Abstract
Long-lived memory B cells (BMem) provide an archive of historic Ab responses. By contrast, circulating Abs typically decline once the immunogen is cleared. Consequently, circulating Abs can underestimate the nature of cognate humoral immunity. On the other hand, the BMem pool should provide a comprehensive picture of Ab specificities that arise over the entire course of infection. To test this hypothesis, we compared circulating Ab and BMem from natural virus suppressors who control HIV-1 without therapy and maintain a relatively intact immune system. We found high frequencies of BMem specific for the conserved neutralizing CD4 induced or CD4 binding site epitopes of gp120, whereas low Ab titers to these determinants were detected in contemporaneous plasma. These data suggest that plasma Ab repertoires can underestimate the breadth of humoral immunity, and analyses of BMem should be included in studies correlating Ab specificity with protective immunity to HIV-1.
Keywords: CD4i antibody, CD4bs antibody, human monoclonal antibody, natural viral suppressors, elite controller
Although an HIV-1 vaccine continues to be elusive after more than 20 years of effort, it remains the single best hope to stop the epidemic (1, 2). The recent failure of an Ad5-vectored “CTL” vaccine and the earlier failure of a gp120 subunit vaccine are sobering testaments to the difficulty of this task (2). Because of these failures, there is renewed focus on the identification and characterization of protective humoral responses in groups of HIV-1-infected individuals who control their infections. These groups include long-term nonprogressors, who maintain stable CD4 counts without disease over many years (3), and elite controllers (4) or, in our clinic, natural viral suppressors (NVS) (5, 6), who control viral replication to undetectable levels without antiretroviral therapy. Several recent studies have attempted to characterize the anti-Env Abs found in the plasma or sera of rare HIV-infected humans that exhibit broadly neutralizing activity (7, 8). However, the specificities of circulating Abs are likely to change and/or decline significantly over time, given the high mutability of the HIV envelope (9), particularly under conditions whereby antigenemia is limiting. Thus, circulating Ab specificities in chronically HIV-1-infected persons are unlikely to represent the full spectrum of Abs elicited by the virus from the time of early acute infection. Furthermore, potentially important Ab responses that occur during the critical period of acute infection might not be detected by serologic analyses of samples taken after viral loads have declined to setpoint. For example, it is known that elite controllers have lower titers of HIV-1-specific Abs than chronic progressors (10, 11).
One way to overcome this limitation is to census HIV Env-directed Ab specificities in the memory B cell (BMem) compartment. It is well established that long-lived BMem provide a historical archive of Ab specificities that have occurred over much of the host lifespan (12). For example, BMem can persist for 50 years after vaccination with vaccinia (13). By contrast, circulating Abs usually decline after antigen clearance. For instance, up to half of vaccinees lose protective Abs within a few years after vaccination with the hepatitis B virus (HBV) vaccine (14). On the other hand, HBV-specific BMem persist in the absence of Ab and set the stage for rapid protective Ab responses upon exposure to HBV or the vaccine (15–18). These studies provide strong collective evidence that BMem are a highly stable record of prior Ab responses.
Analyses of the BMem compartment in HIV-1-infected persons are inherently difficult because HIV-1 pathogenesis includes significant immune dysfunction that extends to humoral immunity (19, 20). Fortunately, elite controllers or NVS individuals who maintain a relatively intact immune system provide an opportunity to census the archive of humoral immunity elicited by the HIV-1 Env protein during infection. Accordingly, we used a local NVS cohort (5, 6) to compare and contrast archived BMem specificities with contemporaneous plasma Ab repertoires directed against 2 highly conserved, cross-reactive neutralization targets on gp120: the CD4 binding site (CD4bs) and the coreceptor binding domain, which includes a subset of CD4-induced (CD4i) epitopes (21–24). Analyses of the BMem compartment indicate a magnitude of prior Ab response that is not apparent in the contemporaneous plasma samples. These results strongly support the use of BMem analyses to complement serologic studies in attempts to correlate Ab specificities and protective immunity against HIV-1. Furthermore, these studies suggest a means for identifying novel targets for broadly cross-reactive anti-Env neutralizing Abs.
Results
NVS Volunteers.
The clinical characteristics are shown in Table 1 for each NVS volunteer (6). The times since diagnosis were 5 years for NVS9 (53-year-old African American male; risk factor, i.v. drug use), 13 years for NVS10 (57-year-old African American female; risk factor, sex), and 17 years for NVS5 (50-year-old African American female; risk factor, sex). None of these individuals received antiretroviral therapy during this time. Total B cell and BMem frequencies are in the normal range (20), indicating the lack of global immune dysregulation in these individuals (Table 1). NVS9 and NVS10 suppressed viral replication to undetectable levels (<75 copies/mL of plasma) at all times tested (supporting information Fig. S1). By contrast, NVS5 had transient spikes of low-level viremia of up to ≈300 copies/mL of plasma during the first 3 years of observation, followed by ≈4 years of control to undetectable levels (Fig. S1). Interestingly, a viral spike of ≈100 copies/mL of plasma was observed shortly after specimens were obtained for the studies reported here. Importantly, all 3 donors had stable CD4+ T cell counts in the normal range throughout the time of observation (Fig. S1). Taken together, these data show that the 3 NVS volunteers maintain undetectable viral loads over many years in the absence of antiretroviral therapy and that in terms of gross phenotypes, their lymphocyte subsets are normal.
Table 1.
Characterization of 3 NVS donors
| Donor | Year of diagnosis | Sampling date | B cell(PBMC %) | BMem cell(B cell %) | Plasma IgG(mg/mL) |
|---|---|---|---|---|---|
| NVS5 | 1991 | July 2006 | 10.5 | 32.7 | 34.2 |
| NVS9 | 2003 | July 2006 | 10.8 | 52.8 | 13.6 |
| NVS10 | 1995 | July 2006 | 10.6 | 36.0 | 18.4 |
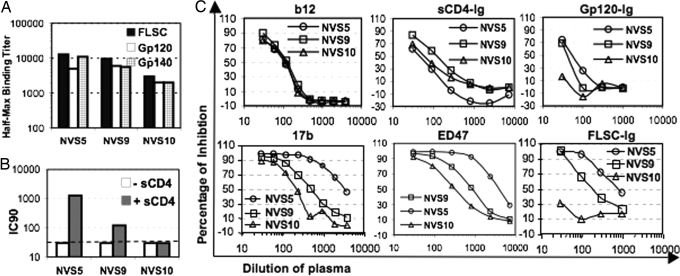
Despite apparent strong control of viral replication, all 3 NVS volunteers remained seropositive for Env epitopes. This was determined by 3 ELISA formats using gp120Ba-L, a full-length single-chain (FLSC) fusion protein of gp120Ba-L and CD4-D1D2, or gp140Ba-L, all based on the HIV-1Ba-L Env protein, as described in Materials and Methods. As shown in Fig. 1A, each volunteer had plasma Abs that recognize each of the 3 antigens, with titers in the range of 10−3 to 10−4. These ELISA formats do not permit epitope-specific discrimination, but they do show that the 3 NVS volunteers are seropositive for Env epitopes at low levels despite undetectable viral loads at the time of specimen collection.
Fig. 1.
Characterization of plasma Abs to HIV-1 Env protein. (A) Plasma Abs against FLSC, gp120Ba-L, and gp140Ba-L were detected by ELISA, and half-maximum binding titers are shown on a log scale. (B) CD4i Abs in plasma of NVS donors were detected by enhanced neutralization of HIV-27312A-V434M in the presence of a subinhibitory concentration of sCD4 (9 nM). IC90 titers are shown; the dashed line is the detection limit. (C) Top shows results of CD4bs Abs in plasma detected by competition ELISA against b12 mAb (Left), competition ELISA against sCD4-Ig (Middle), and inhibition of gp120-Ig binding to cell surface CD4 (Right). Bottom shows results of CD4i Abs in plasma detected by competition ELISA against 17b mAb (Left), competition ELISA against ED47 mAb (Middle) and inhibition of FLSC-Ig binding to cell surface CCR5 (Right). Results of NVS5, NVS9, and NVS10 are shown as circles, squares, and triangles, respectively.
Plasma CD4bs and CD4i Ab Titers.
Of the known conserved neutralization epitopes of HIV-1, those associated with the CD4 and coreceptor binding sites of gp120 are the most consistently immunogenic during infection (21, 25). For this reason, our early analyses focused on the plasma and BMem responses to these epitopes in NVS volunteers. We probed the NVS plasmas for CD4bs Abs using 3 independent competition assays. First, serial 0.5 log plasma dilutions were evaluated for their ability to block binding of a limiting concentration of biotinylated mAb b12 to gp120Ba-L captured on ELISA plates. mAb b12 recognizes a highly conserved neutralization epitope associated with the CD4bs of gp120 (26). Second, serial 0.5 log plasma dilutions were evaluated by capture ELISA for their ability to block binding of soluble (s)CD4-Ig to plates captured with gp120Ba-L. Third, serial 0.5 log plasma dilutions were evaluated by flow cytometry for their ability to block the binding of Allophycocyanin (APC)-tagged gp120-Ig to the CD4+ T cell line, CEM-NKr. As shown in Fig. 1C (Top), the plasma Ab responses to CD4bs epitopes were nil to marginal in all 3 NVS volunteers, with the strongest competitions in any assay format being only 0.5 log above the limit of detection. For example, the half-maximum competition values for each of the plasmas for blocking the binding of b12 were ≈10−2, whereas the background competition is 10−1.5. These marginal titers in each of the 3 independent assay formats strongly suggest that the NVS volunteers studied here have little in the way of ongoing plasma Ab responses to CD4bs epitopes. Similar analyses were also carried out for broadly cross-reactive CD4i Abs that are found in most HIV-1 infected individuals at titers in the 10−3 to 10−5 range by a CD4-triggered neutralization assay using an HIV-2 indicator virus that selectively detects these Abs (21). Using this neutralization assay (Fig. 1B), NVS5 had CD4i-neutralizing Abs, with an IC90 titer of ≈1.5 × 10−3. As expected for CD4i Abs, neutralization was observed only in the presence of limiting concentrations (9 nM) of sCD4 (Fig. 1B). By contrast, NVS9 and NVS10 showed low and negative titers, respectively, in this assay. Thus, the rank order of neutralization in the CD4-triggered assay is NVS5 > NV9 > NVS10.
The CD4i neutralization rank order was confirmed by blocking studies in which the NVS plasmas were probed for competition with the binding of 2 biotinylated CD4i mAbs, 17b and ED47, to FLSC in ELISA and with the binding of fluorescent FLSC-Ig to CCR5 on CfT2h-CCR5 cells. As shown in Fig. 1C (Bottom), NVS5 plasma had higher competition titers in all 3 assays than NVS9 and NVS10. The differences in competition titers for 17b or ED47 were less apparent between NVS9 and NVS10, although the titers were slightly higher for NVS9. By contrast, plasma from NVS9 blocked the binding of fluorescent FLSC-Ig to CfT2h-CCR5 cells at a titer of ≈2 × 10−2, whereas no competition was observed for plasma from NVS10. Taken together, the competition data confirm that the rank order of CD4i Ab responses is NVS5 > NVS9 > NVS10.
Neutralizing Abs in NVS Plasmas.
NVS plasmas were also evaluated in 2 independent “conventional” neutralization formats to further assess their rank order of activity. In the first format, plasma samples from the 3 NVS volunteers were evaluated using a peripheral blood mononuclear cell (PBMC)-based assay. As shown in Table 2, despite the absence of anti-CD4bs Abs, the NVS5 plasma exhibited broad cross-reactivity and neutralized all 10 isolates tested, with IC50 titers ranging from 1.8 × 10−2 to 6.8 × 10−3. By contrast, plasmas from NVS9 and NVS10 neutralized 2 of 10 isolates and 1 of 10 isolates, respectively, at marginal titers in the range 4 × 10−1 to 6 × 10−1.
Table 2.
Neutralization in PBMC format
| Donor | IC50 (dilution of plasma) for HIV-1 strain |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BaL | ADA | 92BR020 | IIIB | 2044 | 2005 | 89.6 | SF2 | 92HT594 | 92HT599 | |
| NVS5 | 380 | 520 | 1,040 | 320 | 240 | 180 | 520 | 6,800 | 320 | 320 |
| NVS9 | <30 | <30 | <30 | <30 | 40 | <30 | <30 | 60 | <30 | <30 |
| NVS10 | <30 | <30 | <30 | <30 | <30 | <30 | <30 | 60 | <30 | <30 |
Similar plasma neutralizing activity was seen in a second format of a cell line-based pseudovirus assay (data not shown). To determine whether the neutralizing activity of plasma is derived from Ab, whole IgG was purified from NVS plasma and evaluated in the second assay format. In accordance with the plasma neutralizing activity, NVS5 IgG neutralized 11 of the 12 pseudoviruses, with IC90 at IgG concentrations ranging from ≈10 μg/mL to 200 μg/mL (Table 3). On the other hand, IgG from NVS9 and NVS10 neutralized 5 of 12 and 3 of 12 pseudoviruses, respectively, at titers in the range of ≈10 μg/mL to 290 μg/mL (Table 3). It should be noted that in both assays (Tables 2 and 3) plasmas or IgGs from NVS9 and NVS10 selectively neutralized X4 viruses or relatively sensitive R5/dual-tropic viruses. Taken together, these data show that the rank order for neutralization is NVS5 ≫ NVS9 ≥ NVS10 in 2 independent assay formats, indicating that NVS5 has an ongoing Ab response that is broadly neutralizing, whereas NVS9 and NVS10 have very weak ongoing neutralizing Ab responses that are narrow in specificity.
Table 3.
Neutralization in Env-pseudotyped virus format
| Donor | IC90 (μg/mL of total IgG) for HIV-1 strain |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6535 | APV6 | APV14 | APV17 | APV19 | BaL | BX08 | MN | NSC | SF162 | JRCSF | NL4–3 | VsVg | |
| NVS5 | 134 | 125 | >300 | 144 | 128 | 193 | 112 | 67 | 12 | 32 | 203 | 90 | >300 |
| NVS9 | >300 | >300 | >300 | 260 | >300 | >300 | >300 | 80 | 9 | 25 | >300 | 243 | >300 |
| NVS10 | >300 | >300 | >300 | >300 | >300 | >300 | >300 | 201 | 80 | 136 | >300 | >300 | >300 |
Natural Viral Suppressors Preserve High Frequency of BMem Specific for Conserved Epitopes of the HIV-1 Env Protein Independent of Serologic Status.
The above results show that although there are only small differences in total circulating Ab titers to gp120 or gp140 epitopes among the NVS subjects, there are marked differences in Ab fine specificity. The Ab responses to CD4bs epitopes were weak to nil in all 3 subjects, and the rank order for circulating Abs to CD4i epitopes was NVS5 > NVS9 > NVS10. Collectively, these data provide no apparent relationship between plasma Ab specificity and control of infection in the 3 NVS subjects. However, given that antigen loads are likely low in these individuals, because they control their infections, it is possible that significant Ab responses have occurred and waned as antigen burdens decrease. In this case, circulating Ab repertoires would provide a poor indication of the initial repertoires that might correlate with the control of infection. Because BMem persist for much of the host lifespan, they should provide a record of these repertoires and might offer a new window through which Ab specificities can be correlated with viral control in our NVS cohort.
To test this hypothesis, we evaluated the NVS subjects for the presence of BMem specific for Env epitopes discordant with those recognized by plasma. First, we established culture conditions and assays to detect anti-Env Abs secreted by single BMem precursors. Our culture conditions were based on those described to polyclonally activate BMem to divide and secrete Abs (27). Preliminary studies showed that culturing 50–100 enriched BMem for 7–14 days was sufficient to induce IgG anti-Env Abs to levels permitting ready analysis of specificity by ELISA. Each culture supernatant was tested in 3 ELISA formats using gp120Ba-L, FLSC, or gp140Ba-L, as described in Materials and Methods. This approach enabled us to differentiate Abs specific for CD4bs and CD4i epitopes from each other and from other HIV-1 Env epitopes. This strategy was validated using human mAbs specific for CD4bs, CD4i, and other HIV-1 Env epitopes. The CD4bs mAbs b12 and M14, as well as CD4-Ig, reacted selectively with gp120, confirming that reactivity with gp120 but not FLSC is indicative of Abs specific for CD4bs epitopes (Fig. S2 A, Middle). By contrast, the CD4i mAbs 17b, ED47, and A32 reacted with FLSC but only poorly when gp120 was used in lieu of FLSC, confirming that reactivity with FLSC but not gp120 detects CD4i-specific mAbs (Fig. S2 A, Left). We were also able to detect Abs specific for other epitopes that are expressed on gp120 as shown by reactivity with mAb 2G12, which binds to carbohydrate epitopes (Fig. S2 A, Right). In this case, there was no difference in binding whether FLSC or gp120 was captured on the plate. Finally, we also evaluated each culture supernatant for reactivity with gp140Ba-L. Because we are comparing binding of single supernatants to multiple Env preparations, it was important to standardize the ELISAs using known mAbs to detect specific binding at levels expected for Abs in BMem culture supernatants. The data shown in Fig. S2 B are indicative of the degrees of binding for different mAbs to gp120, FLSC, or gp140 when their IgG concentrations are at levels typical of those found in supernatants from activated BMem. In the studies that follow, selective reactivity with FSLC is taken as putative CD4i specificity, selective reactivity with gp120 is taken as putative CD4bs specificity, and reactivity with all antigen preparations or gp140 alone is denoted as “Other” Abs. We have confirmed this strategy by mAb isolation from activated BMem (Fig. S3 and unpublished data) and by control studies using either BMem isolated from HIV-1-negative individuals or from CD19+ CD27− cells (naïve B cell) from these NVS subjects. In both cases, no Env-specific precursors were observed.
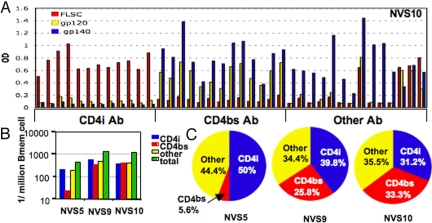
Using these assays, we censused BMem for precursors that recognize CD4bs, CD4i, and “Other” Env epitopes. An example of the type of ELISA data generated is shown in Fig. 2A for NVS10. In this subject, the BMem precursors were approximately equal for CD4i, CD4bs, and “Other” specificities in the range of 200–300 precursors per 106 BMem (Fig. 2 B and C). Further subdivision within the “Other” category is apparent in Fig. 2A, with ≈70% (10 of 14) of the BMem precursors selectively recognizing the gp140 oligomer. We are hesitant at this point to read too much into the apparent specificity for oligomer because the gp140 preparations are heterogeneous in size and perhaps in native structures once they are adsorbed to plastic. This caveat does not change the conclusion that CD4i and CD4bs BMem precursors are prevalent in NVS10, which is not true for the specificities of circulating anti-Env Abs in this individual. A similar picture was found for NVS9 (Fig. 2 B and C), who also had approximately equal frequencies of BMem specific for CD4i, CD4bs, and “Other” Env epitopes in the range of 400–600 precursors per 106 BMem. Again, the prevalence of BMem precursors specific for CD4i and CD4bs epitopes was discordant with the titers of circulating Abs to these epitopes. BMem precursor analysis for NVS5 presented a different and potentially important picture. This individual had very low levels of CD4bs-specific BMem, at ≈20 precursors per 106 BMem. This corresponds to 5.6% of the total Env-specific BMem precursors for this individual and is barely above our limit of detection in this assay format of 10 precursors per 106 BMem. By contrast, the BMem precursor pool was dominated by BMem specific for CD4i and “Other” Env epitopes, corresponding to 50% and 44.4% of detectable Env-specific precursors, respectively (Fig. 2 B and C). The absence of CD4bs-specific plasma Abs and very low levels of CD4bs-specific BMem strongly indicates that the broad neutralizing Ab response for NVS5 is not due to Abs specific for CD4bs epitopes.
Fig. 2.
Identification of HIV-1 Env-specific BMem. One hundred BMem per well were stimulated for 2 weeks, and supernatants were screened for total CD4i, CD4bs, and “Other” HIV-1 Env protein-specific BMem by ELISA using a mixture of anti-κ and anti-λ Abs. (A) Representative ELISA data for supernatants of NVS10. (B) HIV-1 Env-specific BMem in 3 NVS donors are shown as frequency per 106 total BMem. The detection limit is 10 precursors per 106 BMem. (C) Percentages of BMem of CD4i (blue), CD4bs (red), and “Other” specificities (yellow) in each donor (NVS5, NVS9, and NVS10) are shown in a pie chart.
Because our conclusions are based on analysis of supernatants of BMem activated under limiting dilution conditions, it was important to confirm that the Ab specificities found in the supernatants match those of the BMem themselves. This was addressed by mAb isolation using a new algorithm developed for this purpose. A more detailed description of the method will be published elsewhere, but as proof of principle 3 mAbs (N5-I1, N5-I2, and N5-I3) were cloned from BMem in wells whose supernatants were positive for CD4i antibodies. As shown in Fig. S3, all 3 IgG1 mAbs showed strong reactivity with FLSC and low reactivity with gp120 in ELISA. Furthermore, each mAb was encoded by the VH1–69 gene segment (data not shown) that is found in the majority of CD4i mAbs reported to date (28). This is also confirmed by an ongoing analysis of 30 additional Env-specific mAbs isolated from BMem cultures in our NVS cohort (unpublished data). Taken together, the data described above strongly suggest that the specificities of BMem should be evaluated as a component of studies aimed toward correlating Ab specificity and control of HIV-1 infection.
Discussion
The present study tests the hypothesis that analyses of BMem provides a facile and informative method for dissecting past Ab responses against cross-reactive HIV-1 Env epitopes in clinical cohorts in which these responses might not be evident in the circulating Ab pool. Our NVS cohort (5), which comprises HIV-1 infected individuals who control viremia to undetectable levels for many years without antiretroviral therapy, is one such population. These individuals are expected to have low antigen burdens consequent to viral control, and this should be reflected by simultaneously low steady-state Ab responses to certain HIV-1 epitopes. Results from our serologic analyses (Fig. 1) of reactivity with conserved receptor binding sites on gp120 are consistent with this prediction. Our data also agree with recent studies showing that elite controllers and highly active antiretroviral therapy-treated patients with undetectable viremia have lower levels of both Env-binding and neutralizing Abs compared with viremic chronic progressors (10, 11). Nevertheless, our studies show that specificities to conserved neutralizing targets (CD4bs and CD4i epitopes) are clearly evident in the BMem pool of 2 NVS subjects (NVS9 and NVS10) who had little or no cognate Ab titers at the time of plasma collection. Moreover, the high frequencies of these BMem suggest that responses to CD4i and CD4bs epitopes were robust at some point during an earlier stage of infection. Using larger cohorts, it should now be possible to determine whether these archived repertoires correlate with NVS status or some form of transient immunologic control that eventually wanes as viremia is cleared.
Our third NVS subject, NVS5, had broadly neutralizing Abs in the circulation as determined in the CD4-triggered and 2 conventional neutralization assay formats (Fig. 1B, Table 2, and Table 3). This individual had the strongest-binding Ab responses to CD4i epitopes of the NVS subjects, whereas significant responses to CD4bs epitopes were not apparent. This pattern was also reflected in the BMem pool, where ≈50% of the precursors were specific for CD4i epitopes and only a marginal 5.6% were specific for CD4bs epitopes.
It has recently been posited that the broadly neutralizing activity seen in some HIV-positive individuals is attributed to Abs directed against the CD4bs (7, 8) and that natural or vaccine-induced control of infection must rely on such specificity. Because one mAb specific for a CD4bs epitope is broadly neutralizing (26), there is intense interest in identifying such responses in vivo as a correlate of protection (7, 8). Our data showing that NVS5 developed little or no circulating Ab response or BMem precursors specific for CD4bs epitopes over the course of infection yet harbored broadly neutralizing Ig suggest that there are additional and equally important targets for vaccine design. On the basis of our data, it seems unlikely that CD4bs-specific Abs contribute to viral control in NVS5.
NVS5 was also distinct in that there was a good specificity match between the circulating Ab response and the BMem precursor pool. Notably, NVS5 was the individual who exhibited transient, low-level viremias in the range of 100–400 copies during the first 3 years of observation and again in the 8th year of observation, shortly after the specimens studied above were collected (Fig. S1). It is possible that the neutralizing Abs observed in the plasma of NVS5 were elicited in response to the increase in viral load that occurred around the time that the test specimens were collected. Because subsequent analyses of viral loads (Fig. S1) indicate that this rebound is being controlled, NVS5 offers a unique opportunity to implicate particular Ab specificities in the control of infection.
Although our data do not allow us to establish a firm relationship between viral control and Ab specificities, it is interesting to note that CD4i-specific BMem were present at high frequencies in all 3 NVS subjects. Circulating Abs specific for CD4i epitopes were nil to low in the 2 NVS subjects who exhibited tight control of viremias. By contrast, NVS5 exhibited much higher titers of Abs specific for CD4i epitopes in each of the assay formats. This observation is consistent with boosting of these responses by transient viremia and possibly implicates CD4i Abs in the dampening of viral replication in NVS5. At this point it is impossible to establish causality between viral control and the presence of CD4i-specific Abs in NVS5; however, they are consistent with our recent demonstration of a correlation between viral control and circulating Abs specific for CD4i epitopes in rhesus macaques immunized with a version of FLSC in which the CD4 component was derived from rhesus macaques (rhFLSC) (29). It is interesting to note that most HIV-1-infected individuals mount Ab responses to CD4i epitopes and that these responses appear around the time of initial viral control in people (30) and in our rhFLSC vaccine model in rhesus macaques (29).
Overall, our data show that comprehensive analysis of BMem specificity pools will allow for more precise characterization of anti-Env humoral responses than are possible with serologic analyses of plasma Ab responses alone. This view is obvious for subjects NVS9 and NVS10, in whom plasma CD4bs and CD4i responses are low to negative but both sets of specificities are well represented in the BMem pool. Thus, in at least some individuals who strictly control viremia, serologic responses underrepresent the true Ab responses made by that individual at times that the virus was being brought under control. In addition, we have developed a new algorithm to isolate mAbs from BMem precursors that obviates many of the problems encountered with conventional methods, such as hybridomas, EBV transformation, and phage display. This method, coupled with BMem precursor analysis in well-defined controller cohorts, greatly increases the probability that Ab specificity can be correlated with viral control if a causal relationship exists.
Materials and Methods
Subjects and Reagents.
Blood was obtained from NVS donors (5, 6) and normal healthy volunteers under approval of the University of Maryland Institutional Review Board. Plasma was collected from blood after centrifugation and was kept at −80 °C. PBMCs were isolated by Ficoll-Hypaque (Histopaque-1077, Sigma-Aldrich) centrifugation and were stored in liquid nitrogen after resuspension in freezing medium [90% (vol/vol) FBS, DMSO].
Recombinant gp120Ba-L from the HIV-1Ba-L isolate and an FLSC fusion protein of gp120Ba-L and human CD4 D1D2 were produced in the laboratory as described previously (31). An HIV-1Ba-L gp140 oligomer protein (gp140Ba-L) with a chimeric N-terminal gp41 of simian immunodeficiency virus was prepared at Advanced BioScience Laboratories according to published methods (32). Soluble human CD4-D1D2 fused to the Fc domain of human IgG1 (sCD4-Ig) was isolated from 293T cells stably transfected with a sCD4-Ig construct generously provided by Dr. Brian Seed (Massachusetts General Hospital, Boston, MA). Soluble human CD4-D1D4 protein was provided generously by Biogen. An IgG chimera of FLSC (FLSC-Ig) was prepared in our laboratory as described previously (33). An IgG chimera of gp120Ba-L (gp120-Ig) was derived from the FLSC-Ig construct by removal of the CD4-D1D2 coding region and fusing the gp120 region plus the flexible serine-glycine linker of FLSC with the human IgG Fc fragment of the chimera. The resulting gp120-Ig protein was isolated in the laboratory from a stably transfected 293T cell using protein-A Sepharose columns (Sigma-Aldrich) (31). An affinity-purified goat Ab (D7324) specific for the C-terminal peptide of HIV-1 gp120 was purchased from Cliniqa. HIV-1 CD4i mAbs 17b, 19e, ED47, C11, and A32 were purified by protein A affinity chromatography from hybridoma cells or 293T supernatants prepared by transfecting heavy- and light-chain genes encoding the Abs. These mAbs were kindly provided by Dr. James Robinson of Tulane University (New Orleans, LA). CD4bs mAb M14 (34) was kindly provided by Dr. Dimiter Dimitrov of the National Cancer Institute (Frederick, MD). CD4bs mAb b12 (26) was made from 293T cells transfected with synthesized b12-IgG1 expression vectors. The broadly neutralizing mAb 2G12 was purchased from Polymun Scientific.
ELISA.
ELISA was performed using a modified protocol (31, 35). All incubations were performed at 37 °C and used a 50-μL-per-well volume format. Blotto buffer [Tris-buffered saline (TBS; 10 mM Tris and 100 mM NaCl; pH 8.0) with 10% dry milk and 0.1% Nonidet P-40] was used as blocking solution and diluting solution for sample and detecting Abs. TBS-T buffer (TBS with 1% Tween-20) was used as washing solution. Briefly, plates were coated with D7324 (2 μg/mL) in TBS at 4 °C overnight. Recombinant gp120Ba-L (1 μg/mL) or FLSC (1 μg/mL) were captured onto the plates by incubation for 1 h, and then Ab or supernatant of B cell culture diluted 4 times in blotto was incubated in plate for 1 h. Bound Abs were then incubated 1 h with 1:1,000-diluted alkaline phosphatase (AP)-goat antihuman IgG (for detection of mAb) or antihuman λ-chain and/or κ-chain Abs (for detection of total reactive Ab and determination of light chain) (Southern Biotech; catalog no. 2040–04) and detected with Blue Phos Microwell Phosphatase Substrate System (KPL 50–88-00). The gp140 ELISA was performed as above, except gp140Ba-L was directly coated on the plate at 1 μg/mL overnight at 4 °C. Similarly, an in-house quantitative human total IgG ELISA was set up by coating with unconjugated goat antihuman λ and antihuman κ Abs (all from Southern Biotech), detecting with AP-goat antihuman IgG.
Competition ELISA was used as described previously (36) to determine whether plasma samples contain CD4i Abs that block CD4i mAbs (17b and ED47) binding to FLSC or CD4bs Abs that block CD4bs mAbs (b12 and sCD4-Ig) binding to gp120. Briefly, we captured FLSC or gp120 to 96-well plates as above. After washing, captured Env was incubated with the indicated concentrations of plasma samples premixed with a biotinylated mAb of half-maximum binding concentration for 1 h and then detected with AP-conjugated Streptavidin (Southern Biotech) as described above. Data were normalized to the percentage of reactivity seen in the presence of normal serum, and IC50 titer was calculated.
Inhibition of HIV-1 Env Proteins Binding to CD4 or CCR5.
The abilities of Abs to block the binding of gp120-Ig to CD4 or FLSC-Ig to CCR5 were determined by flow cytometry using CEM-NKr cells (for CD4 assays) or C2fTh/CCR5 cells (for CCR5 assays). Briefly, FLSC-Ig or gp120-Ig was labeled with Zenon-APC reagent (Invitrogen) according to the manufacturer's instructions and used in preliminary experiments to confirm specific saturable binding of the ligands to their receptors. The abilities of CD4bs or CD4i Abs in plasma to block binding of the ligands was determined by preincubating limiting concentrations of APC-tagged gp120-Ig or FLSC-Ig with plasma dilutions before incubation with the indicator cells and measurement of bound ligand by flow cytometry using a FACSCalibur flow cytometer (BD Biosciences).
Neutralization Assays.
We tested plasma samples in neutralization assays of conventional Phytohemagglutnin (PHA)-PBMC format (29, 37) performed by our group and IgG purified from plasma in an Env-pseudotyped virus format (9, 38) performed by Monogram Biosciences. As an additional measure of CD4i Abs, plasma was tested in the “CD4-triggered” neutralization assay that uses TZM-bl reporter cells and HIV-27312A/V434M, as described previously (21). This format specifically detects CD4i Abs by markedly enhanced neutralization of the virus in the presence of subinhibitory quantities (9 nM) of sCD4 (21, 29).
Detection of HIV-1 Env-Specific BMem cells.
We adapted an in vitro B cell stimulation method (27) to activate human BMem to become Ab-secreting cells. In brief, B cells were first purified from PBMC by negative bead sorting (StemCell), and BMem were further enriched by positive sorting using anti-CD27 mircrobeads (Miltenyi Biotec). B cell populations depleted of BMem were also recovered to provide negative controls. One hundred BMem per well were cultured in 96-well round-bottom plates, as described previously (27). Supernatants were collected after 2 weeks' incubation and screened for total anti-HIV-1 Env Abs by using a mixture of antihuman λ-chain and κ-chain Abs. Anti-Env-positive supernatants were subsequently evaluated for individual light-chain specificity and heavy-chain isotype. Total IgG in the supernatants was also quantified by capture ELISA. Total human IgG concentrations were typically in the range 0.5–2 μg/mL. Initial studies established the linear relationship between cell concentration and cultures positive for anti-Env Abs as predicted by the Poisson distribution. We chose to use a seeding density of 100 BMem per well because this resulted in fewer than 10% of the wells being positive for Abs specific for Env epitopes. In a typical experiment >95% of the culture wells were IgG positive. Thus, there is a high probability that the anti-Env Ab found in a single well is derived from a single Env-specific BMem precursor. Frequencies of HIV-1 Env-specific BMem precursors were estimated from the fraction of wells containing Env-specific Abs relative to the total wells and normalized as per 106 BMem after correction for BMem purity. These frequencies are in good agreement with results from other studies measuring the frequencies of human BMem for a variety of antigens (39).
Isolation of Env-Specific mAbs from BMem Cultures.
To determine whether the specificity of BMem is reflected accurately by the analysis of culture supernatants, we isolated mAbs from the cultures. Initially we attempted to use a modified EBV transformation method (27) but had no success in isolating stable cell lines secreting Env-specific mAbs in a number of attempts. For this reason we developed a new algorithm to identify heavy chain variable region (VH) and Vκ/λ genes that encode Env-specific Abs in “mini-libraries” prepared directly from the BMem cultures. Briefly, cells from Env Ab-positive wells were harvested without passage, and total RNA was isolated. Human VH and Vκ or Vλ genes were then amplified by RT-PCR and cloned into IgG1 and κ or λ expression vectors with modifications of a previously reported method (40). Individual VH or Vκ/Vλ mini-libraries were prepared for each positive well by pooling all of the VH plasmid clones and, separately, all of the Vκ or Vλ plasmid clones. The VH and Vκ or Vλ mini-libraries were mixed and used to transfect 293T cells, followed by analysis of culture supernatants by ELISA for Env-specific Abs. Modeling studies using cloned VH and Vκ/Vλ genes from existing CD4i and CD4bs mAbs showed that this step could detect the correct VH and Vκ/λ pair if it was present in the mix at a frequency of ≈1% or greater (Fig. S4), which is in a workable range when the mini-libraries are made from 50 to 100 total B cells. Next, functional VH genes were identified by cotransfecting 293T cells with individual VH plasmid clones mixed with either a Vκ or Vλ mini-library and screening culture supernatants by ELISA. Typically, screening 12–25 individual VH clones is sufficient to identify at least 1 that encodes a functional anti-Env Ab. Functional Vκ or Vλ genes are identified by cotransfecting individual clones with the functional VH gene identified in the first step. Finally, corresponding mAb was then produced by transient transfection of 293T cells with the identified VH and Vκ/λ pair and purification by protein A affinity chromatography.
Supplementary Material
Acknowledgments.
We thank Dr. Pal, Advanced BioScience Laboratories, for kindly providing BaL gp140 recombinant protein; and Mrs. Robin Flinko, Christine Obreicht, and Karla Godfery for their technical help. This work was supported by grants from the Bill and Melinda Gates Foundation and from the National Institutes of Health. M.M.S. is supported by National Institutes of Health Grant 1K12RR023250-1.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813392106/DCSupplemental.
References
- 1.Gallo RC. The end or the beginning of the drive to an HIV-preventive vaccine: A view from over 20 years. Lancet. 2005;366:1894–1898. doi: 10.1016/S0140-6736(05)67395-3. [DOI] [PubMed] [Google Scholar]
- 2.Fauci AS, et al. HIV vaccine research: The way forward. Science. 2008;321:530–532. doi: 10.1126/science.1161000. [DOI] [PubMed] [Google Scholar]
- 3.Braibant M, et al. Antibodies to conserved epitopes of the HIV-1 envelope in sera from long-term non-progressors: Prevalence and association with neutralizing activity. AIDS. 2006;20:1923–1930. doi: 10.1097/01.aids.0000247113.43714.5e. [DOI] [PubMed] [Google Scholar]
- 4.Walker BD. Elite control of HIV infection: Implications for vaccines and treatment. Top HIV Med. 2007;15:134–136. [PubMed] [Google Scholar]
- 5.Sajadi MM, Heredia A, Le N, Constantine NT, Redfield RR. HIV-1 natural viral suppressors: Control of viral replication in the absence of therapy. AIDS. 2007;21:517–519. doi: 10.1097/QAD.0b013e328013d9eb. [DOI] [PubMed] [Google Scholar]
- 6.Sajadi MM, et al. Epidemiologic characteristics and natural history of HIV-1 natural viral suppressors. J AIDS. doi: 10.1097/QAI.0b013e3181945f1e. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhillon AK, et al. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J Virol. 2007;81:6548–6562. doi: 10.1128/JVI.02749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 10.Bailey JR, et al. Neutralizing antibodies do not mediate suppression of human immunodeficiency virus type 1 in elite suppressors or selection of plasma virus variants in patients on highly active antiretroviral therapy. J Virol. 2006;80:4758–4770. doi: 10.1128/JVI.80.10.4758-4770.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereyra F, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis. 2008;197:563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 12.Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science. 2002;298:2199–2202. doi: 10.1126/science.1076071. [DOI] [PubMed] [Google Scholar]
- 13.Crotty S, et al. Cutting edge: Long-term B cell memory in humans after smallpox vaccination. J Immunol. 2003;171:4969–4973. doi: 10.4049/jimmunol.171.10.4969. [DOI] [PubMed] [Google Scholar]
- 14.Jilg W, Schmidt M, Deinhardt F. Decline of anti-HBs after hepatitis B vaccination and timing of revaccination. Lancet. 1990;335:173–174. doi: 10.1016/0140-6736(90)90050-f. [DOI] [PubMed] [Google Scholar]
- 15.Banatvala J, Van Damme P, Oehen S. Lifelong protection against hepatitis B: The role of vaccine immunogenicity in immune memory. Vaccine. 2000;19:877–885. doi: 10.1016/s0264-410x(00)00224-3. [DOI] [PubMed] [Google Scholar]
- 16.Bauer T, Jilg W. Hepatitis B surface antigen-specific T and B cell memory in individuals who had lost protective antibodies after hepatitis B vaccination. Vaccine. 2006;24:572–577. doi: 10.1016/j.vaccine.2005.08.058. [DOI] [PubMed] [Google Scholar]
- 17.Wainwright RB, Bulkow LR, Parkinson AJ, Zanis C, McMahon BJ. Protection provided by hepatitis B vaccine in a Yupik Eskimo population—Results of a 10-year study. J Infect Dis. 1997;175:674–677. doi: 10.1093/infdis/175.3.674. [DOI] [PubMed] [Google Scholar]
- 18.West DJ, Calandra GB. Vaccine induced immunologic memory for hepatitis B surface antigen: Implications for policy on booster vaccination. Vaccine. 1996;14:1019–1027. doi: 10.1016/0264-410x(96)00062-x. [DOI] [PubMed] [Google Scholar]
- 19.Moir S, et al. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J Exp Med. 2008;205:1797–1805. doi: 10.1084/jem.20072683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Titanji K, et al. Loss of memory B cells impairs maintenance of long-term serologic memory during HIV-1 infection. Blood. 2006;108:1580–1587. doi: 10.1182/blood-2005-11-013383. [DOI] [PubMed] [Google Scholar]
- 21.Decker JM, et al. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med. 2005;201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizzuto CD, et al. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 24.Wyatt R, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 25.Robinson JE, Elliott DH, Martin EA, Micken K, Rosenberg ES. High frequencies of antibody responses to CD4 induced epitopes in HIV infected patients started on HAART during acute infection. Hum Antibodies. 2005;14:115–121. [PubMed] [Google Scholar]
- 26.Burton DR, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 27.Traggiai E, et al. An efficient method to make human monoclonal antibodies from memory B cells: Potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang CC, et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci USA. 2004;101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeVico A, et al. Antibodies to CD4-induced sites in HIV gp120 correlate with the control of SHIV challenge in macaques vaccinated with subunit immunogens. Proc Natl Acad Sci USA. 2007;104:17477–17482. doi: 10.1073/pnas.0707399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray ES, et al. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J Virol. 2007;81:6187–6196. doi: 10.1128/JVI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fouts TR, et al. Expression and characterization of a single-chain polypeptide analogue of the human immunodeficiency virus type 1 gp120-CD4 receptor complex. J Virol. 2000;74:11427–11436. doi: 10.1128/jvi.74.24.11427-11436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Center RJ, Lebowitz J, Leapman RD, Moss B. Promoting trimerization of soluble human immunodeficiency virus type 1 (HIV-1) Env through the use of HIV-1/simian immunodeficiency virus chimeras. J Virol. 2004;78:2265–2276. doi: 10.1128/JVI.78.5.2265-2276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vu JR, et al. An immunoglobulin fusion protein based on the gp120-CD4 receptor complex potently inhibits human immunodeficiency virus type 1 in vitro. AIDS Res Hum Retroviruses. 2006;22:477–490. doi: 10.1089/aid.2006.22.477. [DOI] [PubMed] [Google Scholar]
- 34.Zhang MY, et al. Identification and characterization of a new cross-reactive human immunodeficiency virus type 1-neutralizing human monoclonal antibody. J Virol. 2004;78:9233–9242. doi: 10.1128/JVI.78.17.9233-9242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore JP, Wallace LA, Follett EA, McKeating JA. An enzyme-linked immunosorbent assay for antibodies to the envelope glycoproteins of divergent strains of HIV-1. AIDS. 1989;3:155–163. doi: 10.1097/00002030-198903000-00006. [DOI] [PubMed] [Google Scholar]
- 36.Turbica I, Posner M, Bruck C, Barin F. Simple enzyme immunoassay for titration of antibodies to the CD4-binding site of human immunodeficiency virus type 1 gp120. J Clin Microbiol. 1995;33:3319–3323. doi: 10.1128/jcm.33.12.3319-3323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fouts T, et al. Crosslinked HIV-1 envelope-CD4 receptor complexes elicit broadly cross-reactive neutralizing antibodies in rhesus macaques. Proc Natl Acad Sci USA. 2002;99:11842–11847. doi: 10.1073/pnas.182412199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amanna IJ, Carlson NE, Slifka MK. Duration of humoral immunity to common viral and vaccine antigens. N Engl J Med. 2007;357:1903–1915. doi: 10.1056/NEJMoa066092. [DOI] [PubMed] [Google Scholar]
- 40.Tiller T, et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.