Summary
We used a quantitative, high-density genetic interaction map, or E-MAP (Epistatic MiniArray Profile), to interrogate the relationships within and between RNA processing pathways. Due to their complexity, and the essential roles of many of the components, these pathways have been difficult to functionally dissect. Here we report the results for 107,155 individual interactions involving 552 mutations, 166 of which are hypomorphic alleles of essential genes. Our data enabled the discovery of links between components of the mRNA export and splicing machineries and Sem1/Dss1, a component of the 19S proteasome. In particular, we demonstrate that Sem1 has a proteasome-independent role in mRNA export as a functional component of the Sac3-Thp1 complex. Sem1 also interacts with Csn12, a component of the COP9 signalosome. Finally, we show that Csn12 plays a role in pre-mRNA splicing, which is independent of other signalosome components. Thus, Sem1 is involved in three separate and functionally distinct complexes.
Introduction
Following the initial identification of yeast genes by the genome sequencing project (Goffeau et al., 1996), the phenotypic analysis of gene deletion mutations (Giaever et al., 2002), and the characterization of cellular protein complexes (Gavin et al., 2006; Krogan et al., 2006), an important next step toward a comprehensive understanding of cellular function is to place these complexes within specific in vivo pathways. Genetic interactions, which report on the extent to which the function of one gene depends on the presence of a second one, can be used to place physically independent proteins and complexes into functional pathways. In this study, we used the Epistatic Mini-Array Profile (E-MAP) approach (Collins et al., 2007b; Roguev et al., 2008; Schuldiner et al., 2005), which quantitatively measures both negative (e.g. synthetic lethality) and positive (e.g. suppression) interactions, to generate a high density, genetic interaction map for a large set of genes (~10% of the genome) in S. cerevisiae that are collectively involved in all aspects of RNA metabolism, including regulating and/or processing rRNAs, mRNAs, snoRNAs, snRNAs and tRNAs. This dataset provided a global view of the epistatic behavior within and between RNA-related processes as well as the genetic “cross-talk” between individual protein complexes that function within these processes. Our analysis allowed us to place the 19S proteasome subunit, Sem1/Dss1, into two other functionally and physically distinct complexes: one involved in mRNA export and the other linked to mRNA splicing. We anticipate that this genetic map will be a useful resource for the RNA community.
Results and Discussion
Generation of an RNA Processing E-MAP
To genetically interrogate RNA processing in Saccharomyces cerevisiae, we created an E-MAP comprised of 552 mutations, each within a different gene involved in one of various RNA-related processes, including mRNA splicing and export, tRNA modification, translation, rRNA processing and RNA degradation (Figure 1A, Supplemental Figure 1, Supplemental Table 1). To facilitate comparison with other E-MAPs, we also included sets of genes involved in other processes, including transcription, chromatin remodelling, and ubiquitin/protein degradation. Finally, we included a number of previously uncharacterized genes that have been functionally linked to RNA-related processes through unbiased, large-scale screens (Gavin et al., 2006; Krogan et al., 2006; Krogan et al., 2004; Tong et al., 2004). This includes 166 hypomorphic alleles of essential genes generated using the DAmP strategy (Schuldiner et al., 2005) (Figure 1A). In all, this genetic map contains 107,155 distinct, pair-wise genetic interaction measurements (Supplemental Table 2 and http://interactome-cmp.ucsf.edu).
Figure 1. Description of the RNA processing E-MAP.
A) Composition of the RNA processing E-MAP. The dark red portions of the pie chart represent the proportions of the processes that correspond to essential genes. B) Comparison of genetic and physical interactions. The graph compares pairs of proteins that are physically associated (PE > 1.0, (Collins et al., 2007a)) and the genetic interaction scores from the corresponding mutants (S-score, (Collins et al., 2006)). C) Comparisons of ratios of negative (S ≤−2.5) to positive (S ≥2.0) (N to P) genetic interactions. The graph is divided into two parts: 1) all genetic interactions from the RNA processing E-MAP and 2) only those from pairs of genes whose corresponding proteins are physically associated (PE > 1.0, (Collins et al., 2007a)). Red bars correspond to genetic interactions derived entirely from deletions of pairs of non-essential genes (n= 58,879 total (left), 110 physically interacting (right)), while yellow bars are derived from pairs including one (n=48,741 total, 73 physically interacting) or two (n= 639 total, 4 physically interacting) DAmP alleles of essential genes. The overrepresentation of negative genetic interactions among pairs of genes that include an essential gene and have physical interactions was found to be highly significant using Fisher’s exact test, with a two-tailed p-value of 3×10−4. The trend was not strongly dependent on using various different thresholds for defining negative and positive interactions (data not shown). GIs, genetic interactions.
We subjected the entire dataset to hierarchical clustering, an approach that groups genes with similar patterns of genetic interactions. Several known protein complexes (e.g. nuclear pore and Prefoldin) are accurately represented as distinct clusters in the E-MAP (Supplemental Figure 1). Similarly, all six known components of the Elongator complex, which was initially linked to transcriptional elongation (Otero et al., 1999), cluster together. Components of Elongator also cluster next to TRM10, a tRNA methyltransferase (Jackman et al., 2003), and display strong negative interactions with several other tRNA modifying enzymes (e.g. ARC1 and DEG1), consistent with work implicating this complex in the regulation of tRNA modification (Esberg et al., 2006). Interestingly, the ubiquitin-like modifier URM1 (Furukawa et al., 2000) also clusters with the components of Elongator, and recent work suggested that Urm1 functions as a thiol group carrier important for thiol modification of tRNAs (Nakai et al., 2008; Schmitz et al., 2008) (Supplemental Figure 1).
Components of known spliceosomal subcomplexes, including the U1 snRNP (MUD1, NAM8, PRP42 and SNU56) and the Prp19 associated Nineteen Complex (NTC) (NTC20, ISY1, SYF2, PRP19) (Jurica and Moore, 2003) satisfyingly cluster together (Supplemental Figure 1). Two largely uncharacterized factors, CWC21 and CWC27, also cluster with the NTC, consistent with work demonstrating that they physically associate with the NTC component Cef1 (Ohi and Gould, 2002). Additionally, we identified many positive genetic interactions between the NTC and the U1 snRNP (Supplemental Figure 1), perhaps reflecting the fact that the U1 snRNP departs the spliceosome approximately when the NTC joins (Chan et al., 2003). Further investigation will be required to understand the implications of these and the many other genetic interactions contained within this E-MAP (Supplemental Table 2, http://interactome-cmp.ucsf.edu).
Global Analysis of Genetic and Physical Interaction Data
While inspection of the E-MAP genetic interaction datasets in isolation have already yielded many interesting observations, analysis of these data in conjunction with large-scale protein-protein interaction datasets is especially powerful. We have shown in other E-MAPs that genes exhibiting positive genetic interactions are more likely to encode proteins that are physically associated (Collins et al., 2007b; Roguev et al., 2008), and inspection of the data from this E-MAP revealed a similar trend (Figure 1B). However, in contrast to what has been observed in other E-MAPS, there is also an enrichment of Protein-Protein Interactions (PPIs) among factors displaying negative genetic interactions (Figure 1B). Notably, the RNA processing E-MAP greatly expands the number of components of essential complexes that have been interrogated by the E-MAP technique, both by including many non-essential factors that participate in several large essential complexes, and by including a large number of hypomorphic alleles of essential factors using the DAmP (Decreased Abundance by mRNA Perturbation) strategy (Schuldiner et al., 2005). This difference from previous E-MAPs may at least in part account for the more prevalent negative genetic interactions observed among physically interacting factors. Indeed, a recent analysis of a previously published E-MAP focused on chromosome biology showed that complexes dominated by negative genetic interactions are more likely to contain at least one essential gene (Bandyopadhyay et al., 2008).
Interestingly, the hypomorphic alleles of essential genes that are included in this E-MAP do not generally have more negative genetic interactions than the non-essential genes when each of these subsets is considered as a whole. We compared the ratio of negative (Genetic Interaction Score (S) ≤−2.5) to positive (S ≥2.0) genetic interactions (N to P) involving either pairs of non-essential genes or pairs where at least one gene is essential (Figure 1C, see Experimental Procedures). In the entire dataset, we found an N to P ratio of ~1, regardless of whether the mutant pairs contained at least one essential gene (1.00) or not (1.29). However, when we restricted the analyses to genes whose corresponding proteins are physically associated (as defined in Figure 1B legend), we found that the genetic interactions observed with essential genes are over three times as likely to be negative (N to P =2.84, p=0.0003) (Figure 1C; Supplemental Table 2) compared to interactions between null mutations of non-essential genes (N to P =0.93).
These observations suggest that, while two mutations in a non-essential complex often result in a positive genetic interaction, the introduction of two mutations in the same essential complex is very likely to have a synthetic negative effect on growth. Thus, a positive interaction observed between two factors that physically interact within an essential complex may suggest that these factors form a non-essential module within the complex. One example of this is the positive interaction between IST3 and BUD13, which have been suggested to physically interact with both the non-essential RES complex and with the essential SF3b complex of the spliceosome (Dziembowski et al., 2004; Wang et al., 2005). Our data support the model that IST3 and BUD13 function as a non-essential module in these complexes.
While factors with protein-protein interactions are overrepresented among the strong genetic interactions observed in the RNA processing E-MAP, there are also certainly many genetic interactions between factors that have functional, but not physical, interactions. Indeed, these interactions comprise a powerful tool for discovering previously uncharacterized pathways connecting protein complexes. To help identify these connections, we used an approach we have recently described (Bandyopadhyay et al., 2008) that classifies protein pairs as either operating within the same module or between functionally related modules given their genetic and physical interactions (see Figure 2A legend for a more detailed description).
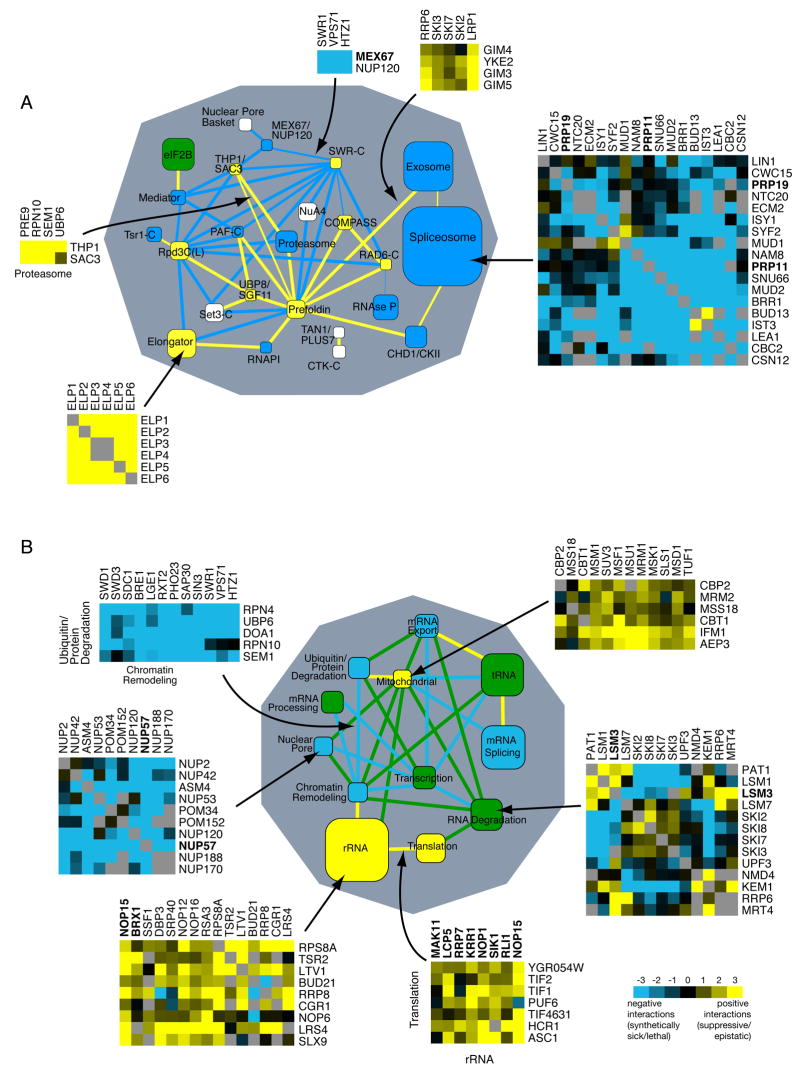
Figure 2. Functional cross-talk between biological processes and protein complexes.
Global views of the genetic cross-talk between different RNA-related protein complexes (A) and processes (B). Blue and yellow represent a statistically significant enrichment of negative and positive interactions, respectively, whereas green corresponds to cases where there are roughly equal numbers of positive and negative genetic interactions. White corresponds to a lack of significant interactions or lack of data. Nodes (boxes) correspond to distinct protein complexes (A) or functional processes (B) whereas edges (lines) represent how the complexes and processes are genetically connected. Line thickness represents the significance of the connection. Node size is proportional to the number of genes in the process or complex. Essential genes (DAmP alleles) are in bold. Representative genetic interactions which contributed to the overall enrichment for interactions are shown for sample nodes and edges, according to the scale shown, with grey boxes representing missing data points.
See Experimental Procedures for a description of how the networks are generated.
Several interesting connections become evident when the data are analyzed in this way. For example, there are negative genetic interactions (blue line) between the chromatin remodeling complex, SWR-C, which incorporates the histone H2A variant, Htz1, into chromatin (Kobor et al., 2004; Krogan et al., 2003; Mizuguchi et al., 2004), and the nuclear pore factors Mex67 and Nup120, consistent with recent work demonstrating that Htz1 is involved in tethering genes to the nuclear periphery (Brickner et al., 2007). Also, we found positive genetic interactions (yellow line) between the complex containing both casein kinase II (CKII) and the chromodomain-containing protein Chd1, and the spliceosome. Consistent with this finding, Reinberg and colleagues found a physical and functional connection in human cells between Chd1 and the spliceosome (Sims et al., 2007). There are many testable hypotheses that can be extracted from this data set in this format and we have created an interactive website that allows for searching of individual interactions as well as the connections between complexes (http://www.cellcircuits.org/complexes/RNAProcessing/html/).
In an effort to gain insight into the functional organization of RNA-related processes at an even broader level, we created a map that highlights strong genetic trends both within and between these processes, without respect to known physical interactions (Figure 2B). Nodes (boxes) now correspond to the interactions within distinct functional processes (defined by a combination of GO term analysis and manual annotation, see Supplemental Table 1) whereas edges (lines) represent the genetic cross-talk between processes.
A striking finding is that several sets of genes that are known to function in the same biochemical processes contain predominantly positive or predominantly negative interactions, as was also observed in an analysis of the computed epistatic behavior among yeast metabolic genes (Segre et al., 2005). For example, genes classed as involved in cytoplasmic rRNA biogenesis and mitochondrial rRNA biogenesis are significantly enriched in positive genetic interactions (Figure 2B, yellow nodes). In contrast, factors involved in mRNA splicing, mRNA export and the nuclear pore have strongly negative interactions (blue nodes). Finally, not all groups show consistent patterns of interactions; for instance, RNA degradation, tRNA biogenesis and transcription show mixed but still significant genetic interactions (green nodes). The broad trends suggest that the patterns of genetic interactions are influenced not only by direct physical interactions between factors and direct functional interactions between complexes, but also probably by higher-level interactions within and between processes.
The Sem1/Dss1 Component of the Proteasome is Involved in mRNA Export Via the Sac3-Thp1 Complex
Our E-MAP uncovered positive genetic interactions between the proteasome and the Sac3-Thp1 (TREX-2) complex involved in mRNA export (Figure 2A), suggesting that a direct functional relationship exists between these two complexes. We confirmed these positive interactions represented suppressive relationships through tetrad analysis and dilution growth assays at 16°C (Supplemental Figure 2). The proteasome is a large molecular machine that degrades ubiquitylated substrates (Collins and Tansey, 2006) and is concentrated at the nuclear periphery in yeast (Enenkel et al., 1998). During the process of mRNA export, multiple components of the Mex67-mediated export pathway interact with the nuclear pore basket via the Sac3-Thp1 complex (Luna et al., 2008).
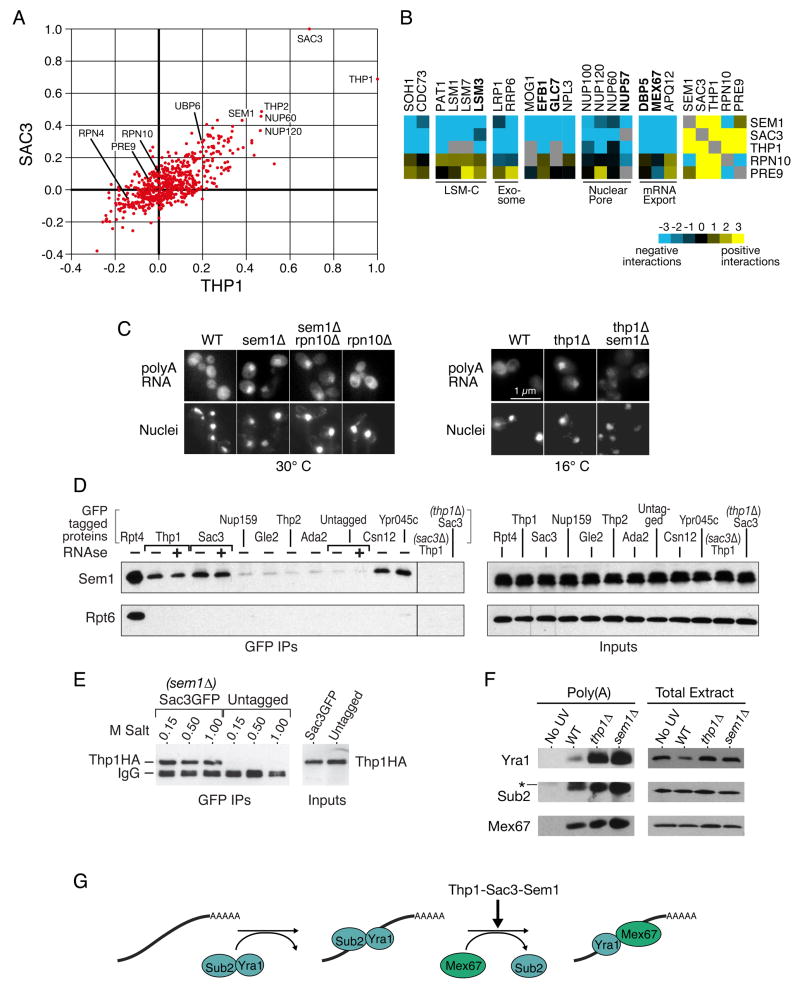
To investigate the significance of the genetic connections between the proteasome and the Sac3-Thp1 mRNA export complex, we compared their global genetic interaction patterns from the E-MAP. The genetic interaction profiles we obtained for SAC3 and THP1 are extremely similar (correlation coefficient = 0.7), consistent with their known biochemical and functional interactions (Fischer et al., 2002; Gallardo et al., 2003). After each other, the genetic interactions of SAC3 and THP1 are most similar to those of the nuclear pore components NUP60 and NUP120, THP2 (a component of the THO/TREX complex that co-transcriptionally associates with the mRNP), and, interestingly, the proteasome lid component SEM1 (Figure 3A and http://interactome-cmp.ucsf.edu/). In contrast, the genetic profiles of other components of the proteasome (e.g. RPN10, PRE9 and RPN4) do not correlate with these export factors. SEM1 differs from other proteasome mutants in sharing many genetic interactions with THP1-SAC3, including negative ones with the exosome (LRP1, RRP6), nuclear pore (NUP100, NUP120, NUP60, NUP57) and other factors implicated in mRNA export (such as MEX67, APQ12, DBP5) (Figure 3B and Supplemental Figure 1).
Figure 3. Sem1 is involved in mRNA export via the Sac3-Thp1 complex.
A) Scatter plot of correlation coefficients for each mutant compared to the profiles generated from SAC3 (y-axis) and THP1 (x-axis). B) Representative genetic interactions from the E-MAP that differentiate SEM1, SAC3, and THP1 from RPN10 and PRE9. Blue and yellow indicate negative and positive genetic interactions, respectively. C) In situ hybridizations with a dT50 probe to detect accumulation of poly(A) RNA (top row). The bottom row is DAPI staining to detect nuclei. Cells were either kept at permissive temperature (30°C, left) or shifted to 16°C for 2 hours before fixation (right). D) Co-immunoprecipitations of Sem1 and Rpt6 with GFP-tagged proteins. The indicated strains were immunoprecipitated with a monoclonal GFP antibody, either with or without prior RNAse A treatment of the extract, and the blot was cut and probed with polyclonal antibodies against Sem1 or Rpt6. The right panel is 1/200th of the sample for the GFP IPs exposed identically to the left. E) Co-immunoprecipitations of Thp1-6HA with Sac3GFP in a sem1 Δ background. The immunoprecipitations were washed at room temperature with buffers containing the indicated amount of salt. The right panel represents1/20th the sample for the GFP IPS. F) In vivo UV crosslinking of proteins to poly(A) RNA. Each lane of the poly(A) eluates contains equal amounts of purified poly(A) RNA. The “no UV” strain contains Thp1-6HA. The * in the Sub2 blot represents a nonspecific cross-reacting band, which is resolved when the gels are run farther (see Supplemental Figure 4). The left and right panels were exposed differently, and the Yra1 poly(A) blot was overexposed to allow visualization of the Yra1 band in the wild-type strain. G) Depiction of the export block identified by the poly(A) crosslinking in part F. Sac3-Thp1-Sem1 could facilitate an exchange of Sub2 for Mex67. See text for more details.
As has been previously reported, deletion of either SAC3 or THP1 results in a substantial increase of polyadenylated mRNA in the nucleus (Fischer et al., 2002; Gallardo et al., 2003; Lei et al., 2003) (Figure 3C). We found that deletion of SEM1 also results in a moderate mRNA export defect at 25°C, 30°C and 37°C (Figure 3C and data not shown), suggesting a direct role for Sem1 in mRNA export, consistent with previous work in fission yeast (Mannen et al., 2008; Thakurta et al., 2005). Interestingly, we found that deletion of SEM1 in the thp1Δbackground not only partially suppresses its growth defect at 16°C (Supplemental Figure 2) but also reduces the intensity of the mRNA export phenotype compared to thp1Δalone (Figure 3C), consistent with a functional link between Sem1 and the Sac3-Thp1 export pathway. However, deletion of proteasome factors other than SEM1 (RPN10, PRE9, UBP6, DOA1, RPN4), or inhibition of the proteasome by the drug MG132 in a pdr5Δbackground (which allows for more efficient uptake of the drug), does not cause an mRNA export defect in this assay (Figure 3C and data not shown). Furthermore, although RPN10 deletion exacerbates the growth, proteasome integrity and DNA damage response defects of a sem1Δstrain (Funakoshi et al., 2004; Sone et al., 2004), we found no significant effect on sem1Δ’s moderate mRNA export defect (Figure 3C). Together, these data argue that Sem1 is directly involved in mRNA export, but other components of the proteasome are not.
We next asked whether Sem1 is physically associated with the Sac3-Thp1 complex. Since we found that introducing a tag on the C-terminus of Sem1 results in a defect in mRNA export (data not shown), we used a polyclonal antibody (Funakoshi et al., 2004) to immunoprecipitate or detect Sem1 from cellular extracts of a panel of GFP-tagged strains. Both Thp1-GFP and Sac3-GFP reciprocally co-immunoprecipitate with Sem1 (Figure 3D and Supplemental Figure 3). Consistent with previous results, GFP-tagged Rpt4, a 19S proteasome component, co-immunoprecipitates both Sem1 and Rpt6, another known component of the proteasome (Funakoshi et al., 2004; Krogan et al., 2004; Sone et al., 2004). In contrast, Sac3-GFP and Thp1-GFP are not associated with Rpt6 (Figure 3D), strongly suggesting that Sem1 interacts with Sac3-Thp1 independent of the rest of the proteasome. Previous work in S. pombe suggested physical associations between Dss1 (S. cerevisiae Sem1) and other mRNA export factors, including Nup159, Rae1 (S. cerevisiae Gle2), and Mlo3 (S. cerevisiae Yra1) (Thakurta et al., 2005); however, we do not detect these physical connections in budding yeast (Figure 3E and data not shown). We also do not find Sem1 associated with TREX (e.g. Thp2) or the transcriptional activation complex SAGA (e.g. Ada2), both of which are complexes that have been reported to interact with Sac3-Thp1 (Fischer et al., 2002; Rodriguez-Navarro et al., 2004). Finally, the physical interaction between Sac3-Thp1 and Sem1 is insensitive to RNAse A treatment (Figure 3D and data not shown), suggesting that this interaction is not mediated by RNA.
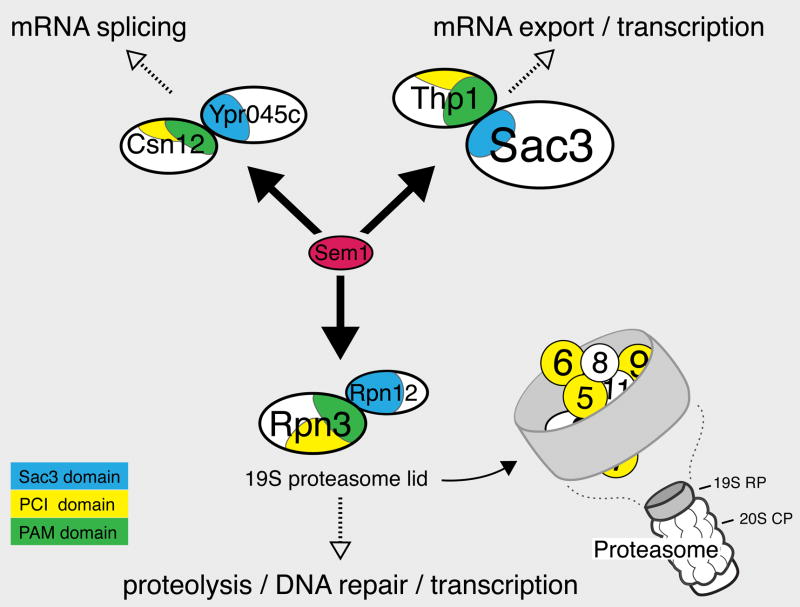
To gain further insight into the physically interacting partners of Sem1, we analyzed data from our earlier large-scale protein-protein interaction project that used a systematic affinity tagging, purification and mass spectrometry approach to identify protein complexes (Krogan et al., 2006). After components of the proteasome, the proteins that most significantly immunoprecipitated Sem1 as prey were Csn12, a component of the CSN (COP9 Signalosome), another large complex with structural similarity to the proteasome lid), and Ypr045c, an uncharacterized protein, both of which also co-purified with each other (http://tap.med.utoronto.ca/). We confirmed the Sem1-Csn12 and Sem1-Ypr045c interactions by co-immunoprecipitation (Figure 3D). Together with our Sac3-Thp1-Sem1 results, these physical interactions suggest that Sem1 may be part of at least three functionally distinct complexes in S. cerevisiae: the 19S proteasome (Funakoshi et al., 2004; Krogan et al., 2004; Sone et al., 2004), Sac3-Thp1, and a complex containing Csn12 and Ypr045c (see Figure 5). This is consistent with previous data showing that some cellular Sem1 fractionates with complexes smaller than the proteasome (Funakoshi et al., 2004). These complexes are all likely to be conserved, as human Dss1 interacts with two metazoan-specific complexes and components of all three S. cerevisiae complexes that we describe here (Baillat et al., 2005). Furthermore, S. cerevisiae or human Sem1/Dss1 can complement a dss1Δ strain for mRNA export in S. pombe (Thakurta et al., 2005).
Figure 5. Model for three Sem1-containing complexes.
19S RP= Regulatory Particle of the proteasome, which includes both the lid (diagrammed here), and the base. 20S CP= Core Particle of the proteasome. The schematic of protein organization in the proteasome lid is based on the model from Sharon et al (Sharon et al., 2006). The “SAC3” domains (PFAM PF03399) are amino acids: Rpn12 20–211, Ypr045c 205–408, and Sac3 248–443, and the PAM domains (Ciccarelli et al., 2003): Rpn3 204–378, Csn12 136–343, and Thp1 151–332. Also shown are the PCI domains (PFAM PF01399) associated with the PAM domains: Rpn3 343–447, Csn12 298–415, and Thp1 300–430.
Examining the domain structure of Sem1-interacting proteins suggests a model for how Sem1 associates with each of these complexes. Rpn3, Thp1, and Csn12 are the only three proteins in S. cerevisiae that harbor the PAM (PCI-Associated Module) domain (Ciccarelli et al., 2003), a highly conserved region upstream of a subset of PCI domains (protein interaction domains named after three complexes rich in these domains: the proteasome, the CSN, and eIF3) (Hofmann and Bucher, 1998; Scheel and Hofmann, 2005). Similarly, Rpn12, Sac3, and Ypr045c are the only three proteins in S. cerevisiae that contain a highly conserved variant of the PCI domain, called the SAC3 domain (PFAM database # PF03399 (Finn et al., 2006)) (Figure 5). Indeed, the region of Sac3 that interacts with Thp1 was mapped to a small section containing the SAC3 domain (Fischer et al., 2002). Furthermore, a number of studies have placed Rpn3, Rpn12, and Sem1 in a subcomplex within the proteasome lid (Fu et al., 2001; Isono et al., 2005; Sharon et al., 2006). Sem1 crosslinks directly to Rpn3 in vivo (Sharon et al., 2006), and binds to Rpn3 in vitro (Wei et al., 2008). Thus, these domains suggest a model that Sem1 may be interacting with PAM and/or SAC3 domains in multiple large protein complexes.
Indeed, our results raise the possibility that the SAC3 and PAM domains bind to one another, and in turn, recruit Sem1. Consistent with this notion, we found that both Sac3 and Thp1 are required for the other to interact with Sem1 (Figure 3D and Supplemental Figure 3). Previous studies have suggested that Sem1 is involved in stabilizing protein complexes.One of the metazoan-specific Sem1/Dss1 containing complexes, that containing BRCA2, is unstable unless Dss1 is co-purified (Li et al., 2006; Yang et al., 2002). The proteasome is also unstable in cells lacking Sem1 (Funakoshi et al., 2004; Sone et al., 2004), although a recent study suggests that this instability does not stem from abrogation of the PAM-domain (Rpn3)- SAC3-domain (Rpn12) interaction (Wei et al., 2008). In the presence of 1M salt, the proteasome falls apart in a sem1Δstrain (Sone et al., 2004). In contrast, we find that in the absence of Sem1, Sac3-GFP still remains associated with immunoprecipitated Thp1-HA in the presence of at least 1 M NaCl (Figure 3E). Consistent with these data, we find that the localization of Thp1 to the nuclear envelope is not strongly affected by sem1Δ, in contrast to sac3Δ, which results in its complete mislocalization ((Fischer et al., 2002) and data not shown). Thus, these data suggest that Sem1 has roles other than simply stabilizing protein complexes.
The Sac3-Thp1 mRNA export complex is implicated in the pathway that loads the export receptor Mex67 (or TAP in higher eukaryotes) onto mRNAs, and our genetic, functional, and physical data suggest that Sem1 may act as part of the Sac3-Thp1 complex. Sub2 and Yra1 are co-transcriptionally loaded onto mRNAs as part of the THO/TREX complex, and Yra1 acts as an adaptor protein to load Mex67 onto the mRNP, making it competent for export (Luna et al., 2008). Because Yra1 can interact with either Sub2 or Mex67, but not both simultaneously, Sub2 may be exchanged for Mex67 (Strasser and Hurt, 2001) (Figure 3G). To investigate the role of Sac3-Thp1-Sem1 in RNP formation, we UV crosslinked proteins to RNA in vivo, made cell extracts, purified the poly(A) RNA over an oligo dT cellulose column, and then examined protein components in the resulting poly(A) eluates in wild-type and mutant backgrounds. In a wild-type strain, low levels of Sub2, Yra1, and Mex67 can be detected crosslinked to poly(A) mRNA in a UV-dependent manner (Figure 3F). Deletion of SEM1, THP1 or SAC3 all result in a significant accumulation of Sub2 and Yra1 on polyA mRNA, whereas Mex67 binding increases only modestly in all three cases (Figure 3F and data not shown). In contrast, large quantities of Mex67, but not Yra1, accumulate on mRNA when it is blocked at the nuclear periphery in a dbp5,rrp6 double mutant (Lund and Guthrie, 2005). These data suggest that the Thp1-Sac3-Sem1 complex facilitates the exchange of Sub2 for Mex67 (Figure 3G). The modest increase in Mex67 association may reflect a partial block during Mex67 loading, or may reflect aberrant binding of Mex67 to the particularly large amounts of Yra1 that appear to be accumulating on the mRNA. Although further work will be required to identify the exact role that this complex plays in the export pathway, our data argue that Sem1 is acting in concert with Sac3-Thp1 to load Mex67 onto polyadenylated mRNA.
The Csn12 Component of the COP9 Signalosome is Involved in mRNA Splicing
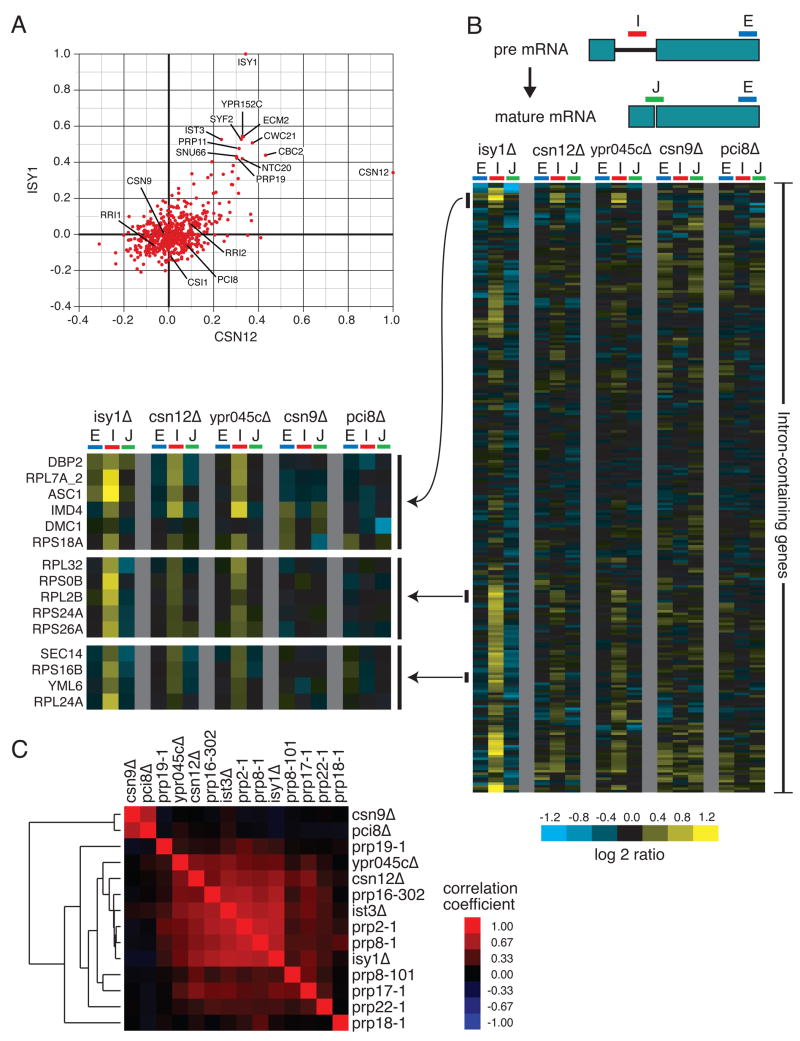
The functions of two of the three Sem1-containing complexes (the proteasome and Sac3-Thp1) were known, so we used the E-MAP, in conjunction with physical interaction data, to gain insight into the function of the Csn12-Ypr045c complex. Csn12 was previously described as a subunit of the COP9 signalosome (CSN), a complex responsible for the removal of the Rub1 modifier (known as Nedd8 in higher eukaryotes) from cullin proteins, which are components of various E3 ubiquitin ligase complexes (Maytal-Kivity et al., 2003). However, deletion of CSN12 fails to cause accumulation of neddylated Cdc53, a phenotype observed in all other mutants of CSN subunits (Wee et al., 2002). Inspection of the quantitative genetic interaction data revealed that CSN12 is most similar genetically to components of the spliceosome. For example, the profile of genetic interactions of csn12Δis highly correlated with that of isy1Δand also several other mRNA splicing factor mutants, including syf2Δ, prp11-DAmP, ist3Δ, snu66Δand prp19-DAmP, whereas deletions of other components of the CSN display very different genetic interaction profiles (Figure 4A). Consistent with a functional link between Csn12 and the spliceosome, large-scale protein TAP studies identified a physical link between only this component of the CSN and many factors involved in mRNA splicing (Collins et al., 2007b; Gavin et al., 2006).
Figure 4. Csn12 is involved in mRNA splicing.
(A) Plot of correlation coefficients generated from comparison of the genetic interaction profiles from csn12Δ or isy1Δ to all other profiles in the E-MAP. B) Splicing-specific microarray profiles for several mutant strains. The schematic displays the positions of the microarray probes that report specifically on the levels of pre-mRNA (in the Intron), mature mRNA (at the Junction), and total mRNA (in the second Exon) for each intron-containing transcript. The relative levels of exon, intron, and junction for a single intron-containing gene are displayed as log2 ratios for the indicated mutant strains compared to a wild type strain, across each row. The ordering of genes was determined by hierarchical clustering. For selected clusters of genes, the splicing profiles across the mutants tested are displayed at higher resolution to the left of the full splicing profiles. C) Pairwise Pearson correlation coefficients were calculated between each of the mutants tested, as well as between these mutants and several previously characterized splicing mutants. The matrix of correlations was subjected to hierarchical clustering.
To test whether Csn12 plays a functional role in mRNA splicing, we used a splicing-specific microarray to measure the levels of pre-mRNA, mature mRNA, and total mRNA for most of the intron-containing genes in the yeast genome (Pleiss et al., 2007). In general, a decrease in splicing efficiency for a given transcript results in an accumulation of its pre-mRNA, and often in a measurable loss of mature mRNA (Clark et al., 2002; Pleiss et al., 2007). We examined the splicing efficiencies of isy1Δ, csn12Δ, and ypr045cΔ (the SAC3 domain-containing protein that physically interacts with Csn12) as well as deletions of CSN9 and PCI8, two other components of the CSN. As expected, deletion of ISY1 causes a substantial accumulation of pre-mRNA for many intron-containing genes (Figure 4B). Interestingly, deletion of either CSN12 or YPR045C causes a pre-mRNA accumulation in many of the same transcripts adversely affected in the isy1Δstrain (Figure 4B). In contrast, csn9Δand pci8Δstrains fail to show a splicing defect (Figure 4B). We also failed to detect a splicing defect in the sem1Δstrain (data not shown), suggesting that, at least under the conditions tested, the loss of Sem1 does not significantly impair the function of the Csn12-Ypr045c complex in splicing.
In order to more quantitatively assess the similarity of the splicing microarray profiles observed with csn12Δand ypr045cΔto those obtained from well characterized splicing factors, we calculated pairwise Pearson correlation coefficients between a number of mutant pairs using the ratios from all of the microarray features reporting on intron-containing genes (Figure 4C). As previously reported, various splicing factor mutations result in a wide range of splicing phenotypes (Pleiss et al., 2007). The splicing microarray profiles of the csn12Δand ypr045cΔstrains are highly correlated with each other as well as with the profiles of a number of well-characterized splicing factor mutants (e.g. prp2-1, prp16-302, prp8-1, ist3Δ, isy1Δ), whereas csn9Δand pci8Δexhibit distinct behavior on the splicing array (Figure 4C). Thus, Csn12 and Ypr045c, but not other components of the CSN, are involved in maintaining normal levels of mRNA splicing.
Perspective
In this study, we generated an E-MAP in budding yeast that is focused on virtually all aspects of RNA processing and includes factors implicated in the generation and regulation of tRNAs, mRNAs, rRNAs, snoRNAs and snRNAs. We have applied innovative analytical tools that allow for the visualization of the genetic cross-talk between the different RNA-related processes and the protein complexes that function within these processes. Using these representations and other data exploration tools such as hierarchical clustering, we predicted and verified several previously uncharacterized biological connections. Along with the examples explored here, we also found a strong link between Npl3, an SR domain-containing protein involved in mRNA export, and mRNA splicing (see Kress et al., this issue).
The genetic data from the E-MAP is even more useful when analyzed in combination with other large data sets, including other E-MAPs (Collins et al., 2007b; Schuldiner et al., 2005) and physical interaction data sets such as the TAP database (Collins et al., 2007a; Gavin et al., 2006; Krogan et al., 2006) (http://interactomecmp.ucsf.edu). This approach allowed us to identify two previously uncharacterized protein complexes containing the 19S proteasome component Sem1, one of which (Sac3-Thp1) acts in mRNA export, and one of which (Csn12-Ypr045c) acts in mRNA splicing (Figure 5). We failed to detect an mRNA export defect in either csn12Δor ypr045cΔ, or a splicing defect in either sac3Δor thp1Δ (data not shown), suggesting that these two complexes are functionally distinct, despite sharing Sem1 as a common component.
The links reported here between Sem1 and multiple mRNA processes suggest a model whereby Sem1 acts to modulate access of nucleic acids to each of these complexes. The proteasome has been suggested to play roles in a number of nucleic acid-dependent processes that could also be facilitated by Sem1, including transcription activation, DNA repair, and RNA degradation (Collins and Tansey, 2006; Gautier-Bert et al., 2003; Krogan et al., 2004). Indeed, inspection of a co-crystal of human Dss1 with the metazoan-specific breast cancer protein BRCA2 and ssDNA led to a similar model, in which Dss1 might modulate access of ssDNA to BRCA2 during double strand break repair. Sem1/Dss1 is a small (10 kD) extremely acidic protein, with physical characteristics strikingly similar to those of nucleic acids (Yang et al., 2002). It has recently been shown that a small patch of conserved acidic residues in Sem1 is necessary and sufficient to mediate interaction with a PAM domain-containing protein (Rpn3) (Wei et al., 2008). Therefore, it is tempting to speculate that Sem1/Dss1 could act as both an RNA and DNA mimic, regulating the association of nucleic acid with several complexes.
Finally, combining the unbiased genetic map with physical interaction data suggests functions for several previously uncharacterized proteins. For example, YNR004W encodes a small protein that has no identifiable domains or homology, but deletion of this non-essential gene causes a similar genetic interaction profile to that seen with tgs1Δ, a protein that trimethylates snoRNA and spliceosomal snRNA caps (Mouaikel et al., 2002). Both TGS1 and YNR004W cluster with components of the spliceosome (Supplemental Figure 1), and a two-hybrid interaction has been reported for the corresponding pair of proteins (Uetz et al., 2000). Strikingly, we have found that deletion of YNR004W causes a nearly identical splicing defect to that observed with tgs1Δ (Supplemental Figure 5). These data suggest that Ynr004w functions directly with Tgs1 to ensure proper tri-methylation of snRNA caps. A number of other currently unnamed open reading frames included in the E-MAP display strong patterns of genetic interactions suggestive of roles in specific processes, providing numerous directions for further experimentation.
In order to facilitate further data-mining by the RNA community, we have developed an interactive and searchable web-based database (Supplemental Figure 6, http://interactome-cmp.ucsf.edu). One can query single mutants for individual genetic interactions or for other factors that share similar genetic interaction profiles. This database is integrated with the results from previous E-MAPs, in both budding (Collins et al., 2007b; Schuldiner et al., 2005) and fission yeast (Roguev et al., 2008), allowing a wealth of information from different fields to be cross-referenced. Additionally, an interface for navigating the higher-level analysis of interactions between complexes has been developed, and is linked to from within the database (Supplemental Figure 7, http://www.cellcircuits.org/complexes/RNAProcessing/html/). We anticipate that this dataset will be used as a resource by others to launch more detailed investigations into the biology of RNA processing.
Experimental Procedures
E-MAP analysis
Strains were constructed and E-MAP experiments were performed as previously described (Collins et al., 2007b; Collins et al., 2006; Schuldiner et al., 2005).
Functional connections between processes and complexes
For analysis of connections between related processes, 446 genes were classified with one of 12 functions, using a combination of GO annotations and manual curation. The median genetic interaction score of protein pairs spanning two processes or within the same process was compared to equal sized random samples of genetic interactions. A cutoff of p<0.001 was used for the between-process connections and a cutoff of p<0.05 was used for connections within a process. Enrichments for strong (both positive and negative) interactions were computed by comparison of the median of the absolute values of the S-scores to the absolute value of a random distribution with a p<0.001. Complexes involved in RNA processing were identified using previously described methods (Bandyopadhyay et al., 2008). This analysis categorizes interactions supported by both strong genetic and physical evidence as operating within a module, or complex. Interactions with a strong genetic but weak physical signal are better characterized as operating between two functionally related modules. Given within- and between-module likelihoods for individual interactions, an agglomerative clustering procedure seeks to merge these interactions into increasingly larger modules and to identify pairs of modules interconnected by many strong genetic interactions. Pairs of functionally related complexes and genetic interactions within a complex were then categorized through analysis of their pairwise genetic interaction distributions as described above.
Yeast Genetics
Yeast were grown and double deletions made according to standard methods (Guthrie and Fink, 2002). For growth tests, cells were grown to log phase and diluted to OD 0.1, and then serially diluted by one fifth incubated at 16°C for 7 days onYPD.
In Situ Hybridizations and Microscopy
In situ hybridizations were performed as described previously with a few modifications (Duncan et al., 2000). Briefly, cells were grown to OD 0.15–0.25 in YEPD, and fixed for one hour in 5% formaldehyde, spheroplasted with zymolyase 100T (Seikagaku Corporation), fixed briefly with paraformaldehyde and permeabilized with cold methanol. Bulk polyadenylated RNA was detected by in situ hybridization with approximately 10 ng of digoxigenin-11-dUTP (Roche) labeled oligo dT50 probe followed by staining with anti-digoxigenin-fluroescein Fab fragments (Roche) (1:25). In parallel, cells were treated with secondary antibody and no probe. Cells were mounted in SlowFade Gold antifade reagent with DAPI (Invitrogen). Images were collected on an Olympus BX-60 microscope with a 100x Olympus UPlanFI NA 1.30 objective and appropriate filters with a Sensys CCD camera (Photometrics) and analyzed using iVision imaging Software (Biovision Technologies). All strains were analyzed at least three times, and all images within a set were treated identically.
Immunoprecipitations
Log phase cultures of cells from the GFP tagged collection (Huh et al., 2003) were washed and resuspended in lysis buffer containing 50 mM Tris HCl pH 7.5, 150 mM NaCl, 0.1 % NP40 and protease inhibitors, and lysed with glass beads. Extracts were clarified by centrifugation at 10,000 g, and immunoprecipitated overnight with 1 μL polyclonal antibody to Sem1 (Funakoshi et al., 2004) (generous gift of Hideki Kobayashi), or 2 μg monoclonal antibody to GFP (Roche) followed by a one hour incubation with 30 μL of a 50% slurry of IgG beads (Amersham). The beads were washed 5 times with lysis buffer and resuspended in 1X SDS sample buffer. Western blots were then probed for Sem1 (1:1000), GFP (1:2000), and Rpt6/Sug1 (1:1000) (generous gift of Thomas Kodadek), or HA (12CA5 Roche). For RNAse A treatment, either 25 μg of RNAse A was added to extracts for 10 min at room temperature, or the extracts were mock treated, and a total RNA sample was analyzed by agarose gel electrophoresis to check degradation of total RNA.
UV Crosslinking Assay
Cells were grown to OD 0.8 and subjected to UV crosslinking, extract preparation, and poly(A) RNA purification as in (Gilbert et al., 2001). Western blots were then probed for Mex67 (1:10000, generous gift of Catherine Dargement), Sub2 (1:2000, polyclonal antisera specific to Sub2 produced in the lab), and Yra1 (1:2000, generous gift of Douglas Kellogg).
Splicing Microarrays
Splicing microarray sample collection, data analysis, and arrays are as previously described with the following modifications (Pleiss et al., 2007). Briefly, total RNA was purified from wild type and mutant yeast cultures growing in log phase at 30°C, and converted to animo-allyl labeled cDNA, which was then coupled to dyes and competitively hybridized to splicing microarrays. Values represent averaged data collected from dye-flipped technical replicates performed on two biological replicates for the csn12Δ, ypr045cΔ, csn9Δ, and pci8Δ arrays. The isy1Δ values are derived from a single representative biological replicate. Similarities between splicing microarray profiles were measured by Pearson correlation, calculated on the complete observations (Becker et al., 1988). Experimental factors were then symmetrically arranged along the X- and Y-axis of Figure 4C using hierarchical clustering (de Hoon et al., 2004) of euclidean distances between correlation values with unweighted centroid linkages. Array data has been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE11634.
Supplementary Material
Acknowledgments
We would like to thank H. Kobayashi, T. Kodadek, D. Kellogg, and C. Dargement for antibodies, D. Cameron for strains, A. Chan, E. Cheng, L. Jieying, M. Dinglasin, and C. Wong for technical assistance, M. Schuldiner for help with the screens, P. Kemmerman for help with the website, L. Booth for work on ynr004wΔ, and V. Panse for sharing unpublished data. We would like to thank members of the Guthrie lab, A. Frankel and J. Gross for reading and advice on the manuscript. G.M.W. was supported by postdoctoral fellowships from the American Cancer Society and the Sandler Foundation. M.B. was supported by an HHMI predoctoral fellowship. C.G. is an American Cancer Society Research Professor of Molecular Genetics. This research was funded by NIH grant GM21119 (C.G.) and from Sandler Family Funding (N.J.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Kelley R, Krogan NJ, Ideker T. Functional maps of protein complexes from quantitative genetic interaction data. PLoS Comput Biol. 2008;4:e1000065. doi: 10.1371/journal.pcbi.1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker RA, Chambers JM, Wilks AR. The new S language: a programming environment for data analysis and graphics. Pacific Grove, Calif.: Wadsworth & Brooks/Cole Advanced Books & Software; 1988. [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Izaurralde E, Bork P. The PAM domain, a multi-protein complex-associated module with an all-alpha-helix fold. BMC Bioinformatics. 2003;4:64. doi: 10.1186/1471-2105-4-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TA, Sugnet CW, Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- Collins GA, Tansey WP. The proteasome: a utility tool for transcription? Curr Opin Genet Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007a;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007b;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Duncan K, Umen JG, Guthrie C. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr Biol. 2000;10:687–696. doi: 10.1016/s0960-9822(00)00527-3. [DOI] [PubMed] [Google Scholar]
- Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprevote O, Seraphin B. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. Embo J. 2004;23:4847–4856. doi: 10.1038/sj.emboj.7600482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enenkel C, Lehmann A, Kloetzel PM. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelopeER network in yeast. Embo J. 1998;17:6144–6154. doi: 10.1093/emboj/17.21.6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esberg A, Huang B, Johansson MJ, Bystrom AS. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Strasser K, Racz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, Lechner J, Hurt E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. Embo J. 2002;21:5843–5852. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. Embo J. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi M, Li X, Velichutina I, Hochstrasser M, Kobayashi H. Sem1, the yeast ortholog of a human BRCA2-binding protein, is a component of the proteasome regulatory particle that enhances proteasome stability. J Cell Sci. 2004;117:6447–6454. doi: 10.1242/jcs.01575. [DOI] [PubMed] [Google Scholar]
- Furukawa K, Mizushima N, Noda T, Ohsumi Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J Biol Chem. 2000;275:7462–7465. doi: 10.1074/jbc.275.11.7462. [DOI] [PubMed] [Google Scholar]
- Gallardo M, Luna R, Erdjument-Bromage H, Tempst P, Aguilera A. Nab2p and the Thp1p-Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J Biol Chem. 2003;278:24225–24232. doi: 10.1074/jbc.M302900200. [DOI] [PubMed] [Google Scholar]
- Gautier-Bert K, Murol B, Jarrousse AS, Ballut L, Badaoui S, Petit F, Schmid HP. Substrate affinity and substrate specificity of proteasomes with RNase activity. Mol Biol Rep. 2003;30:1–7. doi: 10.1023/a:1022261925117. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gilbert W, Siebel CW, Guthrie C. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. Rna. 2001;7:302–313. doi: 10.1017/s1355838201002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;274:546–563. 547. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Part B. San Diego, Calif.: Academic Press; 2002. Guide to yeast genetics and molecular and cell biology. [Google Scholar]
- Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci. 1998;23:204–205. doi: 10.1016/s0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Isono E, Saito N, Kamata N, Saeki Y, Toh EA. Functional analysis of Rpn6p, a lid component of the 26 S proteasome, using temperature-sensitive rpn6 mutants of the yeast Saccharomyces cerevisiae. J Biol Chem. 2005;280:6537–6547. doi: 10.1074/jbc.M409364200. [DOI] [PubMed] [Google Scholar]
- Jackman JE, Montange RK, Malik HS, Phizicky EM. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. Rna. 2003;9:574–585. doi: 10.1261/rna.5070303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, Datta N, Cagney G, Buratowski S, Emili A, Greenblatt JF. Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell. 2004;16:1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- Lei EP, Stern CA, Fahrenkrog B, Krebber H, Moy TI, Aebi U, Silver PA. Sac3 is an mRNA export factor that localizes to cytoplasmic fibrils of nuclear pore complex. Mol Biol Cell. 2003;14:836–847. doi: 10.1091/mbc.E02-08-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zou C, Bai Y, Wazer DE, Band V, Gao Q. DSS1 is required for the stability of BRCA2. Oncogene. 2006;25:1186–1194. doi: 10.1038/sj.onc.1209153. [DOI] [PubMed] [Google Scholar]
- Luna R, Gaillard H, Gonzalez-Aguilera C, Aguilera A. Biogenesis of mRNPs: integrating different processes in the eukaryotic nucleus. Chromosoma. 2008 doi: 10.1007/s00412-008-0158-4. [DOI] [PubMed] [Google Scholar]
- Lund MK, Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell. 2005;20:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Mannen T, Andoh T, Tani T. Dss1 associating with the proteasome functions in selective nuclear mRNA export in yeast. Biochem Biophys Res Commun. 2008;365:664–671. doi: 10.1016/j.bbrc.2007.11.024. [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V, Pick E, Piran R, Hofmann K, Glickman MH. The COP9 signalosome-like complex in S. cerevisiae and links to other PCI complexes. Int J Biochem Cell Biol. 2003;35:706–715. doi: 10.1016/s1357-2725(02)00378-3. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonne R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol Cell. 2002;9:891–901. doi: 10.1016/s1097-2765(02)00484-7. [DOI] [PubMed] [Google Scholar]
- Nakai Y, Nakai M, Hayashi H. Thio-modification of Yeast Cytosolic tRNA Requires a Ubiquitin-related System That Resembles Bacterial Sulfur Transfer Systems. J Biol Chem. 2008;283:27469–27476. doi: 10.1074/jbc.M804043200. [DOI] [PubMed] [Google Scholar]
- Ohi MD, Gould KL. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. Rna. 2002;8:798–815. doi: 10.1017/s1355838202025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park HO, Hayles J, et al. Conservation and Rewiring of Functional Modules Revealed by an Epistasis Map in Fission Yeast. Science. 2008 doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel H, Hofmann K. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinformatics. 2005;6:71. doi: 10.1186/1471-2105-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J, Chowdhury MM, Hanzelmann P, Nimtz M, Lee EY, Schindelin H, Leimkuhler S. The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry. 2008;47:6479–6489. doi: 10.1021/bi800477u. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Segre D, Deluna A, Church GM, Kishony R. Modular epistasis in yeast metabolism. Nat Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- Sharon M, Taverner T, Ambroggio XI, Deshaies RJ, Robinson CV. Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol. 2006;4:e267. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone T, Saeki Y, Toh-e A, Yokosawa H. Sem1p is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. J Biol Chem. 2004;279:28807–28816. doi: 10.1074/jbc.M403165200. [DOI] [PubMed] [Google Scholar]
- Strasser K, Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413:648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- Thakurta AG, Gopal G, Yoon JH, Kozak L, Dhar R. Homolog of BRCA2-interacting Dss1p and Uap56p link Mlo3p and Rae1p for mRNA export in fission yeast. Embo J. 2005;24:2512–2523. doi: 10.1038/sj.emboj.7600713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Wang Q, He J, Lynn B, Rymond BC. Interactions of the yeast SF3b splicing factor. Mol Cell Biol. 2005;25:10745–10754. doi: 10.1128/MCB.25.24.10745-10754.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee S, Hetfeld B, Dubiel W, Wolf DA. Conservation of the COP9/signalosome in budding yeast. BMC Genet. 2002;3:15. doi: 10.1186/1471-2156-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SJ, Williams JG, Dang H, Darden TA, Betz BL, Humble MM, Chang FM, Trempus CS, Johnson K, Cannon RE, Tennant RW. Identification of a Specific Motif of the DSS1 Protein Required for Proteasome Interaction and p53 Protein Degradation. J Mol Biol. 2008 doi: 10.1016/j.jmb.2008.08.044. [DOI] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.