Abstract
The TEM-1 β-lactamase confers bacterial resistance to penicillin antibiotics and has acquired mutations that permit the enzyme to hydrolyze extended spectrum cephalosporins or avoid inactivation by β-lactamase inhibitors. However, many of these substitutions have been shown to reduce activity against penicillin antibiotics and/or result in a loss of stability for the enzyme. In order to gain more information concerning the trade-offs associated with active site substitutions, a genetic selection was used to find second site mutations which partially restore ampicillin resistance levels conferred by an R244A active site TEM-1 β-lactamase mutant. An L201P substitution distant from the active site was identified that enhanced ampicillin resistance levels and increased protein expression levels of the R244A TEM-1 mutant. The L201P substitution also increases the ampicillin resistance levels and restores expression levels of a poorly expressed TEM-1 mutant with a core-disrupting substitution. In vitro thermal denaturation of purified protein indicated that the L201P mutation increases the Tm of the TEM-1 enzyme. The X-ray structure of the L201P TEM-1 mutant was determined to gain insight into the increase in enzyme stability. The proline substitution occurs at the N-terminus of an α-helix and may stabilize the enzyme by reducing the helix dipole as well as lowering the conformational entropy cost of folding due to the reduced number of conformations available in the unfolded state. Collectively the data suggest that L201P promotes tolerance of some deleterious TEM-1 mutations by enhancing protein stability of these mutants.
Introduction
The most prevalent plasmid encoded β-lactamase in Gram-negative bacteria, TEM-1, provides resistance to penicillins and early generation cephalosporins 1. The TEM-1 β-lactamase has evolved in response to the introduction of the extended spectrum β-lactams and β-lactamase inhibitors by acquiring substitutions which modify the substrate profile of the enzyme to match the new selection environment. Many of these active site substitutions alter the substrate profile of the TEM-1 enzyme at the cost of reduced hydrolysis of penicillin β-lactams such as ampicillin, amoxicillin and benzylpenicillin 2; 3; 4; 5; 6; 7; 8; 9.
In addition to sacrificing high-levels of catalytic efficiency against penicillin β-lactams, substitutions near the active site of TEM-1 frequently result in a loss of stability of the enzyme 10. For example, a G238S substitution confers extended-spectrum β-lactamase (ESBL) activity but reduces the protein’s melting temperature (ΔTm = −4.5 relative to wild-type TEM-1) translating to a 1.94 kcal/mol loss of stability 10. Likewise, the R164S ESBL mutation results in a reduced melting temperature (ΔTm = −1.7) and corresponding loss of stability (ΔΔGm = −0.73 kcal/mol) 10. The stability of M69 mutants conferring inhibitor resistance is dependent upon the substituting residue. The M69I substitution results in a loss of stability (ΔΔGm = −1.3), while the M69L TEM-1 mutant is actually more thermodynamically stable than wild type TEM-1 (ΔΔGm = +1.0) 10.
One substitution distant from the active site, M182T, has arisen among antibiotic resistant isolates repeatedly in the context of active site substitutions that reduce stability (http://www.lahey.org/studies). A genetic selection for suppressors of a destabilized TEM-1 β-lactamase led to the identification of the M182T mutation as a general suppressor of folding and/or stability defects 11; 12. The M182T substitution has since been repeatedly identified in clones isolated from directed evolution experiments of TEM-1, which is consistent with its importance in correcting enzyme stability defects associated with other substitutions 13; 14; 15; 16; 17; 18; 19. In restoring stability to TEM-1 variants, the M182T substitution mitigates stability trade-offs associated with amino acid substitutions that provide for extended spectrum cephalosporin hydrolysis and inhibitor resistance 10.
Several TEM-1 variants with substitutions at position 244 have been found in clinical isolates resistant to certain β-lactam/inhibitor combinations 6; 7; 20; 21; 22; 23; 24; 25. The arginine at position 244, however, also plays an important role in substrate binding and catalysis. The X-ray structure of TEM-1 β-lactamase with an acylated penicillin in the active site indicates that the positive charge of this arginine residue interacts, via a bridging water molecule, with the C3 carboxylate group that is common to all classes of β-lactam antibiotics 26. Moreover, substitutions at position 244 increase the Km for hydrolysis of penicillin and, to a lesser extent, cephalosporin antibiotics 7; 9. Yet, substitutions that remove the positive charge from position 244 reduce inactivation by inhibitors as a result of the missing interaction with the C3 carboxylate group 6; 7; 20; 21; 22; 23; 24; 25. Thus, the normally high level of catalytic efficiency towards penem substrates is sacrificed in these mutants in exchange for inhibitor resistance, due to the increase in Km for hydrolysis of β-lactam antibiotics associated with these substitutions.
TEM-1 β-lactamase is a member of a gene family called class A β-lactamases based on primary amino acid sequence homology 27; 28. The class A β-lactamases include a large number of enzymes from both Gram-positive and Gram-negative bacteria 29. Several class A β-lactamases do not encode an arginine at position 244 but rather provide the positive charge that interacts with the C3 carboxylate from another position near the active site 30. For example, the SME-1 and S. albus G β-lactamases provide positive charge from R220 while the Toho-1 β-lactamase has an arginine at position 276. The structures of these enzymes indicate that the guanidinium group from the arginine occupies a similar position in the active site whether it is provided by R220, R244 or R276 31; 32; 33; 34. Therefore, it appears there is structural and evolutionary plasticity in the placement of the arginine that provides the positive charge for substrate binding. To further investigate the loss of catalytic activity associated with substitutions at position 244 and the plasticity with regard to the positive charge, an R244A mutant of TEM-1 β-lactamase was generated. A selection for mutations that increased the ampicillin resistance of E. coli containing the active site mutant R244A was designed to identify second site mutations that restore the positive charge, and thereby the catalytic efficiency, of the enzyme for ampicillin hydrolysis. The selection, however, resulted in the isolation of an L201P substitution that improves fitness by increasing stability and steady-state protein levels of the R244A enzyme. Suppressor function was compared between the L201P substitution and the established, stabilizing, M182T substitution by combining each with other primary substitutions having decreased function. Both L201P and M182T compensate for several but not all primary substitutions and exhibit an overlapping but not identical pattern of suppression. Therefore, these suppressors are "global" in that they act on many different primary substitutions, but they may not act universally to compensate for all defects associated with primary substitutions.
Results and Discussion
Genetic selection of a second site suppressor of the TEM-1 R244A mutant
In TEM-1 β-lactamase, the side chain of arginine at position 244 is directed towards the active site pocket were it interacts with the C3 carboxylate group of a β-lactam substrate 9; 10; 26. In order to initiate study of this position, the arginine side chain was replaced with alanine via site-directed mutagenesis in the pET-TEM-1 expression vector 35. The pET-TEM-1 vector expresses large quantities of soluble TEM-1 β-lactamase via transcription from a T7 promoter and encodes an ompA leader fused to the mature portion of the TEM-1 enzyme 35. This blaTEM-1 gene has subsequently been shown to contain a E28G substitution in mature TEM-1 near the signal cleavage site 35; 36. The mature ompA-TEM-1 E28G β-lactamase is properly processed as determined by N-terminal protein sequencing and does not exhibit measurable changes in catalytic properties relative to wild type TEM-1 with its native signal sequence 36. The R244A substitution results in a 4−8 fold loss of ampicillin resistance relative to TEM-1 E28G β-lactamase as determined by the minimum inhibitory concentration of this β-lactam antibiotic. The R244C mutant of TEM-1 has been shown to have a similar effect on amoxicillin resistance levels 20. The loss of activity seen with these arginine 244 substitutions could be the result of displacement of an active site water molecule that normally interacts with the β-lactam carboxylate, as suggested by the crystal structure of the R244S mutant of TEM-1 10.
To study the plasticity of the positive charge in this region of the active site, a search was made for mutations which partially restore ampicillin resistance to the R244A mutant. This was accomplished by constructing a random library of point mutations of the TEM-1 E28G:R244A gene encoded in the pET-TEM-1 vector by error-prone PCR. From the E28G:R244A library, a suppressor distant from the active site was isolated with a single nucleotide substitution resulting in a L201P mutation. Interestingly, the L201P substitution has been observed previously upon selection for inhibitor-resistant TEM-1 β-lactamases from an error-prone library of TEM-1 enzymes 37. In addition, the L201P substitution was recently identified as a suppressor among populations of highly mutagenized TEM-1 mutants propagated under conditions of neutral drift 38.
In order to ensure separation of function from potential mutations found elsewhere in the pET-TEM-1 plasmid or from the E28G substitution, the L201P:R244A double and R244A single mutants were reconstructed in the original pET-TEM-1 plasmid and the E28G substitution was reverted to wild type by site-directed mutagenesis. The E28G:R244A and R244A mutants in pET-TEM-1 exhibited the same ampicillin minimum inhibitory concentration (MIC = 512 µg/ml). In addition, the pBG66 plasmid that encodes wild type TEM-1 β-lactamase was used to create both the R244A and L201P:R244A TEM-1 mutants by site-directed mutagenesis. Unlike the T7 promoter driven expression of blaTEM-1 from the pET-TEM-1 expression vector, blaTEM-1 is constitutively transcribed by its native promoter in the pBG66 plasmid 39. The reconstructed pET-TEM-1 L201P:R244A in E. coli BL21(DE3) exhibits similar ampicillin resistance (MIC = 512 µg/ml) relative to the R244A mutant as determined by 2-fold antibiotic dilutions. However, a difference in resistance levels was apparent when additional ampicillin concentrations were examined near the minimum inhibitory concentration (MIC) value where R244A exhibited an MIC of 358 µg/ml while the L201P:R44A had an MIC of 512 µg/ml. Similarly, the ampicillin MIC for pBG66-R244A and pBG66-L201P:R244A was 256 µg/ml as determined by 2-fold ampicillin dilutions (Table 1). MIC determinations using commercially available E-test strips containing amoxicillin and a β-lactamase inhibitor, however, indicated that the pBG66-L201P:R244A mutant exhibits an increased level of resistance relative to the pBG66 encoded R244A mutant (Table 1).
Table 1.
Influence of L201P substitution on TEM-1 β-lactamase mediated MIC levels.
| Ampa | Amox+CVb | |
|---|---|---|
| TEM-1 | 2048 | 24 |
| L76N | 32 | 6 |
| M182T | 2048 | 24 |
| L201P | 2048 | 24 |
| R244A | 256 | 32 |
| M182T:L201P | 2048 | 16 |
| L201P:R244A | 256 | 64 |
| L76N:M182T | 512 | 16 |
| L76N:L201P | 64 | 12 |
| L76N:M182T:L201P | 1024 | 16 |
µg/ml ampicillin using broth dilution, pBG66 plasmid
µg/ml amoxicillin with clavulanic acid in 2:1 ratio using E-test strip, pBG66 plasmid
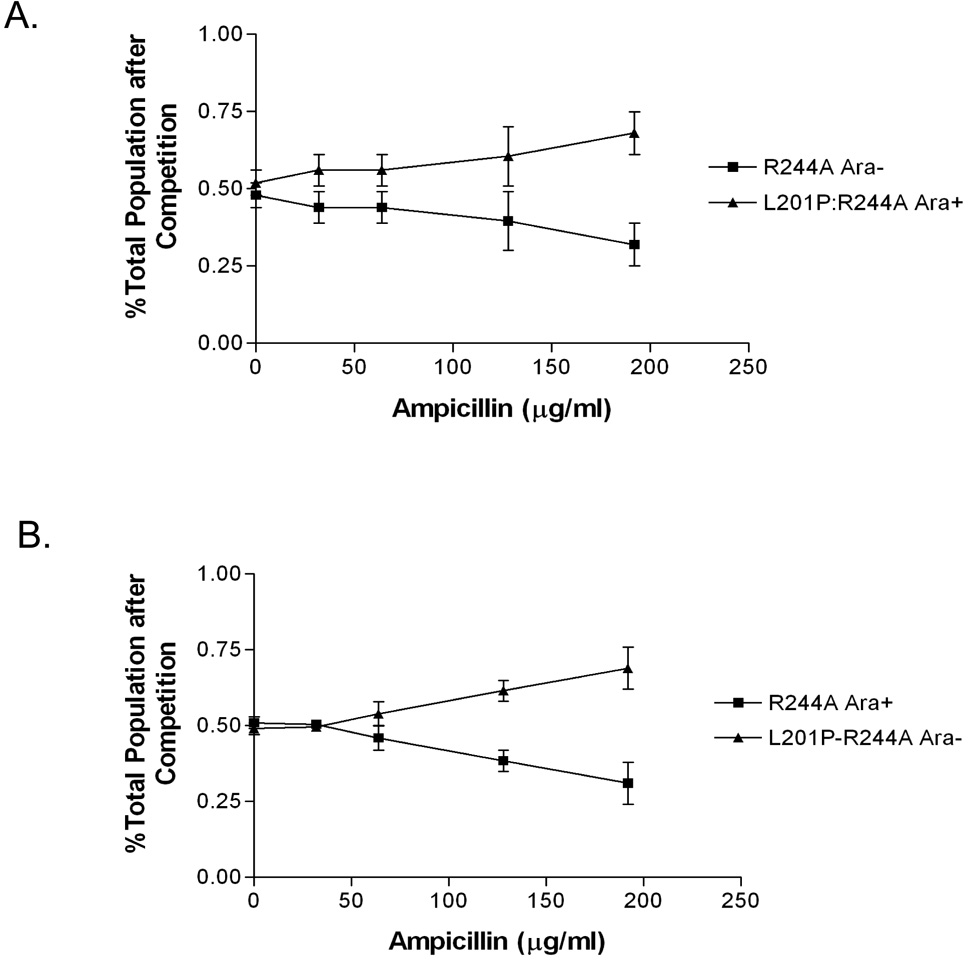
In order to further examine the fitness advantage conferred upon R244A by the L201P substitution, competition experiments were performed between E. coli strains containing the pBG66 plasmid with either the R244A or L201P:R244A β-lactamase genes. These experiments made use of otherwise isogenic Ara+ and Ara− E. coli B strains that, when grown on tetrazolium agar (TA) plates yield pink and red colonies, respectively 40. The R244A and L201P-R244A encoding plasmids were introduced into the Ara+ and Ara− strains and a culture of each was grown to saturation in the absence of ampicillin. Equal volumes of the cultures were then diluted into fresh media containing increasing concentrations of ampicillin and grown to saturation. The cultures were then spread on TA plates and the number of pink and red colonies was determined. The results in Figure 1 clearly indicate that the L201P:R244A mutant out competes the R244A mutant as ampicillin concentrations are increased indicating the L201P substitution provides a fitness advantage.
Figure 1.
Competition experiment between E. coli B cells containing pBG66-R244A and pBG66-L201P:R244A plasmids. A. Equal volumes of overnight cultures of E. coli Ara+ pBG66 R244A and E. coli Ara− pBG66 L201P:R244A were mixed and allowed to compete for growth overnight in LB medium containing the concentration of ampicillin indicated on the X-axis. The Y-axis indicates the percentage of the total culture of E. coli Ara+ pBG66 R244A and E. coli Ara− pBG66 L201P:R244A cells after the competition. B. Reciprocal competition experiment of E. coli Ara− pBG66 R244A and E. coli Ara+ pBG66 L201P:R244A strains.
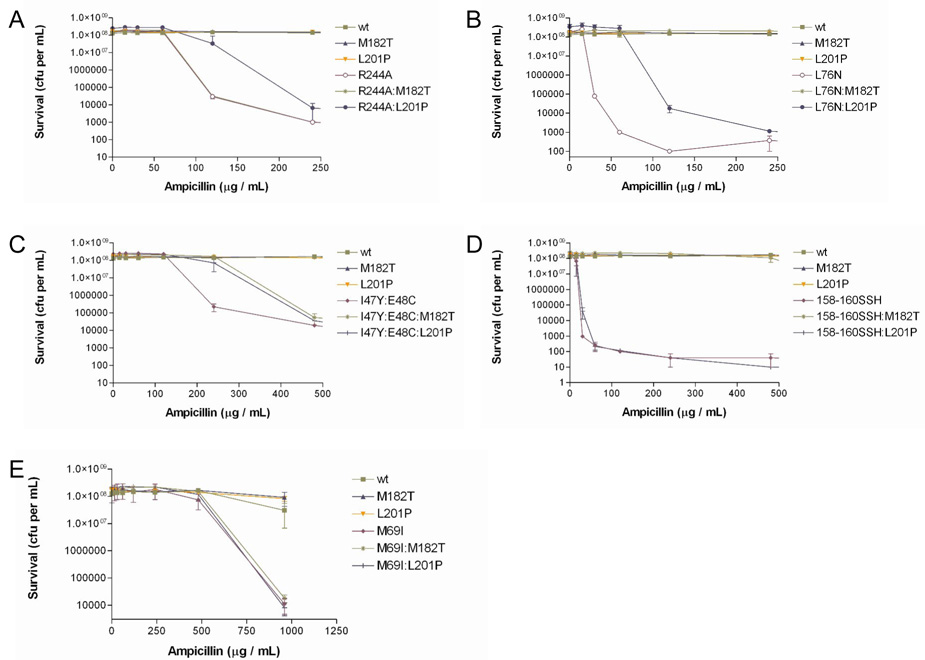
As a further test of the relative fitness of the R244A versus the L201P:R244A mutant, bacterial cultures harboring each mutant on the pBG66 plasmid were spread on agar plates containing increasing concentrations of ampicillin and the number of colonies under each condition were counted to generate a survival curve for each mutant (Fig. 2A). Data from the survival curve experiments were also used to assign ampicillin IC90 values to each TEM-1 mutant, defined here as the concentration of ampicillin required to reduce viability (colony forming units) of TEM-1 expressing bacteria by ≥90% (Table 2). The results indicate the L201P:R244A mutant exhibits enhanced survival relative to the R244A mutant at increasing ampicillin concentrations. Expanded survival curves to an ampicillin concentration of 950 µg/ml are shown in Supplementary Fig. 1A. Taken together, the MIC determinations, competition experiments, and survival curves indicate that the L201P substitution confers enhanced ampicillin resistance to the R244A TEM-1, independent of plasmid and promoter context.
Figure 2.
Survival curves of E. coli with pBG66 plasmid encoding TEM-1 β-lactamase mutants. A. Colony forming units (cfu) on agar plates containing increasing concentrations of ampicillin for E. coli containing R244A, M182T, L201P single and double mutants. B. Cfu for E. coli containing L76N, M182T, L201P single and double mutants. C. Cfu for E. coli containing I47Y:E48C, M182T, L201P mutant combinations. D. Cfu for E. coli containing H158S:V159S:T160H, M182T, L201P mutant combinations. E. Cfu for E. coli containing M69I, M182T, L201P single and double mutants.
Table 2.
Effect of stabilizer substitutions on TEM-1 β-lactamase mediated IC90 levelsa.
| Amp. (µg / mL) | |
|---|---|
| TEM-1 | >960 |
| M182T | >960 |
| L201P | >960 |
| L76N | 30 |
| L76N:L201P | 120 |
| L76N:M182T | 960 |
| R244A | 120 |
| L201P:R244A | 240 |
| M182T:R244A | 120 |
| I47Y:E48C | 240 |
| I47Y:E48C:L201P | 480 |
| I47Y:E48C:M182T | 480 |
| H158S:V159S:T160H | 30 |
| H158S:V159S:T160H:L201P | 30 |
| H158S:V159S:T160H:M182T | 960 |
IC90 , defined as the concentration of ampicillin that reduces cfu per mL of culture by ≥90%, is derived directly from data measurements reported in Figure 2.
It is noteworthy that the M182T suppressor mutation was not identified among the suppressors of the R244A substitution, despite the fact that M182T has been identified previously as a suppressor of folding and/or stability defects of several different primary mutations 13; 14; 15; 16; 17; 18; 19. One explanation is that the library of mutants constructed by PCR contained 1.8 × 105 clones, and the M182T mutant may not have been in this collection. Another possibility is that the M182T substitution does not enhance ampicillin resistance of the R244A primary mutant. To test this possibility, the M182T:R244A double mutant was constructed, and an ampicillin survival curve was generated. As seen in Fig. 2A, the survival curves of the R244A and M182T:R244A are superimposable indicating that, at the resolution of this experiment, M182T does not enhance the ampicillin resistance of the R244A mutant. Therefore, the M182T mutation was not found among the R244A suppressor mutants because it does not alter the ampicillin resistance of R244A.
That the M182T suppressor substitution apparently does not act on the R244A enzyme suggests that the L201P mutation may be allele-specific, i.e., the mutation may increase ampicillin resistance only in combination with R244A. In order to investigate this possibility, double mutant constructs were made with the β-lactamase L76N substitution 11. In TEM-1 β-lactamase, the completely buried leucine at position 76 is part of a hydrophobic core of the enzyme 26; 41. The L76N mutation has a modest effect on the catalytic properties of the enzyme in vitro but results in poor protein expression in vivo. This translates to a significant loss of resistance towards ampicillin 12. Because of these qualities, the L76N mutant can be utilized as a sensitive test of the ability of second site amino acid substitutions to correct protein folding and/or stability defects 11; 12. In addition, L76N was the original primary mutant used to identify M182T as a suppressor of folding and/or stability defects 11.
Both MIC and survival curve experiments were performed with E. coli containing the wild-type, M182T, L201P, L76N, L76N:M182T and L76N:L201P mutants encoded on the pBG66 plasmid. The addition of the M182T substitution restores the ampicillin resistance (MIC) of the pBG66 encoded L76N mutant to high levels (Table 1). The pBG66-L76N:L201P mutant also displays a 2-fold increase of ampicillin resistance relative to pBG66-L76N alone (Table 1). Moreover, the results in Fig. 2B and Table 2 show that the L201P substitution increases the survival of the L76N mutant in ampicillin. These data indicate that L201P can act on substitutions other than R244A. Because the L76N mutant enzyme is unstable, the result also suggests L201P acts to increase stability or improve folding of primary mutants. In addition, the M182T substitution acted as a suppressor of L76N, consistent with previous observations (Fig. 2B) 11.
Several additional mutant combinations were constructed to examine the specificity of action of the M182T and L201P substitutions. The I47Y:E48C mutant was isolated from a library of random substitutions encompassing residues 46–48 of β-lactamase 39. This mutant exhibits reduced ampicillin resistance and lower protein expression levels relative to wild-type TEM-1, and was used previously to demonstrate that M182T can suppress the folding and/or stability defects of mutations other than L76N 11. As seen in Fig. 2C and Table 2, E. coli containing either the I47Y:E48C: M182T or I47Y:E48C:L201P mutants exhibited increased survival on agar plates containing ampicillin compared to the I47Y:E48C parent mutant. This finding indicates both M182T and L201P improve the activity of the I47Y:E48C enzyme in vivo.
The H158S:V159S:T160H TEM-1 β-lactamase mutant was isolated from a library of random substitutions encompassing residues 158–160 39. This mutant was also chosen to test the suppression specificity of M182T and L201P based on its reduced ampicillin resistance and low protein expression levels. It was found that the M182T substitution strongly increased the ampicillin resistance levels of E. coli containing the H158S:V159S:T160H mutant, whereas the L201P substitution had no detectable effect on resistance levels of E. coli containing the mutant (Fig. 2D, Table 2).
Finally, neither M182T nor L201P increased the ampicillin resistance levels of the M69I mutant (Fig. 2E). It was previously shown that the effect of the M182T substitution on the M69I enzyme could only be detected by measuring the specific activity of enzyme lysates at increasing temperatures 11.
The results of the double mutant experiments described above indicate that the TEM-1 L201P substitution can increase the ampicillin resistance of E. coli containing TEM-1 with primary mutations at different locations on the enzyme. This finding is consistent with L201P acting as a global suppressor mutant. However, it was also found that the addition of L201P did not result in detectable changes in ampicillin resistance for the H158S:V159S:T160H or M69I mutants. Similarly, the addition of the M182T substitution increased the ampicillin resistance conferred by several β-lactamase mutants but not M69I or R244A. The failure to observe a change in resistance levels when M182T or L201P are combined with certain mutants could be due to the failure of the assay to detect subtle effects. For example, E. coli containing either M69I:M182T or M69I:L201P did not exhibit increased resistance relative to the M69I parent (Fig. 2E). However, previous studies examining temperature sensitivity have shown that the M69I:M182T enzyme is more active than the M69I parent enzyme at elevated temperatures 11. Therefore, the failure to observe a change in ampicillin resistance levels does not rule out an effect on the stability of the enzyme. Nevertheless, the patterns of suppression of the M182T and L201P substitutions display differences. In general, the M182T substitution is a stronger suppressor than L201P given that M182T confers higher levels of ampicillin resistance in most of the β-lactamase mutants tested. The notable exception is the failure of M182T to enhance the ampicillin resistance of the R244A mutant.
Immunoblot analysis of steady-state levels of β-lactamase mutants
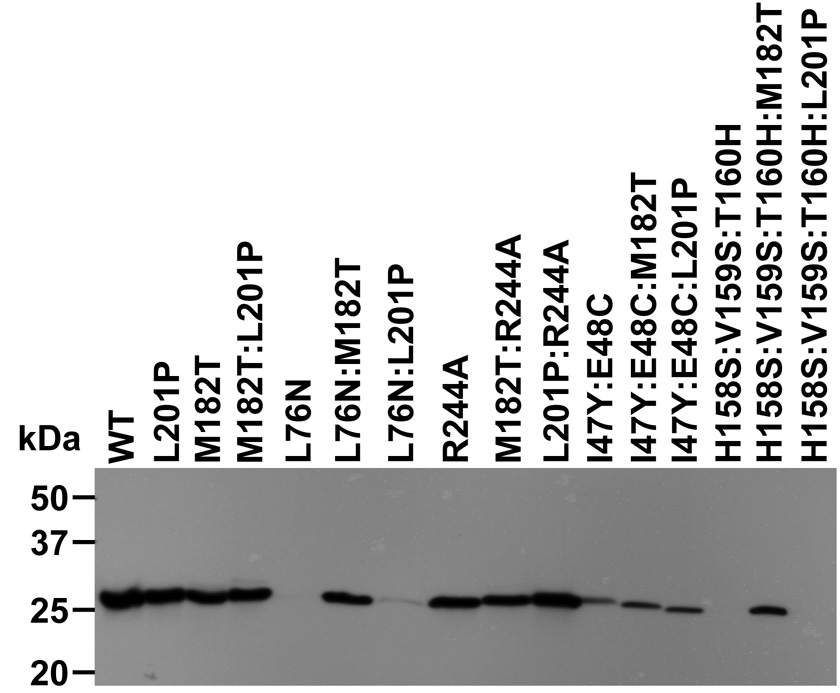
The effect of the L201P substitution on steady-state protein levels of several TEM-1 mutants was quantified by immunoblot analysis of the perplasmic contents of the cell. The protein core-disrupting L76N substitution greatly reduces expression of TEM-1 β-lactamase (Fig. 3 and reference 11 ). A previous study found that the reduced expression level of the L76N mutant is due to periplasmic degradation in that the L76N mutant expression levels are similar to wild-type TEM-1 when expressed in an E. coli strain deficient in four periplasmic proteases 11. Because unstable proteins are rapidly proteolyzed in E. coli, steady-state levels of expression relative to wild type can be used as an indicator of the effect of a mutation on protein stability 42.
Figure 3.
Steady-state protein levels of wild-type and mutant TEM-1 β-lactamases as determined by immunoblot of E. coli periplasmic protein. Positions are: 1, wt TEM-1; 2,L201P; 3, M182T; 4, M182T:L201P; 5, L76N; 6, L76N:M182T; 7, L76N:L201P; 8, R244A; 9, M182T:R244A; 10, L201P:R244A; 11, I47Y:E48C; 12, I47Y:E48C:M182T; 13, I47Y:E48C:L201P; 14, H158S:V159S:T160H; 15, H158S:V159S:T160H:M182T; 16, H158S:V159S:T160H:L201P.
The levels of protein expression of TEM-1 β-lactamase wild type and mutants was monitored by immunoblots of the periplasmic contents of E. coli expressing the enzymes from the pBG66 plasmid using anti-β-lactamase polyclonal antisera 11. Consistent with previous results, the introduction of the M182T substitution rescues steady-state protein levels of the L76N mutant (Fig. 3) 11. The L201P substitution also improves steady-state L76N protein expression levels, which is consistent with it acting as a stability enhancing mutation similar to M182T (Fig. 3) and is also consistent with the increased ampicillin resistance observed in the survival curve (Fig. 2B). The R244A mutation had little effect upon steady-state protein levels suggesting that any effect upon protein folding associated with this active site substitution is modest in comparison to the effect of the L76N mutation (Fig. 3). However, the L201P:R244A clone displayed a higher level of protein expression relative to the R244A clone (Fig. 3, lanes 10 versus 8) while the M182T:R244A clone displayed similar levels as the R244A clone (Fig. 3, lanes 9 versus 8). These observations are consistent with the results of the survival curve experiments with M182T:R244A and L201P:R244A. The higher level of protein expression of L201P:R244A may explain the increased ampicillin resistance which resulted in the isolation of this mutant in the second site mutation selection. The ability of L201P to increase expression levels of both the R244A and L76N mutants of TEM-1 β-lactamase provides evidence that L201P can act on more that one primary mutation in the enzyme.
The results of the survival curve experiments in Fig. 2 suggest that L201P can suppress defects associated with the I47Y:E48C mutant enzyme but not the H158S:V159S:T160H enzyme while the M182T substitution can act on both mutants (Fig. 2C and D, Table 2). The immunobloting results correlate well with these findings in that the I47Y:E48C:L201P enzyme exhibits increased expression relative to the parent enzyme while the H158S:V159S:T160H:L201P enzyme levels are not altered relative to the parent enzyme (Fig. 3). In addition, the expression levels of the I47Y:E48C:M182T and H158S:V159S:T160H:M182T enzymes are higher than the parent enzymes (Fig. 3). Taken together, the survival curve and immunoblotting analyses indicate that both the M182T and L201P substitutions can act on more than one primary mutation, and that the set of mutants that can be suppressed by each overlap but is not identical.
Finally, the M182T and L201P substitutions both increase protein expression levels of other mutants and therefore it was of interest to examine the effect of the combination of M182T and L201P in a double mutant. It was observed that the M182T:L201P double mutant has similar expression levels as the wild type or the M182T or L201P single mutants (Fig. 3).
Enzyme kinetic parameters of TEM-1 β-lactamase mutants
The M182T substitution rescues β-lactamases containing destabilizing mutations but has little influence on enzyme catalytic activity 10; 12. Although ~25 Å distant from the active site, the L201P suppressor could increase enzymatic activity by altering the active site via a long-range structural change propagated down helix α9. To address this issue, the R244A and L201P:R244A enzymes were expressed and purified for kinetic analysis with ampicillin as substrate. No major differences in kcat or Km were found between R244A and L201P:R244A (Table 3). However, as stated above, the L201P substitution is able to increase in vivo ampicillin or amoxicillin-clavulanic resistance levels of the R244A TEM-1 mutant in both the pET-TEM-1 and pBG66 genetic contexts. The finding that the L201P substitution does not enhance enzyme activity via a change in catalytic efficiency but does restore function of both the L76N and R244A mutants in vivo, supports the hypothesis that the L201P substitution acts by increasing the stability of the TEM-1 enzyme and thereby increases steady-state enzyme levels.
Table 3.
Enzyme kinetic parameters for TEM-1 β-lactamase and mutant derivatives for ampicillin hydrolysis.
| TEM-1 | L201P | R244A | R244A-L201P | |
|---|---|---|---|---|
| Km (µM) | 55.2±5.0 | 58.9±7.0 | 2704±492 | 3076±649 |
| kcat (s−1) | 641±51 | 844±8 | 1747±359 | 1706±383 |
| kcat/Km (µM−1 · s−1) | 11.735±2.091 | 14.480±1.890 | 0.645±0.029 | 0.554±0.016 |
Thermodynamic stability of TEM-1 β-lactamase mutant enzymes
To directly address the effect of the L201P substitution upon TEM-1 stability, the L201P, R244A, L201P:R244A and M182T:L201P TEM-1 enzymes were purified for the purpose of obtaining thermal denaturation curves. Measuring the helical signal by circular dichroism, all mutant proteins denatured reversibly in an apparently two state manner, as previously observed for the wild-type enzyme 43. The TEM-1 L201P enzyme exhibited an increased melting temperature relative to wild type TEM-1, by 1.9 °C, indicating increased stability at higher temperatures 10; 43 (Table 4). Typically such a substantial change in the Tm may be interpreted as an increase in the stability of the protein across its folded temperature range. Using the method of Schellmann 44, which analyzes stability changes in the area of the Tm, L201P would be 0.8 kcal/mol more stable than the wild type enzyme. However, it is worth noting that the van’t Hoff enthalpy of denaturation for L201P (111.4 kcal/mol) is lower than that of the wild type enzyme (139.5 kcal/mol), which usually cautions against extrapolating changes in stability around the Tm back to lower temperatures where the enzyme is maximally stable. What we can certainly say is that L201P is more stable at higher temperatures than wild type TEM-1.
Table 4.
Thermodynamic parameters for TEM-1 β-lactamase mutations affecting stability
| Tma | ΔTm | ΔH | ΔΔGu | ΔSu | |
|---|---|---|---|---|---|
| (°C) | (°C) | (kcal/mol) | (kcal/mol) | (kcal/mol*K) | |
| TEM-1b | 51.5±0.1 | — | 139.5±7.9 | — | 0.43 |
| M182Tc | 57.7±0.1 | 6.2±0.2 | 160.3±4.3 | 2.67±0.13 | 0.48 |
| L201P | 53.4±0.1 | 1.9±0.2 | 111.4±0.7 | 0.80±0.08 | 0.34±0.002 |
| L201P:M182T | 58.0±0.2 | 6.5±0.3 | 96.9±5.9 | 2.80±0.13 | 0.29±0.01 |
| R244A | 54.0±0.3 | 2.5±0.4 | 78.1±5.5 | 1.08±0.17 | 0.24±0.02 |
| L201P:R244A | 54.7±0.3 | 3.2±0.4 | 85.8±1.7 | 1.38±0.17 | 0.26±0.004 |
We also determined the Tm of the R244A enzyme, which was 2.5 °C higher than wild type, and therefore even more stable than L201P, at least at the temperature of melting. However, the van’t Hoff enthalpy of denaturation for R244A (78.1 kcal/mol) is much more depressed than was that of L201P, suggesting that this increase of stability can only be trusted at the temperature of melting, and that at lower temperatures the relative stabilities might well be reversed. Moreover, the L201P:R244A double mutant has a Tm that is 0.7 degrees higher than R244A and so the L201P substitution does further stabilize the R244A enzyme. In this double mutant, moreover, the van’t Hoff enthalpy of denaturation is partly restored towards its wild type value, consistent with a stabilization throughout the temperature range of folded protein, and not only at higher temperatures, relative to R244A. This result is consistent with the MIC and survival curve results, and with increased steady state periplasmic expression levels of the L201P:R244A enzyme compared to the R244A enzyme. The reduced hydrolytic activity associated with the R244A substitution results in a significant reduction in ampicillin resistance. The reduction in resistance levels leads to ampicillin MICs being more sensitive to small changes in β-lactamase stability and expression levels 45. This may have provided the sensitivity necessary for the ampicillin selection to identify a suppressor that enhanced steady state expression levels via an increase in the stability of the enzyme.
The M182T and L201P each stabilize TEM-1 β-lactamase. Therefore it was of interest to assess the effects on enzyme stability of a double mutant containing both substitutions. The M182T:L201P double mutant exhibits a 6.5 degree increase in Tm relative to wild type and is marginally more stable than M182T alone, at least around the temperature of melting. Still, the increase in stability of the double relative to the single mutants is much less than additive, which is surprising given how distant the two substitutions are in space in the structure. On closer inspection, residues 201 and 182 both sit at the N-termini of sequential helices, which are connected by a four residue loop. Thus, whereas each of these substitutions can act independently as stabilizers of destabilizing substitutions, together the two substitutions exhibit negative additivity. This in turn suggests that the two helices on which they reside may affect one another. To better understand this observation, and to investigate the effects of Leu201 → Pro at atomic resolution, we determined the structure of L201P by x-ray crystallography.
Structure determination of L201P β-lactamase
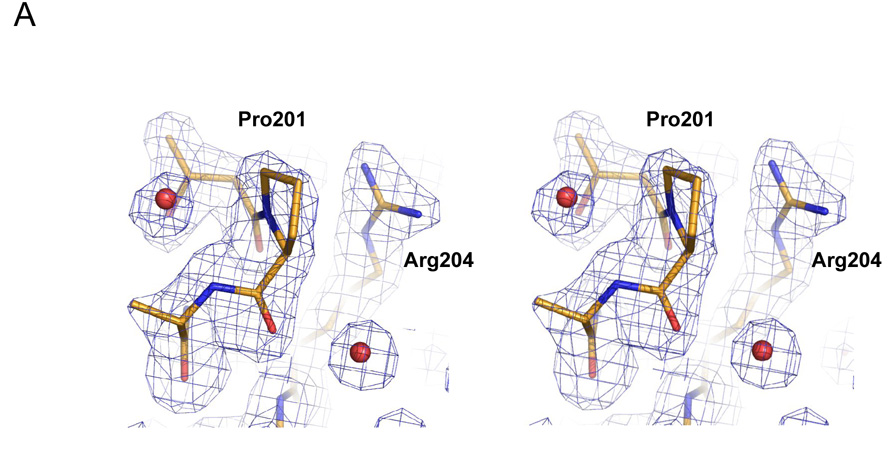
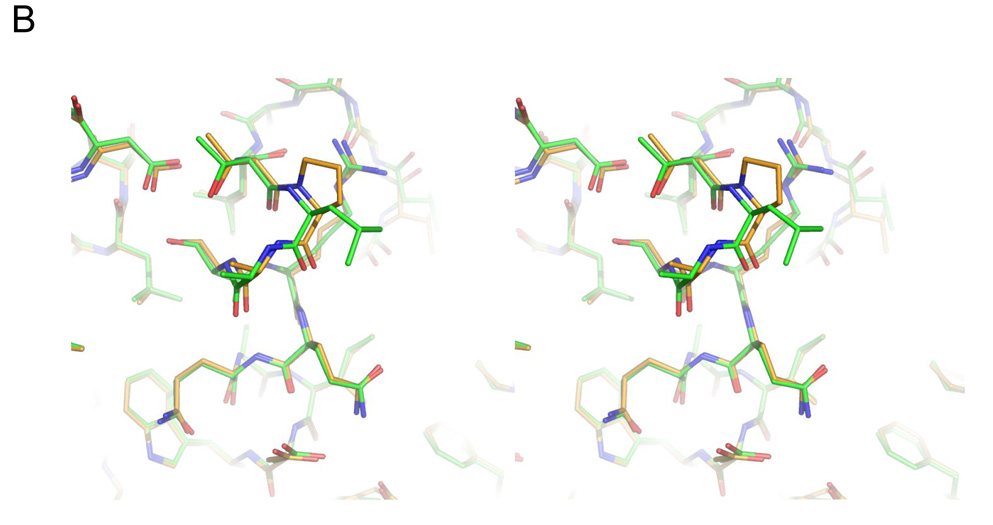
The L201P TEM-1 enzyme was crystallized in conditions similar to that of native TEM-1. Its structure was determined to 1.92 Å by x-ray, and the structure was modeled and refined with good statistics (Table 5). The electron density at and around the point of substitution, Leu201 → Pro, was unambiguous, allowing us to compare the protein environment in the native and mutant enzymes (Fig. 4A). Residue 201 is at the N-terminus of the α9 helix in TEM-1; in both the native and mutant structures the residue is surface exposed, making only van der Waals contacts with Arg204 in both structures. We compared the structure of the L201P mutant to two native structures, (pdb ids 1AXB and 1XPB). For both, the mutant overlapped overlap closely in the region of substitution (Fig. 4B for the superposition with 1AXB), with the overall Cα RMSD between the mutant and 1AXB being only 0.31 Å and in the local region of the substitution (residues 197–205) it was 0.15 Å; the values for the superposition with 1XPB were 0.34 Å and 0.18 Å, respectively. Thus the increased temperature stability of the mutant enzyme is not easily explained by changes in interactions in the folded form of the enzyme. Rather, the proline substitution removes the uncompensated main chain amide proton of Leu201, lying as it does at the N-terminus of helix α9, replacing it with a non-polar carbon of the proline. This reduces the cost of desolvating this group on folding. Consistent with such a role, there is a high propensity for proline at the first (N1) position of α helices 46. Also, the reduced flexibility of the proline side and main chain will increase the free energy of the unfolded form, further increasing the relative stability of the folded enzyme. This entropic effect is thought to be responsible for a ~ 1 kcal/mol increase in stability observed with proline substitutions in T4 lysozyme and ribonuclease Sa 47. Thus, the L201P substitution may enhance the stability of TEM-1 β-lactamase via both helix dipole and entropic mechanisms.
Table 5.
Data collection and refinement statistics.
| Data collection | |
|---|---|
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 41.218 59.140 87.802 |
| α, β, γ (°) | 90.00 90.00 90.00 |
| Resolution (Å) | 50.00-1.92 (1.99-1.92)a |
| Rmerge (%) | 6.1 (35.4) |
| I / σI | 14.0 (2.48) |
| Completeness (%) | 92.0 (83.3) |
| Refinement | |
| Rwork / Rfree | 19.6 % / 24.2% |
| R.m.s deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.473 |
| Ramanchandran plot b: | |
| Most favored region (%) | 93.0 |
| Additionally allowed (%) | 6.6 |
| Generously allowed (%) | 0.4 |
Values in parenthesis represent highest resolution shells.
Calculated by PROCHECK, excluding glycine and proline 54.
Figure 4.
Structure of the L201P mutant of TEM-1 β-lactamase. A. 2Fo-Fc electron density map, contoured at 1σ, shows the mutated residue 201 and surrounding environment, including two ordered water molecules (red spheres) that interact with the protein backbone. Carbon atoms are colored in gold, oxygen atoms in red and nitrogen atoms in blue. B. Overlay of the TEM-1 WT structure (carbons in green) and the L201P mutant (carbons in gold) in the region of the Leu201 → Pro substitution. C. Wild-type TEM-1 (1AXB) helix 9 is shown next to α9 of the L201P structure. The residue at position 201 is presented in stick format and the solvent exposed surface associated with residue 201 is presented in each structure.
Conclusions
The frequent appearance of the M182T substitution in combination with destabilizing active site mutations in TEM-1 β-lactamase variants from clinical isolates provides a medically relevant example of evolutionary pressures acting upon protein stability (http://www.lahey.org/studies) 10; 11; 16. The L201P substitution was previously identified during an in vitro selection experiment for inhibitor resistant TEM-1 mutants37 and it was also recently identified as a suppressor from selection experiments with populations of highly mutagenized TEM-1 genes 38. Unlike the M182T substitution, however, the L201P change has not been identified in natural isolates. The L201P substitution exhibits properties similar to M182T in that it increases stability and steady state protein expression levels and is not strongly allele specific, i.e., it can increase the stability of enzymes with different primary substitutions. The results also indicate, however, that both the M182T and L201P display some specificity with regard to their ability to stabilize enzymes containing destabilizing primary mutations. For example, the M182T substitution had no detectable impact on the R244A mutant, and the L201P substitution had no detectable effect on the H158S:V159S:T160H triple mutant.
Both the M182T and L201P increase the thermodynamic stability of the wild type enzyme. Therefore, M182T and L201P could generally increase stability (−ΔΔG) and act additively with the primary mutations such that the double mutant has increased stability compared to the enzyme containing the primary mutation. The cases where either the M182T or L201P substitutions do not enhance stability could be explained as non-additivity with the primary substitution. By this view, the degree of allele-specificity of a suppressor mutation (substitution) is a function of its additivity relationships with various primary mutations. The exact mechanisms by which M182T and L201P substitutions act and how they communicate with other residue positions in terms of additivity are currently unknown. It is possible that global suppressors actually act on subsets of residue positions and these subsets may overlap completely, partially (as for the targets of M182T and L201P), or not at all. The exact physical mechanism by which the substitutions act on the enzyme would determine additivity relationships. For example, the global stabilizer substitutions could differ with regards to acting upon the native folded protein or certain intermediates on the β-lactamase folding pathway which, in turn, could influence proteolysis or aggregation of the intermediates in vivo. If stabilizer substitutions acted in the same way, they would exhibit a similar pattern of suppression of primary mutations whereas a different pattern of suppression would be observed for stabilizer substitutions that act by different mechanisms. Further experiments are required clarify these questions. Nevertheless, it is clear from these experiments and other recent observations 38 that the Leu201 → Pro substitution provides an important additional example of a stabilizer in TEM β-lactamase that acts on multiple primary mutations and suggests that multiple, single-point restabilizing substitutions may be accessible as a protein evolves new activity, increasing its ability to do so without catastrophic loss of stability.
Materials and Methods
Bacterial strains and plasmids
E. coli XL1-Blue strain (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F'proAB lacIqZΔM15 Tn10 (Tetr)]) was obtained from Stratagene (La Jolla, CA) and utilized in site-directed mutagenesis and determination of steady-state protein expression levels. The E. coli BL21(DE3) strain (F− ompT gal [dcm] [lon] hsdSB (rB − mB −) (λ DE3)) was employed in MIC determination and protein purification. The pET-TEM-1 vector encodes an ompA leader-TEM-1 fusion gene driven by the T7 promoter 35 . The ompA-TEM-1 fusion in pET-TEM-1 also contains an E28G substitution in TEM-1 near the signal cleavage site. The pET-TEM-1 E28G construct was used as template for library construction and for selection of the L201P suppressor mutation. Although no phenotypic changes have been described for the E28G substitution, the substitution was reverted by site-directed mutagenesis prior to subsequent purification of proteins used in enzyme kinetics as well as thermal denaturation experiments and crystallization of the L201P mutant. The pBG66 plasmid encodes wild type TEM-1 β-lactamase under the transcriptional control of its native promoter. Mutations were made in the pBG66 background by site-directed mutagenesis and used in MIC determinations and immunoblot analysis of steady-state protein levels.
Site-directed mutagenesis
All TEM-1 β-lactamase site-directed mutants were created using Stratagene’s (La Jolla, CA) QuickChange kit according to the manufacturer’s protocol. Primers were obtained from Integrated DNA Technologies (Coralville, IA). The following top strand primers were used to introduce mutations (underlined) into the TEM-1 gene: M182T (5’-CGAGCGTGACACCACGACGCCTGCAGCAATGGC-3’), L201P (5’-GGCGAACTACTTACTCCGGCTTCCC-3') and R244A (5’-GCGTGGGTCTGCTGGTATCATTGCAGCACTGGG-3’). The L201P and R244A containing pBG66 TEM-1 mutants were constructed upon the previously described wild-type, M69I, I47Y:E48C, H158S:V159S:T160H, L76N, M182T and L76N:M182T mutants 11. Constructs were sequenced in-house using an Applied Biosystems Instruments (Foster City, CA) Prism Big Dye DNA sequencing kit with an ABI 3100 automated sequencing instrument or by Lonestar Labs (Houston, TX). The pET-TEM-1 E28G mutation was corrected using the top strand primer 5’-CGTAGCGCAGGCCCACCCAGAAACGCTGGTGAAAGTAAAAGATGC-3’.
Library construction and selection
A library of random point mutations was constructed by error-prone PCR using the pET-TEM-1 E28G:R244A gene as template. Mutagenic PCR via inclusion of MnCl2 was performed with 5U Taq polymerase (Promega, Madison, WI), 0.2 mM dATP, 0.2 mM dCTP, 0.4 mM dGTP, 0.4 mM dTTP (dNTPs from Bioline, Randolph, MA), 1.5 mM MgCl2, 1X Taq polymerase buffer (Promega, Madison, WI), 100 ng of outside primers and 45 ng of pET-TEM-1 E28G:R244A plasmid as template in a 100 µl reaction. A series of mutagenic PCR reactions were performed utilizing either 0.0625, 0.125 or 0.25 mM MnCl2 (three reactions performed in triplicate). The PCR reaction parameters were as follows: 95 °C for 1 min; followed by 30 cycles of 95 °C for 1 min, 50 °C for 2 min, and 72 °C for 3 min; ending with 72 °C for 10 min. All 9 mutagenic PCR reactions were pooled and column purified using a Qiagen PCR purification kit (Qiagen Inc., Valencia, CA) before digestion with restriction enzymes XhoI/NdeI/DpnI obtained from New England Biolabs (Ipswich, MA). Insert DNA was gel purified and ligated into pET-TEM-1 digested with XhoI/NdeI that was also treated with calf intestinal alkaline phosphatase (CIP) obtained from New England Biolabs (Ipswich, MA). The DNA in the ligation reaction was extracted with phenol/chloroform, transformed into electrocompetent E. coli BL21(DE3) and spread onto LB plates containing kanamycin. A total of 1.8 × 105 cfu’s were pooled. The library was subjected to selection on 350 µg/ml ampicillin. DNA was isolated from individual clones and sequenced to reveal a t → c mutation resulting in the L201P substitution. The L201P:R244A mutant was recovered and sequenced 6 times. It is not known if the 6 mutants were independent, or if they represent clonal expansion. In addition, the P62L:R244A, A86T:R244A and I208L:R244A mutants were recovered one time each. Only the P62L:R244A mutant (and L201P:R244A) reproducibly improved MIC relative to R244A. The L201P:R244A mutant was pursued because it was detected in multiple clones and it exhibited the most significant increase in ampicillin resistance.
Minimum inhibitory concentration (MIC) determinations
MIC determinations for pET-TEM-1 plasmids
E. coli BL21(DE3) was utilized as the host strain for determining the ampicillin MIC of the pDM122 (pET-TEM-1-R244A), pDM123 (pET-TEM-1-R244A:L201P) pDM124 (pET-TEM-1-L201P), and pET-TEM-1 (wt) constructs. Briefly, overnight cultures were grown at 37°C in LB broth containing 25 µg/ml kanamycin to maintain the plasmid and 100 µM IPTG to induce expression from the T7 promoter. The overnight cultures were diluted 1:104 (~105 bacteria/ml) and 90 µl was inoculated into 1.75 ml fresh LB broth containing 25 µg/ml kanamycin, 100 µM IPTG, and various concentrations of ampicillin. The concentrations of ampicillin tested were 179, 230, 256, 282, 307, 333, 358, 384, 410, 435, 461, 486, 512, 563, 666, 860, 1024, 1106 µg/ml. The cultures were then incubated at 37°C with shaking for ~18 hours and the MIC was determined by examining the minimum inhibitory concentration of ampicillin for bacterial growth.
MIC determinations for pBG66 plasmids
Overnight cultures were grown in LB broth containing 12.5 µg/ml chloramphenicol to maintain the pBG66 plasmid. The overnight cultures were diluted 1:104 (~105 bacteria/ml) and 90 µl was inoculated into 1.75 ml fresh LB broth containing 12.5 µg/ml chloramphenicol and various concentrations of ampicillin. The concentrations of ampicillin tested were 4, 8, 16, 32, 64, 128, 256, 512, 1024, 1536, 2048, 4096 and 8192 µg/ml. The cultures were then incubated at 37°C with shaking for ~18 hours and examined to determine the minimum inhibitory concentration of ampicillin for bacterial growth. For MIC determination using amoxicillin + clavulanic acid E-test strips (AB Biodisk, Solna, Sweden), a one tenth dilution of an overnight culture of E. coli XL1-Blue transformed with a pBG66 construct was spread upon LB plates before placement of the E-test strip on the bacterial lawn. Plates were allowed to incubate overnight at 37°C before scoring for growth. Each assay was conducted in at least duplicate using independent overnight cultures.
Competition Experiments using E. coli B Ara+ and Ara− strains
Competition experiments were performed between E. coli B Ara+ and Ara− cells containing the pBG66 plasmid encoding either the TEM-1 R244A or L201P:R244A β-lactamases. For these experiments, pBG66-TEM-1 R244A and pBG66-TEM-1 L201P:R244A plasmids were transformed into both the E. coli B Ara+ and Ara− strains. Reciprocal experiments were performed, i.e., Ara+ R244A versus Ara− L201P:R244A and Ara+ L201P:R244A versus Ara− R244A for each ampicillin concentration. For simplicity, the methods described below are for the Ara+ R244A versus Ara− L201P:R244A experiment but they were the same for the reciprocal experiment. A culture of each strain was grown overnight in LB medium containing 12.5 µg/ml chloramphenicol to select for the presence of the pBG66 plasmid. Equal volumes of each overnight culture were mixed and diluted to a final concentration of 1 × 104 cells per ml in LB medium containing 12.5 µg/ml chloramphenicol with or without ampicillin. A zero time point was taken from this culture and spread on tetrazolum (TA) agar plates containing 12.5 µg/ml chloramphenicol. After overnight growth at 37°C, Ara+ R244A colonies are white/pink and Ara− L201P:R244A are red. By counting pink versus red colonies it was possible to estimate the number of Ara+ R244A and Ara− L201P:R244A cells in the culture. Competition experiments were performed by adding various concentrations of ampicillin to the Ara+:Ara− mixed starting culture and allowing the cells to compete for growth overnight at 37°C. The cultures were then spread on TA agar plates and pink and red colonies were counted after overnight growth at 37°C.
Bacterial cell survival on ampicillin agar
E.coli XL1-Blue containing the pBG66 plasmid that encodes TEM-1 or a TEM-1 β-lactamase mutant were grown overnight in LB broth containing 12.5 µg/mL chloramphenicol. Overnight cultures were diluted 1:100 into LB broth containing 12.5 µg/mL chloramphenicol and incubated for 4 hours at 37°C to mid-log phase (OD600 = 0.4–0.6). Ten-fold serial dilutions of each culture were made, and 100 µl of each dilution were spread onto LB agar plates containing 0 µg/mL, 15 µg/mL, 30 µg/mL, 60 µg/mL, 120 µg/mL, 240 µg/mL, 480 µg/mL, or 960 µg/mL of ampicillin. After incubation for 24 hours at 37°C, colony forming units (cfu) on each plate were counted. This data was used to calculate the cfu per mL of culture, or survival on a given concentration of ampicillin.
Immunoblot analysis
The effect of the M182T and L201P substitutions upon steady-state expression levels of TEM-1 β-lactamase was determined by immunoblot analysis of E. coli XL1-Blue periplasmic contents as previously described 48. Briefly, overnight cultures transformed with pBG66 encoding wild type TEM-1 or a mutant thereof were diluted 1/50 in fresh LB media containing chloramphenicol and allowed to grow to an OD600 of ~0.3. 1.5 mL of culture was pelleted by centrifugation, resuspended in 200 µl of spheroplast buffer (50 mM Tris-HCl [pH 8], 1mM EDTA, 20% sucrose) and incubated on ice for 10 minutes. The cells were again pelleted by centrifugation, and resuspeded in 200 µl of cold H20 to release the periplasmic contents of the culture. The concentration of soluble protein in each sample was measured using the Bio-Rad Bradford protein assay reagent (Hercules, CA), and approximately 15 µl of sample (adjusted according to protein concentration to ensure equal loading between wells) was resolved on a 12% SDS-PAGE gel. E. coli proteins were electro-transferred to a nitrocellulose membrane (Amersham, GE Heathcare, Piscataway, NJ) using a Bio-Rad (Hercules, CA) semi-dry transfer apparatus. The membrane was blocked overnight in 5% milk before probing with rabbit polyclonal anti-TEM-1 antibody. A donkey anti-rabbit horseradish peroxidase conjugate was utilized with the Amersham ECL chemiluminescent detection reagent (GE Healthcare, Piscataway, NJ) to visualize TEM-1 proteins. This procedure was performed in duplicate using independent cultures.
Protein purification
The L201P, R244A, M182T:L201P and L201P:R244A β-lactamases were purified to ~90% homogeneity. E. coli BL21(DE3) cells transformed with the relevant mutant construct were grown in LB broth with 300 mM sorbitol, 250 mM betaine and 25 µg/ml kanamycin to OD600 0.8, and transcription was induced with 0.4 mM IPTG. The induced culture was grown overnight with shaking at room temperature. Cells were harvested by centrifugation and the periplasmic contents were obtained by osmotic shock. The periplasmic fraction was dialyzed overnight at 4°C before loading onto a Hi-Trap zinc chelating column (Amersham, GE Heathcare, Piscataway, NJ) charged with ZnCl2 5; 49. The β-lactamase containing fraction was eluted by a pH gradient and purity was determined by SDS-PAGE gel electrophoresis. Buffer exchange into 50 mM PO4 pH 7.0 was facilitated by use of a Centricon centrifugal filter (Millipore, Billerica, MA). Mutant β-lactamases L201P, R244A, M182T:L201P and L201P:R244A used in thermal denaturation and crystallization procedures were further purified to ~99% purity by size-exclusion chromatography. Protein concentrations were determined with the Bio-Rad Bradford protein assay reagent (Hercules, CA).
Enzyme kinetics
Michaelis-Menten steady-state kinetic parameters were determined on a Beckman-Coulter spectrophotometer model DU 800 (Fullerton, CA). Ampicillin hydrolysis was monitored at 235 nm. Reactions were performed at 30°C in 50 mM phosphate buffer (pH 7.0). Km and kcat values were determined by fitting initial velocity rates over a range of substrate concentrations to a Michaelis-Menten curve using SigmaPlot. Measurements were performed in triplicate.
Thermal denaturation
Thermal denaturation was carried out in 200 mM potassium phosphate, pH 7.0, as described 43. The enzymes were denatured by raising the temperature in 0.1°C increments at a ramp rate of 2°C/min on a Jasco J-715 spectropolarimeter with a Jasco PTC-348WI Peltier-effect temperature controller and an in-cell temperature probe and stir bar were used, as described. All Tm and ΔHVH values were calculated with the program EXAM 50; the change in heat capacity upon denaturation (ΔCp) was set to 3.8 kcal/mol•K for each enzyme10; 43. Denaturation was marked by an obvious transition in the far-UV CD signal, monitored at 223 nm. Reversibility was measured by the return of folded CD signal divided by the amount of signal lost on unfolding: all enzymes showed greater than 90% reversibility after denaturation. Values of ΔΔGu were determined by the method of Schellman 44, using the entropy of unfolding of the wild-type (WT) enzyme, as determined previously 10; 43.
| (1) |
Structure determination of L201P β-lactamase
L201P was crystallized in 1.6M potassium phosphate buffer (pH 8.7) using a hanging drop method 10. Diffraction data were collected at ALS (Advanced Light Source, Lawrence Berkeley National Lab, Berkeley, CA) Beamline 8.3.1 to 1.92 Å resolution and processed with HKL2000 51. An initial model was obtained through molecular replacement based on the structure of and a TEM-1 M182T mutant structure (1JWP) using EPMR 52. Refinement was performed with CCP4 to a final R value of 19.6% and Rfree 24.2% 53. Full statistics are provided in Table 5.
Supplementary Material
Expanded survival curves of E. coli with pBG66 plasmid encoding TEM-1 β-lactamase mutants, showing survival up to an ampicillin concentration of 950 µg/mL. A. Colony forming units (cfu) on agar plates containing increasing concentrations of ampicillin for E. coli containing R244A, M182T, L201P single and double mutants. B. Cfu for E. coli containing L76N, M182T, L201P single and double mutants. C. Cfu for E. coli containing I47Y:E48C, M182T, L201P mutant combinations. D. Cfu for E. coli containing H158S:V159S:T160H, M182T, L201P mutant combinations.
Acknowledgments
This project was supported by NIH grants AI32956 to TP and GM63815 to BKS. We thank Marvin Makinen for providing the pET-TEM-1 plasmid.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession code:
The coordinates have been deposited in the Protein Data Bank with access code 3CMZ.
References
- 1.Shah AA, Hasan F, Ahmed S, Hameed A. Characteristics, epidemiology and clinical importance of emerging strains of Gram-negative bacilli producing extended-spectrum beta-lactamases. Res. Microbiol. 2004;155:409–421. doi: 10.1016/j.resmic.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Canica MM, Caroff N, Barthelemy M, Labia R, Krishnamoorthy R, Paul G, Dupret JM. Phenotypic study of resistance of beta-lactamase-inhibitor resistant TEM enzymes which differ by naturally occurring variations and by site-directed substitution at Asp276. Antimicrob. Agents Chemother. 1998;42:1323–1328. doi: 10.1128/aac.42.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantu C., III The role of residue 238 of TEM-1 β-lactamase in the hydrolysis of extended-spectrum antibiotics. J. Biol. Chem. 1998;273:26603–26609. doi: 10.1074/jbc.273.41.26603. [DOI] [PubMed] [Google Scholar]
- 4.Cantu C, III, Huang W, Palzkill T. Selection and characterization of amino acid substitutions at residues 237–240 of TEM-1 β-lactamase with altered substrate specificity for aztreonam and ceftazidime. J. Biol. Chem. 1996;271:22538–22545. doi: 10.1074/jbc.271.37.22538. [DOI] [PubMed] [Google Scholar]
- 5.Cantu C, III, Huang W, Palzkill T. Cephalosporin substrate specificity determinants of TEM-1 b-lactamase. J. Biol. Chem. 1997;272:29144–29150. doi: 10.1074/jbc.272.46.29144. [DOI] [PubMed] [Google Scholar]
- 6.Chaibi EB, Sirot D, Paul G, Labia R. Inhibitor-resistant TEM beta-lactamases: phenotypic, genetic and biochemical characteristics. J. Antimicrob. Chemother. 1999;43:447–458. doi: 10.1093/jac/43.4.447. [DOI] [PubMed] [Google Scholar]
- 7.Delaire M, Labia R, Samama JP, Masson JM. Site-directed mutagenesis at the active site of Escherichia coli TEM-1 b-lactamase. Suicide inhibitor-resistant mutants reveal the role of arginine-244 and methionine-69 in catalysis. J. Biol. Chem. 1992;267:20600–20606. [PubMed] [Google Scholar]
- 8.Thomas VL, Golemi-Kotra D, Kim C, Vakulenko SB, Mobashery S, Shoichet BK. Structural consequences of the inhibitor-resistant Ser130Gly substitution in TEM beta-lactamase. Biochemistry. 2005;44:9330–9338. doi: 10.1021/bi0502700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zafaralla G, Manavathu S, Lerner A, Mobashery S. Elucidation of the role of arginine-244 in the turnover processes of class A beta-lactamases. Biochemistry. 1992;31:3847–3852. doi: 10.1021/bi00130a016. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Minasov G, Shoichet BK. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J. Mol. Biol. 2002;320:85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 11.Huang W, Palzkill T. A natural polymorphism in β-lactamase is a global suppressor. Proc. Natl. Acad. Sci. USA. 1997;94:8801–8806. doi: 10.1073/pnas.94.16.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sideraki V, Huang W, Palzkill T, Gilbert HF. A secondary drug resistance mutation of TEM-1 beta-lactamase that suppresses misfolding and aggregation. Proc. Natl. Acad. Sci. USA. 2001;98:283–288. doi: 10.1073/pnas.011454198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barlow M, Hall BG. Predicting evolutionary poteintial: In vitro evolutions accurately reproduces natural evolution of the TEM β-lactmase. Genetics. 2002;160:823–832. doi: 10.1093/genetics/160.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barlow M, Hall BG. Experimental prediction of the evolution of cefepime resistance from the CMY-2 AmpC β-lactamase. Genetics. 2003;164:23–29. doi: 10.1093/genetics/164.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hecky J, Muller KM. Structural perturbation and compensation by directed evolution at physiological temperature leads to thermostabilization of beta-lactamase. Biochemistry. 2005;44:12640–12654. doi: 10.1021/bi0501885. [DOI] [PubMed] [Google Scholar]
- 16.Orencia MC, Yoon JS, Ness JE, Stemmer WP, Stevens RC. Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat. Struct. Biol. 2001;8:238–242. doi: 10.1038/84981. [DOI] [PubMed] [Google Scholar]
- 17.Osuna J, Perez-Blancas A, Soberon X. Improving a circularly permuted TEM-1 beta-lactamase by directed evolution. Protein Eng. 2002;15:463–470. doi: 10.1093/protein/15.6.463. [DOI] [PubMed] [Google Scholar]
- 18.Stemmer W. Rapid evolution of a protein in vitro by DNA shuffling. Nature (London) 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 19.Zaccolo M, Gherardi E. The effect of high-frequency random mutagenesis on in vitro protein evolution: A study on TEM-1 β-lactamase. J. Mol. Biol. 1999;285:775–783. doi: 10.1006/jmbi.1998.2262. [DOI] [PubMed] [Google Scholar]
- 20.Bret L, Chaibi EB, Chanal-Claris C, Sirot D, Labia R, Sirot J. Inhibitor-resistant TEM (IRT) beta-lactamases with different substitutions at position 244. Antimicrob. Agents Chemother. 1997;41:2547–2549. doi: 10.1128/aac.41.11.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bret L, Chanal C, Sirot D, Labia R, Sirot J. Characterization of an inhibitor-resistant enzyme IRT-2 derived from TEM-2 beta-lactamase produced by Proteus mirabilis strains. J. Antimicrob. Chemother. 1996;38:183–191. doi: 10.1093/jac/38.2.183. [DOI] [PubMed] [Google Scholar]
- 22.Imtiaz U, Manavathu E, Mobashery S, Lerner S. Reversal of clavulanate resistance conferred by a ser-244 mutant of TEM-1 beta-lactamase as a result of a second mutation (arg to ser at position 164) that enhances activity against ceftazidime. Antimicrobial Agents and Chemotherapy. 1994;38:1134–1139. doi: 10.1128/aac.38.5.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemozy J, Sirot D, Chanal C, Huc C, Labia R, Dabernat H, Sirot J. First characterization of inhibitor-resistant TEM(IRT) beta-lactamases in Klebsiella pneumoniae strains. Antimicrob. Agents Chemother. 1995;39:2580–2582. doi: 10.1128/aac.39.11.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stapleton PD, Shannon KP, French GL. Construction and characterization of mutants of the TEM-1 beta-lactamase containing amino acid substitutions associated with both extended-spectrum resistance and resistance to beta-lactamase inhibitors. Antimicrob. Agents Chemother. 1999;43:1881–1887. doi: 10.1128/aac.43.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vedel G, Belaaouaj A, Gilly L, Labia R, Philippon A, Nevot P, Paul G. Clinical isolates of Escherichia coli producing TRI β-lactamases: novel TEM-enzymes conferring resistance to β-lactamase inhibitors. J. Antimicrob. Chemother. 1992;30:449–462. doi: 10.1093/jac/30.4.449. [DOI] [PubMed] [Google Scholar]
- 26.Strynadka NCJ, Adachi H, Jensen SE, Johns K, Sielecki A, Betzel C, Sutoh K, James MNG. Molecular structure of the acyl-enzyme intermediate in b-lactam hydrolysis at 1.7 A resolution. Nature. 1992;359:700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- 27.Ambler RP. The structure of beta-lactamases. Phil. Trans. Ry. Soc. London, Ser. B. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 28.Ambler RP, Coulson FW, Frere J-M, Ghuysen J-M, Joris B, Forsman M, Levesque RC, Tiraby G, Waley SG. A standard numbering scheme for the class A β-lactamases. Biochem. J. 1991;276:269–272. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for b-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matagne A, Lamotte-Brasseur J, Frere JM. Catalytic properties of class A beta-lactamases: efficiency and diversity. Biochem. J. 1998;330:581–598. doi: 10.1042/bj3300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dideberg O, Charlier P, Wery JP, Dehotty P, Dusart J, Erpicum T, Frere J-M, Ghuysen J-M. The crystal structure of the beta-lactamase of Streptomyces albus G at 0.3nm resolution. Biochemistry Journal. 1987;245:911–913. doi: 10.1042/bj2450911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibuka AS, Ishii Y, Galleni M, Ishiguro M, Yamaguichi K, Frere JM, Matsuzawa H, Sakai H. Crystal structure of extended-spectrum beta-lactamase Toho-1: Insights into the molecular mechanism for catalytic reaction and substrate specificity expansion. Biochemistry. 2003;42:10634–10643. doi: 10.1021/bi0342822. [DOI] [PubMed] [Google Scholar]
- 33.Matagne A, Misselyn-Baudin A-M, Joris B, Erpicum T, Granier B, Frere J-M. The diversity of the catalytic properties of class A beta-lactamases. Biochemistry Journal. 1990;265:131–146. doi: 10.1042/bj2650131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sougakoff W, L'Hermite G, Pernot L, Naas T, Guillet V, Nordmann P, Jarlier V, Delettre J. Structure of the imipenem-hydrolyzing class A β-lactamase SME-1 from Serratia marcescens. Acta Cryst. 2002;D58:267–274. doi: 10.1107/s0907444901019606. [DOI] [PubMed] [Google Scholar]
- 35.Sosa-Peinado A, Mustafi D, Makinen MW. Overexpression and biosynthetic deuterium enrichment of TEM-1 beta-lactamase for structural characterization by magnetic resonance methods. Protein Expr. Purif. 2000;19:235–245. doi: 10.1006/prep.2000.1243. [DOI] [PubMed] [Google Scholar]
- 36.Savard PY, Gagne SM. Backbone dynamics of TEM-1 determined by NMR: evidence for a highly ordered protein. Biochemistry. 2006;45:11414–11424. doi: 10.1021/bi060414q. [DOI] [PubMed] [Google Scholar]
- 37.Vakulenko SB, Geryk B, Kotra LP, Mobashery S, Lerner SA. Selection and characterization of beta-lactam-beta-lactamase inactivator-resistant mutants following PCR mutagenesis of the TEM-1 beta-lactamase gene. Antimicrob. Agents Chemother. 1998;42:1542–1548. doi: 10.1128/aac.42.7.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bershtein S, Goldin K, Tawfik DS. Intense neutral drifts yield robust and evolvable consensus proteins. J. Mol. Biol. 2008;379:1029–1044. doi: 10.1016/j.jmb.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 39.Huang W, Petrosino J, Hirsch M, Shenkin PS, Palzkill T. Amino acid sequence determinants of β-lactamase structure and activity. J. Mol. Biol. 1996;258:688–703. doi: 10.1006/jmbi.1996.0279. [DOI] [PubMed] [Google Scholar]
- 40.Lenski RE, Simpson SC, Nguyen TT. Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J. Bacteriol. 1994;176:3140–3147. doi: 10.1128/jb.176.11.3140-3147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minasov G, Wang X, Shoichet BK. An ultrahigh resolution structure of TEM-1 beta-lactamase suggests a role for Glu166 as the general base in acylation. J. Am. Chem. Soc. 2002;124:5333–5340. doi: 10.1021/ja0259640. [DOI] [PubMed] [Google Scholar]
- 42.Mayer S, Rudiger S, Ang HC, Joerger AC, Fersht AR. Correlation of levels of folded recombinant p53 in Escherichia coli with thermodynamic stability in vitro. J. Mol. Biol. 2007;372:268–276. doi: 10.1016/j.jmb.2007.06.044. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Minasov G, Shoichet BK. Noncovalent interaction energies in covalent complexes: TEM-1 β-lactamase and β-lactams. Proteins: Struc. Func. Genet. 2002;47:86–96. [PubMed] [Google Scholar]
- 44.Becktel WJ, Schellman JA. Protein stability curves. Biopolymers. 1987;26:1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- 45.Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444:929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- 46.Penel S, Hughes E, Doig AJ. Side-chain structures in the first turn of the alpha-helix. J. Mol. Biol. 1999;287:127–143. doi: 10.1006/jmbi.1998.2549. [DOI] [PubMed] [Google Scholar]
- 47.Matthews BW, Nicholson H, Becktel WJ. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc. Natl. Acad. Sci. USA. 1987;84:6663–6667. doi: 10.1073/pnas.84.19.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palzkill T, Le Q-Q, Venkatachalam KV, LaRocco M, Ocera H. Evolution of antibiotic resistance: several different amino acid substitutions in an active site loop alter the substrate profile of β-lactamase. Mol. Microbiol. 1994;12:217–229. doi: 10.1111/j.1365-2958.1994.tb01011.x. [DOI] [PubMed] [Google Scholar]
- 49.Bowden GA, Paredes AM, Georgiou G. Structure and morphology of protein inclusion bodies in Escherichia coli. Biotechnology. 1991;9:725–730. doi: 10.1038/nbt0891-725. [DOI] [PubMed] [Google Scholar]
- 50.Kirchhoff W. EXAM: A two-state thermodynamic analysis program. Gaithersburg, MD: 1993. [Google Scholar]
- 51.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology: Macromolecular Crystallography Part A. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 52.Kissenger CR, Smith BA, Gehlhaar DK, Bouzida D. Molecular replacement by evolutionary search. Acta Crystallographica. 2001;D57:1474–1479. doi: 10.1107/s0907444901012458. [DOI] [PubMed] [Google Scholar]
- 53.Collaborative Computational. Project N. The CCP4 suite: programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 54.Laskowski RA, MacArther MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Applied Crystall. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded survival curves of E. coli with pBG66 plasmid encoding TEM-1 β-lactamase mutants, showing survival up to an ampicillin concentration of 950 µg/mL. A. Colony forming units (cfu) on agar plates containing increasing concentrations of ampicillin for E. coli containing R244A, M182T, L201P single and double mutants. B. Cfu for E. coli containing L76N, M182T, L201P single and double mutants. C. Cfu for E. coli containing I47Y:E48C, M182T, L201P mutant combinations. D. Cfu for E. coli containing H158S:V159S:T160H, M182T, L201P mutant combinations.