Abstract
A given protein generally has only one native tertiary fold, which is the conformation with the lowest Gibbs free energy. Mad2, a protein involved in the spindle checkpoint, however, has two natively folded states with similar Gibbs free energies. Through binding to its target Cdc20, Mad2 inhibits the multisubunit ubiquitin ligase, the anaphase-promoting complex or cyclosome (APC/C), and delays the onset of anaphase until all sister chromatids achieve bipolar attachment to the mitotic spindle. Without ligand binding or covalent modifications, Mad2 adopts two topologically and functionally distinct native folds in equilibrium under physiological conditions. The transition between the two Mad2 states is regulated by multiple mechanisms and is central to the activation and inactivation of the spindle checkpoint. This review summarizes recent structural and biochemical studies on the two-state behavior of Mad2 and discusses the generality and implications of structural malleability of proteins.
Introduction
During the cell cycle, genomic DNA is duplicated once-and-only-once in S phase. The duplicated sister genomes are physically tethered through the process of sister-chromatid cohesion (Nasmyth, 2002; Onn et al., 2008). In mitosis, the two opposing kinetochores of each sister-chromatid pair attach to microtubules originating from the two opposite spindle poles. This bipolar attachment enables the alignment of sister chromatids at the metaphase plate (Cleveland et al., 2003; Tanaka and Desai, 2008) (Figure 1). At the metaphase-to-anaphase transition, a ubiquitin ligase complex called the anaphase-promoting complex or cyclosome (APC/C) ubiquitinates securin and cyclin B (Peters, 2006; Yu, 2007). Degradation of securin and cyclin B activates separase, which cleaves the cohesin complex that maintains sister-chromatid cohesion (Nasmyth, 2002; Onn et al., 2008). Dissolution of sister-chromatid cohesion then allows microtubule-dependent movement and equal partition of separated chromatids between the two daughter cells. Premature sister-chromatid separation prior to the establishment of proper kinetochore-microtubule attachment leads to loss or gain of chromosomes and aneuploidy in daughter cells. Recent evidence from mouse and human genetic studies has established aneuploidy as a causal factor of tumorigenesis (Hanks et al., 2004; Jeganathan et al., 2007; Michel et al., 2001; Weaver et al., 2007). Aneuploidy is also a major cause of birth defects in humans (Hunt and Hassold, 2008).
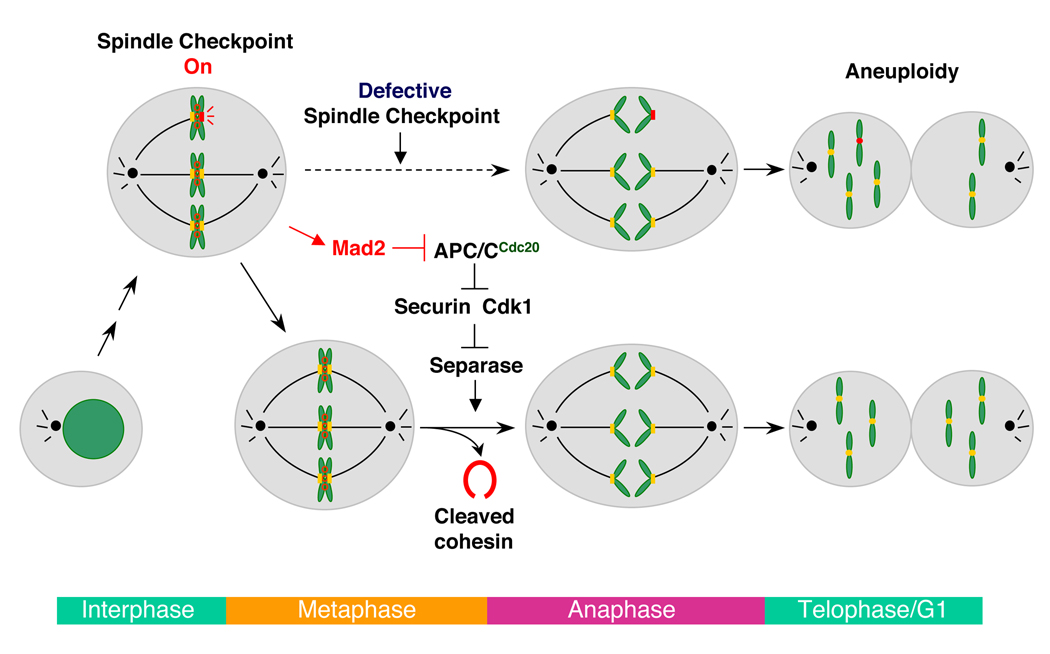
Figure 1. Mitosis and the Spindle Checkpoint.
In mitosis, unattached kinetochores activate the spindle checkpoint. Mad2 is a critical component of the spindle checkpoint and inhibits the ubiquitin ligase APC/C through binding to its activator, Cdc20. Inhibition of APC/C stabilizes its substrates, including securin and cyclin B (the positive regulatory subunit of Cdk1). High levels of securin and Cdk1 block the activation of separase, delaying the cleavage of cohesin and anaphase onset. When the spindle checkpoint is compromised, sister chromatids separate prematurely before all chromatids achieve bipolar attachment to the mitotic spindle. The unattached chromatid (marked with a red kinetochore) segregates randomly into the two daughter cells, resulting in abnormal numbers of chromosomes or aneuploidy.
A cell-cycle surveillance system called the spindle checkpoint monitors the proper attachment of microtubules to kinetochores (Bharadwaj and Yu, 2004; Musacchio and Salmon, 2007). Unattached kinetochores activate this checkpoint, which inhibits APC/C and stabilizes securin and cyclin B, thereby delaying separase activation and the onset of anaphase (Figure 1). The mitotic arrest deficiency 2 (Mad2) protein is a critical molecular component of the spindle checkpoint. Throughout the cell cycle, Mad2 forms a constitutive core complex with Mad1, a coiled-coil protein that is an upstream regulator of Mad2. During mitosis, Mad2 is recruited to unattached kinetochores in a Mad1-dependent manner. Mad1 also promotes the binding of Mad2 to Cdc20, the mitosis-specific activator of APC/C and the molecular target of Mad2 (Yu, 2007). In conjunction with other checkpoint proteins, Mad2 inhibits the ability of Cdc20 to activate APC/C (Yu, 2007), although the detailed mechanism by which Mad2 inhibits APC/C remains unresolved.
A decade ago, Kirschner and coworkers showed that bacterially expressed Mad2 had monomeric and dimeric forms (Fang et al., 1998). Only the dimeric form of Mad2 was functionally active, i.e. it inhibited APC/C-dependent degradation of cyclin B in Xenopus egg extracts. The Mad2 dimers at high concentrations further aggregated into tetramers, which were also active. The Mad2 monomer appeared to be folded, and it blocked the function of the Mad2 dimer in a dominant-negative fashion. This original finding led to the hypothesis that Mad2 is a two-state protein.
The dogma in protein science is that a polypeptide chain with a defined amino acid sequence has a single native fold that is most thermodynamically stable. The thermodynamic hypothesis of Christian Anfinsen states that “the three-dimensional structure of a native protein in its normal physiological milieu (solvent, pH, ionic strength, presence of other components such as metal ions or prosthetic groups, temperature, and other) is the one in which the Gibbs free energy of the whole system is lowest; that is, …the native conformation is determined by the totality of inter-atomic interactions and hence by the amino acid sequence, in a given environment” (Anfinsen, 1973). The possibility that Mad2 had two folded states thus motivated a decade-long structural and functional studies on Mad2 (Mapelli and Musacchio, 2007; Yu, 2006). Collectively, these studies have established that Mad2 indeed has two topologically and functionally distinct native folds in equilibrium at physiological conditions. Furthermore, the transition between the two states of Mad2 is central for its biological function and is regulated by multiple mechanisms in human cells. In this review, we summarize our current understanding of the remarkable two-state behavior of Mad2 and discuss the generality of this behavior.
The Mechanics and Energetics of the Two-State Behavior of Mad2
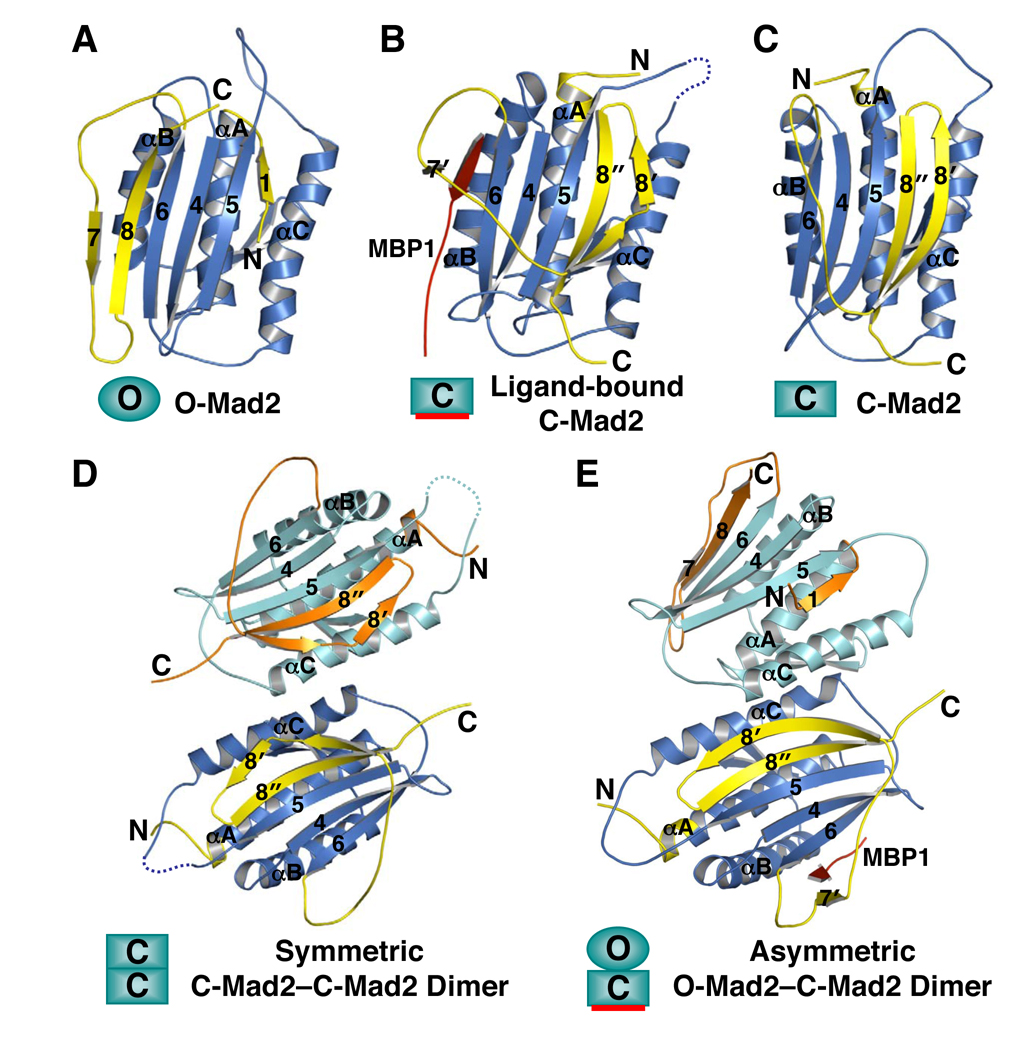
Because purified recombinant Mad2 consisted of multiple conformers, structural studies on this protein relied on conformation-specific mutants that favored a particular conformation. The first structure of Mad2 determined was the solution structure of Mad2ΔC, a mutant of Mad2 with its C-terminal 10 residues deleted that exclusively adopted the monomeric conformation (referred to as open/N1-Mad2 or simply O-Mad2; Figure 2A) (Luo et al., 2000). Using nuclear magnetic resonance (NMR) spectroscopy, the same study also showed that binding of the Mad2-binding motif of Cdc20 triggered a large conformational change on Mad2 and disrupted its oligomerization. The structures of Mad2 bound to a peptide ligand called MBP1 and to Mad2-binding domain of Mad1 were subsequently determined (Luo et al., 2002; Sironi et al., 2002). The ligand-bound conformation of Mad2 (referred to as closed/N2-Mad2 or simply C-Mad2; Figure 2B) is strikingly different from that of O-Mad2. In C-Mad2, the C-terminal β-hairpin and the flexible tail of O-Mad2 rearranges into a new β-hairpin that has a hydrogen bonding network completely different from that of the original one (see Figure 4 below). This new β-hairpin translocates from one side of the Mad2 protein to the other and displaces the original N-terminal β-strand, which forms an additional turn in the central helix. The ligands, such as MBP1, Mad1, or Cdc20, are topologically trapped by the C-terminal β-hairpin of Mad2 and the preceding loop reminiscent of how car passengers are restrained by seat belts. Ligand dissociation requires partial unfolding of Mad2 (the detachment of β8’/8’’ from β5) and is thus expected to be exceedingly slow.
Figure 2. The Many Faces of Mad2.
Ribbon diagrams of N1/open Mad2DC (O-Mad2; PDB ID 1DUJ) (A), N2/closed Mad2-bound to MBP1 (C-Mad2; PDB ID 1KLQ) (B), unliganded C-Mad2R133A (PDB ID 1S2H) (C), the symmetric C-Mad2L13A–C-Mad2L13A dimer (PDB ID 2VFX) (D), and the asymmetric O-Mad2LL –C-Mad2 dimer (PDB ID 2V64) (E). MBP1 is a non-natural high-affinity Mad2-binding peptide identified through phage display (Luo et al., 2002). Loop-less Mad2 (Mad2LL) is a Mad2 mutant with the β5-αC loop shortened (Mapelli et al., 2007).
Figure 4. A Schematic Model Explaining Why O-Mad2 Is an Autoinhibited Conformer.
In O-Mad2, the C-terminal β7/8 hairpin blocks the access of Cdc20 to β6. In C-Mad2, β6 is exposed and can allow edge-on interactions with he Mad2-binding motif of Cdc20. The β8’/8’’ hairpin can then detach from β5, wrap around Cdc20, and re-attach to β5. In the Mad2–Cdc20 complex, Cdc20 is trapped by Mad2, similar to the way that car passengers are restrained by seat-belts. Note that both the original β7/8 hairpin and the corresponding regions in C-Mad2 are colored green. The hydrogen-bonding networks in the β7/8 and β8’/8’’ hairpins are different from one another.
Because the Mad2 dimer was more active in APC/C inhibition, it likely had a conformation that was distinct from O-Mad2 and favored Cdc20 binding. The structural studies of the Mad2 dimer had proved to be difficult, however. Luckily, mutation of a conserved residue in the αC helix, R133, to alanine disrupted the dimerization of Mad2 (Sironi et al., 2001). Strikingly, Mad2R133A had two monomeric conformers at equilibrium that were separable on an anion exchange column (Luo et al., 2004). Based on NMR spectroscopy, the conformer that eluted in the low-salt fractions was O-Mad2. Structure determination of the conformer in the high-salt fractions revealed that it was unliganded C-Mad2, which resembled Mad1- or Cdc20-bound C-Mad2 but had an empty ligand-binding site (Luo et al., 2004) (Figure 2C). The unliganded C-Mad2R133A was more active in APC/C inhibition as compared to O-Mad2R133A. Therefore, Mad2R133A had two topologically and functionally distinct monomeric native folds at equilibrium under physiological conditions.
After extensive efforts, two recent studies finally revealed the structures of the Mad2 dimer (Mapelli et al., 2007; Yang et al., 2008). Yang et al. showed that Mad2 formed both a symmetric dimer that consisted of two unliganded C-Mad2 monomers and an asymmetric dimer that consisted of an O-Mad2 monomer and an unliganded C-Mad2 monomer (Yang et al., 2008) (Figure 2D and 2E). This conformational heterogeneity explained the difficulty of previous structural analyses. The L13A mutant of Mad2 mainly formed the symmetric C-Mad2–C-Mad2 dimer. Structure determination of the Mad2L13A dimer revealed a symmetric dimer interface that centered around the C-terminal end of the αC helix (Figure 2D). With the shortening a flexible loop in O-Mad2 and the addition of MBP1, Mapelli et al. managed to crystallize and determine the structure of the asymmetric O-Mad2–C-Mad2 dimer (Mapelli et al., 2007) (Figure 2E). Collectively, these two structures explained why O-Mad2 could not form an O-Mad2–O-Mad2 symmetric dimer and provided the basis for the ability of Mad2 to form two types of dimers. Yang et al. also showed that Mad2L13A was more active than the wild-type Mad2 in APC/C inhibition in vitro and in eliciting mitotic arrest in cells (Yang et al., 2008). Therefore, unliganded C-Mad2, either as a monomer (as in the case of Mad2R133A) or a dimer, is the functionally more active conformer of Mad2. We emphasize that, despite their obvious structural similarities, ligand-bound C-Mad2 and unliganded C-Mad2 differ in significant ways. Though both can form asymmetric dimers with O-Mad2, only unliganded C-Mad2 can form symmetric dimers.
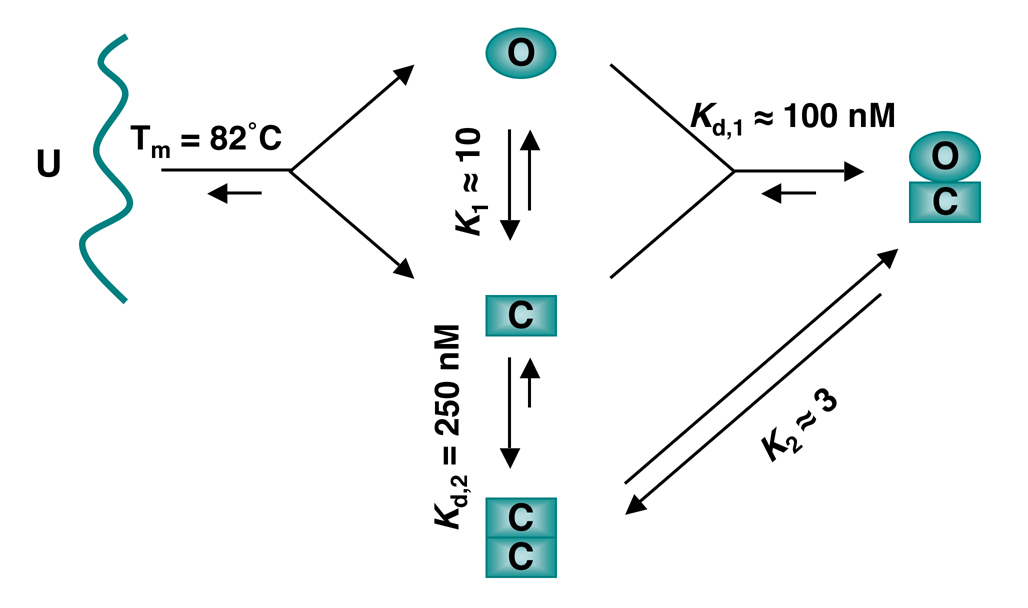
The cartoon diagram in Figure 3 summarizes the equilibria and interconversions of the multiple conformers of Mad2 in vitro. During protein folding, the unfolded Mad2 folds into either O-Mad2 or unliganded C-Mad2. At the physiological temperature (37°C), O-Mad2 spontaneously converts to C-Mad2 with slow kinetics (t1/2 = 9 hrs) (Luo et al., 2004), indicating that O-Mad2 is the high-energy state and that a large energy barrier separates the two states. In the case of Mad2R133A, the molar ratio of O-Mad2:C-Mad2 at equilibrium is about 1:10, i.e. the ΔG of this reaction is about 1.4 kcal/mol (Luo et al., 2004). In the case of the wild-type Mad2, O-Mad2 and C-Mad2 readily associate to form an asymmetric O–C dimer. C-Mad2 also forms a symmetric C–C dimer. The C–C dimer is less stable than the O–C dimer; the molar ratio of the two at equilibrium is about 1:3 (Yang et al., 2008). The conversion rates between the C–C and O–C dimers are slow with a half-life of tens of minutes (our unpublished results), indicating that this structural transition also needs to overcome a high-energy barrier.
Figure 3. The Equilibria and Transitions of Mad2 Conformers.
The unfolded (U) Mad2 chain can fold into either the open (O) Mad2 or closed (C) Mad2 conformers. O-Mad2 and C-Mad2 are at equilibrium with slow interconversion rates. C-Mad2 forms a symmetric C-Mad2–C-Mad2 dimer or an asymmetric O-Mad2–C-Mad2 dimer, which are also at equilibrium. The melting temperature (Tm) of Mad2 is determined by circular dichroism. The equilibrium constants K1 and K2 are estimated from peak intensities of each conformer in NMR spectra (Luo et al., 2004; Yang et al., 2008). Kd,2 is determined for Mad2L13A using equilibrium sedimentation (Yang et al., 2008). Kd,1 is estimated from dissociation constant of O-Mad2 binding to Mad1-bound C-Mad2 (De Antoni et al., 2005).
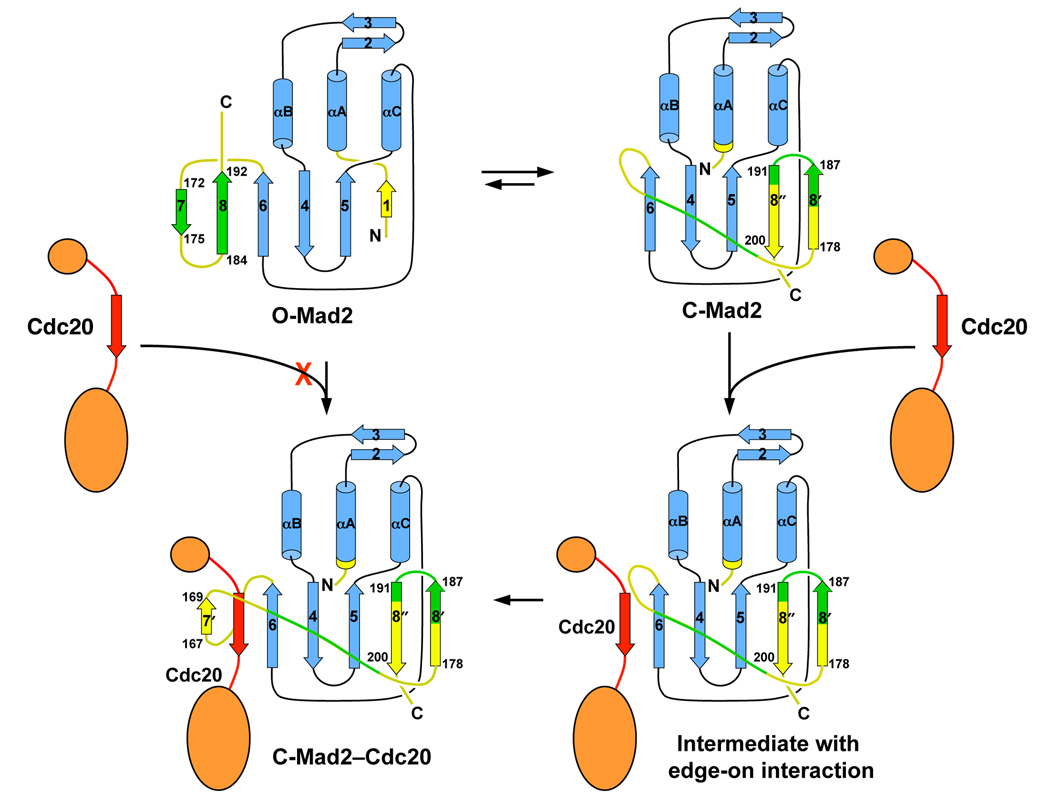
Both the C-Mad2 monomer and the C–C Mad2 dimer are more active in Cdc20 binding and APC/C inhibition as compared to the O-Mad2 monomer and the O–C Mad2 dimer. Why is C-Mad2 more active in Cdc20 binding and APC/C inhibition? Binding of either O-Mad2 or C-Mad2 to Cdc20 produces the same C-Mad2–Cdc20 complex, which has the same dissociation kinetics (off-rate). Therefore, the functional differences between O-Mad2 and C-Mad2 must be caused by differences in the association kinetics (on-rate). In O-Mad2, a vital structural element for Cdc20 binding, strand β6, is blocked by the C-terminal region of Mad2 (Figure 4). It is not accessible for Cdc20 binding. O-Mad2 is thus an autoinhibited conformation. By contrast, β6 is exposed in C-Mad2 and is available for forming edge-on interactions with the Mad2-binding motif of Cdc20. The β8’/8’’ hairpin (the “seat belt”) can then detach from β5, wrap around Cdc20, and reattach to β5, trapping Cdc20 topologically. Thus, we propose that unliganded C-Mad2 binds to Cdc20 with a faster on-rate and a higher affinity as compared to O-Mad2.
In the O–C asymmetric Mad2 dimer, the β8’/8’’ hairpin is a major structural element of the dimer interface (Figure 2E). O-Mad2 thus impedes the movement of this hairpin in C-Mad2 and prevents it from encircling Cdc20, explaining the dominant-negative effects of O-Mad2 over C-Mad2 in the in vitro assays. In the C–C symmetric Mad2 dimer, the β8’/8’’ hairpin is not directly involved in dimerization and can complete the event of Cdc20 binding (Figure 2D).
Because O-Mad2 is a kinetically trapped high-energy state, binding of O-Mad2 to Cdc20 is not a simple equilibrium, as outlined in the following equation: O-Mad2 + Cdc20 <==> C-Mad2–Cdc20. Dissociation of C-Mad2–Cdc20 will mainly produce Cdc20 and unliganded C-Mad2, which is the thermodynamically stable state. Mathematical modeling and quantitative analysis of the spindle checkpoint need to take into account the O-Mad2 <==> C-Mad2 equilibrium when O-Mad2 binding to Cdc20 is considered. The correct binding reaction between Mad2 and Cdc20 at equilibrium is: O-Mad2 + Cdc20 <==> C-Mad2–Cdc20 <==> C-Mad2 + Cdc20.
Function and Regulation of the Two-State Behavior of Mad2
Physiological Relevance of the Different Conformers of Mad2
Is the two-state behavior of Mad2 relevant to its cellular function? Because O-Mad2 is the high-energy state, it is imperative to demonstrate its existence in cells. Based on gel filtration chromatography, the majority of soluble Mad2 in interphase HeLa cells (human ovarian cancer cells) is monomeric (Luo et al., 2004). As discussed above, a remarkably simple yet reliable method for distinguishing O-Mad2 and C-Mad2 is anion exchange chromatography: O-Mad2 elutes in low-salt fractions whereas C-Mad2 elutes in high-salt fractions on anion exchange columns. Mad2 isolated from HeLa cells fractionates in low-salt fractions on anion exchange columns, demonstrating that most of Mad2 in interphase human cells adopts the O-Mad2 conformation (Luo et al., 2004). The functional relevance of O-Mad2 is further underscored by the fact that O-Mad2 is the thermodynamically stable conformation of budding yeast Mad2 in vitro (our unpublished results).
The existence of ligand-bound C-Mad2 is also unequivocal, as the Mad1–Mad2 complex exists throughout the cell cycle and the Mad2–Cdc20-containing complexes are enriched in mitosis (Chen, 2002; Chen et al., 1998; Chung and Chen, 2002; Hwang et al., 1998; Kim et al., 1998). The importance of the O-Mad2–C-Mad2 asymmetric dimerization event is inferred from studies using the Mad2ΔC mutant. Introduction of Mad2ΔC into human cells inactivates the spindle checkpoint (Canman et al., 2002), indicating that Mad2ΔC inhibits the function of endogenous Mad2 in a dominant-negative manner. Mad2ΔC exclusively adopts the O-Mad2 conformation and binds to C-Mad2 to form O–C asymmetric Mad2 dimers in vitro (Luo et al., 2004). Because Mad2ΔC does not interact with Mad1, Cdc20, and other known Mad2-binding proteins, the dominant-negative effect of Mad2ΔC is most likely attributable to its ability to interact with C-Mad2 in cells. Furthermore, Mad2ΔC localizes to kinetochores in mitotic human cells (De Antoni et al., 2005). The Mad2ΔC,R133A mutant that fails to dimerize with C-Mad2 is defective in kinetochore localization (De Antoni et al., 2005). These results demonstrate the functional importance of the O-Mad2–C-Mad2 interaction in human cells.
The existence of unliganded C-Mad2, either as a monomer or a dimer, in human cells has not been established. Because C-Mad2 is the activated conformation, it is expected to bind to O-Mad2, Cdc20, and other Mad2-binding proteins with fast kinetics. Unliganded C-Mad2 is unlikely to be a significantly populated species in vivo. Nevertheless, overexpression of Mad2L13A, a Mad2 mutant that favors the C-Mad2 conformation, is more efficient than the wild-type Mad2 in eliciting mitotic arrest in human cells (Yang et al., 2008), indicating that unliganded C-Mad2 is functional in vivo. Taken together, there is solid evidence supporting the importance of each of the structurally characterized conformers of Mad2 in human cells.
Conformational Activation of Mad2 by Mad1
The majority of Mad2 in interphase human cells is the O-Mad2 conformer, which is in an autoinhibited conformation. How is Mad2 activated conformationally during checkpoint signaling? Mad1 plays a pivotal role in this process. Mad1 forms a constitutive core complex with Mad2 throughout the cell cycle (Chen et al., 1998). Inactivation of Mad1 in various organisms abrogates the spindle checkpoint (Chen et al., 1998; Hardwick and Murray, 1995; Luo et al., 2002), indicating that Mad1 is required for the proper function of Mad2, i.e. binding to Cdc20. On the other hand, because Mad1 and Cdc20 bind to the same site on Mad2, they compete with each other for Mad2 binding (Luo et al., 2002). Indeed, an excess amount of Mad1 inactivates the spindle checkpoint in cells and in Xenopus egg extracts (Canman et al., 2002; Chung and Chen, 2002). Therefore, Mad1 is a double-edged sword in checkpoint signaling and needs to be present at proper amounts. Direct competition between Mad1 and Cdc20 in Mad2 binding provides a straightforward explanation for why too much Mad1 inactivates Mad2 and the spindle checkpoint. The stimulatory effect of Mad1 on Mad2 function is counterintuitive, however.
Then, how does Mad1 promote Mad2 binding to Cdc20? Clues to this mystery first came from fluorescence recovery after photobleaching (FRAP) studies on kinetochore-bound Mad1 and Mad2 (Howell et al., 2004; Shah et al., 2004). The Mad1–Mad2 complex is sequestered at the nuclear pores during interphase and only localizes to kinetochores during mitosis (Campbell et al., 2001; Iouk et al., 2002). Mad1 is a relatively resident kinetochore protein that exchanges slowly with its cytosolic pools (Howell et al., 2004). Intriguingly, there are two different pools of Mad2 at the kinetochores (Shah et al., 2004). About 50% of the kinetochore-bound Mad2 molecules exchange rapidly with cytosolic Mad2 while the other 50% of Mad2 molecules are more resident at the kinetochores. It has been proposed that the resident kinetochore pool of Mad2 is the Mad1-bound C-Mad2 whereas the fast-exchanging pool of Mad2 is O-Mad2 recruited to kinetochores through its interaction with Mad1-bound C-Mad2 (De Antoni et al., 2005). Consistent with this notion, C-Mad2 dissociates from Mad1 with extremely slow kinetics (De Antoni et al., 2005). Furthermore, the kinetics of O-Mad2 binding to the purified recombinant Mad1–Mad2 core complex resembles that of the fast-exchanging kinetochore pool of Mad2 (Vink et al., 2006). Therefore, one Mad1 molecule binds to two molecules of Mad2: the tightly bound C-Mad2 and the loosely bound O-Mad2. Only the loosely bound O-Mad2 is capable of being passed on to Cdc20.
Cdc20 is enriched at kinetochores in mitosis (Howell et al., 2004; Kallio et al., 1998; Kallio et al., 2002). The kinetochore recruitment of O-Mad2 by the Mad1–Mad2 complex increases the local concentration of Mad2. This proximity effect can in theory explain the role of Mad1 in promoting the Mad2–Cdc20 interaction. On the other hand, two main lines of evidence suggest that, in addition to recruiting O-Mad2 to kinetochores, the Mad1–Mad2 core complex also activates Mad2 conformationally. First, the NMR spectra of O-Mad2 bound to C-Mad2 were drastically different from that of free O-Mad2, suggesting that O-Mad2 underwent a major conformational change upon binding to C-Mad2 (Mapelli et al., 2006). Despite the large differences in NMR spectra, the crystal structure of the O-Mad2–C-Mad2 dimer revealed that, surprisingly, O-Mad2 bound to C-Mad2 had virtually the same conformation as free O-Mad2 (Mapelli et al., 2007). Thus, subtle but important structural differences exist between C-Mad2-bound O-Mad2 and free O-Mad2 (Mapelli et al., 2007), although it is unclear at present why the NMR spectra of C-Mad2-bound O-Mad2 are so different from free O-Mad2. Second, the purified recombinant Mad1–Mad2 core complex stimulates the conversion of O-Mad2 to unliganded C-Mad2 in the absence of Cdc20 (Yang et al., 2008). The Mad1–Mad2 core complex thus has the inherent ability to lower the energetic barrier between O-Mad2 and C-Mad2 and promotes the transition between these two Mad2 states.
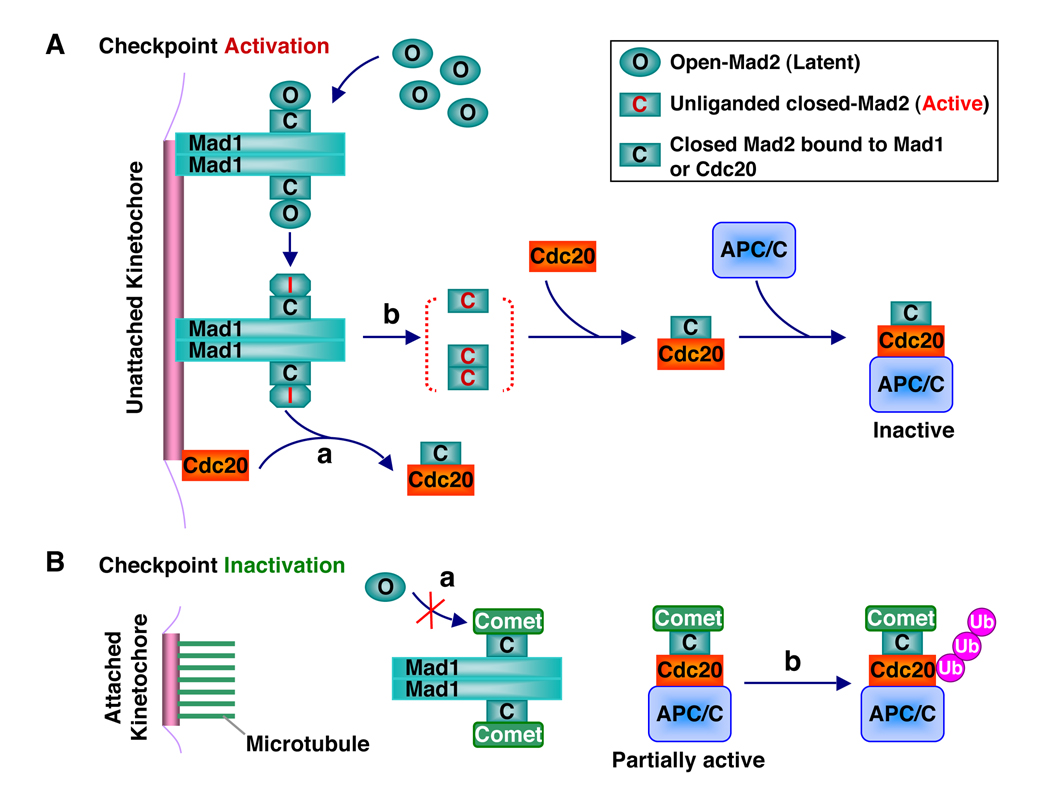
The current data support the following conformational activation model of Mad2 by the Mad1–Mad2 core complex (Figure 5A). In this model, cytosolic O-Mad2 is recruited to kinetochores through its binding to Mad1-bound C-Mad2. Upon binding to C-Mad2, O-Mad2 undergoes subtle conformational changes, which enable it to more readily access a high-energy, transient intermediate Mad2 (I-Mad2) conformation that presumably involves partial unfolding of β7/8 in O-Mad2 to expose β6. Cdc20 may directly bind to I-Mad2. The resulting C-Mad2–Cdc20 complex then dissociates from the Mad1–Mad2 core complex. Alternatively, I-Mad2 completes the O-Mad2–C-Mad2 structural transition on its own and dissociates from the core as unliganded C-Mad2. The unliganded C-Mad2, either as a monomer or a C–C symmetric dimer, then binds to Cdc20 in the vicinity. Several aspects of the conformational activation model need to be further clarified by future experiments. For example, the existence and nature of I-Mad2 need to be established. Furthermore, it has not been shown that the Mad1–Mad2 core complex actually stimulates the binding of O-Mad2 to full-length Cdc20. It is also unclear whether and to what extent I-Mad2 or unliganded C-Mad2 contribute to Cdc20 binding.
Figure 5. A Model for the Conformational Activation of Mad2 by the Mad1–Mad2 Core Complex.
(A) During checkpoint activation, the Mad1–Mad2 core complex recruits cytosolic O-Mad2 to kinetochores through asymmetric O-Mad2–C-Mad2 dimerization and converts O-Mad2 to the transient high-energy I-Mad2 conformer. In pathway a, I-Mad2 binds directly to Cdc20 to form the C-Mad2–Cdc20 complex, which then dissociates from the Mad1–Mad2 core complex. In pathway b, I-Mad2 first dissociates from the Mad1–Mad2 core complex to form unliganded C-Mad2, which exists as a monomer-dimer equilibrium. Unliganded C-Mad2 then binds to Cdc20. The Mad2–Cdc20–APC/C ternary complex is inactive.
(B) During checkpoint inactivation, microtubule binding to kinetochores displaces the Mad1–Mad2 core complex, which is then capped by p31comet to prevent the binding and activation of O-Mad2 (pathway a). p31comet also binds to the Mad2–Cdc20–APC/C ternary complex and promotes the autoubiquitination of Cdc20 and the disassembly of existing Mad2–Cdc20 complexes (pathway b).
This conformational activation model unites the two-state Mad2 model proposed by Luo et al. (Luo et al., 2004) and the template model proposed by De Antoni et al. (De Antoni et al., 2005). However, it differs from the template model in two important ways. First, the conformational activation model emphasizes the roles of unliganded C-Mad2 and the transient high-energy I-Mad2 conformers as activated Mad2 species for binding to Cdc20. Unliganded C-Mad2 is not included in the template model. Second, the template model proposes that Cdc20-bound C-Mad2 in the cytosol can activate O-Mad2 in a mechanism similar to that of the Mad1–Mad2 core complex (De Antoni et al., 2005). In this way, the conformational activation of Mad2 can be propagated away from the kinetochores. Though feasible, there is currently no experimental evidence supporting this hypothesis. The current conformational activation model does not include this propagation mechanism.
Inactivation of Mad2 by p31comet
The spindle checkpoint ensures the proper progression of mitosis during each and every cell division of human cells, even in the absence of spindle damage (Meraldi et al., 2004). It is only activated during mitosis, not during interphase. In addition, when all kinetochores have achieved proper attachment to the mitotic spindle and are under tension, the checkpoint needs to be inactivated to allow sister-chromatid separation. What prevents the Mad1–Mad2 core complex from conformationally activating O-Mad2 during interphase and during checkpoint silencing? An important part of the answer lies with p31comet (formerly known as Cmt2), a Mad2-binding protein initially discovered in a yeast two-hybrid screen (Habu et al., 2002). Depletion of p31comet sensitizes human cells to mitotic arrest exerted by spindle damaging agents, such as Taxol (Xia et al., 2004). p31comet-deficient cells also experience a delay in mitotic exit during the recovery from the spindle damage-induced mitotic arrest. Thus, p31comet plays a role in setting the threshold for checkpoint activation and in checkpoint inactivation. Mechanistically, p31comet selectively associates with ligand-bound or unliganded C-Mad2, but not with O-Mad2 (Xia et al., 2004) (Figure 5B). Through binding to Mad1-bound C-Mad2, p31comet occupies the O-Mad2-binding surface on C-Mad2 and prevents O-Mad2 from binding to the Mad1–Mad2 core complex (Mapelli et al., 2006; Yang et al., 2007). Through binding to Cdc20-bound C-Mad2, p31comet neutralizes the APC/C-inhibitory activity of Mad2 and stimulates the autoubiquitination of Cdc20, which promotes the disassembly of the Mad2–Cdc20-containing checkpoint complexes (Reddy et al., 2007; Stegmeier et al., 2007; Xia et al., 2004). Both mechanisms may contribute to the inhibition of Mad2 by p31comet.
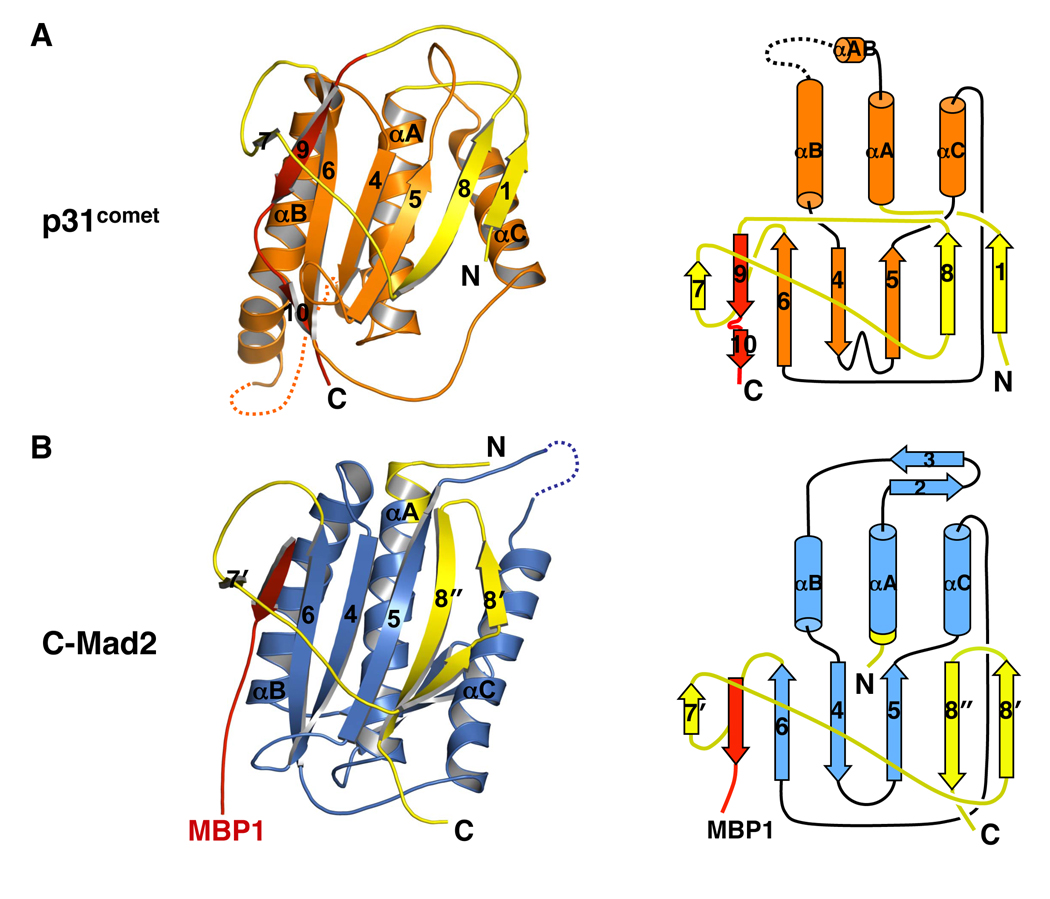
In an unexpected twist, the crystal structure of p31comet bound to C-Mad2 revealed that p31comet had a tertiary fold highly related to Mad2 (Yang et al., 2007) (Figure 6). Structure-based sequence alignment then detected conservation of key residues between Mad2 and p31comet that specified their folds. Mad2 and p31comet might thus share a common ancestor. Mad2 is conserved from yeast to man whereas homologs of p31comet have so far only been found in metazoans. p31comet might have evolved from a Mad2 paralogue to become an “anti-Mad2”. Being a structural mimic of Mad2, p31comet exploits the two-state behavior of Mad2 to block its activation and to neutralize the function of Cdc20-bound Mad2. The story of Mad2 and p31comet provides a stunning example of how two proteins with a similar fold can play antagonistic roles in the same signaling pathway.
Figure 6. p31comet Has a Mad2-like Fold.
(A) Ribbon (left panel) and topology (right panel) diagrams of p31comet. The intramolecular pseudo-ligand is colored red. The seat-belt-like C-terminal region is colored yellow. Despite the insertion of β9/10 into the opening formed by β7, β8, and their connecting loop, p31comet does not contain a knot.
(B) Ribbon (left panel) and topology (right panel) diagrams of MBP1-bound C-Mad2. MBP1 is colored red. The seat-belt-like C-terminal region is colored yellow.
The C-terminal tail of p31comet occupies a site in p31comet that corresponds to the ligand-binding site of Mad2 (Yang et al., 2007) (Figure 6A). It is trapped by β7, β8 and the loop between them in a manner similar to the encirclement of Mad2 ligands by the “seat belt” in Mad2. The C-terminal tail of p31comet is thus an intramolecular pseudo-ligand. It occupies a potential ligand-binding site of p31comet and prevents the binding of exogenous ligands. This important feature might also restrict the conformational malleability of p31comet, making it refractory to potential conformational activation by the Mad1–Mad2 core complex. It is worth noting that the C-terminal region of p31comet does not contain a knot, although at first glance it appears to do so. The main chain of p31comet can be stretched into a straight line when both its N- and C-termini are held and pulled toward opposite directions.
Because p31comet is an inhibitor of Mad2, it logically needs to be inactivated in mitosis when the spindle checkpoint is turned on. It is unclear whether and how p31comet might be regulated during mitosis. Possible regulatory mechanisms include posttranslational modifications, mitosis-specific binding proteins, and subcellular localization.
Generality of the Two-State Behavior of Mad2
Conformational malleability of proteins is rather common. The so-called natively unfolded proteins only adopt defined tertiary folds upon binding to their targets (Dyson and Wright, 2005; Fink, 2005). There are also examples of certain protein domains adopting different folds when bound to different targets (Kim et al., 2000). In these cases, the binding energy between two partners is used to drive or disrupt protein folding. By contrast, Mad2 has two topologically and functionally distinct folds without the binding of cofactors. Other proteins that exhibit such two-state behavior include human plasminogen activator inhibitor-1 (PAI-1), human lymphotactin (Ltn), and the mammalian and fungi prion proteins (Chien et al., 2004; Mottonen et al., 1992; Nar et al., 2000; Perrett and Jones, 2008; Prusiner, 1998; Tuinstra et al., 2008). In this section, we discuss the similarities and differences between Mad2 and other two-state proteins.
Comparison with serpins and Ltn
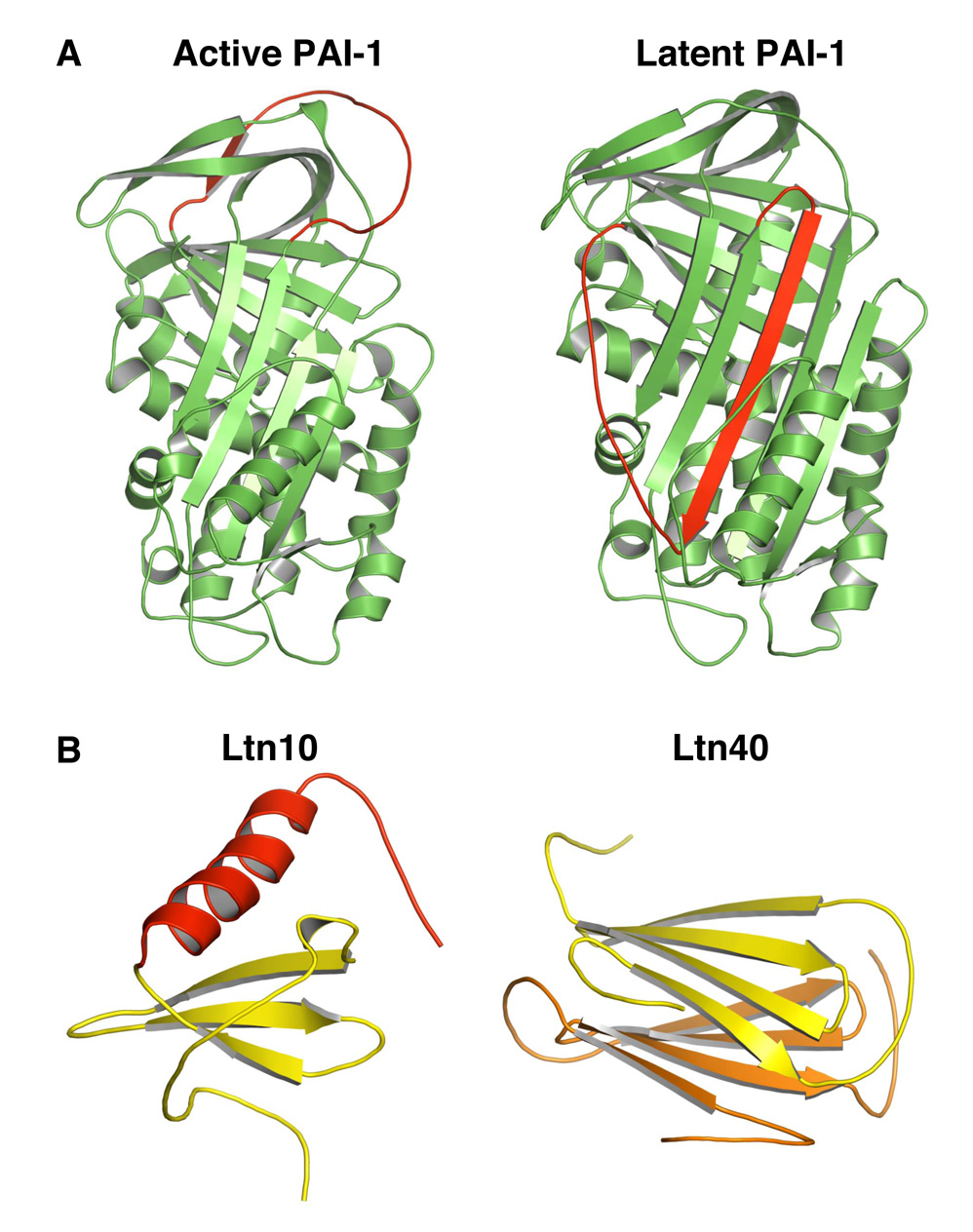
Serine protease inhibitors (serpins) represent one of the first examples of two-state proteins. After cleavage by proteases, serpins undergo a large conformational change that involves the insertion of residues adjacent to the cleavage site as a central strand in a main β sheet (Ye and Goldsmith, 2001). One such serpin protein, active human plasminogen activator inhibitor-1 (PAI-1) spontaneously (without cleavage) folds into a stable latent state, which resembles the cleaved state (Mottonen et al., 1992) (Figure 7A). Unlike Mad2, the structural transition of PAI-1 is irreversible. That is, the active and latent states of PAI-1 are not at equilibrium.
Figure 7. Serpins and Ltn Are Two-state Proteins.
(A) Ribbon diagrams of active (left panel; PDB ID 1DB2) and latent (right panel; PDB ID 1DVN) PAI-1. The region that undergoes a dramatic conformational change is colored red.
(B) Ribbon diagrams of the Ltn10 (left panel; PDB ID 2HDM) and Ltn40 (right panel, PDB ID 2JP1) conformers of Ltn. The two monomers in Ltn40 are colored yellow and orange, respectively.
The human chemokine Ltn has a striking two-state behavior that is more similar to that of Mad2 (Tuinstra et al., 2008) (Figure 7B). At low-temperature (10°C) and high-salt (200 mM NaCl) conditions, Ltn adopts a monomeric canonical chemokine fold, termed Ltn10. At high-temperature (40°C) and no-salt conditions, Ltn has a dimeric fold termed Ltn40, which has a topology completely unrelated to Ltn10. Under physiological conditions (37°C and 150 mM NaCl), both Ltn10 and Ltn40 conformers are equally populated and undergo rapid interconversion. Therefore, Ltn has two unrelated native folds with about the same Gibbs free energy. The two conformers are believed to carry out different subsets of Ltn’s functions. Ltn and Mad2 are thus both two-state proteins that share many common features. Perhaps the most important common feature is that they both have two topologically and functionally distinct native folds at equilibrium under physiological conditions.
There are, however, key differences between the two-state behaviors of Mad2 and Ltn. First, the two conformers of Ltn interconvert much more rapidly than the two conformers of Mad2 do (Tuinstra et al., 2008). The extremely slow rate of interconversion between the two states of Mad2 affords an opportunity for the regulation of this process through cofactor binding and other cellular mechanisms. Second, in the case of Ltn, dimerization is required for the formation of Ltn40, as the dimer interface constitutes the hydrophobic core of this conformer (Tuinstra et al., 2008). By contrast, dimerization is not required for the formation of C-Mad2, because Mad2 mutants deficient in dimerization are capable of folding into both conformers. This property of Mad2 enables it to form two types of dimers: the symmetric C-Mad2–C-Mad2 dimer and the asymmetric O-Mad2–C-Mad2 dimer. The existence of the O-Mad2–C-Mad2 dimer further endows Mad2 with the amazing ability of conformational self-propagation, i.e. C-Mad2 catalyzes the conversion of O-Mad2 to C-Mad2. This unique defining feature of Mad2 is reminiscent of prions.
Comparison with prions
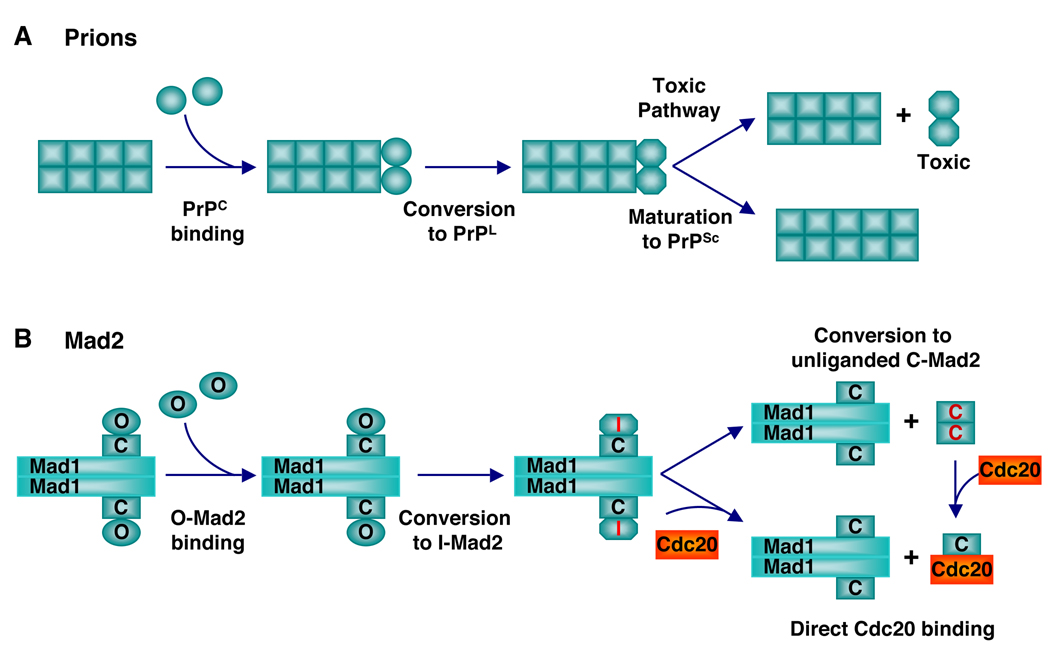
Prions are the protein-only infectious agents that cause neurodegenerative diseases in mammals, including scrapie in sheep and goat, bovine spongiform encephalopathy (BSE or “Mad cow disease”), human Creutzfeldt-Jakob disease (CJD), and human variant CJD (vCJD) (Collinge and Clarke, 2007; Prusiner, 1998). The prion protein has two conformers: the normal monomeric cellular prion protein (PrPC) and the stable amyloid aggregates of misfolded prion (PrPSc). Formation of PrPSc is an irreversible process and involves nucleation. PrPC spontaneously converts to PrPSc with exceedingly slow rates. Several PrPSc molecules can aggregate to form a seeding nucleus. Once formed, this nucleus of PrPSc further recruits PrPC and incorporates it as PrPSc, resulting in the formation of long PrPSc amyloid fibers. Fragmentation of these fibers creates more seeds to catalyze the conversion of additional PrPC. The stability and the ability to self-propagate are believed to underlie the transmissibility of PrPSc. Prion-like proteins have been found in yeast and other fungi (Chien et al., 2004; Perrett and Jones, 2008). For example, the aggregated prion forms of Sup35p in budding yeast account for the genetic trait, [PSI+], which is inherited in a non-Mendelian fashion (Chien et al., 2004; Perrett and Jones, 2008). In contrast to Mad2, the normal and amyloid forms of prions are not at equilibrium, which is a prerequisite for the heritability of prion traits. Neither form of Mad2 encodes a heritable or transmissible trait. Mad2 is clearly not a prion.
On the other hand, the ability of C-Mad2 to promote its own formation through asymmetric O-Mad2–C-Mad2 dimerization bears striking conceptual resemblance to prions (Figure 8). Because PrPSc or the loss of normal PrPC function does not appear to directly cause neuronal cell death (Bueler et al., 1993; Bueler et al., 1992), it has recently been proposed that an intermediate conformation of prions (termed lethal prion or PrPL) might be responsible for the neuronal toxicity (Collinge and Clarke, 2007) (Figure 8A). In this model, PrPSc-catalyzed conversion of PrPC to PrPSc involves the formation of an intermediate, PrPL, which can either convert to PrPSc or dissociate from PrPSc to trigger cell death. This proposed scheme of prion action is conceptually similar to the proposed scheme of Mad2 conformational activation by the Mad1–Mad2 core complex. In the conformational activation model of Mad2 (Figure 8B), an intermediate during the interconversion of the two states termed I-Mad2 is believed to either bind directly to Cdc20 or dissociate to become unliganded C-Mad2, which then binds to Cdc20.
Figure 8. Conceptual Similarities between Prions and Mad2.
(A) A model for the neuronal toxicity of prions. In this model, PrPSc converts PrPC to a transient but lethal intermediate called PrPL, which is toxic to neurons.
(B) The conformational activation model of Mad2. In this model, the Mad1-bound C-Mad2 converts O-Mad2 into a transient high-energy intermediate called I-Mad2, which is active in Cdc20 binding.
Perspectives
Anfinsen’s thermodynamic hypothesis is the foundation for modern protein science. This hypothesis states that the primary structure of a polypeptide chain as specified by its amino acid sequence is sufficient to determine its native fold, which is the conformation with the lowest Gibbs free energy. Both Mad2 and Ltn have two conformers of roughly equal free energy. This two-state behavior appears to be at odds with the thermodynamic hypothesis. The Anfinsen hypothesis does not, however, explicitly state that a single polypeptide chain can only fold into a single native conformation, although this is implied and is generally true for most proteins. The main concept of the thermodynamic hypothesis is the sufficiency of amino acid sequence alone to specify the native fold of a protein in a thermodynamically driven process. Even in the cases of Mad2 and Ltn, the folding and the relative abundance of each of the two states are still governed by thermodynamics. Therefore, the essence of the thermodynamic hypothesis of Anfinsen holds true.
Proteins are dynamic entities. The ground state of a protein is an ensemble of highly related native conformations at fast equilibrium, not a single conformer (Lange et al., 2008). Proteins can also access sparsely populated excited states that may be important for their functions (Hansen et al., 2008). In most cases, the native conformers and excited states of proteins do not differ in overall fold or topology, but rather exhibit local conformational variations. Mad2 and Ltn represent striking examples of proteins with a topologically distinct excited state that has a free energy similar to that of the ground state. Their excited states are thus significantly populated under physiological conditions.
Conformational heterogeneity of proteins poses serious challenges to structural studies. Structural analysis typically starts with efforts to minimize conformational heterogeneity of proteins through trimming their flexible regions and purifying monodispersed species. These efforts may have hindered the observation of functionally relevant two-state behaviors of proteins. Given the relatively small changes in free energy during protein folding reactions and the wide-spread structural malleability of proteins, more proteins with two-state behaviors undoubtedly exist in nature. The combination of X-ray crystallography and other biophysical methods that can better characterize conformational heterogeneity, such as NMR spectroscopy, will be needed to uncover additional examples of two-state proteins and to monitor their metamorphosis.
ACKNOWLEDGMENTS
We thank members of our laboratories for helpful discussions and apologize to colleagues whose primary studies are not cited due to space limitations. Our research on Mad2 is supported by National Institutes of Health (GM61542 to H.Y. and GM085004 to X.L.) and the Welch Foundation (I-1441). H.Y. is an Investigator at the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23:2016–2027. doi: 10.1038/sj.onc.1207374. [DOI] [PubMed] [Google Scholar]
- Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- Bueler H, Fischer M, Lang Y, Bluethmann H, Lipp HP, DeArmond SJ, Prusiner SB, Aguet M, Weissmann C. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- Campbell MS, Chan GK, Yen TJ. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 2001;114:953–963. doi: 10.1242/jcs.114.5.953. [DOI] [PubMed] [Google Scholar]
- Canman JC, Salmon ED, Fang G. Inducing precocious anaphase in cultured mammalian cells. Cell Motil. Cytoskeleton. 2002;52:61–65. doi: 10.1002/cm.10032. [DOI] [PubMed] [Google Scholar]
- Chen RH. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J. Cell Biol. 2002;158:487–496. doi: 10.1083/jcb.200204048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu. Rev. Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- Chung E, Chen RH. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol. Biol. Cell. 2002;13:1501–1511. doi: 10.1091/mbc.02-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, Musacchio A. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink AL. Natively unfolded proteins. Curr. Opin. Struct. Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Habu T, Kim SH, Weinstein J, Matsumoto T. Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 2002;21:6419–6428. doi: 10.1093/emboj/cdf659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks S, Coleman K, Reid S, Plaja A, Firth H, Fitzpatrick D, Kidd A, Mehes K, Nash R, Robin N, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat. Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- Hansen DF, Vallurupalli P, Kay LE. Using relaxation dispersion NMR spectroscopy to determine structures of excited, invisible protein states. J. Biomol. NMR. 2008;41:113–120. doi: 10.1007/s10858-008-9251-5. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Murray AW. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon ED. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- Hunt PA, Hassold TJ. Human female meiosis: what makes a good egg go bad? Trends Genet. 2008;24:86–93. doi: 10.1016/j.tig.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 2002;159:807–819. doi: 10.1083/jcb.200205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio M, Weinstein J, Daum JR, Burke DJ, Gorbsky GJ. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J. Cell Biol. 1998;141:1393–1406. doi: 10.1083/jcb.141.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio MJ, Beardmore VA, Weinstein J, Gorbsky GJ. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J. Cell Biol. 2002;158:841–847. doi: 10.1083/jcb.200201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KF, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- Luo X, Fang G, Coldiron M, Lin Y, Yu H, Kirschner MW, Wagner G. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat. Struct. Biol. 2000;7:224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Rizo J, Yu H. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat. Struct. Mol. Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- Mapelli M, Filipp FV, Rancati G, Massimiliano L, Nezi L, Stier G, Hagan RS, Confalonieri S, Piatti S, Sattler M, Musacchio A. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 2006;25:1273–1284. doi: 10.1038/sj.emboj.7601033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 2007;131:730–743. doi: 10.1016/j.cell.2007.08.049. [DOI] [PubMed] [Google Scholar]
- Mapelli M, Musacchio A. MAD contortions: conformational dimerization boosts spindle checkpoint signaling. Curr. Opin. Struct. Biol. 2007;17:716–725. doi: 10.1016/j.sbi.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VV, Benezra R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- Mottonen J, Strand A, Symersky J, Sweet RM, Danley DE, Geoghegan KF, Gerard RD, Goldsmith EJ. Structural basis of latency in plasminogen activator inhibitor-1. Nature. 1992;355:270–273. doi: 10.1038/355270a0. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nar H, Bauer M, Stassen JM, Lang D, Gils A, Declerck PJ. Plasminogen activator inhibitor 1. Structure of the native serpin, comparison to its other conformers and implications for serpin inactivation. J. Mol. Biol. 2000;297:683–695. doi: 10.1006/jmbi.2000.3604. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister Chromatid Cohesion: A Simple Concept with a Complex Reality. Annu. Rev. Cell Dev. Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- Perrett S, Jones GW. Insights into the mechanism of prion propagation. Curr. Opin. Struct. Biol. 2008;18:52–59. doi: 10.1016/j.sbi.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- Sironi L, Mapelli M, Knapp S, De Antoni A, Jeang KT, Musacchio A. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a 'safety belt' binding mechanism for the spindle checkpoint. EMBO. J. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L, Melixetian M, Faretta M, Prosperini E, Helin K, Musacchio A. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J. 2001;20:6371–6382. doi: 10.1093/emboj/20.22.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, McDonald ER, 3rd, Li MZ, Hannon GJ, Sorger PK, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- Tanaka TU, Desai A. Kinetochore-microtubule interactions: the means to the end. Curr. Opin. Cell Biol. 2008;20:53–63. doi: 10.1016/j.ceb.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink M, Simonetta M, Transidico P, Ferrari K, Mapelli M, De Antoni A, Massimiliano L, Ciliberto A, Faretta M, Salmon ED, Musacchio A. In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Curr. Biol. 2006;16:755–766. doi: 10.1016/j.cub.2006.03.057. [DOI] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Xia G, Luo X, Habu T, Rizo J, Matsumoto T, Yu H. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 2004;23:3133–3143. doi: 10.1038/sj.emboj.7600322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Li B, Liu CJ, Tomchick DR, Machius M, Rizo J, Yu H, Luo X. Insights into mad2 regulation in the spindle checkpoint revealed by the crystal structure of the symmetric mad2 dimer. PLoS Biol. 2008;6:e50. doi: 10.1371/journal.pbio.0060050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Li B, Tomchick DR, Machius M, Rizo J, Yu H, Luo X. p31comet blocks Mad2 activation through structural mimicry. Cell. 2007;131:744–755. doi: 10.1016/j.cell.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Goldsmith EJ. Serpins and other covalent protease inhibitors. Curr. Opin. Struct. Biol. 2001;11:740–745. doi: 10.1016/s0959-440x(01)00275-5. [DOI] [PubMed] [Google Scholar]
- Yu H. Structural activation of Mad2 in the mitotic spindle checkpoint: the two-state Mad2 model versus the Mad2 template model. J. Cell Biol. 2006;173:153–157. doi: 10.1083/jcb.200601172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol. Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]