Abstract
Sapje-like (sapcl100) was one of eight potential zebrafish muscle mutants isolated as part of an early-pressure screen of 500 families. This mutant shows a muscle tearing phenotype similar to sapje (dys−/−) and both mutants fail to genetically complement suggesting they have a mutation in the same gene. Protein analysis confirms a lack of dystrophin in developing sapje-like embryos. Sequence analysis of the sapje-like dystrophin mRNA shows that exon 62 is missing in the dystrophin transcript causing exon 63 to be translated out of frame terminating translation at a premature stop codon at the end of exon 63. Sequence analysis of sapje-like genomic DNA identified a mutation in the donor splice junction at the end of dystrophin exon 62. This mutation is similar to splicing mutations associated with human forms of Duchenne Muscular Dystrophy. Sapje-like is the first zebrafish dystrophin splicing mutant identified to date and represents a novel disease model which can be used in future studies to identify therapeutic compounds for treating diseases caused by splicing defects.
INTRODUCTION
Muscular dystrophy is the one of the most common X-linked human genetic disorders. Mutations in the dystrophin gene account for almost 90% of the human forms of the disease (1–3). Depending on disease severity, these mutations can cause Duchenne (more severe) or Becker (less severe) Muscular Dystrophy (4). While many different types of dystrophin mutations can cause muscular dystrophy, the most common are deletions (65%), mutations (30%) and duplications (5%) (5).
Dystrophin is a large protein (427 kDa) that localizes in healthy muscle to the inside of the sarcolemmal membrane and is thought to function by connecting transmembrane proteins with the actin cytoskeleton inside muscle (6–8). Loss of dystrophin can destabilize the dystrophin-associated protein complex (DAPC) thereby increasing the fragility of the diseased muscle cell membrane. Mutations in many DAPC proteins have also been linked to different forms of muscular dystrophy (9–13). Once the membrane is compromised, calcium infiltrates the muscle and the fiber degenerates, eliciting a large immune response and the regeneration of additional diseased muscle fibers. While specific mutations in dystrophin were identified in 1986 as the cause of Duchenne Muscular Dystrophy (3), new treatments are only recently emerging based on either dystrophin replacement or gene correction. These therapies seem promising and are currently being evaluated (reviewed in 14,15). Despite these advances, approaches to therapy would benefit from the establishment of additional new disease models adapted to high-throughput drug screening.
Over the last eight years, muscular dystrophy has been investigated in zebrafish as a potential disease model (reviewed in 16). Morpholino experiments have shown that dystroglycan, dystrophin, δ-sarcoglycan, titin and caveolin-3 are all required for normal muscle development in fish during the first seven days of development (17–21). In addition, four of the five available zebrafish muscle degeneration mutants isolated from the 1996 Tuebingen screen (22) have now been characterized. Bassett et al. characterized the first of these fish in 2003 and showed that sapje carries a nonsense mutation in exon 4 of the dystrophin gene (23). The second and third mutants (both allelic and designated candyfloss) were shown to carry mutations in laminin α2 (24), whereas the mutation in runzel has been linked to the titin locus (25). In humans, mutations in laminin α2 have been associated with a congenital muscular dystrophy (26), and mutations in titin have been linked with various muscle disorders including Tibial muscular dystrophy, Familial Dilated Cardiomyopathy and limb-girdle muscular dystrophy type 2J (27–31).
All four of the currently characterized zebrafish dystrophy mutants were originally isolated as part of the 1996 Tuebingen screen (22). These dystrophic mutants show a phenotype of muscle degeneration between two and five days post-fertilization (dpf). Since zebrafish embryos are transparent at early stages, Granato et al. used muscle birefringence to assay muscle integrity. Birefringence is a physical property in which light is rotated as it passes through ordered matter and was used in the initial characterizations of muscle in the 1950s (32).
To isolate additional zebrafish dystrophic mutants, an early-pressure screen was completed, looking for mutants showing decreased birefringence over time, decreased activity and bending. Early pressure is a technique in which the haploid genome of the mother can be screened by making gynogenetically diploid embryos (reviewed in 33). In this technique, the final phase of meiosis is inhibited using pressure to prevent the extrusion of the polar body after egg fertilization. Using this technique, offspring from 500 female carriers were screened for defects in muscle birefringence, activity and posture. Eight potential muscle mutants were isolated and one (sapje-like) showed, upon using the birefringence assay, muscle lesions very similar to that of sapje, a known dystrophin mutant. In this report, we show that the sapje-like muscular dystrophy mutant has a mutation within the dystrophin exon 62 donor splice junction causing the resulting mRNA to be translated out of frame and protein translation to be prematurely terminated. Sapje-like (sapcl100) is the first zebrafish dystrophy mutant isolated to date with a dystrophin splice site mutation making it an ideal model for investigating corrective therapies for muscle disorders associated with splicing.
RESULTS
Genetic isolation of zebrafish muscle mutants
An early-pressure screen was performed to genetically isolate zebrafish mutants with symptoms of muscle disease. Briefly, as described in Supplementary Material, Figure S1, the screen was performed by mutating the spermatagonia of 80 adult male zebrafish with ethylnitrosylurea (ENU). Sperm from ENU-treated males were cleared by outcrossing with wild-type females. The mutation rate was then determined to be 1 × 10−3 mutations/allele as assayed by complementation with albino heterozygous mutants. ENU-treated males were outcrossed to wild-type females to generate a stock of heterozygous females. Upon reaching sexual maturity, these females were squeezed, and the eggs were fertilized with irradiated sperm and subjected to high pressure to create gynogenetically diploid embryos. Fertilized eggs were scored at 5 dpf for birefringence defects, decreased activity and a bent posture. The offspring of 500 females were screened and 8 distinct mutants with abnormal birefringence of muscle were isolated. Carrier females were outcrossed to maintain the line and the resultant progeny incrossed to verify phenotype.
Phenotype of the sapje-like mutant
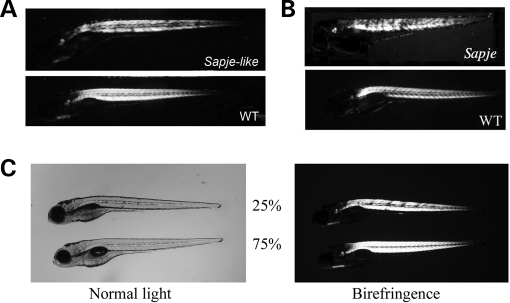
Of the eight isolated muscle mutants, one mutant (sapje-like) showed a muscle birefringence phenotype very similar to sapje, a known dystrophin mutant. Both mutants show normal muscle birefringence at 2 dpf (not shown), but then the muscle degenerates such that by 5 dpf muscle lesions are clearly apparent as assayed using muscle birefringence (Fig. 1A). Since sapje-like showed a muscle phenotype very similar to that of sapje (Fig. 1B), complementation analysis was performed. When sapje-like heterozygous fish were crossed with sapje heterozygous fish, 25% of the offspring showed the muscle birefringence phenotype suggesting that the mutation in each of the mutants resided in the same gene (Fig. 1C). This cross was repeated twice to verify our results. Since sapje was shown previously to carry a nonsense mutation in exon 4 of the zebrafish dystrophin gene (23), this evidence strongly suggested that sapje-like also carried a recessive mutation in the dystrophin gene.
Figure 1.
Sapje-like mutant offspring show muscle defects which can be assayed using birefringence. (A) Muscle degeneration in sapje-like mutants is clearly visible as small dark spots at 5 dpf using birefringence. These spots represent areas of muscle tearing. (B) The sapje-like birefringence is very similar to that seen for sapje, a known zebrafish dystrophin mutant. (C) Sapje and sapje-like fail to complement each other. Heterozygous sapje mutants (±) were crossed with heterozygous sapje-like mutants (±) and the offspring assayed at 4 dpf for defects in muscle using birefringence. From this cross, 25% of the offspring showed muscle lesions (top fish, right) suggesting that the two fish have mutations in the same gene. All panels are shown at approximately 10-fold magnification.
Immunohistochemical analysis confirms loss of dystrophin protein in the sapje-like mutant
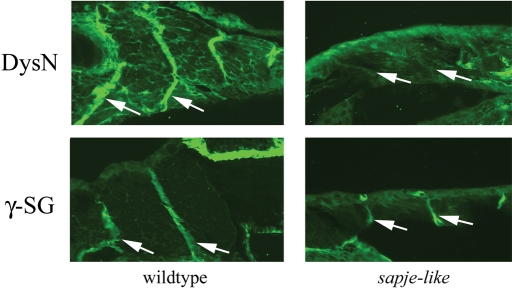
In human Duchenne Muscular Dystrophy patients and in the zebrafish dystrophin morphant (19), dystrophin protein is missing and its associated proteins are often partially down-regulated at the muscle sarcolemma. To assay for overall dystrophin protein expression in the sapje-like mutant, wild-type and sapje-like mutants were analyzed by immunohistochemistry. Sections from 5 dpf embryos were immunostained with antibodies designed specifically against zebrafish dystrophin and in all cases, dystrophin protein was absent at the myosepta of developing sapje-like mutants (Fig. 2, top).
Figure 2.
Immunohistochemical analysis of sapje-like horizontal sections at five days post-fertilization (dpf). Dystrophin protein localizes in unaffected siblings to the muscle myosepta (top left), but is absent in the mutant (top right). γ-Sarcoglycan is also strongly expressed in unaffected siblings (bottom left) and also in the sapje-like mutant (bottom right). From this analysis, it is difficult to determine if γ-sarcoglycan is down-regulated in the mutant. The myoseptas are designated with white arrows. For each antibody, pictures were taken at the same exposure level for both the wild-type and sapje-like mutant. All panels are shown at approximately 80-fold magnification.
To test if the dystrophin-associated proteins were partially down-regulated in the sapje-like mutant, sections from 5 dpf wild-type and sapje-like mutants were stained with antibodies against the following DAPC components: β-sarcoglycan, δ-sarcoglycan and γ-sarcoglycan. Due to similar staining patterns, only the panels from the γ-sarcoglycan staining are shown in Figure 2 (bottom). This analysis showed little difference in the intensities of the sarcoglycan myoseptal staining patterns in the 5 dpf and 4-week-old mutant fish (data not shown).
Western blot analysis shows that the DAPC is partially destabilized in the sapje-like mutant
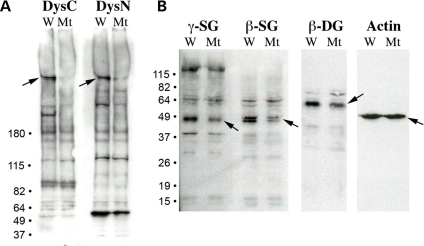
Since protein expression levels are difficult to compare using immunohistochemical analysis, immunoblot analysis was performed to compare expression of dystrophin and dystrophin-associated proteins in the sapje-like mutant. Protein extracts from wild-type and sapje-like mutants at 5 dpf were separated by gel electrophoresis and transferred to nitrocellulose membrane. Using antibodies developed against both the N- and C- terminus of zebrafish dystrophin, immunoblot analysis confirmed the loss of the dystrophin protein specifically in the sapje-like mutant (Fig. 3A, see arrows). While the dystrophin protein appeared to be completely absent in the blot using the C-terminus dystrophin antibody, the N-terminus antibody detected a reduced (although present) smaller dystrophin protein in the mutant. This band likely represents the truncated dystrophin isoform resulting from the splice site mutation (see later).
Figure 3.
Immunoblot analysis shows that dystrophin and the sarcoglycans are reduced in the sapje-like mutant. (A) Antibodies against both the C- and N-terminus of dystrophin (DysC and DysN, respectively) show that dystrophin is strongly down-regulated in the sapje-like mutant (‘Mt’) relative to the unaffected wild-type siblings (‘W’). (B) γ- and β-Sarcoglycan are down-regulated in the sapje-like mutant. β-Dystroglycan also appears to be affected in the sapje-like mutant, but the actin control and background non-specific bands seen in all of the blots show that the lanes were loaded equally. Size markers are shown on the left and each of the proteins is identified with an arrow on the blot.
To determine if expression of the dystrophin-associated proteins was affected in the sapje-like mutant, western blot analysis was also performed for the sarcoglycans. In the sapje-like mutant, the β-, δ- and γ-sarcoglycan proteins were moderately although specifically reduced (Fig. 3B). This data suggest that the sarcoglycan subcomplex is weakly destabilized in the sapje-like mutant, similar to that seen in some human Duchenne patients (sarcoglycan destabilization can be variable). Western analysis was also performed for β-dystroglycan, a member of the DAPC that is not typically reduced in human Duchenne patients. While subtle, β-dystroglycan also appears to be slightly reduced, suggesting the integrity of the entire complex might be partially compromised in the mutant. Equal staining of the actin control in both the wild-type and mutant embryos confirms equal loading across the wells (Fig. 3B).
sapje-like carries a mutation in the donor splice junction of exon 62 of the zebrafish dystrophin gene
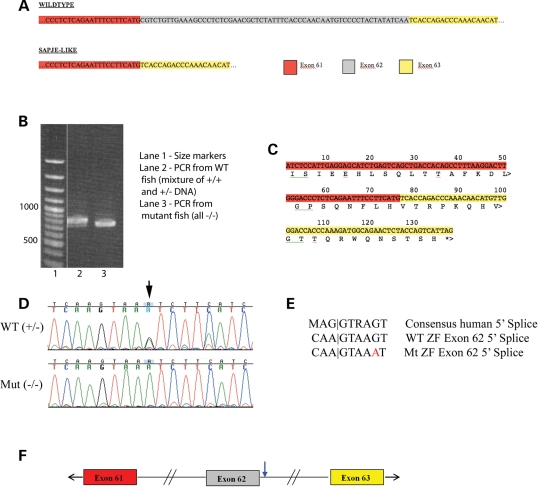
The birefringence phenotype, complementation results and protein analysis strongly suggest that the sapje-like causative mutation resides in the dystrophin gene. In humans, the dystrophin gene spans 2.5 million bases of the human X chromosome and previous work suggests the gene might be equally as large in zebrafish (19). Because the zebrafish dystrophin gene has not been cloned, it was difficult to conclusively identify zebrafish orthologues to the 79 human dystrophin exons. To facilitate sequencing the dystrophin coding sequence, sections of the zebrafish dystrophin gene transcript were PCR amplified in sections from cDNA using primers designed to the more conserved sequences. Resulting PCR products were sequenced from sapje-like mutant fish and compared with the sequence from unaffected siblings. Sequencing these products revealed that sapje-like mRNA was missing dystrophin exon 62 since the sequence went directly from exon 61 to 63 (Fig. 4A). In addition, using primers homologous to exons 57 and 63, a clear size difference is seen in the sapje-like PCR product, suggesting a portion of the cDNA is missing in the mutant transcript (Fig. 4B). The smaller mutant band can still be faintly seen in the unaffected siblings, some of which are heterozygous for the mutation. For both the sequencing analysis (Fig. 4A) and the PCR experiment (Fig. 4B), the same cDNA preparations were used and were made by phenotypically grouping the embryos. The removal of exon 62 results in a translational frameshift yielding an inadvertent termination signal near the end of exon 63 (Fig. 4C).
Figure 4.
Sapje-like has a mutation in the donor splice junction of dystrophin exon 62. (A) Based on sequencing the zebrafish dystrophin cDNA, zebrafish exon 61 is directly adjacent to exon 63 in the sapje-like mutant and missing exon 62. (B) Using primers within zebrafish dystrophin exons 57 and 63, PCR amplification of the zebrafish cDNA templates produces a smaller PCR product in the sapje-like mutant than in the unaffected siblings as assayed by agarose gel electrophoresis. (C) When exon 61 is directly adjacent to exon 63 as in sapje-like, this shifts the reading frame of exon 63 resulting in a premature stop codon at the end of exon 63. (D) Sequencing of the 5′ splice site adjacent to exon 62 reveals a G to A base change (see arrow). (E) Comparison of the human consensus 5′ splice site with the wild-type and mutant zebrafish dystrophin exon 62 5′ splice site. The sapje-like mutation resides at position +5 (G→A) and is highlighted in red. (F) The sapje-like mutation is located in the Exon 62 donor splice junction (blue arrow).
To identify the mutation, splice sites between exons 61-62 and exons 62-63 were sequenced from genomic DNA in mutant and wild-type embryos. A mutation was found in the +5 position of the exon 62 splice donor (Fig. 4D). Based on the human consensus splice site (Fig. 4E), this base is normally present 80% of the time (34). This base change was homozygous in only affected sapje-like embryos and was not found in any of the other strains of mutants that were analyzed (20 individual fish were tested). The location of the mutation within the gene is diagrammatically shown in Figure 4F.
As a means of confirming the nature of the splice mutation, the Leiden Muscular Dystrophy database (http://www.dmd.nl/) (35) was searched to determine if similar mutations were causative in human patient populations. A +5G→A splice site mutation was identified in humans for seven different exons (exons 4, 7, 32, 41, 43, 55 and 67), suggesting that such changes can cause disease. Importantly, the wild-type 5′ exon 62 zebrafish splice junction does not conform to the consensus human splice site at the −1 position (A versus T). This position is present in 80% of the splice sites although A is present 10% of the time (34). It is possible that this difference in the wild-type splice junction at the −1 position may make the zebrafish 5′ exon 62 splice junction more sensitive to the +5G→A mutation. Importantly, the human exon 55 mutation (which has a +5G→A splice site mutation; 36) has the same change at the −1 position and is identical to the zebrafish exon 62 splice donor suggesting that this may be the case.
Sequence conservation of the zebrafish dystrophin gene
As part of identifying the sapje-like mutation, the entire dystrophin coding sequence was determined by designing primers to conserved exons and using PCR amplification of the dystrophin coding sequence from cDNA. Since the sequence was assembled by overlapping ends of PCR products rather than sequencing an entire EST, it is possible (although unlikely) that portions of two highly conserved genes could be assembled together although all coding sequences position on the same genomic sequence suggesting that the entire sequence is part of the same large gene. To fill in gaps in the zebrafish cDNA, five small missing sections were added by including zebrafish EST sequences from the Genbank database. Blast homology results suggest that zebrafish dystrophin is more closely related to human dystrophin (57% identical/74% similar from exons 2–78) than human utrophin (33% identical/53% similar from exons 2–57 and 38% identical/56% similar from exons 34–78). When zebrafish dystrophin was compared with the known sequences of zebrafish utrophin (37), there was no significant homology. Taken together, these data suggest that all of the amplified exons likely belong to the zebrafish dystrophin gene.
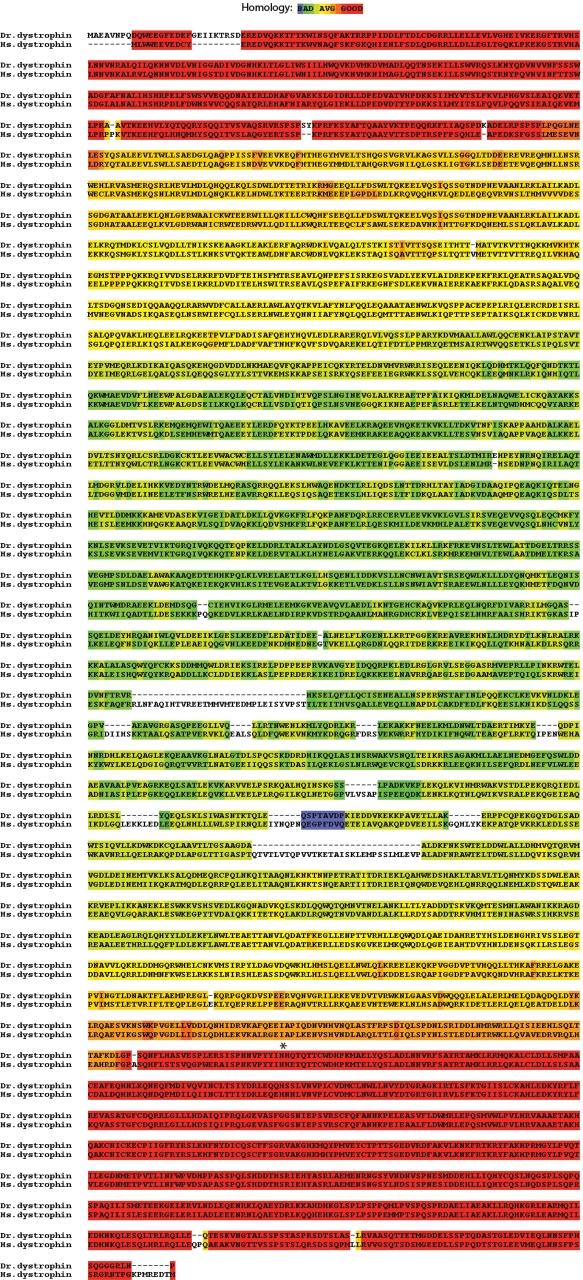
To our knowledge, zebrafish are the most evolutionarily diverged vertebrate from human from which the dystrophin sequence has been fully sequenced. As a means of determining which portions of the dystrophin sequence were most conserved among vertebrates, the zebrafish dystrophin sequence was compared with the human sequence using the Tcoffee algorithm (Fig. 5) (38–42). In this figure, highly conserved sequences are shown in red and less-conserved regions in blue. As is clearly evident, the N- (the actin binding region) and C-terminus (the DAPC interacting region) of the zebrafish dystrophin sequence are highly conserved with the most variability present in the middle spectrin repeat section of the protein. Interestingly, when zebrafish cDNA sequence was compared with the genomic sequence, all intron/exon boundaries found in humans were conserved in zebrafish with the exception that zebrafish exon 79 does not encode protein (data not shown). Given the low degree of conservation of the spectrin repeat region of the protein, it was somewhat unexpected to see that the intron/exon boundaries were maintained even in this genomic region.
Figure 5.
Conservation of the zebrafish dystrophin protein sequence. The zebrafish dystrophin sequence is designated ‘Dr. dystrophin’ and the human sequence is designated ‘Hs. Dystrophin’. The degree of homology has been evaluated by the Tcoffee alignment program (38,39) on a blue (not conserved) to red (highly conserved) scale. The location of the zebrafish sapje-like mutation is shown with an asterisk.
DISCUSSION
Because of their small size, proliferative capacity and transparency, zebrafish are quickly becoming an ideal complementary model to mice for the study of human disease. While further removed from humans than mice, zebrafish offer many advantages for identifying new diseases genes and investigating potential therapies through cell transfer experiments or large-scale chemical screens. Over the last eight years, a number of laboratories have established the zebrafish as a disease model for muscular dystrophy and morpholino experiments have confirmed the phenotype of a dystrophic fish at 5 dpf as inactive, bent and having disorganized muscle (reviewed in 16).
In 1996, a large scale genetic screen was completed in which a small group of muscle degenerative mutants were labeled as potentially dystrophic (22). Bassett et al. characterized the first of these mutants and showed that sapje carries a nonsense mutation in exon 4 of the dystrophin gene (23). Three other mutants have since been characterized. Two carry a mutation in laminin α2 and one carries a mutation in titin (24,25). Because this group of mutants is limited, an early-pressure genetic screen was conducted to identify additional zebrafish muscle degenerative mutants. One of these mutants (sapje-like) has a phenotype similar to sapje and our analysis shows that this recessive mutant carries a splice mutation in exon 62 of the zebrafish dystrophin gene. In addition to being a unique dystrophin mutant, sapje-like also confirms the dystrophin null-mutation described previously for sapje (23).
While mutations in the human muscle dystrophin isoform cause muscular dystrophy, there are many other human forms of dystrophin that are transcribed from alternative promoters (43). In humans, there are four full-length 427 kDa dystrophin isoforms that are transcribed in brain (Dp427c), lymphocyte (Dp427l), purkinje cells (Dp427p) and muscle (Dp427m). In addition, there are many other shorter dystrophin isoforms including Dp260 (retinal dystrophin), Dp140 (brain), Dp116 (Schwann cells), Dp71 (ubiquitous) and Dp40 (ubiquitous). While expression of these isoforms have not yet been reported in zebrafish, the western blot using the C-terminus antibody (recognizes the region conserved in all isoforms) specifically recognizes a 260 kDa dystrophin isoform down-regulated in the sapje-like mutant (Fig. 3A). This suggests that zebrafish express at least full-length and Dp260 dystrophin isoforms. Since the sapje-like mutation lies just upstream of where the DP71 promoter would be (in the intron between exon 62 and 63), all dystrophin isoforms larger than DP71 should be affected. As in human, defects in muscle Dp427m cause the most overt phenotype (muscular dystrophy) although there are additional defects in dystrophic patients including cognitive defects which may be caused by changes in other dystrophin isoforms. It is unclear whether the sapje-like mutation causes cognitive impairment or visual defects although direct physical manifestations resulting from the mutation are almost identical to sapje in which only the largest dystrophin isoform is affected.
While genomic deletions cause the vast majority of the cases of Duchenne Muscular Dystrophy, splicing defects can cause muscular dystrophy and many other diseases. Sapje-like represents the first zebrafish dystrophic mutant characterized to date that has dystrophy due to a splicing defect. As such, future drug screens using sapje-like are likely to uncover therapeutics that work in a completely different manner than drugs which would be able to correct sapje which carries a nonsense mutation in exon 4 of the zebrafish dystrophin gene. Since the muscular dystrophy phenotype in sapje-like can be easily and specifically assayed using birefringence, this makes sapje-like valuable not just for finding drugs capable of correcting muscular dystrophy, but potentially any other disease caused by splicing defects.
The location of the sapje-like splicing defect was identified in the donor splice site of exon 62. The loss of exon 62 was first noted experimentally by sequencing the complete zebrafish dystrophin cDNA in affected and unaffected siblings. The fragment that was initially sequenced was amplified using primers in exons flanking exon 62 (primers located in exon 57 and 63). In this scenario, the only product that could amplify in the splicing mutant was a transcript that skipped exon 62 (since that exon was incapable of splicing and would yield a PCR product too big for amplification and sequencing). While spliced (exon 61 to exon 63) and unspliced (exon 62 to intron to exon 63) transcript may exist in the sapje-like mutant, no full-length dystrophin protein is produced and the muscle degenerates due to the dystrophic phenotype.
Sapje-like is a novel zebrafish dystrophin mutant isolated as part of a large genetic screen. Its characterization has shown that it has symptoms of muscle degeneration similar to that seen in human patients with Duchenne Muscular Dystrophy. We have identified the molecular lesion as a G→A mutation in the donor splice junction of zebrafish dystrophin exon 62. Sapje-like is the first characterized dystrophic mutant which was not isolated as part of the 1996 Tuebingen screen. Since many of the Tuebingen dystrophic mutants have already been characterized and there are at least 30 different forms of muscular dystrophy, this suggests that additional dystrophic mutants remain to be identified and validates our approach to genetically isolate additional mutants as part of our own genetic screen. The identification of sapje-like suggests that our screen was successful and that the characterization of our remaining muscle mutants will help elucidate additional disease mechanisms.
MATERIALS AND METHODS
Early-pressure screen
As outlined in Supplementary Material, Figure S1 and described in (44), 80 male AB strain adult zebrafish were mutagenized with three treatments of 3 mm ENU (one treatment per week). Following treatment, 27 males were found to be fertile. After clearing the males by random outcrosses for a month, the mutation rate was assessed by complementation analysis with heterozygous albino mutants. Of 2400 scored embryos, 2 albinos were identified suggesting a mutation rate of approximately 1.2 × 10−3 mutations per albino locus. This mutation rate is similar to that from previously published reports. Mutagenized males were crossed with wild-type AB females to create a stock of heterozygous carriers which were grown to adulthood.
To perform the early-pressure screen, heterozygous females were separated from the males. The night before squeezing, the females were primed by placing in a tank with males although each sex was separated by a screen barrier. In the morning, the males were sacrificed and the testes were ground in Hanks Buffer to make a sperm stock. The sperm was subjected to UV irradiation for 3 min to crosslink the DNA. The females were anesthetized and systematically squeezed to extract the eggs and the eggs were fertilized with the UV-irradiated sperm. After squeezing, the females were placed in numbered tanks to be later correlated with the clutch of scored eggs. The sperm was activated by adding 1 ml of fish water and after 1 min 24 s, the fertilized eggs were subjected to high pressure (8 000 pounds/inch) for an additional 4 min 36 s to inhibit the second phase of meiosis (polar body extrusion). Pressure was gently released and the eggs were placed in a plate with fish water. The following day, the dead eggs were removed and the clutch was counted if more than five eggs were viable (norm estimated at 10–30 eggs). Gross phenotype was noticed at 1 dpf and muscle structure, activity and bending confirmation assayed at 5 dpf. If the clutch showed any mutants of interest (recombination can make telomeric mutations hard to score by EP), the squeezed mother was outcrossed to wild-type AB males and the resultant progeny incrossed to genetically verify phenotype. The screen was complete when 500 viable clutches were scored. Of these, eight showed a muscle-specific phenotype (not gross abnormalities) including three which show the hallmarks of muscular dystrophy (muscle degeneration over time).
Birefringence
Muscle birefringence was analyzed by placing anesthesized embryos on a glass polarizing filter and covering the embryos with second polarizing filter elevated to prevent the embryos from being harmed. The filters were then placed on an underlit dissecting scope (Leica, model MZ75) and the top polarizing filter twisted until only the light refracting through the striated muscle was visible. Since the degree of birefringence is affected by the horizontal orientation of the fish, the fish were gently oscillated back and forth to account for differences in positioning.
Complementation analysis
Complementation analysis was performed twice. First to assay the mutation rate by crossing mutagenized males with heterozygous albino females. Second, complementation was used to determine if sapje and sapje-like were allelic by crossing heterozygous sapje-like mutants with heterozygous sapje mutants. Resultant progeny were scored using birefringence.
Immunoblot analysis
For separation of large proteins like dystrophin, 3–8% Tris–Acetate gradient gels were used and electrophoresis was at 150 V for ∼2 h. Following separation, proteins were transferred to 0.45 µm nitrocellulose membranes in a submerged transfer apparatus according to the instructions provided by the manufacturer (Invitrogen). For separation of smaller proteins (<100 kDa), 12% Tris–Glycine gradient gels (4% stacking) were used and electrophoresis was at 130 V for ∼2 h. Following separation, the proteins were transferred to 0.45 µm nitrocellulose using a Semi-Dry Transfer Apparatus according to the instructions provided by the manufacturer (Bio-Rad). All antibodies used have either been described previously or are commercially available.
Membranes were incubated with the following antibodies in a cocktail of 1× PBS (phosphate-buffered saline), 0.1% Tween-20 and 5% non-fat dry milk (NFDM): ‘DysC’ (1:333 dilution) (19), ‘DysN’ (1:333 dilution) (18), δ-sarcoglycan (1:300 dilution) (12,19), γ-sarcoglycan ‘γ-SG’ (1:333 dilution) (18), β-sarcoglycan ‘β-SG’ (Novocastra) (1:20 dilution) (19), β-dystroglycan ‘β-DG’ (Novocastra) (1:10 dilution) (19) and actin ‘A2066 ‘(Sigma) (1:300 dilution) (19). Membranes were incubated with primary antibodies for 1 h at 25°C then washed two times quickly and three times for 10 min in a solution of 1× PBS with 0.1% Tween-20.
An anti-rabbit-conjugated goat HRP secondary antibody (1:5000 dilution) (Jackson ImmunoResearch Lab) was used to detect the following primary antibodies: ‘DysC’, ‘DysN’, ‘δ-SG’, ‘γ-SG’ and actin ‘A2066’. An anti-mouse-conjugated goat HRP secondary antibody (1:5000 dilution) (Jackson ImmunoResearch Lab) was used to detect the ‘β-SG’ and ‘β-DG’ primary antibodies. The membranes were incubated with 1:5000 dilution of secondary antibody in a solution of 1× PBS with 0.1% Tween-20 and 5% NFDM for 1 h at 25°C before washing two times quickly and three times for 10 min in a solution of 1× PBS with 0.1% Tween-20. The blots were incubated with Western Lighting Chemiluminescence Reagent Plus (Perkin Elmer) as described by the distributor and exposed to film for up to 5 min.
Immunohistochemistry
Juvenile (5-day-old) zebrafish were anesthetized in 0.3% tricaine solution for 2–10 min, coated in Optimal Cutting Temperature Compound (Sakura) and frozen in liquid nitrogen-cooled 4-methyl butane. 5–10 µm sections (as indicated) were cut at −20°C using a Microm HM 505 E cryostat, transferred to silane-coated microscope slides and fixed by immersing the slides in methanol for 3 min before equilibrating in 1× PBS at 4°C overnight.
To block non-specific binding of the antibodies, sections were incubated for 30 min at room temperature in PBS+10% horse serum. Following blocking, the slides were incubated overnight at 4°C using the same antibodies described for immunoblot analysis. Slides were washed three times in 1× PBS and sections incubated with FITC-conjugated secondary antibodies (Jackson ImmunoResearch Lab) at a 1:100 dilution for 1 h at room temperature. Slides were washed three times in 1× PBS before mounting in 25 µl vectashield. Slides were analyzed using a Zeiss Axioplan II microscope with a triple filter and the images stored using either IP Lab or OpenLab Software.
Sequencing the zebrafish dystrophin gene
Zebrafish cDNA was synthesized for both wild-type and sapje-like 5 dpf embryos by first isolating total RNA with a QIAGEN RNA isolation kit. The poly A containing mRNA was used as a template for cDNA synthesis using a One-Cycle cDNA Synthesis Kit (Affymetrix). To specifically amplify segments of the zebrafish dystrophin gene, PCR primers were designed by finding regions of zebrafish chromosome 1 exon sequences which could encode peptide fragments having high homology to the analogous section of the human dystrophin protein sequence. Using this approach, 20 PCR reactions were designed to amplify the complete dystrophin cDNA in overlapping fragments. Overall, 12 000 bases of the zebrafish dystrophin cDNA were sequenced with a few remaining bases filled in using pre-existing EST sequences available in Genbank. For identification of the splice site mutation, primers were designed from genomic sequence outside the splice regions and the region of interest amplified from wild-type and mutant genomic DNA.
PCR reactions were performed at 35 cycles of 1 min at 94°C, 2 min at 52°C, and 1 min at 72°C using a GC 2 PCR kit (Clontech). Finished PCR reactions were purified with a QIAGEN PCR Cleanup Kit and prepared for sequencing.
All DNA sequencing was performed in The Mental Retardation Research Center Sequencing Core Facility located within Children's Hospital Boston. Sequencing reactions were prepared with ∼50 ng of PCR product and 10 pm of primer in a 12 µl reaction volume. DNA was fluorescently labeled and analyzed on an Applied Biosystems 3730 DNA Analyzer. The sequences were imported into Sequencher and comparisons made between wild-type and sapje-like DNA and cDNA sequence to identify the exon missing in the mutant fish and the causative mutation. In addition, at least 10 linked SNPs were also identified as part of this analysis.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a National Institutes of Health Program Project Grant (grant number 5P50NS040828-07 to L.M.K.); the Bernard F. and Alva B. Gimbel Foundation ( to L.M.K.) and a Developmental Grant awarded by the Muscular Dystrophy Association (to J.R.G). Funding for the Children's Hospital Sequencing Facility was provided by a NIH grant to the Mental Retardation and Developmental Disabilities Research Center (MRDDRC) Molecular Genetics Core Facility at Children's Hospital Boston (grant number 5P30HD018655-26).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge members of the Kunkel and Zon laboratory for their experimental advice, help in performing these experiments and the drafting of this manuscript. Specifically, we would like to thank Dr Yi Zhou and Dr Len Zon (both of Children's Hospital Boston) for their helpful advice and providing necessary fishroom space. In addition, we are grateful to Chris Lawrence and Tim Jones who managed our fish facility. We would also like to thank members of the Children's Hospital Sequencing Facility including Christine Briggs and Hal Schneider for sequencing the zebrafish dystrophin PCR products. L.M.K. is an investigator of the Howard Hughes Medical Institute.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Burghes A.H., Logan C., Hu X., Belfall B., Worton R.G., Ray P.N. A cDNA clone from the Duchenne/Becker muscular dystrophy gene. Nature. 1987;328:434–437. doi: 10.1038/328434a0. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman E.P., Brown R.H., Jr, Kunkel L.M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 3.Monaco A.P., Neve R.L., Colletti-Feener C., Bertelson C.J., Kurnit D.M., Kunkel L.M. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323:646–650. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- 4.Hoffman E.P., Fischbeck K.H., Brown R.H., Johnson M., Medori R., Loike J.D., Harris J.B., Waterston R., Brooke M., Specht L., et al. Characterization of dystrophin in muscle-biopsy specimens from patients with Duchenne's or Becker's muscular dystrophy. N. Engl. J. Med. 1988;318:1363–1368. doi: 10.1056/NEJM198805263182104. [DOI] [PubMed] [Google Scholar]
- 5.Flanigan K.M., von Niederhausern A., Dunn D.M., Alder J., Mendell J.R., Weiss R.B. Rapid direct sequence analysis of the dystrophin gene. Am. J. Hum. Genet. 2003;72:931–939. doi: 10.1086/374176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arahata K., Ishiura S., Ishiguro T., Tsukahara T., Suhara Y., Eguchi C., Ishihara T., Nonaka I., Ozawa E., Sugita H. Immunostaining of skeletal and cardiac muscle surface membrane with antibody against Duchenne muscular dystrophy peptide. Nature. 1988;333:861–863. doi: 10.1038/333861a0. [DOI] [PubMed] [Google Scholar]
- 7.Bonilla E., Samitt C.E., Miranda A.F., Hays A.P., Salviati G., DiMauro S., Kunkel L.M., Hoffman E.P., Rowland L.P. Duchenne muscular dystrophy: deficiency of dystrophin at the muscle cell surface. Cell. 1988;54:447–452. doi: 10.1016/0092-8674(88)90065-7. [DOI] [PubMed] [Google Scholar]
- 8.Zubrzycka-Gaarn E.E., Bulman D.E., Karpati G., Burghes A.H., Belfall B., Klamut H.J., Talbot J., Hodges R.S., Ray P.N., Worton R.G. The Duchenne muscular dystrophy gene product is localized in sarcolemma of human skeletal muscle. Nature. 1988;333:466–469. doi: 10.1038/333466a0. [DOI] [PubMed] [Google Scholar]
- 9.Araishi K., Sasaoka T., Imamura M., Noguchi S., Hama H., Wakabayashi E., Yoshida M., Hori T., Ozawa E. Loss of the sarcoglycan complex and sarcospan leads to muscular dystrophy in beta-sarcoglycan-deficient mice. Hum. Mol. Genet. 1999;8:1589–1598. doi: 10.1093/hmg/8.9.1589. [DOI] [PubMed] [Google Scholar]
- 10.Bonnemann C.G., Modi R., Noguchi S., Mizuno Y., Yoshida M., Gussoni E., McNally E.M., Duggan D.J., Angelini C., Hoffman E.P. Beta-sarcoglycan mutations cause autosomal recessive muscular dystrophy with loss of the sarcoglycan complex. Nat. Genet. 1995;11:266–273. doi: 10.1038/ng1195-266. [DOI] [PubMed] [Google Scholar]
- 11.Lim L.E., Duclos F., Broux O., Bourg N., Sunada Y., Allamand V., Meyer J., Richard I., Moomaw C., Slaughter C., et al. Beta-sarcoglycan: characterization and role in limb-girdle muscular dystrophy linked to 4q12. Nat. Genet. 1995;11:257–265. doi: 10.1038/ng1195-257. [DOI] [PubMed] [Google Scholar]
- 12.Nigro V., Moreira E.d.S., Piluso G., Vainzof M., Belsito A., Politano L., Puca A.A., Passos-Bueno M.R., Zatz M. Autosomal recessive limb-girdle muscular dystrophy (LGMD2F) is caused by a mutation in the delta-sarcoglycan gene. Nat. Genet. 1996;14:195–198. doi: 10.1038/ng1096-195. [DOI] [PubMed] [Google Scholar]
- 13.Noguchi S., McNally E.M., Othmane K.B., Hagiwara Y., Mizuno Y., Yoshida M., Yamamoto H., Bonnemann C.G., Gussoni E., Denton P.H., et al. Mutations in the dystrophin-associated protein gamma-sarcoglycan in chromosome 13 muscular dystrophy. Science. 1995;270:819–822. doi: 10.1126/science.270.5237.819. [DOI] [PubMed] [Google Scholar]
- 14.Odom G.L., Gregorevic P., Chamberlain J.S. Viral-mediated gene therapy for the muscular dystrophies: successes, limitations and recent advances. Biochim. Biophys. Acta. 2007;1772:243–262. doi: 10.1016/j.bbadis.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunkel L.M., Bachrach E., Bennett R.R., Guyon J., Steffen L. Diagnosis and cell-based therapy for Duchenne muscular dystrophy in humans, mice, and zebrafish. J. Hum. Genet. 2006;51:397–406. doi: 10.1007/s10038-006-0374-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guyon J.R., Steffen L.S., Howell M.H., Pusack T.J., Lawrence C., Kunkel L.M. Modeling human muscle disease in zebrafish. Biochim. Biophys. Acta. 2007;1772:205–215. doi: 10.1016/j.bbadis.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Cheng L., Guo X.F., Yang X.Y., Chong M., Cheng J., Li G., Gui Y.H., Lu D.R. Delta-sarcoglycan is necessary for early heart and muscle development in zebrafish. Biochem. Biophys. Res. Commun. 2006;344:1290–1299. doi: 10.1016/j.bbrc.2006.03.234. [DOI] [PubMed] [Google Scholar]
- 18.Guyon J.R., Mosley A.N., Jun S.J., Montanaro F., Steffen L.S., Zhou Y., Nigro V., Zon L.I., Kunkel L.M. Delta-sarcoglycan is required for early zebrafish muscle organization. Exp. Cell Res. 2005;304:105–115. doi: 10.1016/j.yexcr.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 19.Guyon J.R., Mosley A.N., Zhou Y., Davidson A.J., Sheng X., Chiang K., Brien K.F.O., Volinski J.M., Zon L.I., Kunkel L.M. The dystrophin associated protein complex in zebrafish. Hum. Mol. Genet. 2003;12:601–615. [PubMed] [Google Scholar]
- 20.Nixon S.J., Wegner J., Ferguson C., Mery P.F., Hancock J.F., Currie P.D., Key B., Westerfield M., Parton R.G. Zebrafish as a model for caveolin-associated muscle disease; caveolin-3 is required for myofibril organization and muscle cell patterning. Hum. Mol. Genet. 2005;14:1727–1743. doi: 10.1093/hmg/ddi179. [DOI] [PubMed] [Google Scholar]
- 21.Parsons M.J., Campos I., Hirst E.M., Stemple D.L. Removal of dystroglycan causes severe muscular dystrophy in zebrafish embryos. Development. 2002;129:3505–3512. doi: 10.1242/dev.129.14.3505. [DOI] [PubMed] [Google Scholar]
- 22.Granato M., Eeden F.J.v., Schach U., Trowe T., Brand M., Furutani-Seiki M., Haffter P., Hammerschmidt M., Heisenberg C.P., Jiang Y.J., et al. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development. 1996;123:399–413. doi: 10.1242/dev.123.1.399. [DOI] [PubMed] [Google Scholar]
- 23.Bassett D.I., Bryson-Richardson R.J., Daggett D.F., Gautier P., Keenan D.G., Currie P.D. Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development. 2003;130:5851–5860. doi: 10.1242/dev.00799. [DOI] [PubMed] [Google Scholar]
- 24.Hall T.E., Bryson-Richardson R.J., Berger S., Jacoby A.S., Cole N.J., Hollway G.E., Berger J., Currie P.D. The zebrafish candyfloss mutant implicates extracellular matrix adhesion failure in laminin alpha2-deficient congenital muscular dystrophy. Proc. Natl. Acad. Sci. USA. 2007;104:7092–7097. doi: 10.1073/pnas.0700942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steffen L.S., Guyon J.R., Vogel E.D., Howell M.H., Zhou Y., Weber G.J., Zon L.I., Kunkel L.M. The zebrafish runzel muscular dystrophy is linked to the titin gene. Dev. Biol. 2007;309:180–192. doi: 10.1016/j.ydbio.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helbling-Leclerc A., Zhang X., Topaloglu H., Cruaud C., Tesson F., Weissenbach J., Tomé F.M., Schwartz K., Fardeau M., Tryggvason K., et al. Mutations in the laminin alpha 2-chain gene (LAMA2) cause merosin-deficient congenital muscular dystrophy. Nat. Genet. 1995;11:216–218. doi: 10.1038/ng1095-216. [DOI] [PubMed] [Google Scholar]
- 27.Knoll R., Hoshijima M., Hoffman H.M., Person V., Lorenzen-Schmidt I., Bang M.L., Hayashi T., Shiga N., Yasukawa H., Schaper W., et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- 28.Itoh-Satoh M., Hayashi T., Nishi H., Koga Y., Arimura T., Koyanagi T., Takahashi M., Hohda S., Ueda K., Nouchi T., et al. Titin mutations as the molecular basis for dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 2002;291:385–393. doi: 10.1006/bbrc.2002.6448. [DOI] [PubMed] [Google Scholar]
- 29.Gerull B., Atherton J., Geupel A., Sasse-Klaassen S., Heuser A., Frenneaux M., McNabb M., Granzier H., Labeit S., Thierfelder L. Identification of a novel frameshift mutation in the giant muscle filament titin in a large Australian family with dilated cardiomyopathy. J. Mol. Med. 2006;84:478–483. doi: 10.1007/s00109-006-0060-6. [DOI] [PubMed] [Google Scholar]
- 30.Haravuori H., Vihola A., Straub V., Auranen M., Richard I., Marchand S., Voit T., Labeit S., Somer H., Peltonen L., et al. Secondary calpain3 deficiency in 2q-linked muscular dystrophy: titin is the candidate gene. Neurology. 2001;56:869–877. doi: 10.1212/wnl.56.7.869. [DOI] [PubMed] [Google Scholar]
- 31.Hackman P., Vihola A., Haravuori H., Marchand S., Sarparanta J., De Seze J., Labeit S., Witt C., Peltonen L., Richard I., et al. Tibial muscular dystrophy is a titinopathy caused by mutations in TTN, the gene encoding the giant skeletal-muscle protein titin. Am. J. Hum. Genet. 2002;71:492–500. doi: 10.1086/342380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schapira G., Dreyfus J.C., Joly M. Changes in the flow birefringence of myosin as a result of muscular atrophy. Nature. 1952;170:494–495. doi: 10.1038/170494b0. [DOI] [PubMed] [Google Scholar]
- 33.Patton E.E., Zon L.I. The art and design of genetic screens: zebrafish. Nat. Rev. Genet. 2001;2:956–966. doi: 10.1038/35103567. [DOI] [PubMed] [Google Scholar]
- 34.Zhang M.Q. Statistical features of human exons and their flanking regions. Hum. Mol. Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]
- 35.Aartsma-Rus A., Van Deutekom J.C., Fokkema I.F., Van Ommen G.J., Den Dunnen J.T. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- 36.Taylor P.J., Maroulis S., Mullan G.L., Pedersen R.L., Baumli A., Elakis G., Piras S., Walsh C., Prosper-Gutierrez B., De La Puente-Alonso F., et al. Measurement of the clinical utility of a combined mutation detection protocol in carriers of Duchenne and Becker muscular dystrophy. J. Med. Genet. 2007;44:368–372. doi: 10.1136/jmg.2006.047464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bohm S., Jin H., Hughes S.M., Roberts R.G., Hinits Y. Dystrobrevin and dystrophin family gene expression in zebrafish. Gene Expr. Patt. 2008;8:71–78. doi: 10.1016/j.modgep.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 39.Poirot O., O'Toole E., Notredame C. Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Notredame C., Abergel C. Using multiple alignment methods to assess the quality of genomic data analysis. In: Andrade M., editor. Bioinformatics and Genomes: Current Perspectives. Norfolk: Horizon Scientific Press; 2003. pp. 30–50. [Google Scholar]
- 41.Notredame C., Holme L., Higgins D.G. COFFEE: a new objective function for multiple sequence alignmnent. Bioinformatics. 1998;14:407–422. doi: 10.1093/bioinformatics/14.5.407. [DOI] [PubMed] [Google Scholar]
- 42.O'Sullivan O., Suhre K., Abergel C., Higgins D.G., Notredame C. 3DCoffee: Combining protein sequences and structures within multiple sequence alignments. J. Mol. Biol. 2004;340:385–395. doi: 10.1016/j.jmb.2004.04.058. [DOI] [PubMed] [Google Scholar]
- 43.Sadoulet-Puccio H.M., Kunkel L.M. Dystrophin and its isoforms. Brain Pathol. 1996;6:25–35. doi: 10.1111/j.1750-3639.1996.tb00780.x. [DOI] [PubMed] [Google Scholar]
- 44.Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 4th edn. Eugene: University of Oregon Press; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.