Abstract
Transposase domain proteins mediate DNA movement from one location in the genome to another in lower organisms. However, in human cells such DNA mobility would be deleterious, and therefore the vast majority of transposase-related sequences in humans are pseudogenes. We recently isolated and characterized a SET and Transposase domain protein termed Metnase that promotes DNA double strand break (DSB) repair by non-homologous end-joining (NHEJ). Both the SET and Transposase domain were required for its NHEJ activity. In this study we found that Metnase interacts with DNA Ligase IV, an important component of the classical NHEJ pathway. We investigated whether Metnase had structural requirements of the free DNA ends for NHEJ repair, and found that Metnase assists in joining all types of free DNA ends equally well. Metnase also prevents long deletions from processing of the free DNA ends, and improves the accuracy of NHEJ. Metnase levels correlate with the speed of disappearance of γ-H2Ax sites after ionizing radiation. However, Metnase has little effect on homologous recombination repair of a single DSB. Altogether, these results fit a model where Metnase plays a role in the fate of free DNA ends during NHEJ repair of DSBs.
1. Introduction
DNA double-strand breaks (DSBs) are produced by ionizing radiation, replication fork collapse at single-strand DNA lesions, and programmed cleavage by immunologic nucleases. Mammalian cells repair DSBs by two mechanisms, homologous recombination (HR) and non-homologous end-joining (NHEJ). HR restores the broken DNA to its original sequence, using homologous sequences on sister chromatids, homologous chromosomes, or repeated elements, as repair templates [1]. It is more active in S and G2 phases of the cell cycle, where replicated sequences are available to serve as repair templates [1]. NHEJ is the dominant DSB repair pathway in mammalian cells, but it tends to be less accurate than HR, because NHEJ often involves end-processing that reveals microhomologies that direct annealing prior to ligation, producing deletions (and sometimes insertions) of nucleotides at the repair site [2]. Large scale deletions result if annealing takes place far from the DSB. A consequence of this end processing is the production of errors in the joined DNA sequence at the DSB. The ability of the damaged cell to minimize errors at the DSB site yet efficiently ligate free ends is therefore critical for maintaining genomic stability [3,4].
In the classical NHEJ pathway, the Mre11-Rad50-Nbs1 (MRN) complex detects DNA DSBs and this complex recruits the ATM kinase to the DSB site [5]. ATM is activated and it phosphorylates targets that slow the cell cycle to allow repair to occur [6]. The Ku70/80 complex binds to the free DNA ends and recruits the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) [7–9], which activates the DNA Ligase IV/XRCC4/XLF complex, promoting synapsis and then ligation of the free DNA ends [10–18]. An alternative NHEJ pathway has been described that shows greater dependence on annealing of microhomologies flanking the DSB. This is sometimes called microhomology-mediated end-joining, and it always deletes one of the two microhomologous regions and intervening DNA. Several studies have identified factors that regulate the choice of NHEJ pathways. Ku70/80, XRCC4, or histone H1 promote accuracy of NHEJ [19–21], whereas FEN1 promotes error-prone NHEJ [20]. The requirement of Ku70/80 for accurate end joining can be bypassed when joining occurs between sequences with high G/C content [22]. In the absence of DNA-PKcs, blunt-end joining is less efficient but not less accurate [21].
Previously we identified a novel human protein termed Metnase, which promotes DSB repair by NHEJ [23]. Metnase contains two functional domains: a SET domain and a nuclease domain from the Mariner transposase family. The SET domain methylates histone H3 at lysine 36, which is associated with open chromatin. The nuclease domain contains a DDE motif that is common to both retroviral integrase and transposase families. Metnase under-expression decreases resistance to ionizing radiation and reduces repair of DNA DSBs by NHEJ [23]. Both Metnase domains are required for these activities since mutation or deletion of either domain abolishes these activities.
We recently determined that Metnase also functions as a DNA endonuclease [24]. Metnase-mediated DNA cleavage is sequence-independent and non-processive, although Metnase does preferentially cleave 5’ overhangs of partial duplex DNA near the duplex end [24]. We also found that Metnase interacts with the DNA repair component Pso4, and this interaction is required for DSB repair [25]. Based on these results it was postulated that Metnase could regulate the processing of free DNA ends during NHEJ repair. Here we show that Metnase improves equally the efficiency of NHEJ of broken ends with 5’ or 3’ overhangs, or blunt ends, and reduces the frequency of long deletions arising during NHEJ. In addition, we show that Metnase physically interacts with DNA Ligase IV upon DSB induction, placing it within the classical NHEJ pathway. These data lend insight into the function of this novel transposase/nuclease domain protein in human cells.
2. Materials and methods
2.1. Co-immunoprecipitation of Metnase with DNA Ligase IV and XRCC4
HEK-293 cells stably expressing V5-tagged Metnase were transfected with a vector expressing FLAG-tagged DNA Ligase IV using calcium phosphate transfection. Two days later cells were treated with 10 Gy ionizing radiation (IR) or mock treated. Following a 1 hr incubation cells were harvested and lysed with a buffer containing PBS, 10% glycerol, 1% NP-40, and protease and phosphatase inhibitors. Cell extracts (200 µg) were used for immunoprecipitation in 300 µL RIPA buffer with an anti-FLAG antibody, anti-V5 antibody, or anti-XRCC4 antibody in the presence or absence of 1 unit of DNase I. Immunoprecipitates were analyzed by 10% SDS-PAGE and Western blotting for interaction between Metnase and DNA Ligase IV or XRCC4. HEK-293 cells were transfected individually with either V5-tagged Metnase or FLAG-tagged Ligase IV as above. Two days later the cells were harvested and total protein lysates prepared. Cell extracts were immunoprecipitated with either an anti-V5 or an anti-FLAG antibody. Immunoprecipitates were analyzed using anti-DNA Ligase IV or anti-Metnase to determine interaction of the tagged proteins with the endogenous DNA Ligase IV or Metnase.
2.2. Generation of cell lines over- and under-expressing Metnase
Cell lines over-expressing Metnase were generated by stable transfection of HEK-293 cells with pcDNA3.1-Metnase-V5/His, and cells transfected with empty pcDNA3.1-V5/His vector (Invitrogen, Carlsbad, CA) served as control. Transfectants were selected in 800 µg/ml G418 for 14–21 days. After this time individual colonies were picked, expanded and protein lysates extracted. Expression of Metnase was monitored by Western immunoblotting using polyclonal antibodies to Metnase [23].
Cell lines under-expressing Metnase were generated by stable transfection of HEK-293 cells with pRNA-U6.1/Hyg-siRNA-Metnase, and cells transfected with empty pRNA-U6.1/Hyg vector (Genescript, Piscataway, NJ) served as controls as described [23]. Transfectants were selected in 150 µg/ml hygromycin for 10–14 days. After this time individual colonies were picked, expanded and RNA was isolated. Reverse transcription-PCR (RT-PCR) was used as the initial screen for clones with reduced Metnase expression compared to the control (U6) clones, and RT-PCR of 18S rRNA was used to control input RNA levels. The primers for Metnase and 18S rRNA, and the Metnase siRNA target sequences were described previously [23]. Cells under-expressing Metnase grow slowly and can revert to normal expression levels, thus Metnase under-expression was confirmed by RT-PCR prior to all experiments with these cells.
2.3. NHEJ Repair Assays With or Without Coupled Integration
NHEJ repair was assayed using both one and two step assays, where the One Step assay analyzes direct plasmid end-joining without integration, and the Two Step assay analyzes plasmid end-joining coupled to genomic integration. In the one step assay, pBluescript KS+ digested with appropriate restriction enzymes and linear plasmid DNA was separated from any undigested plasmid by agarose gel electrophoresis and purified. HEK-293 cells expressing low, normal, or high levels of Metnase were seeded at 50,000 cells/well in 6 well plates and transfected with 10 µg of the digested plasmid using calcium phosphate [25]. Twenty-four hours post-transfection plasmid DNA was isolated from the cells using the Qiagen mini plasmid kit. DNA was quantified with a NanoDrop spectrophotometer (Fisher Scientific) and 10 ng of extracted plasmid was transformed into E. coli TOP10 cells and blue and white colonies were scored on LB plates with ampicillin, X-Gal and IPTG as described [23]. White and blue colonies were picked for PCR using M13 Forward and M13 Reverse primers. The fraction of colonies that failed to produce PCR products was a measure of the fraction of NHEJ deletions that were longer than 90 nucleotides (the distance from the DSBs to the M13 primer ends). Controls performed with the parent vector in parallel always yielded a PCR product (data not shown). For shorter deletions, PCR products were sequenced to define NHEJ junctions including deletions and insertions at the DSB site.
To analyze plasmid NHEJ coupled with integration (Two Step NHEJ assay), plasmid U6.1 was digested with HindIII or KpnI (5’ and 3’ overhangs, respectively), which cleave the plasmid between U6 promoter and the hygromycin-resistance gene (hygro) [25]. Linearized plasmid was purified and transfected into HEK-293 cells expressing normal or high levels of Metnase and hygromycin resistant colonies were counted to obtain a measure of end-joining followed by genomic integration (the effect of Metnase under-expression was not tested because these cells were already hygromycin-resistant). DNA was isolated from colonies and PCR amplified using T7 and BGH primers located 40 nucleotides from the restriction enzyme sites. The fraction of products that failed to produce PCR products (>40 bp deletions), and NHEJ junction analysis by DNA sequencing was performed as above. All statistical analysis was performed by using unpaired t tests unless otherwise specified.
2.4. DSB repair by Homologous Recombination
Five µg of the pcDNA Metnase expression vector or empty vector was co-transfected into 106 HT256 cells with 5 µg of pCMV(3xNLS)I-SceI to induce DSBs or the negative control vector pCMV(I-SceI−). HR repair of the I-SceI-induced DSB reconstitutes a functional neomycin phosphotransferase (neo) gene, as described [26]. HR was measured as the number of G418-resistant colonies per 200,000 cells; cell viability measured in non-selective medium was not affected by Metnase over-expression (data not shown).
2.5.γ-H2Ax Immunofluorescence
The formation of γ-H2Ax foci correlates with DSB induction, and the rate of γ-H2Ax disappearance correlates with the rate of DSB repair [27–31]. HEK-293 cells over- or under-expressing Metnase and the corresponding vector controls were treated with 2 Gy IR. Cells were harvested from untreated populations at 0.25, 2 and 6 hours after IR. After fixation, rehydration, and blocking, cells were stained with an anti-γ-H2Ax monoclonal mouse antibody (Cal-Biochem, NJ) at 4°C overnight. After washing, an Alexa 488-tagged goat anti-mouse IgG antibody (Invitrogen, Carlsbad, CA) was used for secondary staining for 1 hour at room temperature. After washing, cells were covered in Vectashield mounting media with DAPI (Vector, Burlingame, CA) and sealed. Images were obtained with a Biorad inverted microscope fitted with filter sets specific for DAPI and FITC/Alexa 488. Images were optimized consistently within experiments with ImageJ software (http://rsb.info.nih.gov/ij/).
3. Results
3.1. Metnase Interacts with DNA Ligase IV
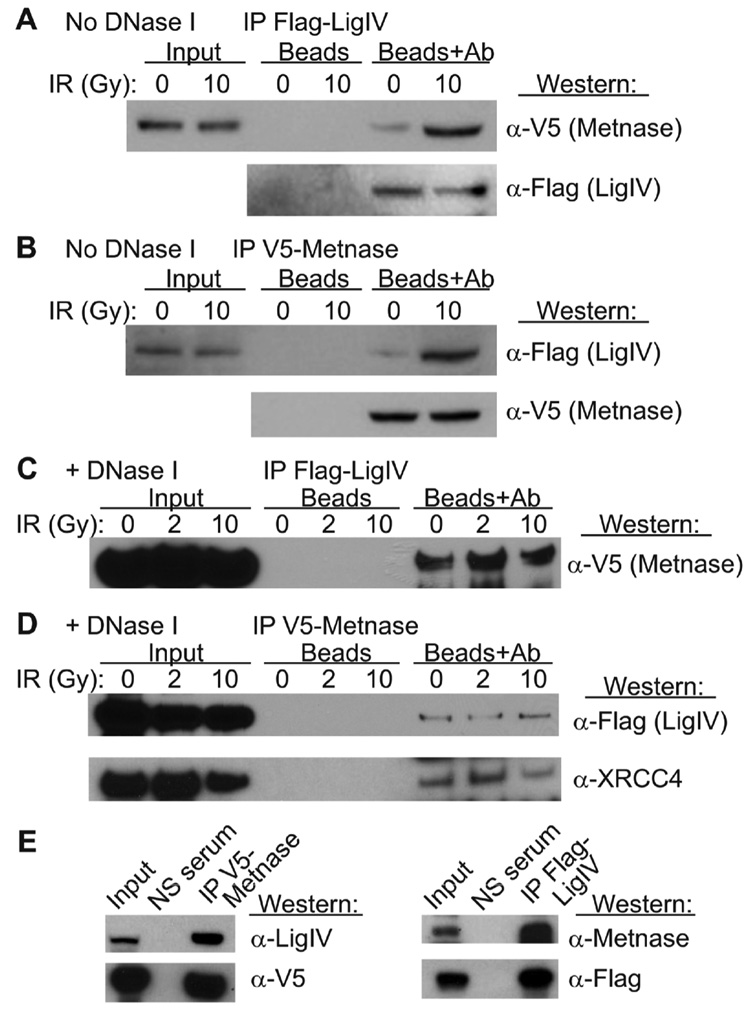
Because Metnase promotes NHEJ and confers resistance to IR [23], we tested whether Metnase interacts with components of the NHEJ system by co-immunoprecipitation. Using co-immunoprecipitation assays, Metnase was found to interact with DNA Ligase IV. Cells expressing V5-tagged Metnase and FLAG-tagged DNA Ligase IV were generated, and protein was isolated before and after DSB induction with IR. When proteins were immunoprecipitated with anti-FLAG and then probed with anti-V5, Metnase was detected (Fig. 1A). This interaction was also seen in the reciprocal reaction in which immunoprecipitation with anti-V5 was probed anti-FLAG, detecting DNA Ligase IV (Fig. 1B). Metnase and DNA Ligase IV interacted weakly in the absence of IR, but upon radiation exposure the interaction was enhanced. When DNase I was added to the reactions, co-immunoprecipitation of Metnase and DNA Ligase IV was still apparent (Fig. 1C, D) indicating that these proteins interact (directly or indirectly) independently of DNA. However, in the absence of DNA, the Metnase-DNA Ligase IV interaction was no longer enhanced by IR, indicating that the damage-enhanced interaction is DNA dependent. XRCC4 is a well-known binding partner of DNA Ligase IV, and we found that XRCC4 also co-precipitated with Metnase in the presence of DNase I, and in irradiated and untreated cells (Fig. 1D).
Fig. 1. Metnase interacts with DNA Ligase IV and XRCC4.
Extracts from HEK-293 cells expressing V5-tagged Metnase and Flag-tagged DNA Ligase IV were immunoprecipitated with Protein-G beads and anti-Flag (panel A) or anti-V5 antibodies (panel B); mock-IP controls without antibodies are indicated by “Beads.” Immunoprecipitates were separated by PAGE and Western blots were probed with anti-V5 or anti-Flag antibodies. IR increases the interaction between Metnase and DNA Ligase IV when DNA is present. (C, D) Immunoprecipitation as above except samples were treated with DNase I. In panel D the anti-V5 immunoprecipitate was also probed with anti-XRCC4 antibodies. (E) Extracts from HEK-293 cells expressing wither V5-tagged Metnase or Flag-tagged DNA Ligase IV were immunoprecipitated with Protein-G beads and anti-V5 or anti-Flag antibodies. Immunoprecipitates were probed with anti-DNA Ligase IV or anti-Metnase.
In order to show that these interactions were not simply the result of co-ordinated over-expression of both proteins, we transfected HEK-293 cells with either the FLAG-tagged Ligase IV or the V5-tagged Metnase, immunoprecipitated with the tag-specific antibodies and then immunoblotted using the antibodies specific for the endogenous proteins. We determined that the FLAG-tagged DNA Ligase IV interacted with the endogenously expressed Metnase protein (Fig. 1E). This interaction was also seen when V5-tagged Metnase immunoprecipitates were immunoblotted for endogenous DNA Ligase IV (Fig. 1E).
3.2. Metnase Promotes Precise and Imprecise NHEJ of Plasmid DNA with Any Type of Overhang in a One Step Assay
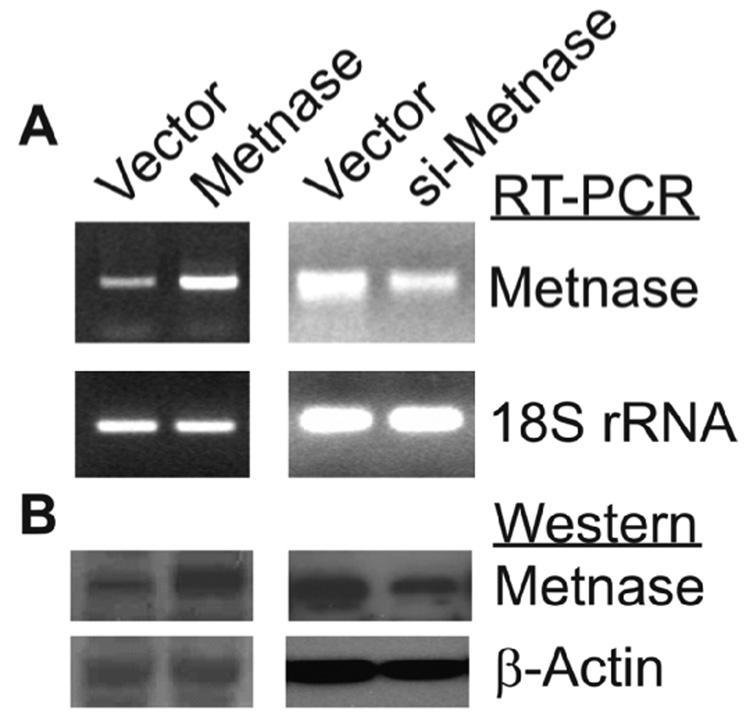
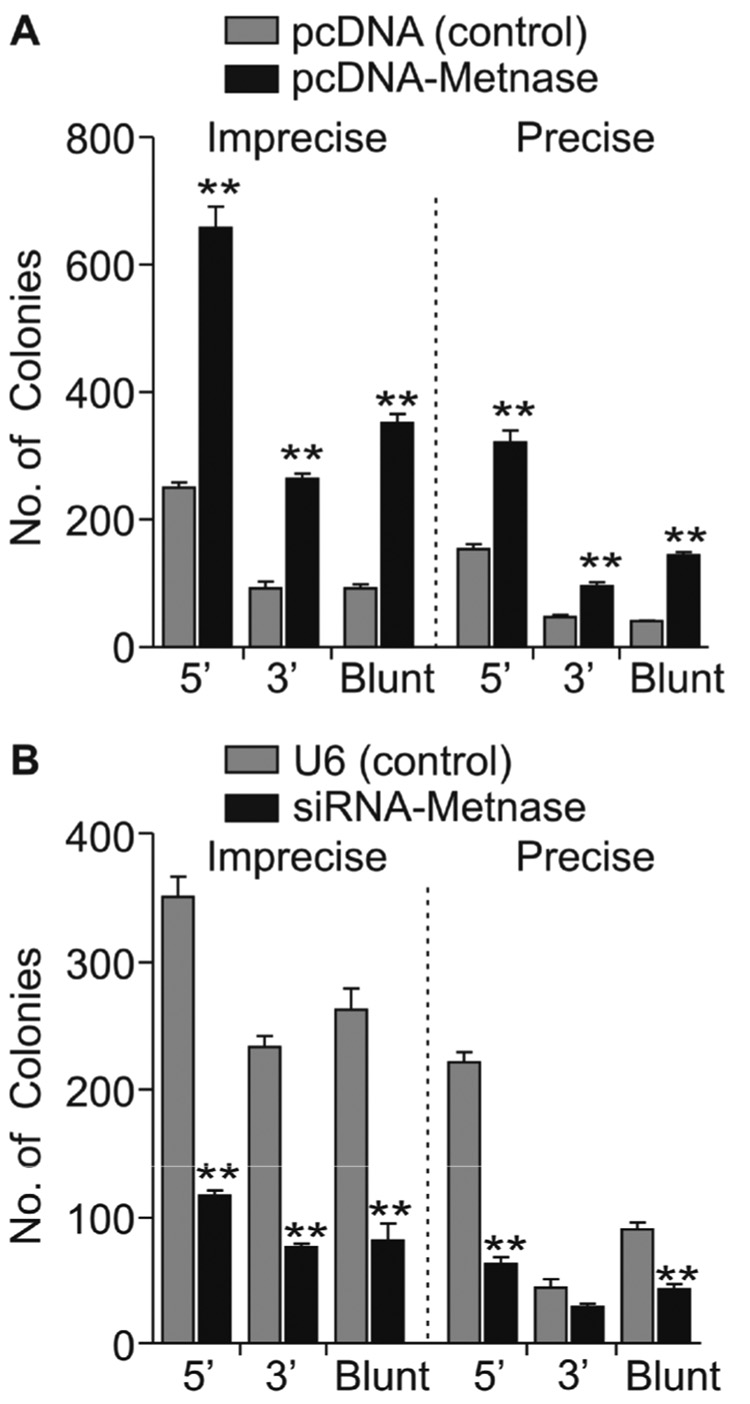
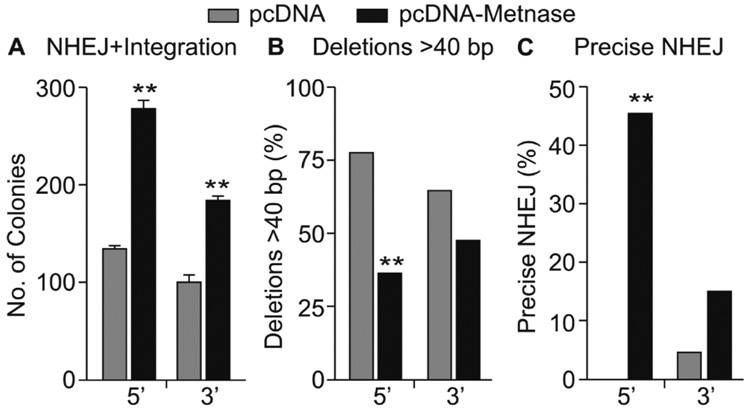
Derivatives of HEK-293 cells stably over- or under-expressing Metnase were generated and Metnase expression levels were determined both by RT-PCR and Western blotting (Fig. 2). Based on Western blotting, over-expression resulted in a 5-fold increase, and under-expression a 3-fold decrease in Metnase. As reported previously [23], Metnase RNA levels measured by RT-PCR correlated well with protein levels by Western blotting (Fig. 2). Plasmid pBluescript was linearized with EcoRI or KpnI to generate 5’ or 3’ overhangs, or with EcoRV to generate blunt ends, transfected into HEK-293 cells over- or under-expressing Metnase, and 24 hr later plasmid DNA was isolated and used to transform E. coli to assay for precise (β-galactosidase positive colonies) and imprecise (β-galactosidase negative) NHEJ. We observed that Metnase over-expression enhanced both precise and imprecise NHEJ (Fig. 3A), and under-expression decreased precise and imprecise NHEJ, and these effects were seen with each type of DNA end (Fig. 3B). Metnase over-expression increased total NHEJ by 2.4-fold with the 5’ overhang, 2.6-fold with the 3’ overhang, and 3.7-fold with blunt ends. Metnase under-expression decreased total NHEJ by 2.2-fold with 5’ overhangs, 2.6-fold with 3’ overhangs, and 2.8-fold with blunt ends. Metnase over- and under-expression resulted in statistically significant differences in both precise and imprecise NHEJ with all three end types (all P < 0.003) except Metnase under-expression did not significantly reduce precise NHEJ of the 3’ overhang (P = 0.13) (Fig. 3). However, Metnase under-expression did result in a significant decrease in total NHEJ of the 3’ overhang (2.6-fold; P = 0.0003). Thus, Metnase expression levels strongly correlate with the efficiency of plasmid NHEJ, but altering Metnase expression has similar effects on each type of end, and does not significantly affect the ratio of precise and imprecise NHEJ. We conclude that Metnase is a general enhancer of NHEJ, able to promote NHEJ with any type of free DNA end.
Fig. 2. Cell lines that over- and under-express Metnase.
(A) RT-PCR using Metnase-specific primers with 18S rRNA as loading control. (B) Western immunoblots with an anti-Metnase antibody and anti-β-actin as loading control. Vector: controls; Metnase: Metnase over-expressing cells; si-Metnase: under-expressing cells.
Fig. 3. Metnase promotes plasmid NHEJ with 5’ and 3’ overhangs and blunt ends in a One Step assay.
(A) Metnase over-expression increases precise and imprecise NHEJ. (B) Metnase under-expression decreases precise and imprecise NHEJ. Values are averages (±SEM) of 3 distinct determinations. **P < 0.01.
In this assay β-galactosidase positive colonies may result from precise NHEJ, or from imprecise NHEJ that deletes (or inserts) 3 bp segments to maintain the β-galactosidase gene reading frame. Therefore, we PCR amplified the sequence surrounding the DSB from 20 blue colonies for each type of end and each Metnase expression level. In all cases, ≥85% of blue colonies resulted from precise NHEJ and the remainder had lost one or two 3 bp segments (data not shown). There were no significant differences in codon loss rates in the different experimental conditions.
3.3. Metnase prevents long deletions during NHEJ in a One Step Assay
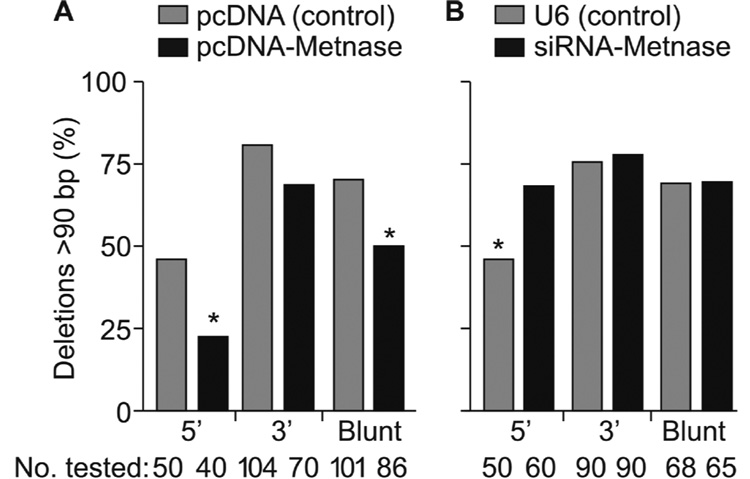
For each type of DSB end and each Metnase expression level, the majority of DSB repair was by imprecise NHEJ (Fig. 3). To determine whether Metnase influenced the extent or type of deletions (or insertions) at repair sites, we analyzed plasmid DNA from imprecise end joining products by PCR using primers that amplify a 90 bp fragment flanking the DSB sites. Large deletions will eliminate one or both primer sites and fail to produce PCR products. Large insertions may also prevent PCR if primer target sites are separated by too great a distance, but these are extremely rare [32,33]. We analyzed 40–104 repair products of plasmid DNA with 5’ or 3’ overhangs or blunt ends in HEK-293 cells expressing low, normal, or high levels of Metnase (Fig. 4). With 5’ overhangs and blunt ends large deletions were significantly less frequent in cells over-expressing Metnase (P = 0.03 and 0.007, respectively), and Metnase under-expression significantly increased the frequency of large deletions with 5’ overhangs (P = 0.02). Under-expression had no effect with 3’ overhangs or blunt ends. To determine whether the lack of a PCR product was due to the loss of one or both primer sites, we designed primers about 200 bp upstream and downstream from the DSB sites. These primers yielded PCR products on all colonies analyzed and sequencing showed that large deletions did indeed remove a primer site and these deletions were 92–98 bp in all cases (data not shown). We sequenced imprecise repair junctions of products with relatively short deletions (β-galactosidase negative colonies that yielded PCR products) and found little difference in deletion size with each free DNA end type and different Metnase expression levels (Table 1; Supplemental Fig. S1). NHEJ may involve alignment of 1 to several bases at processed ends (microhomology) and the use of microhomology (and more extensive stretches of microhomology) is increased in cells with defects in NHEJ proteins like Ku and DNA-PKcs [19,34]. We found that NHEJ of each type of end occurred without use of extensive microhomology regardless of Metnase expression level (Supplemental Fig. S1). Thus, Metnase enhances NHEJ efficiency and accuracy, but it does so without altering the use of microhomology.
Fig. 4. Metnase prevents large deletions during NHEJ.
(A) Metnase over-expression decreases the frequency of deletions >90 bp in length during NHEJ of 5’ overhangs and blunt ends; *P = 0.027; **P = 0.0066. (B) Metnase under-expression increases the frequency of large deletions during NHEJ of 5’ overhangs. Values as in Fig. 3; * indicates P = 0.021. The number of repair products tested is shown below each graph.
Table 1.
Characteristics of NHEJ junctions with normal vs. high Metnase levels
| Metnase Expressiona | Type of Endb | No. of Deletions | Deleted bp Range (Average bp ±SD) | 5’ bp Deleted Average ±SD | 3’ bp Deleted Average ±SD | No. of Insertions | Inserted bp Range (Average bp ±SD) |
|---|---|---|---|---|---|---|---|
| Normal | 5’ | 26 | 1–50 (6.6 ±10) | 2.3 ±3.7 | 4.3 ±9.4 | 15 | 42–402 (192 ±121) |
| High | 5’ | 30 | 1–49 (10.4 ±13.4) | 7.2 ± 11.4 | 3.0 ±4.7 | 18 | 1–426 (124 ±115) |
| Normal | 3’ | 20 | 5–49 (16.5 ±14.8) | 6.0 ±4.2 | 18.3 ±10.7 | 5 | 1–58 (23 ±26.3) |
| High | 3’ | 22 | 7–112 (26.3 ±26.7) | 9.3 ±13.0 | 17.0 ±18.8 | 11 | 1–271 (39 ±76.7) |
| Normal | Blunt | 27 | 2–66 (9.6 ±15.5) | 4.6 ±10.3 | 6.3 ±10.7 | 7 | 32–570 (248 ±179) |
| High | Blunt | 38 | 2–128 (15.6 ±31) | 8.4 ±16.5 | 9.3 ±18.4 | 10 | 1–612 (182 ±211) |
Normal Metnase expression in HEK-293 cells transfected with empty vector, pcDNA; high expression in cells transfected with pcDNA-Metnase.
5’ overhangs produced by EcoRI, 3’ by KpnI, and blunt ends by EcoRV.
3.4. Metnase promotes accurate NHEJ in a Two Step assay
To further investigate Metnase promotion of NHEJ we measured the efficiency of end-joining of a plasmid linearized at a site between a hygro resistance coding sequence and its promoter coupled to integration. This assay scores relatively accurate NHEJ, since large deletions would remove critical promoter elements or part of the hygromycin (hygro) coding sequence, followed by integration into the genome [25]. We performed this assay with U6.1 plasmid DNA linearized with HindIII or KpnI which create 5’ and 3’ overhangs, respectively, and transfected into HEK-293 cells or a derivative that stably over-expressed Metnase. The number of hygro-resistant colonies was significantly enhanced (2-fold) with Metnase over-expression with both the 5’ and 3’ ends (Fig. 5A; both P < 0.0001). Consistent with the One Step NHEJ assay results above, the fraction of hygro-resistant colonies with large deletions (>40 bp in length by PCR assay) was significantly reduced with Metnase over-expression and the 5’ overhang (P < 0.0001). The same trend was seen with 3’ ends but was not statistically significant with these sample sizes (P = 0.1) (Fig. 5B). In addition, Metnase over-expression increased the fraction of precise repair products (Fig. 5C). This increase was statistically significant for 5’ overhangs (P < 0.0001), but not with 3’ overhangs (P = 0.08). Together, the results from One and Two Step NHEJ assays indicate that Metnase promotes precise and relatively accurate NHEJ independently of the type of DNA end, but its strongest effects are seen with 5’ overhangs.
Fig. 5. Over-expression of Metnase enhances NHEJ accuracy and efficiency in a Two Step coupled NHEJ/integration assay.
(A) Over-expression of Metnase increases the number of hygromycin-resistant colonies; averages ± SEM for 9 determinations. (B, C) Metnase over-expression decreases the frequency of large deletions and increases precise repair; 40–104 products tested per condition, **P < 0.0001 by Fisher exact tests.
3.5 Metnase does not influence DSB repair by homologous recombination
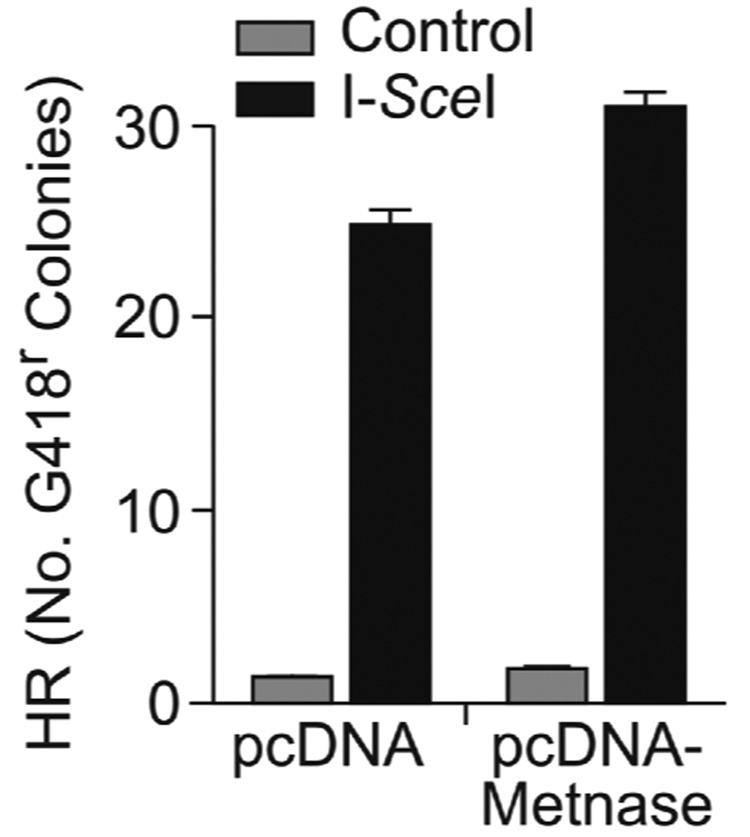
NHEJ and homologous recombination (HR) are competing DSB repair pathways [1], and mutations that decrease NHEJ typically increase HR [35,36]. Because Metnase over-expression enhances plasmid NHEJ (above) and increases resistance to IR (which is largely due to NHEJ)[23], we hypothesized that Metnase overexpression was unlikely to improve HR repair of DSBs. Using a chromosomal substrate in which HR is induced by I-SceI DSBs, we found that HR was not statistically affected by Metnase overexpression (Fig. 6).
Fig. 6. Metnase does not suppress HR.
HT256 co-transfected with control vector or pcDNA-Metnase, and I-SceI sense or control vector and G418-resistant colonies were scored. Values as in Fig. 5A.
3.6. Metnase promotes removal of IR-induced γ-H2Ax foci
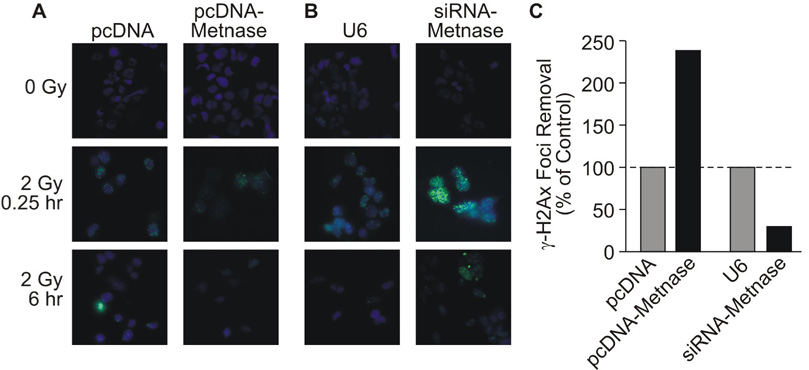
As noted above, cellular resistance to IR primarily reflects NHEJ activity, and IR resistance is proportional to Metnase expression levels [23]. IR-induced DSBs are marked by γ-H2Ax foci [37]. We were interested in whether altering NHEJ activity by modulating Metnase levels would alter the kinetics of γ-H2Ax focus removal. γ-H2Ax foci were induced similarly in cells expressing normal or high levels of Metnase, and consistent with the enhanced NHEJ and IR resistance with over-expressed Metnase (Fig. 3, Fig. 5)[23], cells over-expressing Metnase removed 31% of IR-induced γ-H2Ax foci within 6 hr after IR, whereas control cells removed only 13% of foci (Fig. 7A, B). Similarly, cells in which Metnase was knocked down showed slower removal of γ-H2Ax foci than control cells (Fig. 7C, D). Thus, the speed that IR-induced γ-H2Ax foci are resolved is proportional to Metnase levels, demonstrating that the resistance to IR mediated by Metnase is indeed likely due to repair of DNA damage.
Fig. 7. Metnase promotes resolution of γ-H2Ax foci after IR.
(A) Representative images of γ-H2Ax foci (green) with DAPI (blue) in control (unirradiated) cells, and 0.25 or 6 hr after IR-treated cells with normal and over-expressed Metnase. (B) As in panel A except control and Metnase under-expression are shown. (C) The percentages γ-H2Ax-positive cells were counted with no IR (averaging ~5% regardless of Metnase expression level), and at 0.25 and 6 hr after 2 Gy IR. At 0.25 hr, >75% of cells had γ-H2Ax foci and this number declined after 6 hr. Plotted are the ratios of γ-H2Ax-positive cells at 6 hr divided by 0.25 hr, with control values normalized to 100%.
4. Discussion
We identified Metnase as a SET and transposase domain protein that methylates histone H3, enhances foreign DNA integration, and promotes NHEJ [23]. In this study we further defined its NHEJ activity on different types of DSB ends, and showed that it interacts with the key NHEJ complex DNA Ligase IV-XRCC4. Interestingly, Metnase and DNA Ligase IV interact independently of DNA, but the interaction is enhanced when cells are exposed to IR and DNA is not removed prior to immunoprecipitation. This implies that Metnase and DNA Ligase IV can stably interact, but that IR enhances their interaction on DNA, presumably at sites of damage. This interaction also suggests that Metnase functions in the classical NHEJ repair pathway [2,38,39].
The processing and ligation of different types of DNA ends has important implications for genome instability since these processes directly regulate repair accuracy. Sequence analysis of repair junctions showed that Metnase expression had little effect on processing in the immediate vicinity of any of the broken ends. However, in One Step NHEJ assays and Two Step coupled NHEJ/integration assays, Metnase expression prevented large deletions. Metnase appeared to enhance precise NHEJ only in the coupled NHEJ/integration assay. This may be due to the selection pressure for a functional hygro-resistance expression cassette since even relatively short deletions could inactivate the promoter and/or alter the hygro coding sequence. Thus, the selection for the functional hygro gene may have amplified the effect of Metnase on precise repair.
There are several ways that Metnase might promote accurate repair. First, Metnase may associate with the NHEJ apparatus at DSBs and regulate end-processing. Artemis has 5’-3’ exonuclease activity, and the MRN complex has 3’-5’ exonuclease activity with blunt and recessed 3’ ends that pauses at sites of microhomology [40–43]. Metnase could inhibit these activities to prevent large deletions. Second, Metnase may preferentially modify chromatin at the DSB via its histone methylase activity, which in turn could allow limited processing, but prevent more extensive processing leading to large deletions. Third, Metnase may directly bind to free ends, and protect them against further degradation from other nucleases. These three mechanisms are not mutually exclusive, and one could envision a model where Metnase directly modifies chromatin at a DSB, regulates the processing activity of the NHEJ apparatus, and protects free ends from degradation.
We show here that Metnase promotes NHEJ of 5’ and 3’ overhangs and blunt ends, but the underlying mechanism remains unclear. Metnase may promote NHEJ by binding to free ends, and attracting DNA Ligase IV. Conversely, DNA Ligase IV may attract Metnase to DSBs where it modulates the end-processing activities of MRN or Artemis as suggested above. DNA Ligase IV has been reported to bind to 5’ overhangs reduce end degradation [44]. This activity may be dependent on, or regulated by, interaction with Metnase at broken ends. Metnase promotes NHEJ but does not significantly affect DSB repair by HR. This is not surprising given that these pathways compete for repair of DSBs [1].
We previously reported that Metnase has structure-specific DNA endonuclease activity [24] and others have shown that it has some but not all of the nucleolytic activities typical of transposases [45–47]. These results suggest that Metnase may play a role in end-processing during NHEJ. However, altering Metnase levels did not strongly affect the spectrum of small-scale deletions or relative fractions of deletions vs. insertions (Table 1). Moreover, a role in end-processing is difficult to reconcile with our finding that long deletions are decreased when Metnase is over-expressed. Because Metnase promotes NHEJ of all types of ends, minimizes large-scale deletions, and increases precise repair, we propose that Metnase enhances NHEJ kinetics, which would increase NHEJ accuracy by reducing the time for end-processing. The Metnase nuclease activity may be more important during DNA integration [23] than for NHEJ. Although mutations in the Metnase nuclease domain prevent Metnase from stimulating NHEJ [23], we cannot rule out the possibility that these mutations alter Metnase structure to prevent its interaction with DNA Ligase IV, or that they reduce the ability of Metnase to bind free DNA ends. It is also possible that the nuclease domain is important for DNA end binding, but has little nucleolytic activity at the free DNA ends.
Some components of the NHEJ pathway promote repair of specific types of DNA ends, such as DNA-PKcs and Ku, which preferentially promote repair of 5’ overhangs [19–22]. We found that Metnase promotes NHEJ of 5’ and 3’ overhangs and blunt ends equally well, but its ability to enhance NHEJ accuracy was most evident with 5’ overhangs. This property, along with its ability to interact with DNA Ligase IV, provides further support for the idea that Metnase promotes DSB repair via the classical NHEJ pathway involving DNA-PKcs, Ku, and DNA Ligase IV-XRCC4. Although the precise mechanism by which Metnase promotes NHEJ remains to be determined, it is likely that this activity depends on its interaction with the DNA Ligase IV-XRCC4 complex.
This present study characterizes the end-joining repair properties of Metnase which evolved from a transposase. Transposases catalyze uncontrolled movement of DNA segments within genomes, and this would be deleterious in mammals which have relatively long life spans and large numbers of divisions of individual cells. Such unregulated transposition would result in high rates of apoptosis or malignancy. This is likely why transposase functions have been selected against in mammals, and that the vast majority of transposase sequences in humans are pseudogenes [48]. Metnase, on the other hand, represents an interesting example in which mammalian cells have diverted a transposase sequence to enhance the accuracy of NHEJ, which would promote accurate DSB repair and enhance genome stability.
Supplementary Material
Table 2.
Characteristics of NHEJ junctions with normal vs. low Metnase levels.
| Metnase Expressiona | Type of Endb | No. of Deletions | Deleted bp Range (Average bp ±SD) | 5’ bp Deleted Average ±SD | 3’ bp Deleted Average ±SD | No. of Insertions | Inserted bp Range (Average bp ±SD) |
|---|---|---|---|---|---|---|---|
| Normal | 5’ | 15 | 2–98 (12.8 ±18.4) | 4.1 ±3.8 | 9.2 ±19.6 | 16 | 26–291 (139 ±88.7) |
| Low | 5’ | 17 | 1–62 (11.9 ±16.8) | 3.5 ±5.4 | 55.5 ±11.7 | 12 | 43–402 (135 ±95.7) |
| Normal | 3’ | 22 | 9–77 (26.4 ±18.4) | 5.6 ±5.6 | 20.7 ±19.9 | 5 | 283–625 (424 ±160) |
| Low | 3’ | 20 | 5–130 (15.8 ±16.8) | 7.4 ±20.4 | 8.4 ±10.7 | 2 | 40–113 (76.5 ±51.6) |
| Normal | Blunt | 20 | 2–11 (2.8 ±2.4) | 0.7 ±2.5 | 2.2 ±1.4 | 1 | 150 |
| Low | Blunt | 25 | 2–13 (3.0 ±2.8) | 2.8 ±7.1 | 3.2 ±5.2 | 4 | 1–168 (82.0 ±93.6) |
Normal Metnase expression in HEK-293 cells transfected with empty vector, U6; high expression in cells transfected with U6-siRNA-Metnase.
5’ overhangs produced by EcoRI, 3’ by KpnI, and blunt ends by EcoRV.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that there are no conflicts of interest.
REFERENCES
- 1.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 2.Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson DO, Sekiguchi JM, Chang S, Frank KM, Gao Y, DePinho RA, Alt FW. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl. Acad. Sci. USA. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu YP, Yu JC, Cheng TC, Lou MA, Hsu GC, Wu CY, Chen ST, Wu HS, Wu PE, Shen CY. Breast cancer risk associated with genotypic polymorphism of the nonhomologous end-joining genes: a multigenic study on cancer susceptibility. Cancer Res. 2003;63:2440–2446. [PubMed] [Google Scholar]
- 5.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 6.van den Bosch M, Bree RT, Lowndes NF. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 2003;4:844–849. doi: 10.1038/sj.embor.embor925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammarsten O, Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc. Natl. Acad. Sci. USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smider V, Rathmell WK, Lieber MR, Chu G. Restoration of X-ray resistance and V(D)J recombination in mutant cells by Ku cDNA. Science. 1994;266:288–291. doi: 10.1126/science.7939667. [DOI] [PubMed] [Google Scholar]
- 9.Taccioli GE, Gottlieb TM, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann AR, Alt FW, Jackson SP, Jeggo PA. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 10.Block WD, Yu Y, Merkle D, Gifford JL, Ding Q, Meek K, Lees-Miller SP. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic Acids Res. 2004;32:4351–4357. doi: 10.1093/nar/gkh761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calsou P, Frit P, Humbert O, Muller C, Chen DJ, Salles B. The DNA-dependent protein kinase catalytic activity regulates DNA end processing by means of Ku entry into DNA. J. Biol. Chem. 1999;274:7848–7856. doi: 10.1074/jbc.274.12.7848. [DOI] [PubMed] [Google Scholar]
- 12.Chan DW, Lees-Miller SP. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J. Biol.Chem. 1996;271:8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- 13.Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell. Biol. 2003;23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douglas P, Sapkota GP, Morrice N, Yu Y, Goodarzi AA, Merkle D, Meek K, Alessi DR, Lees-Miller SP. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem. J. 2002;368:243–251. doi: 10.1042/BJ20020973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 16.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 17.Merkle D, Douglas P, Moorhead GB, Leonenko Z, Yu Y, Cramb D, Bazett-Jones DP, Lees-Miller SP. The DNA-dependent protein kinase interacts with DNA to form a protein-DNA complex that is disrupted by phosphorylation. Biochemistry. 2002;41:12706–12714. doi: 10.1021/bi0263558. [DOI] [PubMed] [Google Scholar]
- 18.Reddy YV, Ding Q, Lees-Miller SP, Meek K, Ramsden DA. Non-homologous end joining requires that the DNA-PK complex undergo an autophosphorylation-dependent rearrangement at DNA ends. J. Biol. Chem. 2004;279:39408–39413. doi: 10.1074/jbc.M406432200. [DOI] [PubMed] [Google Scholar]
- 19.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang L, Deng L, Chen Y, Li GC, Shao C, Tischfield JA. Modulation of DNA end joining by nuclear proteins. J. Biol. Chem. 2005;280:31442–31449. doi: 10.1074/jbc.M503776200. [DOI] [PubMed] [Google Scholar]
- 21.van Heemst D, Brugmans L, Verkaik NS, van Gent DC. End-joining of blunt DNA double-strand breaks in mammalian fibroblasts is precise and requires DNA-PK and XRCC4. DNA Repair. 2004;3:43–50. doi: 10.1016/j.dnarep.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Sandoval A, Labhart P. High G/C content of cohesive overhangs renders DNA end joining Ku-independent. DNA Repair. 2004;3:13–21. doi: 10.1016/j.dnarep.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 23.Lee SH, Oshige M, Durant ST, Rasila KK, Williamson EA, Ramsey H, Kwan L, Nickoloff JA, Hromas R. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc. Natl. Acad. Sci. USA. 2005;102:18075–18080. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roman Y, Oshige M, Lee YJ, Goodwin K, Georgiadis MM, Hromas RA, Lee SH. Biochemical characterization of a SET and transposase fusion protein, Metnase: its DNA binding and DNA cleavage activity. Biochemistry. 2007;46:11369–11376. doi: 10.1021/bi7005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beck BD, Park SJ, Lee YJ, Roman Y, Hromas RA, Lee SH. Human Pso4 is a Metnase (SETMAR)-binding partner that regulates Metnase function in DNA repair. J. Biol. Chem. 2008;283:9023–9030. doi: 10.1074/jbc.M800150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu H, Guo X, Meng X, Liu J, Wray J, Allen C, Nickoloff JA, Shen Z. The BRCA2-interacting protein BCCIP functions in RAD51 and BRCA2 focus formation and homologous recombinational repair. Mol. Cell. Biol. 2005;25:1949–1957. doi: 10.1128/MCB.25.5.1949-1957.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kegel P, Riballo E, Kuhne M, Jeggo PA, Lobrich M. X-irradiation of cells on glass slides has a dose doubling impact. DNA Repair. 2007;6:1692–1697. doi: 10.1016/j.dnarep.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 28.Kuhne M, Riballo E, Rief N, Rothkamm K, Jeggo PA, Lobrich M. A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res. 2004;64:500–508. doi: 10.1158/0008-5472.can-03-2384. [DOI] [PubMed] [Google Scholar]
- 29.Lobrich M, Rief N, Kuhne M, Heckmann M, Fleckenstein J, Rube C, Uder M. In vivo formation and repair of DNA double-strand breaks after computed tomography examinations. Proc. Natl. Acad. Sci. USA. 2005;102:8984–8989. doi: 10.1073/pnas.0501895102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacPhail SH, Banath JP, Yu TY, Chu EH, Lambur H, Olive PL. Expression of phosphorylated histone H2AX in cultured cell lines following exposure to X-rays. Int. J. Radiat. Biol. 2003;79:351–358. doi: 10.1080/0955300032000093128. [DOI] [PubMed] [Google Scholar]
- 31.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to γ-H2AX foci. Mol. Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 32.Allen C, Miller CA, Nickoloff JA. The mutagenic potential of a single DNA double-strand break is not influenced by transcription. DNA Repair. 2003;2:1147–1156. doi: 10.1016/s1568-7864(03)00139-3. [DOI] [PubMed] [Google Scholar]
- 33.Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood. 2006;107:777–780. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene. 2005;24:949–961. doi: 10.1038/sj.onc.1208332. [DOI] [PubMed] [Google Scholar]
- 35.Allen C, Kurimasa A, Brennemann MA, Chen DJ, Nickoloff JA. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc. Natl. Acad. Sci. USA. 2002;99:3758–3763. doi: 10.1073/pnas.052545899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delacote F, Han M, Stamato TD, Jasin M, Lopez BS. An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 2002;30:3454–3463. doi: 10.1093/nar/gkf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pilch DR, Sedelnikova OA, Redon C, Celeste A, Nussenzweig A, Bonner WM. Characteristics of γ-H2AX foci at DNA double-strand breaks sites. Biochem. Cell Biol. 2003;81:123–129. doi: 10.1139/o03-042. [DOI] [PubMed] [Google Scholar]
- 38.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lees-Miller SP, Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 40.Assenmacher N, Hopfner KP. MRE11/RAD50/NBS1: complex activities. Chromosoma. 2004;113:157–166. doi: 10.1007/s00412-004-0306-4. [DOI] [PubMed] [Google Scholar]
- 41.Dahm K. Functions and regulation of human artemis in double strand break repair. J. Cell. Biochem. 2007;100:1346–1351. doi: 10.1002/jcb.21226. [DOI] [PubMed] [Google Scholar]
- 42.Drouet J, Frit P, Delteil C, de Villartay JP, Salles B, Calsou P. Interplay between Ku, Artemis, and the DNA-dependent protein kinase catalytic subunit at DNA ends. J. Biol. Chem. 2006;281:27784–27793. doi: 10.1074/jbc.M603047200. [DOI] [PubMed] [Google Scholar]
- 43.Paull TT, Gellert M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc Natl Acad Sci U S A. 2000;97:6409–6414. doi: 10.1073/pnas.110144297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith J, Riballo E, Kysela B, Baldeyron C, Manolis K, Masson C, Lieber MR, Papadopoulo D, Jeggo P. Impact of DNA ligase IV on the fidelity of end joining in human cells. Nucleic Acids Res. 2003;31:2157–2167. doi: 10.1093/nar/gkg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc. Natl. Acad. Sci. USA. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu D, Bischerour J, Siddique A, Buisine N, Bigot Y, Chalmers R. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol. Cell. Biol. 2007;27:1125–1132. doi: 10.1128/MCB.01899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miskey C, Papp B, Mates L, Sinzelle L, Keller H, Izsvak Z, Ivics Z. The ancient mariner sails again: transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends. Mol. Cell. Biol. 2007;27:4589–4600. doi: 10.1128/MCB.02027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.