, progression of disease in human Duchenne muscular dystrophy patients leads to significantly shortened life expectancy, while mdx mice have a near-normal lifespan. As many as 50% of humans with Duchenne muscular dystrophy also develop severe and life-threatening cardiomyopathy, while the mdx model shows minor cardiac pathology (48). Loss of δ-sarcoglycan in the mouse models a rare form of human limb-girdle muscular dystrophy referred to as LGMD2F. However, loss of any of the proteins within the DGC leads to a similar disease mechanism that involves destabilization of the sarcolemma and influx of Ca2+, which induces degeneration of myofibers. Thus, while the scgd−/− mouse employed here corresponds to a rare form of human disease, its use to dissect disease progression is likely relevant to many forms of muscular dystrophy. Indeed, scgd−/− mice show both skeletal muscle and significant cardiac muscle disease, they show fairly widespread pathology in all skeletal muscles examined that is reminiscent of the pathology observed in severe forms of human muscular dystrophy, and they have foreshortened lifespan as seen in many human muscular dystrophies such as Duchenne's (30).
Another possibility is that different membrane alterations occur between the mdx and scgd−/− mouse models such that Cn activity is differentially affected. The hypothesis has been proposed that the DGC and accessory structural proteins can anchor and regulate the activation of various signaling effectors. Hence, it is possible that loss of δ-sarcoglycan and the other sarcoglycans with it, preferentially affects Cn signaling or an accessory signaling pathway that modulates Cn in a manner that is distinct from a loss in dystrophin alone. As a final consideration, it should also be noted that the genetic background of the mdx mouse is C57-based while the scgd−/− mouse is 129Sv-based, which could also account for some variability in phenotype.
The mechanism whereby Cn inhibition protects against skeletal muscle and heart muscle disease in the scgd−/− background is uncertain, although some evidence supports the hypothesis that cell death or limited necrosis is involved. For example, studies conducted in neurons, lymphocytes, and tumor cell lines have shown both pro- and anti-apoptotic roles for Cn activation (49). In the heart, transgenic mice expressing activated CnA were protected from cell death following ischemia-reperfusion injury, while genetic disruption of the CnAβ gene enhanced myocyte death induced by ischemia-reperfusion injury (50,51). In contrast to these studies, isoproterenol or aldosterone stimulation promoted apoptosis in association with Cn activation, suggesting that Cn could also promote cell death in cardiac myocytes (52,53). Mechanistically, Cn can localize to the mitochondria in fibroblasts in association with FK506 binding protein 38, resulting in Bcl-2 and Bcl-xl redistribution and effects on cell death (54). Cn has also been implicated as a direct inducer of apoptosis in primary hippocampal neurons by dephosphorylating the pro-apoptotic factor Bad, resulting in its induction of mitochondrial-dependent cell death (55). More recently, we also observed that Cn could serve a pro-apoptotic role in cardiac myocytes and fibroblasts through a direct regulatory effect on apoptosis signal-regulating kinase 1 (56). While it has yet to be determined whether Cn signaling regulates cell death/necrosis in skeletal muscle myofibers, we did observe less central nucleation and inflammation in scgd−/− mice lacking the CnB1 gene specifically in skeletal muscle. However, direct assessment of apoptosis with hairpin 1 ligation failed to reveal that skeletal muscle fibers die by such a mechanism in scgd−/− mice. Instead, we have observed a prominent increase in necrosis as the potential mechanism underlying myofiber loss in scgd−/− mice2. Thus, it is possible that Cn activation can promote myofiber death by enhancing necrosis in a model of limb-girdle muscular dystrophy, although it may not in the mdx model if myofiber degeneration proceeds through a different mechanism (apoptosis). Regardless of the exact mechanism, our results indicate that Cn inhibition may provide some benefit in select types of muscle disease, such as limb-girdle muscular dystrophy. This interpretation might also suggest that CsA could be potentially of benefit as well (at least some aspects of the disease). However, caution is warranted because CsA is not a pure inhibitor of calcineurin, as it can also alter tissue necrosis by directly regulating mitochondrial permeability pore transition (57). Moreover, previous clinical trials with CsA in humans with Duchenne and Becker muscular dystrophy were equivocal, although most were underpowered. Thus, while Cn may offer some therapeutic value, new inhibitors may be needed or dramatically different time courses used in a preventative scheme rather than for treating more advanced disease.
Acknowledgement
This work was supported by the National Institutes of Health (to J.D.M.). J.D.M. is an Established Investigator of the American Heart Association. S.A.P. was supported by training grants from the National Institutes of Health (5T32HL007752 and 1T32AR053461-01). We would also like to thank Wyeth Research for support in finalizing revisions to this manuscript.
Abbreviations
- Cn
calcineurin
- Cn*
constitutively active Cn
- CnA
calcineurin A subunit
- CnB
calcineurin B subunit
- CsA
cyclosporine A
- DGC
dystrophin-glycoprotein complex
- HW/BW
heart-weight divided by body-weight
- JNK
c-Jun N-terminal kinase
- MAPK
mitogen-activated protein kinase
- MEF2
myocyte enhancer factor 2
- MLC
myosin light chain
- MCK
muscle creatine kinase
- NFAT
nuclear factor of activated T cells
- RLU
relative light units
- Wt
wildtype
Footnotes
Doug P. Millay and Jeffery D. Molkentin, unpublished observation.
REFERENCES
- 1.Crabtree GR. J. Biol. Chem. 2001;276:2313–2316. doi: 10.1074/jbc.R000024200. [DOI] [PubMed] [Google Scholar]
- 2.Klee CB, Ren H, Wang X. J. Biol. Chem. 1998;273:13367–13370. doi: 10.1074/jbc.273.22.13367. [DOI] [PubMed] [Google Scholar]
- 3.Rusnak F, Mertz P. Physiol. Rev. 2000;80:1483–1521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- 4.Buttini M, Limonta S, Luyten M, Boddeke H. Histochem. J. 1995;27:291–299. doi: 10.1007/BF00398971. [DOI] [PubMed] [Google Scholar]
- 5.Jiang H, Xiong F, Kong S, Ogawa T, Kobayashi M, Liu JO. Mol. Immunol. 1997;34:663–669. doi: 10.1016/s0161-5890(97)00054-0. [DOI] [PubMed] [Google Scholar]
- 6.Muramatsu T, Giri PR, Higuchi S, Kincaid RL. Proc. Natl. Acad. Sci. USA. 1992;89:529–533. doi: 10.1073/pnas.89.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takaishi T, Saito N, Kuno T, Tanaka C. Biochem. Biophys. Res. Commun. 1991;174:393–398. doi: 10.1016/0006-291x(91)90533-d. [DOI] [PubMed] [Google Scholar]
- 8.Hogan PG, Chen L, Nardone J, Rao A. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 9.Abbott KL, Friday BB, Thaloor D, Murphy TJ, Pavlath GK. Mol. Biol. Cell. 1998;9:2905–2916. doi: 10.1091/mbc.9.10.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoey T, Sun YL, Williamson K, Xu X. Immunity. 1995;2:461–472. doi: 10.1016/1074-7613(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 11.Bigard X, Sanchez H, Zoll J, Mateo P, Rousseau V, Veksler V, Ventura-Clapier R. J. Biol. Chem. 2000;275:19653–19660. doi: 10.1074/jbc.M000430200. [DOI] [PubMed] [Google Scholar]
- 12.Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM, Wu H, Zhu W, Bassel-Duby R, Williams RS. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delling U, Tureckova J, Lim HW, De Windt LJ, Rotwein P, Molkentin JD. Mol. Cell. Biol. 2000;20:6600–6611. doi: 10.1128/mcb.20.17.6600-6611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCullagh KJ, Calabria E, Pallafacchina G, Ciciliot S, Serrano AL, Argentini C, Kalhovde JM, Lomo T, Schiaffino S. Proc. Natl. Acad. Sci. USA. 2004;101:10590–10595. doi: 10.1073/pnas.0308035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyazaki M, Hitomi Y, Kizaki T, Ohno H, Haga S, Takemasa T. J. Physiol. Pharmacol. 2004;55:751–764. [PubMed] [Google Scholar]
- 16.Naya FJ, Mercer B, Shelton J, Richardson JA, Williams RS, Olson EN. J. Biol. Chem. 2000;275:4545–4548. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- 17.Oh M, Rybkin II, Copeland V, Czubryt MP, Shelton JM, van Rooij E, Richardson JA, Hill JA, De Windt LJ, Bassel-Duby R, Olson EN, Rothermel BA. Mol. Cell. Biol. 2005;25:6629–6638. doi: 10.1128/MCB.25.15.6629-6638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parsons SA, Millay DP, Wilkins BJ, Bueno OF, Tsika GL, Neilson JR, Liberatore CM, Yutzey KE, Crabtree GR, Tsika RW, Molkentin JD. J. Biol. Chem. 2004;279:26192–26200. doi: 10.1074/jbc.M313800200. [DOI] [PubMed] [Google Scholar]
- 19.Parsons SA, Wilkins BJ, Bueno OF, Molkentin JD. Mol. Cell. Biol. 2003;23:4331–4343. doi: 10.1128/MCB.23.12.4331-4343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serrano AL, Murgia M, Pallafacchina G, Calabria E, Coniglio P, Lomo T, Schiaffino S. Proc. Natl. Acad. Sci. USA. 2001;98:13108–13113. doi: 10.1073/pnas.231148598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H, Naya FJ, McKinsey TA, Mercer B, Shelton JM, Chin ER, Simard AR, Michel RN, Bassel-Duby R, Olson EN, Williams RS. EMBO J. 2000;19:1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kegley KM, Gephart J, Warren GL, Pavlath GK. Dev. Biol. 2001;232:115–126. doi: 10.1006/dbio.2001.0179. [DOI] [PubMed] [Google Scholar]
- 24.Horsley V, Friday BB, Matteson S, Kegley KM, Gephart J, Pavlath GK. J. Cell Biol. 2001;153:329–338. doi: 10.1083/jcb.153.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durbeej M, Campbell KP. Curr. Opin. Genet. Dev. 2002;12:349–361. doi: 10.1016/s0959-437x(02)00309-x. [DOI] [PubMed] [Google Scholar]
- 26.Lapidos KA, Kakkar R, McNally EM. Circ. Res. 2004;94:1023–1031. doi: 10.1161/01.RES.0000126574.61061.25. [DOI] [PubMed] [Google Scholar]
- 27.Chakkalakal JV, Harrison MA, Carbonetto S, Chin E, Michel RN, Jasmin BJ. Hum. Mol. Genet. 2004;13:379–388. doi: 10.1093/hmg/ddh037. [DOI] [PubMed] [Google Scholar]
- 28.Stupka N, Gregorevic P, Plant DR, Lynch GS. Acta. Neuropathol. (Berl) 2004;107:299–310. doi: 10.1007/s00401-003-0807-x. [DOI] [PubMed] [Google Scholar]
- 29.St-Pierre SJ, Chakkalakal JV, Kolodziejczyk SM, Knudson JC, Jasmin BJ, Megeney LA. FASEB J. 2004;18:1937–1939. doi: 10.1096/fj.04-1859fje. [DOI] [PubMed] [Google Scholar]
- 30.Hack AA, Lam MY, Cordier L, Shoturma DI, Ly CT, Hadhazy MA, Hadhazy MR, Sweeney HL, McNally EM. J. Cell Sci. 2000;113:2535–2544. doi: 10.1242/jcs.113.14.2535. [DOI] [PubMed] [Google Scholar]
- 31.Coral-Vazquez R, Cohn RD, Moore SA, Hill JA, Weiss RM, Davisson RL, Straub V, Barresi R, Bansal D, Hrstka RF, Williamson R, Campbell KP. Cell. 1999;98:465–474. doi: 10.1016/s0092-8674(00)81975-3. [DOI] [PubMed] [Google Scholar]
- 32.Neilson JR, Winslow MM, Hur EM, Crabtree GR. Immunity. 2004;20:255–266. doi: 10.1016/s1074-7613(04)00052-4. [DOI] [PubMed] [Google Scholar]
- 33.Bothe GW, Haspel JA, Smith CL, Wiener HH, Burden SJ. Genesis. 2000;26:165–166. [PubMed] [Google Scholar]
- 34.Bueno OF, Wilkins BJ, Tymitz KM, Glascock BJ, Kimball TF, Lorenz JN, Molkentin JD. Proc. Natl. Acad. Sci. USA. 2002;99:4586–4591. doi: 10.1073/pnas.072647999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkins BJ, Dai YS, Bueno OF, Parsons SA, Xu J, Plank DM, Jones F, Kimball TR, Molkentin JD. Circ. Res. 2004;94:110–118. doi: 10.1161/01.RES.0000109415.17511.18. [DOI] [PubMed] [Google Scholar]
- 36.Woessner JF., Jr. Arch. Biochem. Biophys. 1961;93:440–447. doi: 10.1016/0003-9861(61)90291-0. [DOI] [PubMed] [Google Scholar]
- 37.Allen DG, Whitehead NP, Yeung EW. J. Physiol. 2005;567:723–35. doi: 10.1113/jphysiol.2005.091694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berchtold MW, Brinkmeier H, Muntener M. Physiol. Rev. 2000;80:1215–1265. doi: 10.1152/physrev.2000.80.3.1215. [DOI] [PubMed] [Google Scholar]
- 39.Talmadge RJ, Otis JS, Rittler MR, Garcia ND, Spencer SR, Lees SJ, Naya FJ. BMC Cell. Biol. 2004;5:28. doi: 10.1186/1471-2121-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molkentin JD. Cardiovasc. Res. 2004;63:467–475. doi: 10.1016/j.cardiores.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 41.Megeey LA, Kablar B, Perry RL, Ying C, May L, Rudnicki MA. Proc. Natl. Acad. Sci. USA. 1999;96:220–225. doi: 10.1073/pnas.96.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolodziejczyk SM, Walsh GS, Balazsi K, Seale P, Sandoz J, Hierlihy AM, Rudnicki MA, Chamberlain JS, Miller FD, Megeney LA. Curr. Biol. 2001;11:1278–1282. doi: 10.1016/s0960-9822(01)00397-9. [DOI] [PubMed] [Google Scholar]
- 43.Lang JM, Esser KA, Dupont-Versteegden EE. Exp. Biol. Med. (Maywood) 2004;229:503–511. doi: 10.1177/153537020422900608. [DOI] [PubMed] [Google Scholar]
- 44.Parsons SA, Millay DP, Sargent MA, McNally EM, Molkentin JD. Am. J. Pathol. 2006;168:1975–1985. doi: 10.2353/ajpath.2006.051316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel K, Amthor H. Neuromuscul. Disord. 2005;15:117–126. doi: 10.1016/j.nmd.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 46.Tanabe Y, Esaki K, Nomura T. Acta Neuropathol. (Berl) 1986;69:91–95. doi: 10.1007/BF00687043. [DOI] [PubMed] [Google Scholar]
- 47.Anderson JE, Ovalle WK, Bressler BH. Anat. Rec. 1987;219:243–257. doi: 10.1002/ar.1092190305. [DOI] [PubMed] [Google Scholar]
- 48.Finsterer J, Stollberger C. Cardiology. 2003;99:1–19. doi: 10.1159/000068446. [DOI] [PubMed] [Google Scholar]
- 49.Baines CP, Molkentin JD. J. Mol. Cell. Cardiol. 2005;38:47–62. doi: 10.1016/j.yjmcc.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 50.Bueno OF, Lips DJ, Kaiser RA, Wilkins BJ, Dai YS, Glascock BJ, Klevitsky R, Hewett TE, Kimball TR, Aronow BJ, Doevendans PA, Molkentin JD. Circ. Res. 2004;94:91–99. doi: 10.1161/01.RES.0000107197.99679.77. [DOI] [PubMed] [Google Scholar]
- 51.De Windt LJ, Lim HW, Taigen T, Wencker D, Condorelli G, Dorn GW, 2nd, Kitsis RN, Molkentin JD. Circ. Res. 2000;86:255–263. doi: 10.1161/01.res.86.3.255. [DOI] [PubMed] [Google Scholar]
- 52.Mano A, Tatsumi T, Shiraishi J, Keira N, Nomura T, Takeda M, Nishikawa S, Yamanaka S, Matoba S, Kobara M, Tanaka H, Shirayama T, Takamatsu T, Nozawa Y, Matsubara H. Circulation. 2004;110:317–323. doi: 10.1161/01.CIR.0000135599.33787.CA. [DOI] [PubMed] [Google Scholar]
- 53.Saito S, Hiroi Y, Zou Y, Aikawa R, Toko H, Shibasaki F, Yazaki Y, Nagai R, Komuro I. J. Biol. Chem. 2000;275:34528–34533. doi: 10.1074/jbc.M002844200. [DOI] [PubMed] [Google Scholar]
- 54.Shirane M, Nakayama KI. Nat. Cell. Biol. 2003;5:28–37. doi: 10.1038/ncb894. [DOI] [PubMed] [Google Scholar]
- 55.Wang HG, Pathan N, Ethell IM, Krajewski S, Yamaguchi Y, Shibasaki F, McKeon F, Bobo T, Franke TF, Reed JC. Science. 1999;284:339–343. doi: 10.1126/science.284.5412.339. [DOI] [PubMed] [Google Scholar]
- 56.Liu Q, Wilkins BJ, Lee YJ, Ichijo H, Molkentin JD. Mol. Cell. Biol. 2006;26:3785–3797. doi: 10.1128/MCB.26.10.3785-3797.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waldmeier PC, Zimmermann K, Qian T, Tintelnot-Blomley M, Lemasters JJ. Curr. Med. Chem. 2003;10:1485–1506. doi: 10.2174/0929867033457160. [DOI] [PubMed] [Google Scholar]