Abstract
Background/Aims
We previously reported that hepatitis C virus (HCV) core protein up regulated transcription of apolipoprotein C-IV (ApoC-IV, 10.7-fold increase), a member of the apolipoprotein family implicated in liver steatosis. Here, we identified host transcription factors regulating the ApoC-IV gene expression.
Methods
Transcriptional regulators were identified by DNA affinity purification and steatosis was detected by oil red staining and triglyceride assay.
Results
We defined a 163-bp ApoC-IV promoter as a core protein responsive element, and identified Ku antigen complex (Ku70 and Ku80) as well as nuclear receptors PPARγ/RXRα as key regulators of ApoC-IV gene expression. Both Ku70 overexpression and PPARγ agonist significantly increased ApoC-IV promoter activity; conversely, Ku70 silencing or mutation of PPARγ binding site diminished the ApoC-IV promoter activity. Interestingly, transient transfection of ApoC-IV cDNA into a human hepatoma cell line was able to trigger moderate lipid accumulation. In agreement with this in vitro study, ApoC-IV transcript level was increased in HCV infected livers which correlated with triglyceride accumulation.
Conclusions
ApoC-IV overexpression may perturb lipid metabolism leading to lipid accumulation. HCV core protein may modulate ApoC-IV expression through Ku antigen and PPARγ/RXRα complex.
Keywords: Apolipoprotein, gene regulation, liver steatosis, viral infection
Introduction
Liver steatosis is defined by cytoplasmic accumulation of lipid droplets. It is triggered by a variety of host and environmental factors including obesity, diabetes, and alcohol consumption. In addition, chronic infection with hepatitis C virus (HCV) is also associated with increased prevalence of liver steatosis from 20–30% found in the general population to 55% [1, 2]. It has been reported that sustained response to antiviral therapy led to disappearance of fat from the liver, whereas a relapse of HCV infection correlated with recurrence of steatosis [1, 3], thus highlighting the role of virus replication in the induction of steatosis. The HCV expresses four structural proteins (core, E1, E2, p7) and six non-structural proteins (NS2, NS3, NS4A, 4B, NS5A and 5B) [4]. Steatosis can be induced experimentally in the mouse model by isolated expression of HCV structural proteins [5], or just the core protein [6].
As a major component of the HCV nucleocapsid, the core protein is required for viral genome packaging and virion formation. Interestingly, it is localized to triglyceride-rich lipid droplets in cytoplasm whether in transfected cell lines or in hepatocytes of HCV-infected chimpanzees [7, 8]. The core protein has been reported to physically interact with apolipoprotein AII [9] and apolipoprotein B containing lipoproteins [10], both implicated in the storage of triglyceride inside hepatocytes. Indeed, transgenic mice expressing core protein were impaired in the function of microsomal triglyceride transfer protein and secretion of very low density lipoprotein (VLDL); they also manifested intrahepatic triglyceride accumulation [11]. Similarly, transfection of the core protein coding sequence into the Huh-7 hepatoma cell line led to intracellular triglyceride accumulation [12].
Recently, we performed microarray analysis of Huh-7 cells transiently transfected with HCV core gene, and observed transcriptional up regulation of a significant number of genes responsible for fat/lipid metabolism [13]. Of particular interest was a marked up regulation of ApoC-IV, a member of apolipoprotein family that is frequently overexpressed in steatotic livers [14] and HCV-associated HCC samples [15]. In the present study, we identified host transcription factors regulating the ApoC-IV gene expression, evaluated effect of HCV core protein on ApoC-IV promoter activity, and measured ApoC-IV transcripts in HCV infected liver samples. We demonstrate that ApoC-IV was regulated by Ku antigen/PPARγ and its overexpression could increase lipid accumulation in Huh-7 cells.
Materials and methods
ApoC-IV promoter and expression constructs
A 1.7-kb DNA fragment upstream of the ApoC-IV coding sequence was amplified from genomic DNA of Huh-7 cells using Expand High Fidelity PCR System (Roche Diagnostics, Indianapolis, IN), and cloned to pGL3-Basic luciferase reporter plasmid (Promega, Madison, WI). From this construct, a panel of 5’ deletion mutants and a site-directed mutant of PPARγ/RXRα binding site was generated by PCR. The 384-bp ApoC-IV coding sequence and 1830-bp Ku70 coding region were amplified by RT-PCR from total RNA extracted from Huh-7 cells and cloned into pcDNA3.1/Zeo−/+ vector (Invitrogen, Carlsbad, CA). All constructs were verified by DNA sequencing.
Cell culture and transfection
Culture of Huh-7 and Huh-7.5 cells and transfection of HCV replicons have been described [13]. Huh-7 Tet-on cells, which harbor HCV core coding sequence of 1b genotype under the transcriptional control of tetracycline regulatory elements (Li et al, unpublished), were cultured in tetracycline-free medium. Core protein expression was induced by 1 µg/ml of doxycycline (Sigma, Saint Louis, Missouri). A reporter plasmid for the secreted alkaline phosphatase (SEAP) was co-transfected to evaluate the transfection efficiency. Polyamine (Mirus, Madison, WI) was used as a transfection reagent for DNA constructs [13], and lipofectamine 2000 for siRNA transfection (Invitrogen, Carlsbad, CA). Differences among the groups were examined by student’s t test.
Electrophoretic mobility shift assay (EMSA) and chromatin immunoprecipitation (ChIP)
EMSA was performed according to User Manual (ProteinOne, Bethesda, MD and Promega, Madison, WI). Chromatin was immunoprecipitated with antibodies against Ku70, Ku80 or irrelevant IgG (Active Motif, Carlsbad, CA). Presence of ApoC-IV promoter sequence in the purified DNA was verified by PCR.
Western blot and IP-Western blot analyses
Experiments were performed using standard procedures or as recommended by manufacturers. Antibodies: Ku70 and Ku80 (NeoMarkers, Fremont, CA), RNA polymerase II (Active Motif, Carlsbad, CA), actin (Sigma, Saint Louis, Missouri), PPARα, β, γ and histone-1 (Santa Cruz Biotechnology, Santa Cruz, CA) respectively.
Detection of ApoC-IV mRNA and triglyceride in liver samples
Liver tissues were obtained from the Tissue Bank at University of Minnesota. Total RNA was extracted (Invitrogen, Carlsbad, CA) followed by reverse transcription (Roche Diagnostics, Indianapolis, IN) and real time PCR detection of ApoC-IV mRNA (Qiagen, Valencia, CA) using primers listed in Table 1. The ApoC-IV mRNA in transfected Huh-7 cells was detected by RT-PCR (see Table 1 for primers). Triglyceride was detected by Oil Red O staining and quantified by enzymatic assay which measures the concentration of glycerol release after hydrolysis of the triglyceride (Zen-Bio, Inc., Research Triangle Park, NC).
Table 1.
Primers used for PCR amplification
| Gene | Sense | Anti-sense |
|---|---|---|
| Real time RT-PCR | ||
| ApoC-IV | CCCCCACCAAAGCTAAAGAT | GTTCACCACTGTCTCCAGCA |
| 18S | CCGCAGCTAGGAATAATGGA | CCCTCTTAATCATGGCCTCA |
| RT-PCR | ||
| ApoC-IV | AGAGGCCCAGGAAGGAACCCTG | GCTGTCTTTGGATTCGAGGAACCA |
| GAPDH | ATCATCAGCAATGCCTCCTGC | GCCATCACGCCACAGTTTCCCG |
| ChIP | ||
| ApoC-IV | ATGTACTGTGCCTCCCACTTTATG | CCTGTTTCTGAGGAGGGACATTTC |
| GAPDH | TACTAGCGGTTTTACGGGCG | TCGAACAGGAGGAGCAGAGAGCGA |
Results
Identification of a core protein responsive element in the ApoC-IV promoter
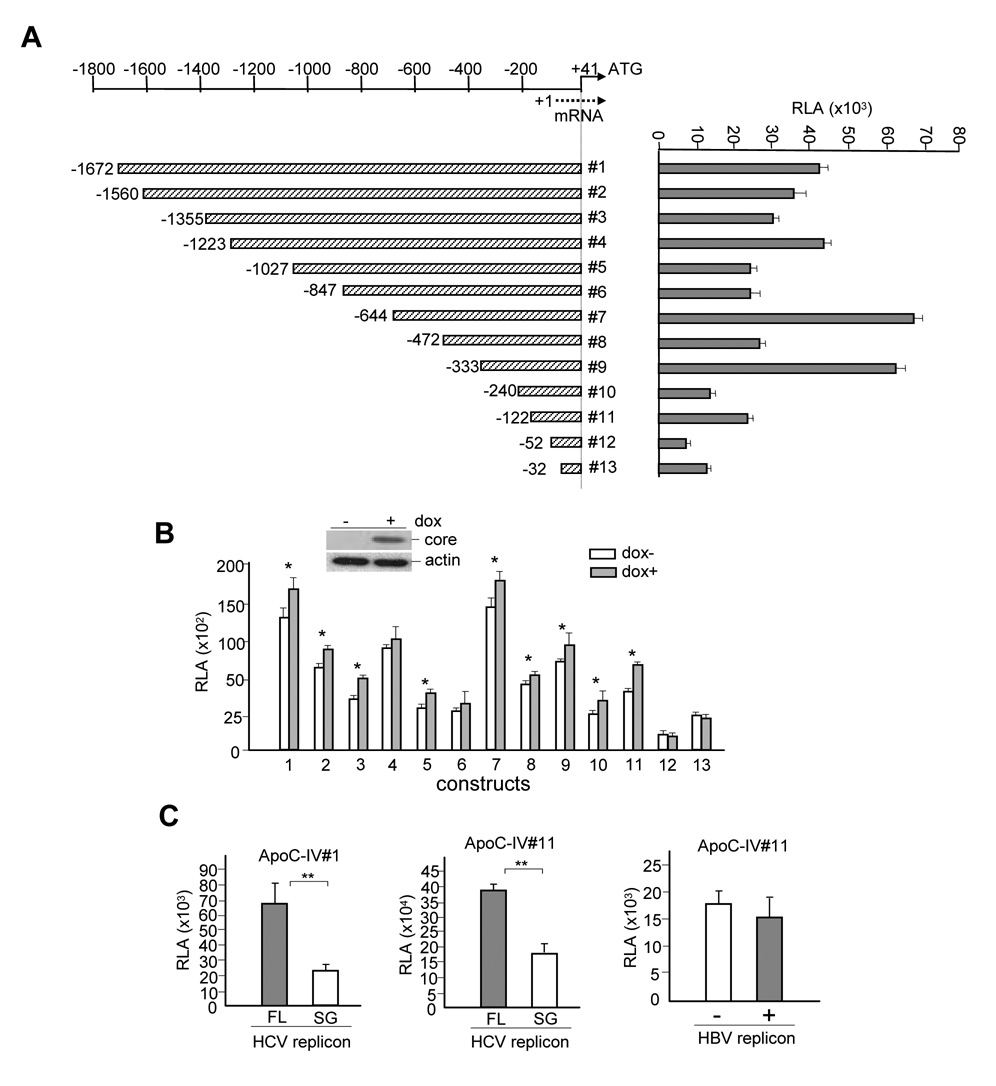
We cloned a 1.7-kb genomic sequence upstream of ApoC-IV gene to examine the molecular mechanism by which HCV core protein up regulates ApoC-IV gene transcription. The promoter was inserted to a luciferase reporter plasmid so as to measure the promoter activity by luciferase assay. Progressive shortening of the 5’ end led to fluctuation of luciferase expression (Fig. 1A) consistent with the presence of multiple positive elements (nt −1672 to −1355, −1223 to −1027, −644 to −472, and −333 to −240, −122 to −52) and negative regulatory sequences (nt −1355 to −1223, −847 to −644, −472 to −333, and −242 to −122). The impact of HCV core protein was determined by transfection of the reporter constructs to Huh-7 Tet-on cells with inducible core protein expression (1b genotype) (Fig. 1B). With the exception of constructs #12 and #13, core protein expression increased luciferase expression from all the promoter constructs (the difference did not reach statistical significance for constructs 4 and 6), suggesting its action through the proximal region of the ApoC-IV promoter.
FIG. 1. ApoC-IV promoter activity and impact of core protein expression.
(A) Schemes of human ApoC-IV promoter constructs (left panel) and relative luciferase activities (RLA) of transfected Huh-7 cells (right panel). Nucleotide positions are relative to the transcription initiation site. (B) Effect of core protein expression on luciferase activities. Transfected Huh-7 Tet-on cells were treated with doxycycline (dox) to induce core protein expression. (C) Effect of HCV and HBV replication on ApoC-IV promoter activity. Left and middle panels: Huh-7.5 cells electroporated with HCV replicons. The cells were further transfected with the ApoC-IV promoter construct #1 or #11, followed by luciferase assay. FL: full-length; SG: subgenomic replicon. Right panel: Huh-7 cells co-transfected with HBV and ApoC-IV promoter construct #11. Luciferase activity was measured at day 5 post-transfection (similar results were obtained at day 3). The replication defective mutant (−) contains a frameshift mutation in the core gene. For all the panels, relative luciferase activities are presented as mean ± SD (n=4) and adjusted by transfection efficiency according to SEAP assay. * P< 0.05, ** P < 0.005.
We further examined the ApoC-IV promoter activity in the context of HCV replication. Huh-7.5 cells were electroporated with the full-length replicon of the 1b genotype or the subgenomic replicon that does not express the structural proteins[13, 16]. Indeed, the luciferase expression from both 1.7-kb (#1) and 163-bp (#11) promoter constructs tested were much higher in association with the full-length HCV genome than with the subgenomic fragment of 1b genotype (Fig. 1C, left and middle panels). In addition, the fold increase of the promoter activity observed in the context of the full-length HCV replication (Fig. 1C) was much higher than those in cells expressing core protein alone (Fig. 1B), which was consistent with the higher expression level of the core protein in replicon transfected cells (data not shown). Similar results were also observed in Huh-7.5 cells infected with HCV of 2a genotype (data not shown). In contrast, replication of hepatitis B virus, another hepatotropic virus that does not induce steatosis, failed to stimulate the ApoC-IV promoter activity (Fig. 1C, right panel). These results are consistent with a role of the core protein in stimulating ApoC-IV promoter activity.
Core protein expression enhanced binding of Ku antigen to the 163-bp promoter fragment
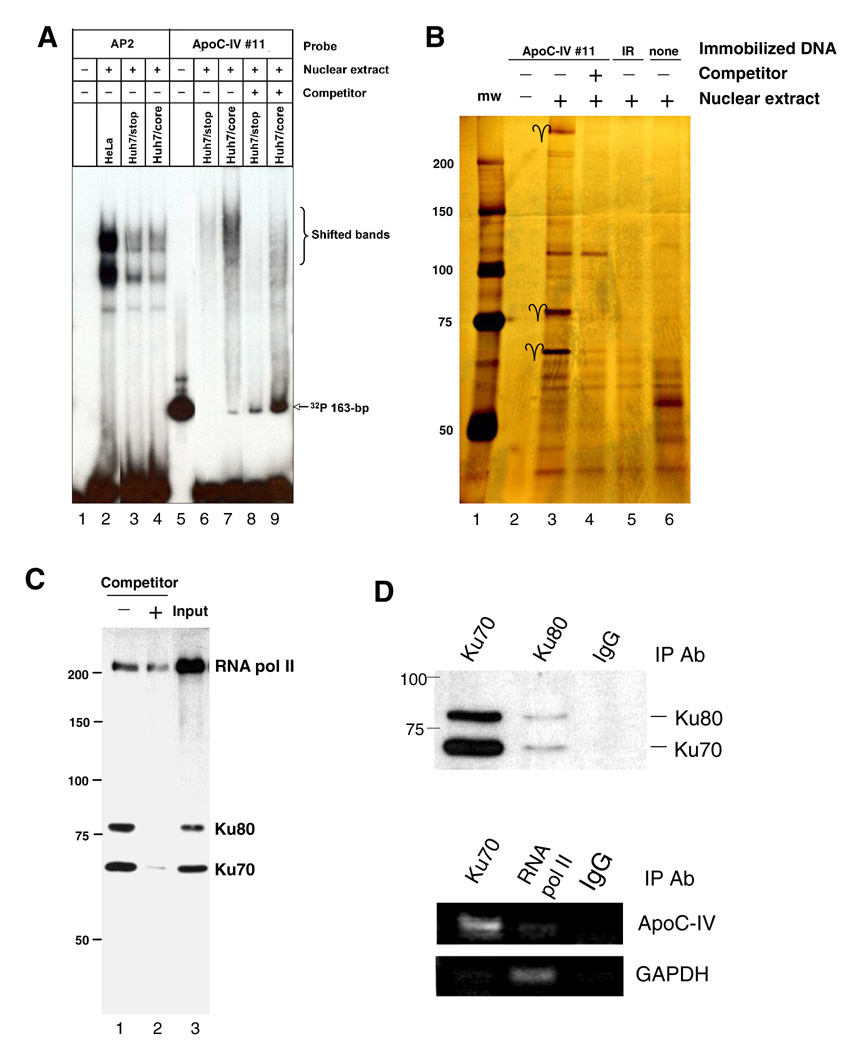
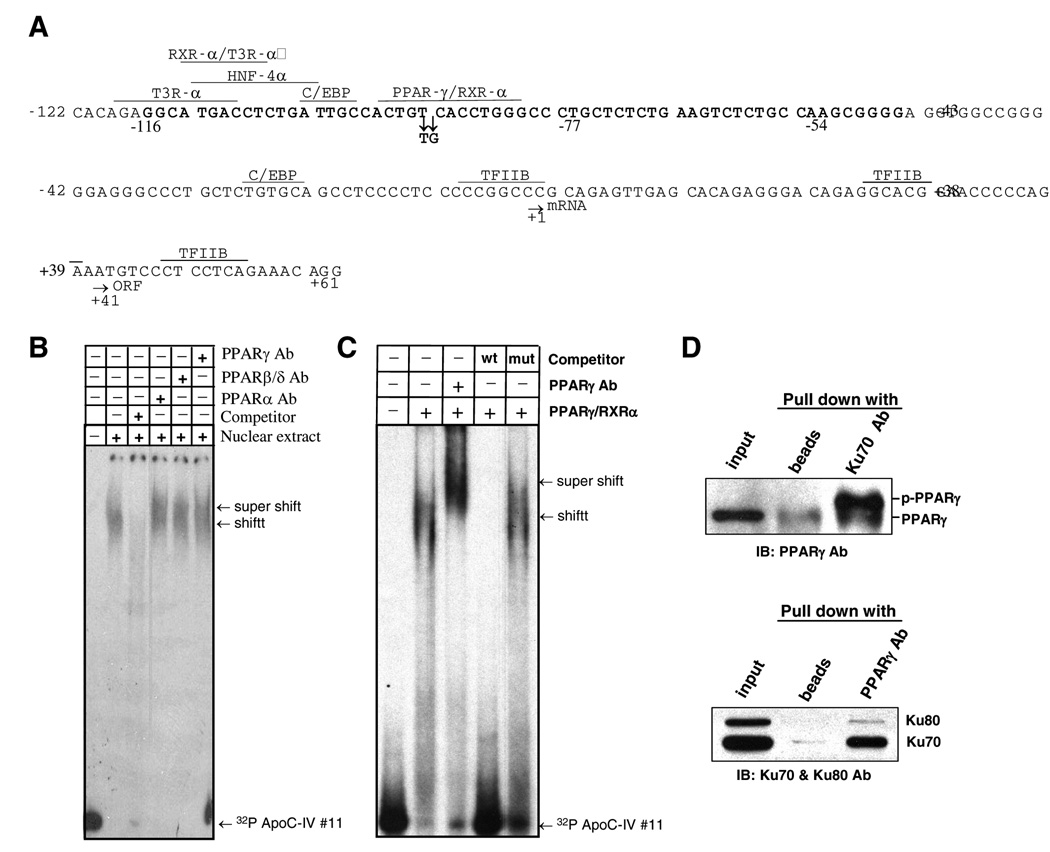
Considering that construct #11 containing a 163-bp sequence was the minimum ApoC-IV promoter that could be activated by HCV core protein, we employed this construct for most of the subsequent experiments. EMSA revealed binding of nuclear proteins, with stronger signals obtained from core expressing cells than from cells transfected with its null mutant (Fig. 2A, compare lanes 7 and 6). Pre-incubation with excess amount of cold 163-bp DNA reduced shifted bands (lanes 9 and 8). To identify the nuclear proteins associated with the proximal ApoC-IV promoter, the promoter was biotinylated and immobilized to agarose beads. SDS-PAGE and silver staining of retained proteins revealed three prominent proteins of 70-kDa, 80-kDa, and > 200-kDa, which were markedly reduced by an excess of unbiotinylated 163-bp promoter fragment (Fig. 2B). Peptide sequencing revealed the 70-kDa species as Ku70, a protein involved in regulation of DNA replication and transcription (Table 2). Interestingly, Ku70 is known to dimerize with Ku80 to form Ku antigen, the regulatory subunit of a 460-kDa DNA-dependent protein kinase. Thus, the 80-kDa molecule could be Ku80, which was confirmed by Western blot analysis (Fig. 2C). Detection of RNA polymerase II in this complex reinforces the 163-bp fragment as a functional promoter engaged in mRNA transcription.
FIG. 2. Interaction of Ku antigen with the 163-bp ApoC-IV promoter (#11).
(A) DNA/protein complex revealed by gel shift assay. The [γ-32P]ATP-labeled 163-bp DNA was incubated with the nuclear extracts from Hela cells, or Huh-7 cells transfected with core gene or its null mutant (Huh7/stop). The [γ-32P]ATP -labeled DNA bearing AP2 binding site was assayed in parallel. (B) Nuclear proteins retained to the immobilized ApoC-IV promoter as revealed by SDS-PAGE and silver staining. Arrows indicate the 70-kDa, 80-kDa, and > 200-kDa proteins. Control binding experiments were performed on empty beads (none) or with an irrelevant oligonucleotide (IR). (C) Western blot to confirm the retained 70-kDa and 80-kDa proteins as Ku70 and Ku80. The blot was incubated with a mixture of antibodies against Ku70, Ku80 and RNA pol II. Input: crude nuclear extracts. For all the three panels, the soluble 163-bp ApoC-IV promoter DNA was used as a competitor. (D) ChIP assay to validate in vivo association of Ku antigen with ApoC-IV promoter. Chromatin was immunoprecipitated with antibodies against Ku70, Ku80, or irrelevant IgG. Presence of Ku antigen in the precipitate was determined by Western blot analysis using anti-ku70 & ku80 antibodies (upper panel), while presence of ApoC-IV and GAPDH promoters was determined by PCR (lower panel).
Table 2.
Peptide Sequences of Ku70
| Sequence | Location in Ku70 |
|---|---|
| DSLIFLVDASK | 36–46 |
| ILELDQFK | 116–123 |
| DIISIAEDEDLR | 219–230 |
The nuclear extracts of Huh-7 cells were preincubated with avidin-agarose beads (Pierce, Rockford, IL), followed by incubation with the biotinylated163-bp ApoC-IV promoter immobilized on avidin-agarose beads. Bound proteins were separated in 0.1% SDS-8% PAGE and revealed by silver staining (Invitrogen, Carlsbad, CA). The protein bands were excised and sequenced at the Proteomic Core Facility of the Rhode Island Hospital.
We next performed ChIP assay to confirm Ku antigen interaction with ApoC-IV promoter as part of chromosomal DNA. DNA was cross-linked to bound nuclear proteins and sheared to fragments of ~500 bp prior to immunoprecipitation. Western blot analysis revealed precipitation of both Ku70 and Ku80 by either Ku70 or Ku80 antibody, but not by irrelevant IgG (Fig. 2D, upper panel). PCR amplification confirmed co-precipitation of ApoC-IV promoter sequence (nt −300 to −21) by the Ku70 antibody, but not the control IgG (Fig. 2D, middle panel). Consistent with active transcription, the ApoC-IV promoter could also be precipitated with an antibody to RNA polymerase II, albeit at reduced efficiency. The GAPDH promoter was immunoprecipitated by an antibody against RNA polymerase II, but not by Ku70 antibody (Fig. 2D, lower panel).
Ku antigen expression was critical for ApoC-IV gene transcription
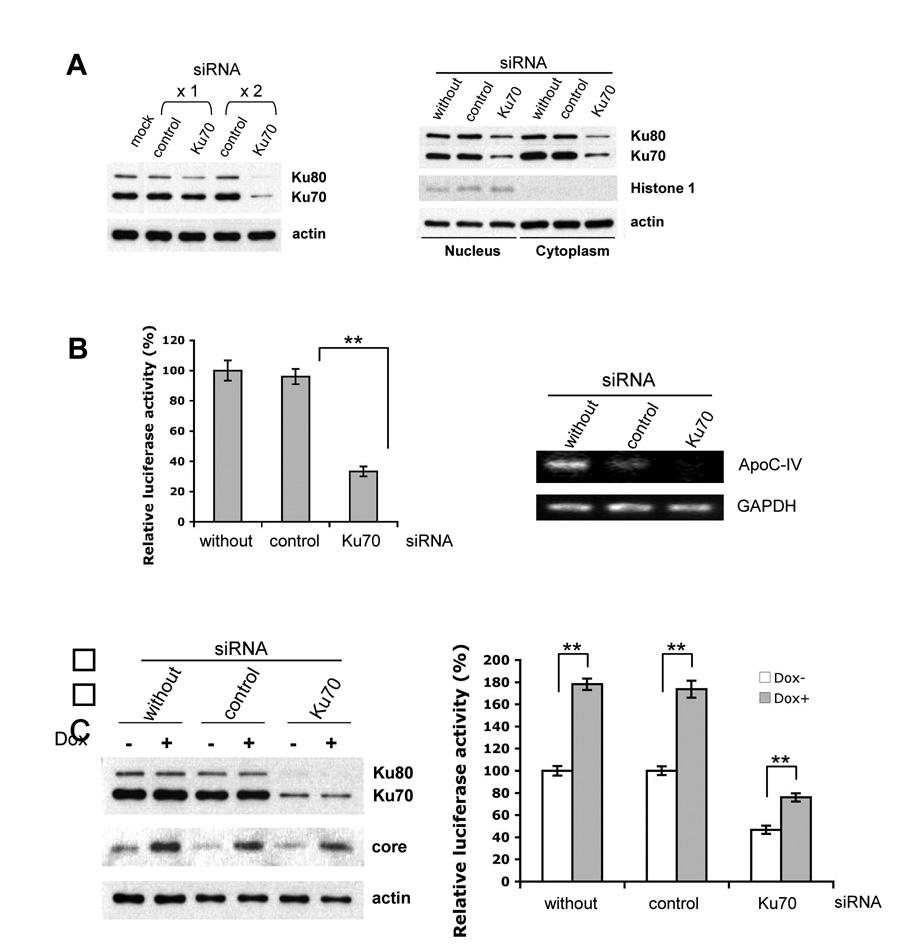
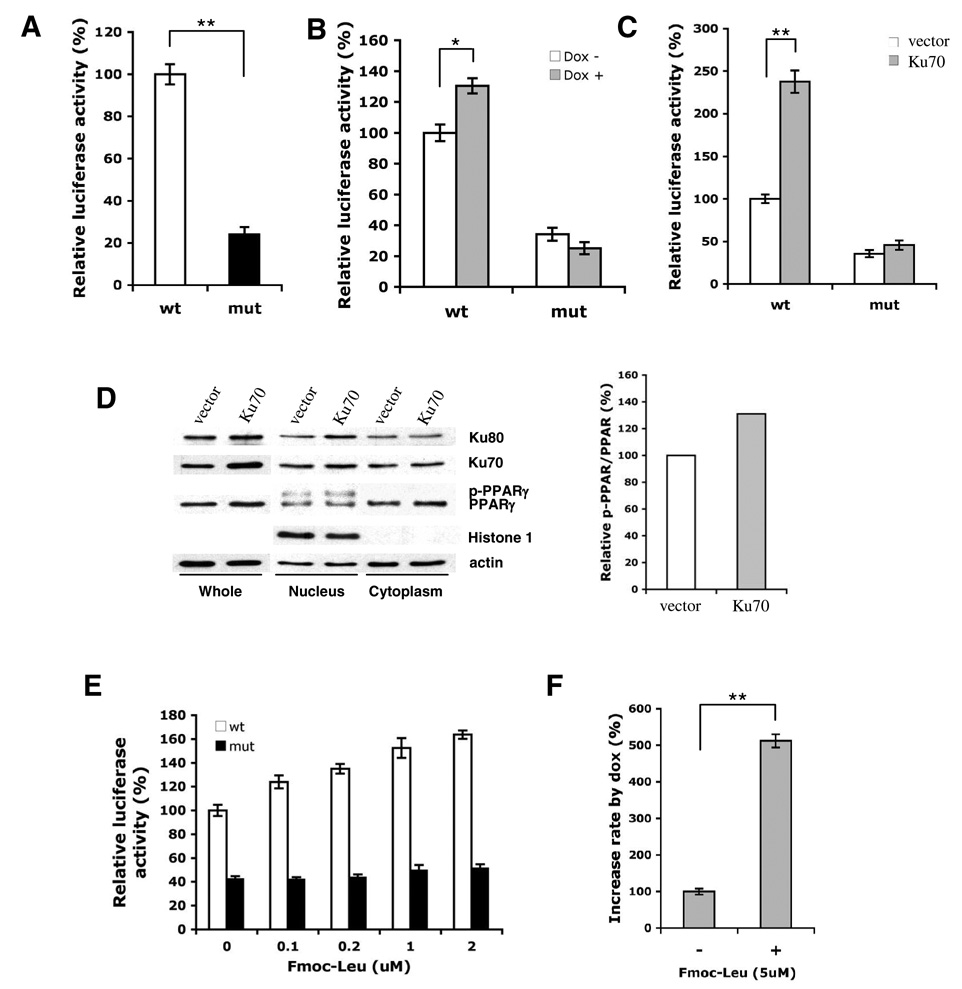
The functional significance of Ku antigen in association with the ApoC-IV promoter was evaluated by gene silencing and overexpression. Twice transfection of Ku70 siRNA (30 nM) reduced Ku70 expression by >70%, while the control siRNA targeting duck carboxypeptidase D [17] was ineffective (Fig. 3A, left panel). Both cytoplasmic and nuclear pools of Ku70 were reduced (Fig. 3A, right panel). Consistent with the role of Ku70 in Ku80 stabilization, silencing of Ku70 also reduced Ku80 level. Importantly, the ApoC-IV promoter activity was reduced by 60% in Ku70 silenced cells in comparison to cells transfected with irrelevant siRNA (Fig. 3B, left panel). Similarly, level of the endogenous ApoC-IV mRNA, but not GAPDH mRNA, was reduced as a consequence of Ku70 silencing (Fig. 3B, right panel). In Huh-7 Tet-on cells, induction of core protein expression by Dox increased ApoC-IV promoter activity, while knockdown of Ku70 expression had the opposite effect (Fig. 3C). Although the promoter activity continued to increase after induction of the core protein expression, the fold increase was much reduced. Apparently, the residual Ku70 protein could still respond to core protein and stimulate ApoC-IV transcription.
FIG. 3. Impact of Ku70 silencing on ApoC-IV transcription and promoter activity.
(A) Western blot analysis of Ku70 and Ku80 in Huh-7 cell lysate. Left panel: total cell lysate following once (x1) or twice (x2) transfection with siRNA. Right panel: nuclear and cytoplasmic fractions after twice transection with siRNA. The control siRNA targeted duck carboxypeptidase D. Actin served as a marker for loading control, while Histone 1 was a marker of nuclear proteins. (B) luciferase expression from the 163-bp ApoC-IV promoter (left) and endogenous ApoC-IV and GAPDH mRNA (right) from Huh-7 cells twice transfected with the siRNA. (C) Huh-7 Tet-on cells transfected twice with the Ku70 or control siRNA, with or without doxycycline (Dox) induction of core protein expression. Left panel: Western blot analysis. Right panel: luciferase expression from the 163-bp ApoC-IV promoter. ** P < 0.005.
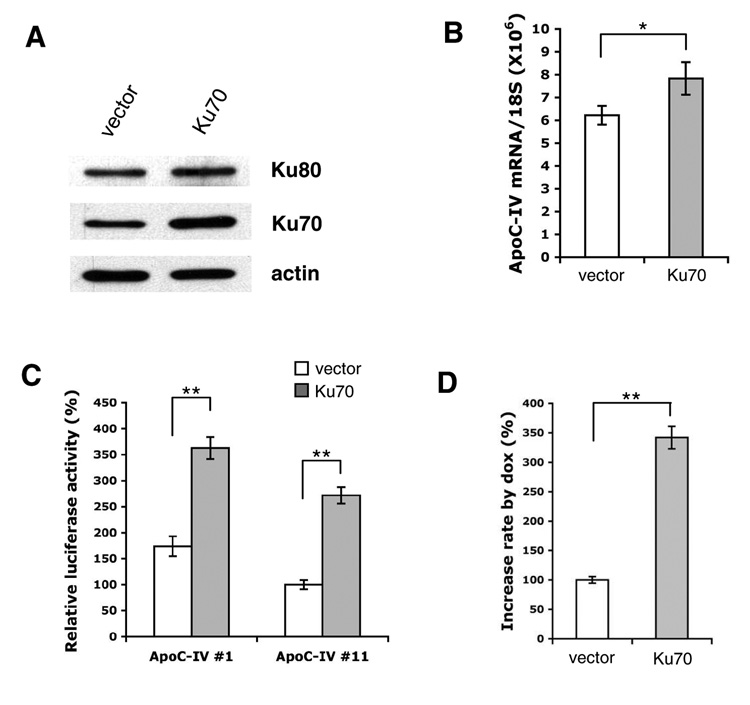
In a complementary approach, transfection of Ku70 cDNA to Huh-7 cells moderately increased endogenous ApoC-IV mRNA and markedly stimulated luciferase expression from the 1.7-kb or 163-bp promoter construct (Fig. 4A–C). In Huh-7 Tet-on cells, overexpression of Ku70 greatly induced ApoC-IV promoter activity in the context of core protein expression (> 3-fold) (Fig. 4D). Taken together with data from Ku70 knockdown, these findings implicate Ku70 as an important mediator of transcriptional activation by core protein.
FIG. 4. Effect of Ku70 overexpression on ApoC-IV transcription and promoter activity in Huh-7 cells (A–C) and Huh-7 Tet-on cells (D).
Cells were transfected with either pcDNA3.1 vector or Ku70 cloned to this vector (Ku70). (A) Western blot analysis of Ku70, Ku80, and actin. (B) RT-PCR detection of endogenous ApoC-IV mRNA. Data are presented as mean ± SD (n=3). * P<0.05. (C) Luciferase expression from the 1.7-kb ApoC-IV promoter (#1) or 163-bp promoter (#11). (D) Luciferase expression from the 163-bp ApoC-IV promoter in the absence or presence of doxycycline. Data are presented as fold increase (Dox+ vs. Dox−) (mean ± SD, n=4). ** P<0.005.
Ku70 formed a complex with nuclear hormone receptors PPARγ and RXRα
Using TRANSFAC [18, 19] and Genomatix database search, we identified other transcription factors/regulators that could potentially bind to the 163-bp ApoC-IV promoter, including PPAR/RXR-α, T3R-α, HNF-4, and C/EBP (Fig. 5A). These factors are important for transcriptional regulation of lipoproteins [20] and their binding sites at −120 to −80 of ApoC-IV promoter overlaps with the tentative Ku antigen binding site (our unpublished observation). EMSA revealed interaction of the 163-bp ApoC-IV promoter with all three members of PPAR family (α, β/δ, and γ) present in the nuclear extracts of Huh-7 cells (Fig. 5B), or as purified proteins (ProteinOne PPAR Triad kit, data not shown). Since PPARγ has been reported to contribute to hepatic steatosis and triglyceride clearance [21], experiments were performed to examine its relevance in ApoC-IV promoter regulation. We found that binding of PPARγ to the ApoC-IV promoter was inhibited by an excess of unlabeled promoter DNA but not by a mutated form with two nucleotide substitutions in the consensus sequence of the PPARγ/RXRα binding site (Fig. 5C, two right lanes). Moreover, immunoprecipitation experiments using lysate of Huh-7 cells transfected with Ku70 expression construct revealed that PPARγ, especially the phosphorylated form involved in transcription [22–24], were efficiently precipitated with the Ku70 antibody (Fig. 5D, upper panel); reciprocal precipitation with an anti-PPARγ antibody pulled down both Ku70 and Ku80 (Fig. 5D, lower panel).
FIG. 5. PPARγ/RXRα association with the 163-bp ApoC-IV promoter and complex formation with Ku70.
(A) Predicted transcription factor binding sites on the 163-bp promoter by TRANSFAC and Genomatix database search. The start sites for transcription (mRNA) and translation (ORF) are indicated by arrows, as well as the two point mutations introduced into the predicted PPARγ/RXRα binding site. Sequence in boldface (−116 to −54) is required for Ku antigen binding. (B) Binding of PPARα, β/δ, and γ to the 163-bp ApoC-IV promoter as indicated by supershift assay using specific antibodies. (C) Gel shift assay to demonstrate binding of purified PPARγ/RXRα to ApoC-IV promoter. Note that PPARγ antibody supershifted DNA band. (C) IP-Western blot analysis to demonstrate association of Ku70 with phosphorylated PPARγ (p-PPARγ). Huh-7 cells were transfected with Ku70 expression plasmid. Cell lysate was immunoprecipitated with Ku70 antibody followed by Western blot detection of PPARγ (upper panel), or immunoprecipitated with PPARγ antibody followed by Western blot analysis of Ku70 and Ku80 (lower panel).
Activation of ApoC-IV promoter by HCV core protein and Ku antigen was mediated by PPARγ/RXRα
Experiments were conducted to determine whether PPARγ/RXRα binding to the ApoC-IV promoter was required for transcription, and whether the core protein activated ApoC-IV promoter through Ku antigen, which in turn relied on complex formation with PPARγ/RXRα. We found that double nucleotide substitution in the core sequence of the PPARγ/RXRα binding site (from ACCT to ATGT, see Fig 5A) markedly impaired ApoC-IV promoter activity (Fig. 6A), and abrogated stimulatory effect of the core protein (Fig. 6B). Moreover, the mutated promoter was no longer activated by Ku70 overexpression (Fig. 6C). Interestingly, overexpression of Ku70 slightly increased PPARγ protein level in the cytoplasm and the phosphorylated form in the nucleus, the active form involved in transcription (Fig. 5D) [22–24]. In addition, application of Fmoc-Leu, a PPARγ activator, increased the ApoC-IV promoter activity in a dose-dependent manner (Fig. 6E). In the presence of Fmoc-Leu, core protein expression increased ApoC-IV promoter activity by 5-fold (Fig. 6F). Thus, PPARγ/RXRα served as a critical transcription factor(s) in ApoC-IV gene expression. It mediated transcriptional up regulation by HCV core protein and Ku antigen.
FIG. 6. Requirement of PPARγ/RXRα for up regulation of ApoC-IV promoter activity by core protein and Ku antigen.
(A) Effect of mutating PPARγ/RXRα binding site on ApoC-IV promoter activity. Huh-7 cells were transfected with #11 ApoC-IV promoter constructs containing the wild-type (wt) sequence or mutated (mut) PPARγ/RXRα binding motif, and luciferase activity was measured two days later. (B) HCV core protein activated ApoC-IV promoter containing the wild-type but not mutated PPARγ/RXRα consensus sequence. Core protein was induced in Huh-7 Tet-on cells by doxycycline. (C) Overexpression of Ku70 activated wild-type but not mutated ApoC-IV promoter. (D) Overexpression of Ku70 increased phosphorylated PPARγ in the nucleus. The relative intensity of the bands of phosphorylated vs. nonphosphorylated PPARγ is quantified in the right panel using KODAK ID image software. (E) Activation of ApoC-IV transcription by a PPARγ agonist. Huh-7 cell were transfected with ApoC-IV promoter containing wild type or mutated PPARγ/RXRα binding. site, and Fmoc-Leu was added in various concentrations followed by luciferase assay two days later. (F) Fold change of ApoC-IV promoter activity in core protein induced (+) vs. non-induced (−) Huh-7 Tet-on cells in the presence of 5 µM Fmoc-Leu. For all experiments, luciferase activity is presented as mean ± SD (n=4). ** P<0.005.
Overexpression of ApoC-IV protein in cell culture led to triglyceride accumulation
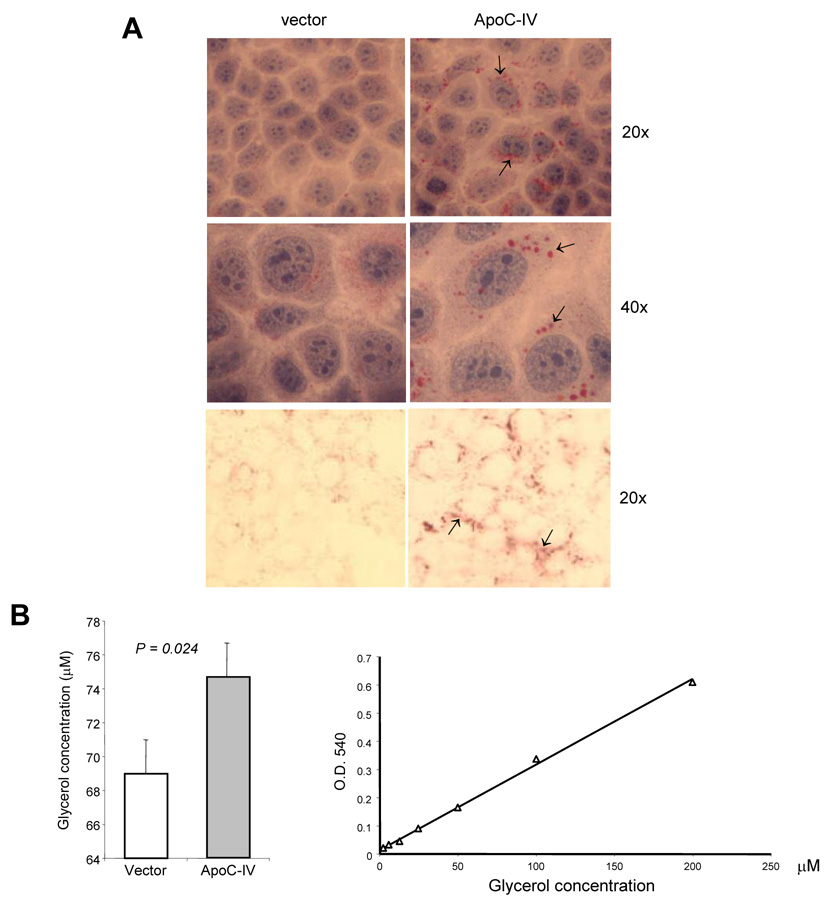
In order to test the role of ApoC-IV protein in lipid accumulation, we cloned its cDNA from Huh-7 cells. Following a transient transfection of the ApoC-IV cDNA, Huh-7 cells were subsequently cultured in serum-free medium or medium containing delipidated FBS to reduce possible influence of lipids from culture medium. With serum-free medium, cells transfected with the ApoC-IV cDNA displayed lipid droplets of granule-like structure in the cytoplasm and perinuclear space, distinct from the weak staining and homogenous distribution observed in cells transfected with the empty vector (Fig. 7A, upper and middle panels). A similar difference was observed in cells cultured with medium containing delipidated FBS (Fig. 7A, lower panel). The enzymatic assay revealed a moderate increase in triglyceride in cells transfected with ApoC-IV gene as compared to mock transfected cells (Fig. 7B, left, p = 0.024). Considering the transfection efficiency of about 40%, the triglyceride content in ApoC-IV transfected cells was approximately 2.7-fold higher than that in vector transfected cells.
FIG. 7. Triglyceride accumulation in transfected Huh-7 cells.
(A) Oil Red O staining of cells transfected with the ApoC-IV cDNA or empty vector. Upper and middle panels: cells cultured in FBS-free medium with additional H&E staining. Lower panel: cells cultured in delipidated medium. Arrows indicate lipid accumulation. (B) Enzymatic assay of triglyceride in transfected cells (left panel) and standard curve (right panel).
Elevation of ApoC-IV transcript in HCV infected livers
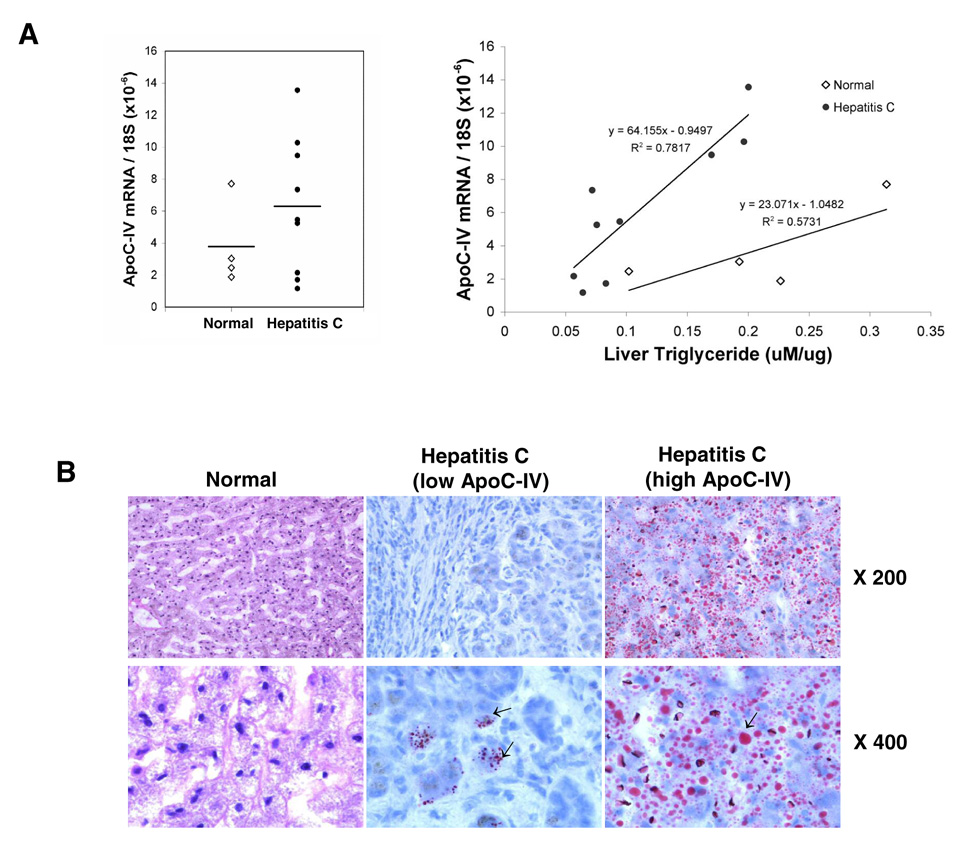
To determine the relevance of ApoC-IV up regulation to lipid accumulation in vivo, we examined ApoC-IV mRNA levels and triglyceride content in nine HCV infected cirrhotic livers and four normal livers. Despite individual variations, the mean ApoC-IV mRNA level was higher in HCV infected livers (6.2±4.2) than in the control group (3.7±2.6) (Fig. 8A, left panel). Importantly, the ApoC-IV transcript levels correlated with the concentration of intracellular triglycerides in the HCV infected livers as revealed by a coefficient of determination (R2 = 0.78) (Fig. 8A, right panel), although no such correlation was found in the normal livers. In the liver tissues with high level of ApoC-IV mRNA, severe steatosis was detected, as indicated by wide intracellular distribution of lipid droplets (Fig. 8B, right panel). In contrast, accumulation of lipid droplets was mild and sporadic in the liver tissues with low level of ApoC-IV mRNA (Fig. 8B, middle panel). Taken together, these results demonstrate that the ApoC-IV gene expression level is highly associated with liver steatosis in vivo in HCV infected livers, and overexpression of this protein alone could trigger triglyceride accumulation (or fusion of lipid droplets) in hepatocytes cultured in vitro.
FIG. 8. Correlation of ApoC-IV mRNA level with triglyceride concentration in HCV infected livers.
Nine HCV infected liver tissues were collected at the time of liver transplantation (age: 50.1±4.2; sex: 8 males, 1 female; HCV viral titer: 2.8×103 to 4.9×105 i.u./ml; genotype: 4 1a, 2 1b, 1 3a and 2 ND). Four normal liver tissues were included as controls (age 24.5±13.8; sex: 1 male, 3 females). (A) ApoC-IV mRNA/18S ribosomal RNA (left panel) and its correlation with triglyceride concentration (right panel). The coefficient of determination (R2) and equation are indicated. Open diamond: normal liver. Dark circle: HCV-infected liver. Horizontal bar: average value. (B) Oil-Red O staining of liver sections. The left, middle, and right panels represent normal liver, HCV infected liver with low and high ApoC-IV mRNA level, respectively. Arrows indicate lipid droplets. Note that the genotype difference in triglyceride accumulation was not observed due to limited samples examined.
Discussion
Hepatitis C virus infection has been recognized as one of the key risk factors for liver steatosis. Accumulating evidence implicates ApoC-IV in the development of liver steatosis. The transcription of ApoC-IV gene has been found up regulated in steatotic liver [14], HCV related HCC samples [15], and human liver cell line expressing HCV core protein [13]. In the present study, we found correlation of ApoC-IV expression with the extent of steatosis in HCV infected liver. We also demonstrated that ApoC-IV overexpression in Huh-7 cells triggered moderate triglyceride accumulation.
The transcriptional control of the human ApoC-IV has not been investigated before. In the present study, we identified both positive and negative elements in the ApoC-IV promoter and established a 163-bp fragment as the minimum promoter. This region also contains a responsive element to HCV core protein. Through EMSA and ChIP technique, we identified Ku antigen (Ku70 and Ku80) as nuclear proteins associated with the minimum ApoC-IV promoter. Ku antigen regulates transcription of a variety of viral and cellular genes such as murine mammary tumor virus, heat shock protein 70, glycophorin B, and NF-κB1/p50 in a positive or negative manner. We found silencing of Ku70 diminished ApoC-IV transcription, whereas transfection of Ku70 cDNA enhanced ApoC-IV transcription. Thus, the Ku antigen is a positive transcriptional regulator of ApoC-IV gene expression.
Transcriptional regulation of apolipoproteins involves both liver-specific transcription factors such as hepatocyte nuclear factors (HNF-1, 3, 4) and hormone nuclear receptors (PPAR/RXRα) [20]. PPARγ plays an essential role in adipogenesis, glucose homeostasis, lipid metabolism as well as development of hepatic steatosis. We found all three members of PPAR in the presence of RXRα could directly bind to the minimum ApoC-IV promoter. Two lines of evidence indicate critical role of PPARγ/RXRα complex in transcriptional activation of the ApoC-IV promoter. First, mutating the PPARγ/RXRα binding site in the minimum ApoC-IV promoter impaired reporter gene expression. Secondly, a PPARγ agonist increased the ApoC-IV promoter activity and augmented core protein’s ability to induce ApoC-IV promoter. Importantly, ApoC-IV promoter with mutated PPARγ/RXRα site was no longer activated by Ku70 overexpression, suggesting the PPARγ/RXRα complex works downstream of Ku antigen. In this regard, phosphorylated PPARγ, the nuclear PPARγ involved in transcription [22–24], could interact with Ku70. Ku70 overexpression increased total PPARγ level and phosphorylated PPARγ in the nucleus (Fig. 6D). It is likely that Ku70 binding to PPARγ leads to its phosphorylation via the DNA-dependent protein kinase associated with the Ku antigen, which has been found to phosphorylate several transcription factors such as p53, c-fos, and Ap1 [25]. Based on our current findings and work reported by others, a working model of ApoC-IV gene regulation by Ku antigen/PPARγ complex was proposed (see Fig. 9) [26, 27].
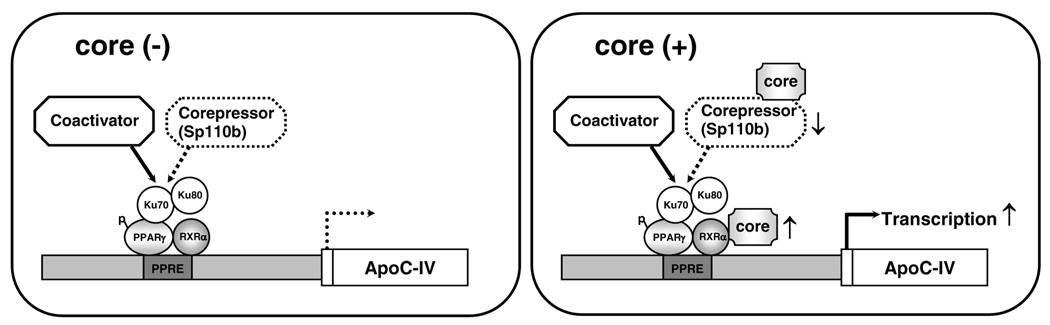
FIG. 9. Proposed model of transcriptional activation of the ApoC-IV gene by the HCV core protein.
In the absence of HCV infection, the ApoC-IV gene is tightly controlled by specific transcription factors/regulators. The heterodimer PPARγ/RXRα binds to the proximal promoter via PPRE site where it recruits other transcriptional regulators including Ku70/Ku80 and coactivator(s), corepressor (e.g. Sp110b). In the presence of HCV infection, viral core protein may enhance Ku70 and PPARγ/RXRα mediated transcription by its direct binding to RXRα in the nucleus thus activating its activity [26]. Alternatively, the core protein can also bind to Sp110b in the cytoplasm to prevent its nuclear translocation and function as a corepressor [27].
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr. Charles Rice, Rockefeller University, New York, for providing replicon constructs and Huh-7.5 cells. This work was supported by grants DK066950, CA109733, CA95490, CA35711, AA08169 and AA20666 from the National Institutes of Health, and RSG 06-059-01-MCB from the American Cancer Society. E. Kim was supported by the Korea Research Foundation Grant from the Korean Government (MOEHRD) (KRF-2005-214-E00052). J. Li was a Liver Scholar of the American Liver Foundation.
Abbreviations
- ApoC-IV
apolipoprotein C-IV
- PPAR
peroxisome proliferators-activated receptor
- RXR
retinoid X receptor
- VLDL
very low density lipoprotein
- SEAP
secreted alkaline phosphatase
- EMSA
electrophoretic mobility shift assay
- ChIP
chromatin immunoprecipitation
- RT-PCR
reverse transcription-polymerase chain reaction
- Dox
doxycycline
- HNF
hepatocyte nuclear factor
- PPRE
PPAR responsive element
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Clark JM, Brancati FL, Diehl AM. Nonalcoholic fatty liver disease. Gastroenterology. 2002;122:1649–1657. doi: 10.1053/gast.2002.33573. [DOI] [PubMed] [Google Scholar]
- 2.Lonardo A, Adinolfi LE, Loria P, Carulli N, Ruggiero G, Day CP. Steatosis and hepatitis C virus: mechanisms and significance for hepatic and extrahepatic disease. Gastroenterology. 2004;126:586–597. doi: 10.1053/j.gastro.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 3.Rubbia-Brandt L, Giostra E, Mentha G, Quadri R, Negro F. Expression of liver steatosis in hepatitis C virus infection and pattern of response to alpha-interferon. J Hepatol. 2001;35:307. doi: 10.1016/s0168-8278(01)00087-3. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager R. Hepatitis C virus molecular clones: from cDNA to infectious virus particles in cell culture. Curr Opin Microbiol. 2006;9:416–422. doi: 10.1016/j.mib.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Lerat H, Honda M, Beard MR, Loesch K, Sun J, Yang Y, et al. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology. 2002;122:352–365. doi: 10.1053/gast.2002.31001. [DOI] [PubMed] [Google Scholar]
- 6.Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, Ishibashi K, Matsuura Y, et al. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J Gen Virol. 1997;78:1527–1531. doi: 10.1099/0022-1317-78-7-1527. [DOI] [PubMed] [Google Scholar]
- 7.Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, et al. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci U S A. 1997;94:1200–1205. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLauchlan J, Lemberg MK, Hope G, Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. Embo J. 2002;21:3980–3988. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabile A, Perlemuter G, Bono F, Kohara K, Demaugre F, Kohara M, et al. Hepatitis C virus core protein binds to apolipoprotein AII and its secretion is modulated by fibrates. Hepatology. 1999;30:1064–1076. doi: 10.1002/hep.510300429. [DOI] [PubMed] [Google Scholar]
- 10.Diaz O, Delers F, Maynard M, Demignot S, Zoulim F, Chambaz J, et al. Preferential association of Hepatitis C virus with apolipoprotein B48-containing lipoproteins. J Gen Virol. 2006;87:2983–2991. doi: 10.1099/vir.0.82033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perlemuter G, Sabile A, Letteron P, Vona G, Topilco A, Chretien Y, et al. Hepatitis C virus core protein inhibits microsomal triglyceride transfer protein activity and very low density lipoprotein secretion: a model of viral-related steatosis. Faseb J. 2002;16:185–194. doi: 10.1096/fj.01-0396com. [DOI] [PubMed] [Google Scholar]
- 12.Abid K, Pazienza V, de Gottardi A, Rubbia-Brandt L, Conne B, Pugnale P, et al. An in vitro model of hepatitis C virus genotype 3a-associated triglycerides accumulation. J Hepatol. 2005;42:744–751. doi: 10.1016/j.jhep.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 13.Fukutomi T, Zhou Y, Kawai S, Eguchi H, Wands JR, Li J. Hepatitis C virus core protein stimulates hepatocyte growth: correlation with upregulation of wnt-1 expression. Hepatology. 2005;41:1096–1105. doi: 10.1002/hep.20668. [DOI] [PubMed] [Google Scholar]
- 14.Chiappini F, Barrier A, Saffroy R, Domart MC, Dagues N, Azoulay D, et al. Exploration of global gene expression in human liver steatosis by high-density oligonucleotide microarray. Lab Invest. 2006;86:154–165. doi: 10.1038/labinvest.3700374. [DOI] [PubMed] [Google Scholar]
- 15.Iizuka N, Oka M, Yamada-Okabe H, Mori N, Tamesa T, Okada T, et al. Comparison of gene expression profiles between hepatitis B virus- and hepatitis C virus-infected hepatocellular carcinoma by oligonucleotide microarray data on the basis of a supervised learning method. Cancer Res. 2002;62:3939–3944. [PubMed] [Google Scholar]
- 16.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong S, Li J, Wands JR. Carboxypeptidase D is an avian hepatitis B virus receptor. J Virol. 1999;73:8696–8702. doi: 10.1128/jvi.73.10.8696-8702.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 20.Zannis VI, Kan HY, Kritis A, Zanni E, Kardassis D. Transcriptional regulation of the human apolipoprotein genes. Front Biosci. 2001;6:D456–D504. doi: 10.2741/zannis. [DOI] [PubMed] [Google Scholar]
- 21.Gavrilova O, Haluzik M, Matsusue K, Cutson JJ, Johnson L, Dietz KR, et al. Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J Biol Chem. 2003;278:34268–34276. doi: 10.1074/jbc.M300043200. [DOI] [PubMed] [Google Scholar]
- 22.Diradourian C, Girard J, Pegorier JP. Phosphorylation of PPARs: from molecular characterization to physiological relevance. Biochimie. 2005;87:33–38. doi: 10.1016/j.biochi.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Lazennec G, Canaple L, Saugy D, Wahli W. Activation of peroxisome proliferator-activated receptors (PPARs) by their ligands and protein kinase A activators. Mol Endocrinol. 2000;14:1962–1975. doi: 10.1210/mend.14.12.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Relic B, Benoit V, Franchimont N, Kaiser MJ, Hauzeur JP, Gillet P, et al. Peroxisome proliferator-activated receptor-gamma1 is dephosphorylated and degraded during BAY 11-7085-induced synovial fibroblast apoptosis. J Biol Chem. 2006;281:22597–22604. doi: 10.1074/jbc.M512807200. [DOI] [PubMed] [Google Scholar]
- 25.Koike M. Dimerization, translocation and localization of Ku70 and Ku80 proteins. J Radiat Res (Tokyo) 2002;43:223–236. doi: 10.1269/jrr.43.223. [DOI] [PubMed] [Google Scholar]
- 26.Tsutsumi T, Suzuki T, Shimoike T, Suzuki R, Moriya K, Shintani Y, et al. Interaction of hepatitis C virus core protein with retinoid X receptor alpha modulates its transcriptional activity. Hepatology. 2002;35:937–946. doi: 10.1053/jhep.2002.32470. [DOI] [PubMed] [Google Scholar]
- 27.Watashi K, Hijikata M, Tagawa A, Doi T, Marusawa H, Shimotohno K. Modulation of retinoid signaling by a cytoplasmic viral protein via sequestration of Sp110b, a potent transcriptional corepressor of retinoic acid receptor, from the nucleus. Mol Cell Biol. 2003;23:7498–7509. doi: 10.1128/MCB.23.21.7498-7509.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.