Abstract
Drought, flood, salinity, or a combination of these limits rice production. Several rice varieties are well known for their tolerance to specific abiotic stresses. We determined genetic relationship among 12 rice varieties including 9 tolerant to drought, flood, or salinity using inter-simple sequence repeat (ISSR) markers. Based on all markers, the nine tolerant varieties formed one cluster distinct from the cluster of three control varieties. The salt-tolerant varieties were closest to two flood-tolerant varieties, and together they were distinct from the drought-tolerant varieties. (GA)8YG was the most informative primer, showing the highest polymorphic information content (PIC) and resolving power (Rp). The drought-, flood-, and salt-tolerant varieties grouped in three distinct clusters within the group of tolerant varieties, when (GA)8YG was used. Sabita was the only exception. The two aus varieties, Nagina22 and FR13A, were separated and grouped with the drought- and flood-tolerant varieties, respectively, but they were together in dendrograms based on other primers. The results show that ISSR markers associated with (GA)8YG delineated the three groups of stress-tolerant varieties from each other and can be used to identify genes/new alleles associated with the three abiotic stresses in rice germplasm.
Keywords: Drought, Submergence, Salinity, Inter-simple sequence repeat-polymerase chain reaction (ISSR-PCR), (GA)8YG, Nagina22 (N22), FR13A, Pokkali
INTRODUCTION
There is concern about low yields in rice growing areas that are prone to drought, submergence/flash floods, salinity, or a combination of these, and the concern has increased with the projected climate change scenarios. The uncertainty of duration, timing and intensity of the abiotic stresses requires tremendous plasticity in adaptation. It is necessary to increase tolerance to such abiotic stresses in fragile ecosystems where only rice can be grown (IRRI, 1995).
There are several well-known rice varieties that are adapted to specific abiotic stresses, e.g., Nagina22 (N22) for drought tolerance, FR13A for flood tolerance, and Pokkali for salinity tolerance, but the genetic relationship among such tolerant varieties is unknown. Even though these are well known as major sources of only one abiotic stress and extensively researched for that (Gorantla et al., 2007; Xu et al., 2006; Mohammadi-Nejad et al., 2008), they have some tolerance to other abiotic stresses as well. For example, FR13A also has moderate drought tolerance (Adkins et al., 1995), Pokkali also has acid tolerance and submergence tolerance in seedling stage (Shylaraj et al., 2006), and N22 also has heat tolerance (Ziska et al., 1996; Jagadish et al., 2008). It is also emerging that the metabolic pathways activated in response to many abiotic stresses and even biotic stresses are common. Transcription factors, such as drought-responsive element binding (DREB) protein, are known to enhance tolerance to more than one abiotic stress, e.g., drought-, salt-, and cold-tolerance, and even biotic stress (Agarwal et al., 2006).
There has been a paradigm shift in breeding for complex traits such as yield and abiotic stress tolerance. In such cases, the phenotype may not always be a good predictor of genotype (Tanksley and McCouch, 1997). Wild species that are poor in yield have contributed genes that increased yield in rice, tomato, and several other crops (McCouch et al., 2007; Swamy and Sarla, 2008). Extending this observation further, it is possible to enhance tolerance to one stress through hybridization and introgression from lines with tolerance to a different stress. As a starting point to test the hypothesis that introgression from one stress-adaptive cultivar may increase tolerance to another stress, it is desirable to have an idea of the genetic relationship among such cultivars.
Molecular markers provide a robust estimate of genetic similarity that is often not obtained using morphological data alone. Inter-simple sequence repeat (ISSR) markers based on AG and GA repeats have been reported to be very informative and cost-effective in determining genetic relationships among diverse accessions of rice germplasm (Olufowote et al., 1997; Garland et al., 1999; Joshi et al., 2000; Davierwala et al., 2000; Sarla et al., 2003; 2005), and have been shown to be very useful in different steps of the breeding process (Reddy et al., 2002). (GATA)n containing loci were reported to be useful in fingerprinting of rice (Davierwala et al., 2001) and tomato (Rao et al., 2006). There is increasing interest in presence of GA repeats in promoters and their role in gene regulation (Santi et al., 2003; Zhang et al., 2006). In a previous study, segregation of 14 ISSR markers specific to the salt-tolerant variety CSR10 was examined in CSR10×Taraori Basmati F3 population (Kaushik et al., 2002). These markers were present at high frequencies (54%~91%) in the selected salt-tolerant plants compared with those (9%~45%) in the sensitive ones. The ISSR markers were amplified using 12 primers, 4 (primers 823, 840, 853, 884) of which were based on GA and AG repeats. This indicated that these repeats are probably linked to genomic DNA sequences with significant effects on salt tolerance (Kaushik et al., 2002). Sequencing of amplicons can allow the identification of alleles of such sequences from different germplasm. The usefulness of ISSR markers in determining diversity within rice varieties tolerant to diverse abiotic stresses has not been previously investigated. We wanted to know the genetic relationship among 9 well-known cultivars tolerant to drought, flood, or salinity and 3 popular cultivars, and whether grouping based on (GA)8-, (AG)8-, and (GATA)4-associated ISSR markers corresponds to their tolerance to a particular abiotic stress.
MATERIALS AND METHODS
The material comprised 9 well-known rice varieties tolerant to drought (Vandana, Rasi, N22), flood/submergence (FR13A, Jalmagna, Sabita), or salinity (CSR30, Pokkali, Nonasail), and 3 popular varieties (IR64, IR28, and Swarna) as controls (Table 1).
Table 1.
Significant details of the four groups of rice varieties and recent references on their use
| Accession No. | Name of variety | Significance | References |
| Control varieties | |||
| 1 | IR64 | Widely grown rainfed shallow-land variety, highly susceptible to drought stress | Ziska et al., 1996; Lafitte et al., 2007 |
| 2 | IR28 | Generally used as a salt-susceptible control in experiments on salinity stress | Ziska et al., 1996; Chao et al., 2005 |
| 3 | Swarna | Rainfed shallow-land variety, wide adaptability, widely planted in India, responsive to low N, high yield | McNally et al., 2006 |
| Drought-tolerant varieties | |||
| 4 | Vandana | Popular drought-tolerant variety, early maturity, deep-rooted, national check for short-duration varieties | Singh, 2006 |
| 5 | Rasi | Drought-tolerant variety, suitable for shallow lowlands and irrigated areas, photoinsensitive, late duration, early maturity, resistant to blast, developed at Directorate of Rice Research (DRR) in 1977 | Prasad et al., 2001 |
| 6 | N22 | Drought- and heat-tolerant variety, early maturity, deep-rooted, adapted to upland conditions, stress-responsive genes identified using expressed sequence tags (ESTs) generated from drought-stressed seedlings. It tolerates drought as it flowers early, thus escaping the day-time high temperature, which may prevent seed set | de Datta et al., 1988; Ziska et al., 1996; Gorantla et al., 2007 |
| Flood-tolerant varieties | |||
| 7 | FR13A | Flood-tolerant variety, surviving up to two weeks of complete submergence, carrying the Sub1 allele near the centromere of chromosome 9, photosensitive | Xu et al., 2006 |
| 8 | Jalmagna | Popular variety for very deep water (>1 m), high iron and zinc contents in grain, photosensitive | Singh, 2004 |
| 9 | Sabita | Popular variety for deep water (41~75 cm deep), predominating in India’s rainfed lowland ecosystem, photosensitive, late maturity, national check in variety trials for semi-deep water (0.5~1 m deep) since 1988, international check for the Eastern India rainfed lowland shuttle breeding program since 1992 | Prasad et al., 2001; Mallik et al., 2006 |
| Salt-tolerant varieties | |||
| 10 | CSR30 (Yamini) | Salt-tolerant variety with basmati qualities, recommended for partially reclaimed sodic soils of Uttar Pradesh and Haryana | DRR Annual Report, 2000~2001 |
| 11 | Pokkali | Highly salt-tolerant landrace variety from Kerala, used in several molecular studies on salt tolerance | Sahi et al., 2006 |
| 12 | Nonasail | Landrace for inland salinity, parent of Lunishree, a well-known salt-tolerant variety | Sarla et al., 2005 |
A total of 17 primers (8 based on (AG)8, 8 on (GA)8, and 1 on (GATA)4) for inter-simple sequence repeat-polymerase chain reaction (ISSR-PCR) were used in the study (Table 2).
Table 2.
Number of bands, polymorphism information content, resolving power, and mean genetic similarity revealed by each of the 17 primers in 12 rice varieties
| No. | Primer No. | PS* | nbp | npl | nb | rb/a | rb/l | PIC | Rp | MGS |
| (AG)n-based primers | ||||||||||
| 1 | 807 | (AG)8T | 21 | 17 | 156 | 13.00 | 7.43 | 0.52 | 9.0 | 0.74 |
| 2 | 808 | (AG)8C | 9 | 8 | 68 | 5.67 | 7.56 | 0.53 | 4.0 | 0.71 |
| 3 | 809 | (AG)8G | 12 | 9 | 109 | 9.08 | 9.08 | 0.36 | 3.8 | 0.82 |
| 4 | 815 | (CT)8G | 10 | 7 | 97 | 8.08 | 9.70 | 0.32 | 3.8 | 0.82 |
| 5 | 834 | (AG)8YT | 11 | 7 | 112 | 9.33 | 10.18 | 0.25 | 3.3 | 0.87 |
| 6 | 835 | (AG)8YC | 15 | 13 | 80 | 6.67 | 5.33 | 0.68 | 5.0 | 0.71 |
| 7 | 836 | (AG)8YA | 18 | 13 | 131 | 10.92 | 7.28 | 0.53 | 7.1 | 0.76 |
| 8 | 884 | HBH(AG)7 | 20 | 18 | 108 | 9.00 | 5.40 | 0.70 | 8.7 | 0.62 |
| Mean | 14.5 | 11.5 | 108 | 8.97 | 7.75 | 0.49 | 5.6 | 0.76 | ||
| (GA)n-based primers | ||||||||||
| 9 | 810 | (GA)8T | 12 | 7 | 128 | 10.67 | 10.67 | 0.18 | 2.7 | 0.91 |
| 10 | 811 | (GA)8C | 9 | 7 | 78 | 6.50 | 8.67 | 0.40 | 2.7 | 0.81 |
| 11 | 812 | (GA)8A | 18 | 9 | 142 | 11.83 | 7.89 | 0.42 | 4.0 | 0.87 |
| 12 | 822 | (TC)8A | 12 | 10 | 70 | 5.83 | 5.83 | 0.63 | 3.7 | 0.74 |
| 13 | 840 | (GA)8YT | 11 | 8 | 88 | 7.33 | 8.00 | 0.44 | 3.3 | 0.82 |
| 14 | 841 | (GA)8YC | 14 | 9 | 108 | 9.00 | 7.71 | 0.46 | 4.0 | 0.83 |
| 15 | 842 | (GA)8YG | 22 | 22 | 120 | 10.00 | 5.45 | 0.75 | 14.3 | 0.50 |
| 16 | 885 | BHB(GA)7 | 19 | 10 | 190 | 15.83 | 10.00 | 0.25 | 4.0 | 0.89 |
| Mean | 14.6 | 10.2 | 115.5 | 9.62 | 8.03 | 0.44 | 4.8 | 0.80 | ||
| 17 | 872 | (GATA)4 | 13 | 10 | 74 | 6.17 | 5.69 | 0.60 | 7.67 | 0.58 |
PS: Primer sequence;
Y: T or C; n bp: number of band positions (loci); n pl: number of polymorphic loci; n b: number of bands; r b/a: mean number of bands/accession; r b/l: mean number of bands/locus; PIC: polymorphism information content; Rp: resolving power; MGS: mean genetic similarity
Nine cultivars belong to the indica group, two cultivars, N22 and FR13A, to the aus group (Garris et al., 2005) or Isozyme Gr II (Glaszmann, 1987), and CSR30 to the aromatic group (Garris et al., 2005) or Isozyme Gr V (Glaszmann, 1987).
Genomic DNA was isolated from freshly harvested young leaves of five plants of each cultivar by Mini prep method. Quality and quantity of DNA were estimated both visually by ethidium bromide staining of 8 mg/ml agarose gel and using a spectrophotometer. DNA was diluted to uniform concentration of about 10 ng/μl. ISSR-PCR was conducted in a reaction volume of 15 μl containing 30 ng template DNA, 0.2 μmol/L primer, 200 μmol/L each dNTP, 10 mmol/L Tris-Cl (pH 8.3), 50 mmol/L KCl, 2.0 mmol/L MgCl2, and 1 U of Taq polymerase. PCR amplification conditions were set as initial denaturation at 94 °C for 5 min, 40 cycles of denaturation at 94 °C for 1 min, annealing at 50 °C for 1 min, extension at 72 °C for 2 min, and a final extension at 72 °C for 7 min. PCR was performed in 96-well plate thermal cycler (Eppendorf, Germany). The amplified products were mixed with loading dye (0.4 g/ml sucrose and 2.5 mg/ml bromophenol blue), resolved on 18 mg/ml agarose gel in 0.5× Tris borate EDTA (TBE) buffer under room temperature at constant voltage of 100 V, and detected by ethidium bromide staining (0.5 mg/ml). The gels were documented under ultraviolet light using gel documentation unit from Pharmacia Biotech, Sweden.
Each amplified product was scored as 1 or 0 depending on its presence or absence. The frequency of microsatellite polymorphism was calculated based on the presence or absence of common bands. The polymorphism information content (PIC) value was calculated as 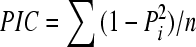 , where n is the number of band positions analyzed in the set of accessions, and Pi is the frequency of ith pattern. The ability of the primers to distinguish between accessions was assessed by calculating their resolving power (Rp) as Rp=∑I
b, where I
b is band informativeness, I
b=1−[2×(0.5−pi)] and pi is the proportion of accessions containing band i (Prevost and Wilkinson, 1999). The genetic associations among varieties were evaluated using Dice similarity coefficient for pair-wise comparisons based on the proportion of shared bands produced by primers. Similarity matrices were generated using ‘SIMQUAL’ sub-program of NTSYS-PC software. Similarity coefficients were used for cluster analysis of varieties using ‘SAHN’ sub-program of NTSYS-PC software and dendrograms were obtained using un-weighted pair-group method with arithmetic average (UPGMA) sub-program of NTSYS-PC software version 2.0 from Exeter software, NY, USA (Rohlf, 1993). Bootstrap analysis was performed using 1000 permutations in Winboot. Bootstrap values over 50 are considered significant and mentioned on the dendrogram.
, where n is the number of band positions analyzed in the set of accessions, and Pi is the frequency of ith pattern. The ability of the primers to distinguish between accessions was assessed by calculating their resolving power (Rp) as Rp=∑I
b, where I
b is band informativeness, I
b=1−[2×(0.5−pi)] and pi is the proportion of accessions containing band i (Prevost and Wilkinson, 1999). The genetic associations among varieties were evaluated using Dice similarity coefficient for pair-wise comparisons based on the proportion of shared bands produced by primers. Similarity matrices were generated using ‘SIMQUAL’ sub-program of NTSYS-PC software. Similarity coefficients were used for cluster analysis of varieties using ‘SAHN’ sub-program of NTSYS-PC software and dendrograms were obtained using un-weighted pair-group method with arithmetic average (UPGMA) sub-program of NTSYS-PC software version 2.0 from Exeter software, NY, USA (Rohlf, 1993). Bootstrap analysis was performed using 1000 permutations in Winboot. Bootstrap values over 50 are considered significant and mentioned on the dendrogram.
RESULTS
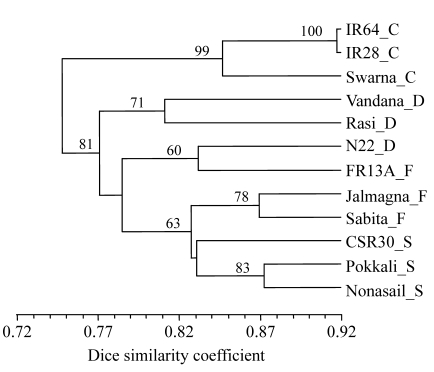
UPGMA dendrogram derived from data of all primers (1859 bands at 246 band positions) showed that the 3 control varieties formed one group and the 9 tolerant varieties formed another group (Fig.1).
Fig. 1.
UPGMA dendrogram of 12 genotypes based on all 17 primers
Suffix after each variety refers to its group: C: control; D: drought tolerant; F: flood tolerant; S: salt tolerant
In the latter group, there were 3 clear clusters, the first consisting of 2 drought-tolerant indica varieties (Rasi and Vandana), the second of 2 aus varieties, FR13A (flood-tolerant) and N22 (drought-tolerant), and the third of 2 flood-tolerant varieties and 3 salt-tolerant varieties in 2 distinct sub-clusters. The aromatic salt-tolerant variety CSR30 grouped with the other non-aromatic salt-tolerant varieties at a low bootstrap value of 41 (not shown in Fig.1). Jalmagna and Sabita, the flood-tolerant varieties grouped together and joined the group of salt-tolerant varieties at 83% similarity.
Grouping based on data of 8 (GA)8-based primers (924 bands at 117 band positions, dendrogram not presented) was almost similar to the grouping based on all primers. The dendrogram showed 2 clusters—a minor cluster consisting of all the 3 control varieties and a major cluster consisting of 2 sub-clusters. FR13A grouped along with all the 3 drought-tolerant varieties in sub-cluster 1. All the 3 salt-tolerant varieties along with other two flood-tolerant varieties formed sub-cluster 2 of the major cluster.
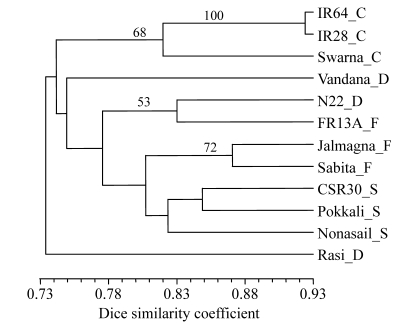
Grouping based on 8 (AG)8-based primers (861 bands at 116 band positions) (Fig.2) showed that Rasi was distinct from all other varieties at 73% similarity. The next to separate were the 3 control varieties as one group. Vandana separated next. The remaining 7 varieties formed one cluster of 2 sub-clusters—a small sub-cluster of N22 and FR13A, the 2 aus varieties, and a large one with 2 small groups, of which one had 2 flood-tolerant varieties and the other had 3 salt-tolerant varieties.
Fig. 2.
UPGMA dendrogram of 12 genotypes based on (AG)n primers
Suffix after each variety refers to its group: C: control; D: drought tolerant; F: flood tolerant; S: salt tolerant
In the dendrogram based on (GATA)4 (dendrogram not presented), two major groups were observed and each group had at least one variety in each of flood-, drought-, and salt-tolerant groups. The control varieties, and Rasi, Jalmagna, Nonasail, and Pokkali were in one cluster and the two aus varieties, FR13A and N22, were in the other cluster along with Vandana, CSR30, and Sabita.
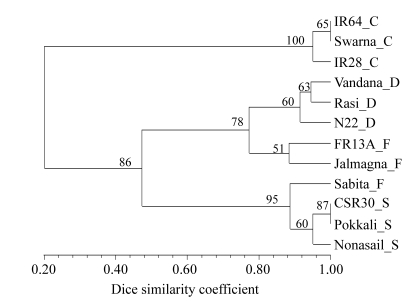
Overall the most informative primer was (GA)8YG, i.e., primer 842 in terms of the highest PIC and Rp values. Markers amplified by (GA)8YG could distinguish all varieties except 2 pairs, IR64-Swarna and CSR30-Pokkali. In the dendrogram based on primer 842 alone, the 3 control varieties grouped with a bootstrap value of 100% and all the tolerant varieties in one group with bootstrap value 86% (Fig.3).
Fig. 3.
UPGMA dendrogram of 12 genotypes based on the most informative primer (GA)8YG
Suffix after each variety refers to its group: C: control; D: drought tolerant; F: flood tolerant; S: salt tolerant
All the nodes had high bootstrap values. Within the tolerant varieties group were two clusters, one consisting of all salt-tolerant varieties and Sabita, and the other cluster consisting of three drought-tolerant varieties in one sub-cluster and two flood-tolerant varieties in the other sub-cluster.
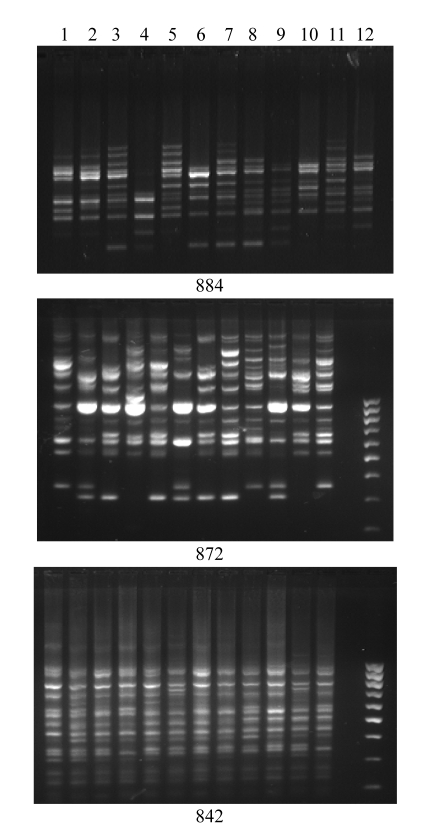
GA-based primers amplified more bands (924) compared with AG-based primers (861), though the numbers of loci amplified by AG- and GA-based primers were about the same. However, the average PIC and Rp of AG-based primers were higher than those of GA-based primers. The number of bands amplified by (GA)n was also greater than that by (AG)n in each group of varieties. The flood-tolerant varieties amplified the maximum number of bands compared with the other 3 groups. Among the 248 band positions amplified, 17 were unique (present only in one variety) and occurred in 6 varieties (Vandana, Rasi, FR13A, N22, Sabita, and Swarna). FR13A showed maximum unique bands at 6 band positions with 4 primers, followed by Swarna at 5 band positions. Bands specific to control varieties were amplified at 8 loci using the primers 842, 884, and 812. Primers 842 and 884 also showed bands specific to drought-tolerant varieties. Likewise primers 836 and 885 showed bands specific to salt- and flood-tolerant varieties. Three primers 809, 812, and 836 showed bands that occur only in the aus varieties N22 and FR13A. The primer 842 showed the highest PIC value (0.75), the highest Rp value (14.33), and the lowest mean genetic similarity among the accessions (Table 2, Fig.4).
Fig. 4.
ISSR-PCR amplification profiles of 12 accessions using primers 884, 872, and 842, respectively
In each gel, lanes 1~12 from left to right correspond to accession Nos. 1~12, respectively, of Table 1
PIC values of AG-based primers ranged from 0.7 in primer 884 to 0.25 in primer 834. Among the AG-based primers, primer 884 was the most informative (Fig.4). It showed high Rp value (8.67) with the lowest mean genetic similarity among all accessions.
DISCUSSION
Knowledge of the genetic relationship between cultivars is useful for choosing superior yet genetically divergent parents for hybridization to optimize genetic variation in subsequent generations. Based on all markers, our study provides evidence that the abiotic stress-tolerant varieties are collectively different from the three standard cultivars used as control.
The two aus varieties, N22 and FR13A, corresponding to Gr II of Glaszmann classification grouped together, consistent with the population structure of rice wherein the aus types (Gr II) were delineated in the indica cluster (Garris et al., 2005; McNally et al., 2006). It is interesting that FR13A, one of the best known flood-tolerant varieties, is genetically close to N22, one of the best known drought-tolerant varieties, both belonging to Gr II (Glaszmann, 1987). N22 is also one of the most heat-tolerant rice varieties (de Datta et al., 1988; Ziska et al., 1996).
Our data also show that the salt-tolerant varieties are genetically closer to the flood-tolerant varieties than to the drought-tolerant varieties, if all the markers are considered. This may well be a reflection of the kinds of varieties chosen. Among the salt-tolerant varieties, Pokkali and Nonasail were closest with 88% similarity and both are tolerant to coastal salinity. CSR30, which is tolerant to inland salinity, joined the two at about 83% similarity. CSR30 belongs to the aromatic group (Gr V), which could be a reason for its joining the indica (Gr I) salt-tolerant varieties at a low bootstrap value. Sabita, a deep-water mega variety popular in coastal areas, was always close to the salt-tolerant varieties. The proximity of flood-tolerant varieties to salt-tolerant varieties or drought-tolerant varieties needs to be analyzed in a larger and diverse rice-germplasm set tolerant to abiotic stresses. For instance, it would be interesting to know if several varieties tolerant to inland salinity as against coastal salinity were used, whether they would still group with the flood-tolerant varieties. Even among varieties generically referred to as flood-tolerant ones here, there are considerable differences in tolerance to extent, duration, salinity, and temperature of water bodies in which rice gets submerged.
It is significant that (GA)8YG-associated markers grouped the varieties according to their tolerance to a specific stress. This has not been reported previously. The separation of the two genetically close aus varieties, N22 and FR13A, and their grouping with other drought-tolerant and flood-tolerant varieties, respectively, clearly show that markers associated with (GA)8YG have a strong positive association with DNA sequences linked to specific stress tolerances and not to the phylogenetic similarities. In all other dendrograms the two aus varieties, N22 and FR13A, group together. (GA)8YG was earlier reported to be one of the most informative primers for studying diversity in rice landraces and wild species (Sarla et al., 2003; 2005). The primer (GATA)4 could help distinguish all the 12 varieties and is suitable for use in fingerprinting as reported earlier for rice and tomato (Davierwala et al., 2001; Rao et al., 2006).
Genomic regions associated with GA repeats are known to be involved in gene regulation. GA repeats (GAGA elements) are known to control gene expression in animals and plants (Sangwan and Brian, 2002; Santi et al., 2003; van Steensel et al., 2003). These are reported to be most abundant in 5′ regions of genes. In Arabidopsis, (GA)8 repeats occur particularly within 1500 bp upstream of gene start codons (Zhang et al., 2004; 2006). In the context of abiotic stress tolerance, it is reported that non-coding microsatellite repeats provide opportunities for rapid adaptive changes in regulatory regions or play specific roles in gene regulation in response to environmental stimuli (Zhang et al., 2006). Our data show that clustered (GA)8 and (AG)8 repeats occur more frequently in the flood-tolerant varieties than in the other 3 groups of varieties, since more bands were obtained in the flood-tolerant varieties. These flood-tolerant varieties are also photosensitive.
Evaluation of varieties for different stresses is required, as very few studies have assessed cross-tolerances. It is likely that FR13A can be a source of new alleles for drought and salt tolerance and N22 for traits other than drought tolerance. In an earlier study on the ability to withstand drought in R2 families of 5 rice cultivars, significant improvement in drought tolerance was found in 6% of the FR13A families compared with parental controls (Adkins et al., 1995). It is suggested that in experiments on salt or drought tolerance if FR13A or Sabita is used as an outgroup, more interesting insights into its drought, flood or salt tolerance would be obtained. Many genes are known to be over-expressed by more than one stress; e.g., the gene OsGGT (glycogenin glucosyltransferase) from FR13A is over-expressed not only in response to submergence but upon drought and salt stresses as well (Qi et al., 2005). In another study, as many as 67 upregulated genes were found to be common to 9 different abiotic stresses (Swindell, 2006).
It is noteworthy that some of the submergence-prone areas are also drought prone, and that rice crop can face drought first and flood later or the reverse. Also, in the field situation there may be considerable overlap of stresses experienced by rice, and each variety may have a good combination of tolerances. For example, RD19 released in Thailand in 1980 for traditional deep-water areas is also fairly drought tolerant (Prasad et al., 2001), and Janki tolerates both drought and submergence (Mackill et al., 1996). Thus, it is possible to have both drought and flood tolerance but maybe at different stages of growth or different modes of tolerance. In our view, crossing of varieties belonging to different groups as shown in this study can lead to useful transgressive segregants with increased tolerance to one stress or overlap of tolerance to more than one stress.
The results reported here have significant implications for rice breeding and basic studies as well. Closely related or moderately distant parents can be chosen based on the overall grouping reported here and depending on the breeding objectives. The two flood-tolerant varieties which grouped with salt-tolerant varieties are likely sources of new alleles for salt tolerance. Sequencing of the ISSR amplicons amplified by (GA)8YG can help identify allelic variation for stress-associated genes from different germplasm.
CONCLUSION
Our results show that there is a link between GA repeats and tolerance to different abiotic stresses. Firstly, the amplified ISSR regions, when used for clustering, grouped the control genotypes (IR64, IR28, and Swarna) separately from the stress-tolerant genotypes. Secondly, among the stress-tolerant genotypes the drought-, flood-, and salt-tolerant genotypes were also clearly delineated when (GA)8YG was used.
Acknowledgments
We are thankful to Department of Biotechnology (DBT), University Grants Commission-Council of Scientific and Industrial Research (UGC-CSIR), and Indian Council of Agricultural Research (ICAR), Government of India for fellowship. We thank Dr. K Muralidharan, Head, Crop Protection, Directorate of Rice Research (DRR) for a critical review of the manuscript and the Project Director, DRR for encouragement and support.
References
- 1.Adkins SW, Kunanuvatchaidach R, Godwin ID. Somaclonal variation in rice. 2. Drought tolerance and other agronomic characters. Aust J Bot. 1995;43(2):201–209. doi: 10.1071/BT9950201. [DOI] [Google Scholar]
- 2.Agarwal PK, Agarwal P, Reddy MK, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006;25(12):1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- 3.Chao DY, Luo YH, Shi M, Luo D, Lin HX. Salt-responsive genes in rice revealed by cDNA microarray analysis. Cell Res. 2005;15(10):796–810. doi: 10.1038/sj.cr.7290349. [DOI] [PubMed] [Google Scholar]
- 4.Davierwala AP, Chowdari KV, Kumar S, Reddy APK, Ranjekar PK, Gupta VS. Use of three different marker systems to estimate genetic diversity of Indian elite rice varieties. Genetica. 2000;108(3):269–284. doi: 10.1023/A:1004160232679. [DOI] [PubMed] [Google Scholar]
- 5.Davierwala AP, Ramakrishna W, Chowdari V, Ranjekar PK, Gupta VS. Potential of (GATA)n microsatellites from rice for inter- and intra-specific variability studies. BMC Evol Biol. 2001;1(1):7. doi: 10.1186/1471-2148-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Datta S, Malabuyoc J, Aragon E. A field screening technique for evaluating rice germplasm for drought tolerance during the vegetative stage. Field Crops Research. 1988;19(2):123–134. doi: 10.1016/0378-4290(88)90050-0. [DOI] [Google Scholar]
- 7.Garland SH, Lewin L, Abedinia M, Henry R, Blakeney A. The use of microsatellite polymorphisms for the identification of Australian breeding lines of rice (Oryza sativa L.) Euphytica. 1999;108(1):53–63. doi: 10.1023/A:1003688612179. [DOI] [Google Scholar]
- 8.Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch SR. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169(3):1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glaszmann JC. Isozymes and classification of Asian rice varieties. Theor Appl Genet. 1987;74(1):21–30. doi: 10.1007/BF00290078. [DOI] [PubMed] [Google Scholar]
- 10.Gorantla M, Babu PR, Lachagari VB, Reddy AM, Wusirika R, Bennetzen JL, Reddy AR. Identification of stress-responsive genes in an indica rice (Oryza sativa L.) using ESTs generated from drought-stressed seedlings. J Exp Bot. 2007;58(2):253–265. doi: 10.1093/jxb/erl213. [DOI] [PubMed] [Google Scholar]
- 11.IRRI (International Rice Research Institute) Fragile Lives in Fragile Ecosystems. Manila, Philippines: 1995. p. 976. [Google Scholar]
- 12.Jagadish SVK, Craufurd PQ, Wheeler TR. Phenotyping parents of mapping population of rice for heat tolerance during anthesis. Crop Sci. 2008;48(3):1140–1146. doi: 10.2135/cropsci2007.10.0559. [DOI] [Google Scholar]
- 13.Joshi SP, Gupta VS, Aggarwal RK, Ranjekar PK, Brar DS. Genetic diversity and phylogenetic relationship as revealed by inter-simple sequence repeat (ISSR) polymorphism in the genus Oryza . Theor Appl Genet. 2000;100(8):1311–1320. doi: 10.1007/s001220051440. [DOI] [Google Scholar]
- 14.Kaushik A, Sani N, Jan S, Singh RK, Jan R. Genetic structure of a segregating CSR10×Taraori Basmati F3 population for salinity tolerance. Rice Genetics Newsletter. 2002;19:85–87. [Google Scholar]
- 15.Lafitte HR, Yongsheng G, Yan S, Li ZK. Whole plant responses, key processes, and adaptation to drought stress: the case of rice. J Exp Bot. 2007;58(2):169–175. doi: 10.1093/jxb/erl101. [DOI] [PubMed] [Google Scholar]
- 16.Mackill DJ, Coffman WR, Garrity DP. Rainfed Lowland Rice Improvement IRRI. Manila, Philippines: 1996. p. 242. [Google Scholar]
- 17.McCouch SR, Sweeney M, Li J, Jiang H, Thomson M, Septiningsih E, Edwards J, Moncada P, Xiao J, Garris A. Through the genetic bottleneck: O. rufipogon as a source of trait-enhancing alleles for O. sativa . Euphytica. 2007;154(3):317. doi: 10.1007/s10681-006-9210-8. [DOI] [Google Scholar]
- 18.McNally KL, Bruskiewich R, Mackill D, Buell CR, Leach JE, Leung H. Sequencing multiple and diverse rice varieties. Connecting whole-genome variation with phenotypes. Plant Physiol. 2006;141(1):26–31. doi: 10.1104/pp.106.077313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohammadi-Nejad G, Arzani A, Rezail AM, Singh RK, Gregorio GB. Assessment of rice genotypes for salt tolerance using microsatellite markers associated with the saltol QTL. African Journal of Biotechnology. 2008;7(6):730–736. [Google Scholar]
- 20.Olufowote JO, Xu Y, Chen X, Park WD, Beachell HM, Dilday RH, Goto M, McCouch SR. Comparative evaluation of within-cultivar variation of rice (Oryza sativa L.) using microsatellite and RFLP markers. Genome. 1997;40(3):370–378. doi: 10.1139/g97-050. [DOI] [PubMed] [Google Scholar]
- 21.Prasad GSV, Muralidharan K, Rao CS, Prasad ASR. Stability and yield performance of genotypes: a proposal for regrouping world rice area into mega environments. Curr Sci. 2001;81:1337–1346. [Google Scholar]
- 22.Prevost A, Wilkinson MJ. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet. 1999;98(1):107–112. doi: 10.1007/s001220051046. [DOI] [Google Scholar]
- 23.Qi Y, Kawano N, Yamauchi Y, Ling J, Li D, Tanaka K. Identification and cloning of a submergence-induced gene OsGGT (glycogenin glucosyltransferase) from rice (Oryza sativa L.) by suppression subtractive hybridization. Planta. 2005;221(3):437–445. doi: 10.1007/s00425-004-1453-9. [DOI] [PubMed] [Google Scholar]
- 24.Rao R, Corrado G, Bianchi M, DiMauro A. (GATA)4 DNA fingerprinting identifies morphologically characterized “San Marzano” tomato plants. Plant Breed. 2006;125(2):173–176. doi: 10.1111/j.1439-0523.2006.01183.x. [DOI] [Google Scholar]
- 25.Reddy PM, Sarla N, Siddiq EA. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica. 2002;128(1):9–17. doi: 10.1023/A:1020691618797. [DOI] [Google Scholar]
- 26.Rohlf FJ. NTSYS-PC Version 2.0. Setauket, New York: State University of New York; 1993. Exeter Software. [Google Scholar]
- 27.Sahi C, Singh A, Kumar K, Blumwald E, Grover A. Salt stress response in rice: genetics, molecular biology, and comparative genomics. Funct Integr Genomics. 2006;6(4):263–284. doi: 10.1007/s10142-006-0032-5. [DOI] [PubMed] [Google Scholar]
- 28.Sangwan I, Brian MRO. Identification of a soybean protein that interacts with GAGA element dinucleotide repeat DNA. Plant Physiol. 2002;129(4):1788–1794. doi: 10.1104/pp.002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santi L, Wang Y, Stile MR, Berendzen K, Wanke D, Roig C, Pozzi C, Muller K, Muller J, Rohde W, et al. The GA octodinucleotide repeat binding factor BBR participates in the transcriptional regulation of the homeobox gene Bkn3 . Plant J. 2003;34(6):813–826. doi: 10.1046/j.1365-313X.2003.01767.x. [DOI] [PubMed] [Google Scholar]
- 30.Sarla N, Bobba S, Siddiq EA. ISSR and SSR markers based on AG and GA repeats delineate geographically diverse Oryza nivara accessions and reveal rare alleles. Curr Sci. 2003;84:683–690. [Google Scholar]
- 31.Sarla N, Neeraja CN, Siddiq EA. Use of anchored (AG)n and (GA)n primers to assess genetic diversity of Indian landraces and varieties of rice. Curr Sci. 2005;89:1371–1381. [Google Scholar]
- 32.Shylaraj KS, Sasidharan NK, Sreekumaran V. VTL 6: a semi-tall, non-lodging, and high yielding rice (Oryza sativa L.) variety for the coastal saline zones of Kerala. Journal of Tropical Agriculture. 2006;44(1-2):48–51. [Google Scholar]
- 33.Singh DN. Participatory plant breeding as a method of rice breeding. International Rice Research Notes. 2006;31(2):48–50. [Google Scholar]
- 34.Singh UP. Farmers participatory diagnosis of flood prone deepwater rice-cropping system in eastern India. International Rice Research Notes. 2004;29(2):85–87. [Google Scholar]
- 35.Swamy BPM, Sarla N. Yield enhancing quantitative trait loci (QTLs) from wild species. Biotechnology Advances. 2008;26(1):106–120. doi: 10.1016/j.biotechadv.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 36.Swindell WR. The association among gene expression responses to nine abiotic stress treatments in Arabidopsis thaliana . Genetics. 2006;174(4):1811–1824. doi: 10.1534/genetics.106.061374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanksley SD, McCouch SR. Seed banks and molecular maps: unlocking genetic potential from the wild. Science. 1997;277(5329):1063–1066. doi: 10.1126/science.277.5329.1063. [DOI] [PubMed] [Google Scholar]
- 38.van Steensel B, Delrow J, Bussemaker HJ. Genome wide analysis of Drosophila GAGA factor target genes reveals context dependent DNA binding. Proc Nat Acad Sci. 2003;100(5):2580–2585. doi: 10.1073/pnas.0438000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442(7103):705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- 40.Zhang LD, Yuan DJ, Yu SW, Li ZG, Cao YF, Miao ZQ, Qian HM, Tang KX. Preference of simple sequence repeats in coding and non-coding regions of Arabidopsis thaliana . Bioinformatics. 2004;20(7):1081–1086. doi: 10.1093/bioinformatics/bth043. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Zuo K, Zhang F, Cao Y, Wang J, Zhang Y, Sun X, Tang K. Conservation of noncoding microsatellites in plants: implication for gene regulation. BMC Genomics. 2006;7(1):323. doi: 10.1186/1471-2164-7-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ziska LH, Manalo PA, Ordonez RA. Intraspecific variation in the response of rice (Oryza sativa L.) to increased CO2 and temperature: growth and yield response of 17 cultivars. J Exp Bot. 1996;47(9):1353–1359. doi: 10.1093/jxb/47.9.1353. [DOI] [Google Scholar]