Abstract
Many genes in eukaryotic genomes produce multiple transcripts through a variety of molecular mechanisms including alternative splicing. Alternatively spliced transcripts often encode functionally distinct proteins, indicating that gene regulation at this level makes an important contribution to organismal complexity. The multilevel splicing cascade that regulates sex determination and sex-specific development in Drosophila is a classical example of the role of alternative splicing in cell differentiation. Recent evidence suggests that a large proportion of genes in the Drosophila genome may be spliced in a sex-biased fashion, raising the possibility that alternative splicing may play a more general role in sexually dimorphic development and physiology. However, the prevalence of sex-specific splicing and the extent to which it is shared among genotypes are not fully understood. Genetic variation in the splicing of key components of the sex determination pathway is known to influence the expression of downstream target genes, suggesting that alternative splicing at other loci may also vary in functionally important ways. In this study, we used exon-specific microarrays to examine 417 multitranscript genes for evidence of sex-specific and genotype-specific splicing in 80 different genotypes of Drosophila melanogaster. Most of these loci showed sex-biased splicing, whereas genotype-specific splicing was rare. One hundred thirty-five genes showed different alternative transcript use in males vs. females. Real-time PCR analysis of 6 genes chosen to represent a broad range of biological functions showed that most sex-biased splicing occurs in the gonads. However, somatic tissues, particularly adult heads, also show evidence of sex-specific splicing. Comparison of splicing patterns at orthologous loci in seven Drosophila species shows that sexual biases in alternative exon representation are highly conserved, indicating that sex-specific splicing is an ancient feature of Drosophila biology. To investigate potential mechanisms of sex-biased splicing, we used real-time PCR to examine the expression of six known regulators of alternative splicing in males vs. females. We found that all six loci are themselves spliced sex specifically in gonads and heads, suggesting that regulatory hierarchies based on alternative splicing may be an important feature of sexual differentiation.
THE pervasive and frequently marked differences in morphology and behavior between females and males evolve when selection favors different phenotypes in the two sexes, imposed by their distinct roles in reproduction and competition (Lande 1980). Intergenomic conflict may arise between sexes with predominantly identical genomes but different fitness requirements, a discord that may be resolved by the evolution of sex-specific gene expression (Connallon and Knowles 2005; Ellegren and Parsch 2007). Advances in high-throughput technologies such as whole-genome sequencing and transcriptome profiling have afforded new insight into the widespread occurrence of sex-biased gene expression in diverse species including Drosophila (Jin et al. 2001; Parisi et al. 2003; Ranz et al. 2003), mice (Yang et al. 2006), nematodes (Reinke et al. 2000; Thoemke et al. 2005), mosquitoes (Iatrou and Biessmann 2008), and birds (Mank et al. 2008). In the best characterized Drosophila model, sexual dimorphism affects not only transcript abundance per se, but also the genetic architecture of transcriptome variation and the mode by which this variation is inherited (Wayne et al. 2007).
An additional level of sexual dimorphism is found at loci that produce multiple transcripts through alternative pre-mRNA splicing (AS) or the use of alternative transcription initiation and termination sites (McIntyre et al. 2006). AS makes an important contribution to organismal complexity by allowing a single gene to produce multiple, functionally distinct proteins (Graveley 2001; Ben-Dov et al. 2008). Alternative splicing is a common phenomenon among eukaryotes (Kim et al. 2007), with estimates in excess of 70% of the genome in humans (Johnson et al. 2003; Gupta et al. 2004; Ben-Dov et al. 2008), up to 40% in Drosophila melanogaster (Stolc et al. 2004), and of 10% in Caenorhabditis elegans (Kim et al. 2007). The biological importance of AS is reflected in its tissue- and stage-specific regulation (Stolc et al. 2004; Thoemke et al. 2005; Blencowe 2006; Clark et al. 2007) and in the preponderance of AS in tissues with diverse cell types and in genes with regulatory functions such as cell adhesion molecules and DNA- and RNA-binding proteins (Pan et al. 2004; Barberan-Soler and Zahler 2008; Ben-Dov et al. 2008).
The best characterized system of alternative splicing regulation is the pathway of somatic sex determination in Drosophila (Nagoshi et al. 1988; Bell et al. 1991; Cline 1993). Three RNA-binding proteins, Sex-lethal (SXL), Transformer (TRA), and Transformer 2 (TRA2), form a multilevel splicing cascade that regulates sex-specific splicing of the downstream targets male-specific lethal-2, fruitless, and doublesex, which in turn control sexually dimorphic cell differentiation and dosage compensation (comprehensive reviews by Baker 1989; Black 2003; Penalva and Sanchez 2003). At the top of the cascade, female-specific splicing is initiated by early zygotic expression of Sxl from the “establishment” promoter in response to the presence of two X chromosomes. This sets up a stable feedback loop in which SXL protein induces female-specific splicing of Sxl mRNA transcribed from the “maintenance” promoter that becomes active later in development (Bell et al. 1988, 1991). Default splicing of Sxl transcript in males yields a truncated, nonfunctional protein. In females, SXL controls the splicing of tra to produce a functional TRA protein, which, together with TRA2, promotes female-specific splicing of the dsx and fru transcription factors that go on to direct female-specific development of somatic tissues. In males, default splicing of tra, dsx, and fru results in the production of male-specific DSX and FRU isoforms, which promote male-specific differentiation. In adult flies, the abundance of splice variants of Sxl, tra2, dsx, ix, and her varies among genotypes (Tarone et al. 2005). This variation may partly explain genetic variation in the expression of downstream target genes such as Yolk protein 1 (Tarone et al. 2005).
The sex determination pathway is conserved within the genus Drosophila (Erickson and Cline 1998; Pomiankowski et al. 2004). Conservation of sex-specific splicing of doublesex in the holometabolous insect orders Diptera, Lepidoptera, and Hymenoptera suggests that the splicing-based mechanism of sex determination may be quite ancient (Pomiankowski et al. 2004; Cho et al. 2007). However, the upstream regulators of doublesex splicing are more evolutionarily labile. Transformer is restricted to certain dipterans (Pomiankowski et al. 2004) and is rapidly evolving in Drosophila (Kulathinal et al. 2003), while Sxl, which is the master regulator of sex determination in Drosophila, is restricted to this genus (Pomiankowski et al. 2004).
Despite the importance of AS in sex determination, the prevalence of sex-specific splicing in Drosophila was not fully appreciated until systematic genomewide analyses became possible. McIntyre et al. (2006), using D. melanogaster microarrays designed to detect over >2700 multitranscript genes, found that between 10 and 22% of these genes showed sex-biased expression of alternative transcripts. Moreover, many known regulators of mRNA splicing (Park et al. 2004) show sex-biased expression, suggesting a possible explanation for the prevalence of sex-biased splicing (McIntyre et al. 2006). Genetic variation in the splicing of several components of the sex determination pathway (Tarone et al. 2005) suggests that the splicing of other multitranscript genes may vary across genotypes, especially if these genes are regulated by the sexual hierarchy. Although McIntyre et al. (2006) found little effect of sex-by-genotype interaction on alternative transcript abundance, this could be due to the limited number of strains examined in that study. A larger number of genotypes are necessary both to determine the generality of sex-specific splicing among genotypes and to test for genetic variation in alternative transcript preference.
Here, we report that a large proportion of multitranscript genes show consistent patterns of sex-biased splicing in 80 crosses derived from 11 strains of D. melanogaster. Although genotype has a large effect on transcript abundance, genotype-specific splicing is rare. RT–PCR analysis suggests that most, but not all, sex-biased splicing occurs in gonads. A comparison of seven Drosophila species separated by up to 60 million years of evolution shows that most sexual biases in alternative exon representation are highly conserved. Finally, we show that several known regulators of mRNA splicing are themselves spliced sex specifically, suggesting that regulatory hierarchies based on alternative splicing may be an important feature of sexual differentiation.
MATERIALS AND METHODS
Drosophila lines and culture:
Recombinant inbred lines (RIL-derived lines):
Four replicates of two D. melanogaster laboratory strains OregonR and Russian 2b, and six randomly chosen recombinant inbred lines derived from these parents were grown from small mass matings on standard dextrose medium. Parents were removed after 3 days at 25°, 12:12-hr light/dark cycle, and 20 virgin females and males were collected within 24 hr and matured separately (64 samples total). At day 3 posteclosion flies were snap frozen in liquid nitrogen and stored at −80° (McIntyre et al. 2006).
Diallel derived F1 heterozygous crosses (DIL-derived lines):
The nine parental D. melanogaster lines used for this experiment were derived from single females collected in the wild (Winters, CA) and inbred under full-sib mating for ∼40 generations (Yang and Nuzhdin 2003). The lines were further isogenized using standard balancer techniques (Nuzhdin et al. 1998) and then crossed in a full diallel breeding scheme with reciprocals, excluding self crosses of the parents for a total of 72 F1 crosses (Wayne et al. 2007). Thus these 72 genotypes represent all the pairwise combinations of the nine parental alleles. Flies were reared as above, two biological replicates for each cross, and two sexes resulted in 288 samples.
D. melanogaster RNA sample preparation, probe selection, microarray hybridization, and signal detection:
Total RNA was isolated from the frozen samples using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions and then purified using standard techniques (RNeasy kit; QIAGEN, Valencia, CA). RNA concentration was determined on a Nanodrop Spectrophotometer (NanoDrop Technologies, Wilmington, DE) and RNA integrity was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
The chip was synthesized on an Agilent platform (bioinformatics.ufl.edu/pages_for_research/Drosophilia_chip.htm, AMADID 012798) (McIntyre et al. 2006). This platform was found to be highly reproducible and reliable for the detection of alternative exons (McIntyre et al. 2006). The 20,772 probe sequences on the array were BLASTed (Altschul et al. 1990) to the D. melanogaster genome, FlyBase version 5.1 (Wilson et al. 2008). There were 158 probe sequences that did not produce a BLAST hit to the current genome build and were not considered further. As this work focuses on identifying sex-specific expression of transcripts with alternative exons, 12,653 probes representing 10,049 genes without alternative exons and 666 probes representing 163 paralogous gene families were not considered further. Alternative exons may arise via the preferential inclusion or skipping of a cassette exon, via mutually exclusive splicing of cassette exons, or from multiple initiation/termination sites (Graveley 2001). For genes with known alternative transcripts, the microarrays included 7060 probes targeting 2777 genes. Probe types were split into two groups, constitutive and alternative. A total of 2724 genes (6851 probes) had at least 1 probe targeting a constitutive exon, and 1651 of these genes had >1 probe targeting constitutive exons. There were 4770 probes targeting constitutive exons only. There were 417 genes (1409 probes) where at least 2 probes for the same gene targeted different exons.
This permits us to attribute differences in the relative abundance of alternative transcripts in males vs. females to sex-biased transcription or sexually dimorphic transcription of specific exons.
Fluorescent cRNA synthesis and microarray hybridization procedures for the recombinant inbred line (RIL) and diallel inbred line (DIL) experiments are described in McIntyre et al. (2006) and Wayne et al. (2007), respectively. Briefly, males and females of the same genotype were labeled in different dyes and hybridized to the same chip. Dye swaps were performed over biological replicates. This design may improve the ability to detect differences among sexes compared to differences among lines. Hybridizations were performed at the Interdisciplinary Center for Biotechnology Research Microarray Core, University of Florida. Slides were scanned by an Agilent Microarray Scanner and spot quantification was performed using Imagene software version 6.0 at the Genomics Database Facility, Purdue University (West Lafayette, IN). Transcript abundance was estimated as the natural log of the spot mean minus the mean of the local background. For the RIL experiment, probes with signal >95% of the negative control spots were considered detected, and a particular line/sex combination was considered detected if >50% of the replicates were detected. Probes were removed from analysis if they were not detected in at least one treatment. There were seven technical failures for the RIL experiment, resulting in the loss of these samples. For the DIL experiment, the 90% value of the negative control spots was set as the detection threshold, and both replicates of each cross/sex combination were required for analysis. All technical failures were repeated until they were successful. To ensure that the results were directly comparable between experiments, only those genes where two or more probes targeting distinct exons detected in both experiments were included in the analysis of alternative transcript expression (n = 243 genes).
Statistical analyses:
The effects of sex and genetic variation are complicated by the structure of the multiple-transcript genes. Probes were classified as belonging to constitutive or alternative exons. For each gene, we initially examined all the probes for that gene, regardless of the type of exon the probe interrogated, using the model
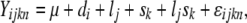 |
where Yijkn is the transcript abundance for dye i, line j, sex k, and replicate n; μ is the overall mean of the transcript abundance for that gene; d is the dye effect; l is the line effect; s is the effect of sex; ls is for line-by-sex interaction; and ɛ is the error. We refer to this model as model 1. Model 1 was also fitted for only the probes representing constitutive exons, for all genes.
In the case where there were two or more alternative exons, the model
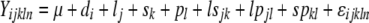 |
was fitted to assess alternative exon representation for each gene, where Yijkln is the transcript abundance for dye i, line j, sex k, probe l, and replicate n; μ is the overall mean of the transcript abundance for that gene; d is the dye effect; l is the line effect; s is the effect of sex; p is the probe effect; ls is for line-by-sex interaction; lp is for line-by-probe interaction; sp is for sex-by-probe interaction; and ɛ is the error. Only probes that interrogated alternative exons were included in this model. This model (model 2) was also fitted with a three-way interaction term for line, probe, and sex. This term was not statistically significant, and the model fit was significantly worse; therefore the model with the three-way interaction term included was not considered further. All effects were considered fixed, since the question was whether “males” were different from “females” and whether particular lines were different from each other in their effects. The main effect of sex or line and the sex-by-line interaction in this model represents the average of probes across alternative exons. The models above were all fitted with an additional random effect for slide. These models gave the same basic inferences as those presented and differences in the number and type of significant effects were negligible.
If males and females produce different exons preferentially, then probes targeting different exons will be detected at different levels between the sexes. Similarly, if different exons are preferentially transcribed or spliced in different genetic backgrounds, then probes targeting different exons will be detected at different levels between genotypes. If the difference is large enough, and variance small enough, then the F test for interaction between the probe and sex (spkl) or the probe and line (lpjl) will be significant. These are the main hypotheses of interest. P-values were corrected for multiple tests, using a false discovery rate (FDR) approach (Benjamini and Hochberg 1995). An FDR level of 0.05 was used for determining whether results were statistically significant. As type I and type II errors are inversely related, analyses were repeated at FDR 0.1 and 0.2. Results were consistent with the 0.05 level and are reported in the supplemental materials. All analyses were performed using SAS software version 9.1 (SAS Institute, Cary, NC).
Real-time PCR:
D. melanogaster tissue dissections:
One D. melanogaster line (line 89) derived from the Winters isofemale strains described above was randomly chosen for real-time PCR to verify the microarray results and to examine sex-specific exon expression at the tissue level. Four biological replicates were reared from small mass matings on standard medium at 25°, 12:12-hr light/dark cycle. From each replicate, virgin males and females were collected within 24 hr and matured separately. At day 3 posteclosion, 15 whole flies of each sex were snap frozen in TRIzol. In addition, the heads, ovaries/testes, and carcasses from 20 females and males, respectively, were dissected in Ringer's solution and snap frozen separately in TRIzol. This resulted in a total of 32 D. melanogaster samples for PCR: 4 samples of whole bodies of each sex and 4 samples of three tissues (heads, gonads, and carcasses) by two sexes.
Drosophila species collections:
Three biological replicates each of D. simulans (Winters, CA), D. yakuba (strain Tai 18, Ivory Coast), D. ananassae (KMJ1, Japan), D. pseudoobscura (Winters, CA), D. willistoni (14030-0811.0, Tucson Drosophila Stock Center), and D. virilis (15010-1051.0, Tucson) were reared on standard media at 25° in uncrowded mass cultures. Nonvirgin males and females 3–4 days of age were separated by sex and stored immediately in TRIzol at −80°. From each species, male and female heads were collected by freezing several hundred 3- to 4-day-old nonvirgin flies on dry ice and shaking them through a series of prechilled wire sieves. Heads were retained in the 230-μm sieve; visual inspection showed that no significant contamination from other body parts was present in the head fraction. A total of 72 Drosophila species samples were collected for real-time PCR, three replicates of whole bodies and heads of 6 species by two sexes.
PCR:
Primers for real-time PCR were designed to flank alternative exon junctions, so that each primer pair amplified only a subset of alternative transcripts and did not detect genomic DNA or unspliced mRNA (supplemental Table 1). Total RNA was isolated and purified as previously described, with the addition of 1 μl of linear polyacrylamide (Ambion, Austin, TX) to the D. melanogaster gonad and all species head samples. TURBO DNase (Ambion) digestion was carried out for 20 min at 37°. Reverse transcription was performed on 300 ng of total RNA, using oligo(dT)16 following manufacturer's instructions (Applied Biosystems, Foster City, CA). Real-time PCR was performed on 1 μl of cDNA product in a total volume of 25 μl, using RT2 SYBR Green/Fluorescein qPCR mastermix (SuperArray, Frederick, MD). PCR was performed on a MyiQ single-color real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA). Cycle thresholds (CTs) were determined using MyiQ v2.0 software.
For each gene, the relative abundance of different exon junctions (and, by implication, different alternative transcripts) was compared in males vs. females for whole bodies, carcasses, gonads, and heads. The ratio of the CT values for the alternative exon junctions amplified by different primers pairs was calculated and tested for sex effects, using the ANOVA model
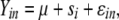 |
where Yin is the ratio of CT values for alternative exons for sex i and replicate n, μ is the overall mean expression ratio for that gene, s is the effect of sex, and ɛ is the error.
RESULTS
Effects of sex and genotype on overall transcript abundance of multitranscript genes:
We examined expression variation at 2777 loci that produce multiple transcripts through the use of alternative exons. Among the 2724 of these genes that had at least one probe targeting a constitutive exon, 2428 showed detectable hybridization of any probe(s) (including constitutive and/or alternative exons) in both the RIL and the DIL experiments (Table 1). In the RIL data, 64.6% of these genes showed significant sex-biased expression, and the expression of 3.7% of the genes was significantly different among genotypes (Table 1, FDR 0.1, 0.2; supplemental Table 2). In the DIL data, 87.3% of the 2428 genes examined showed significant sex bias, and 60.5% showed significant genetic variation. The direction of the sex difference was affected by genotype (line-by-sex interaction) for 4.1% of the genes (Table 1, FDR 0.1, 0.2; supplemental Table 2).
TABLE 1.
Results from ANOVA model by gene for all probes for that gene detected in the RIL and DIL data sets, with significant genes shown at FDR 5%
| Genes | Sex | Line | Line × sex | |
|---|---|---|---|---|
| RIL | 2432 | 1569 | 108 | 0 |
| DIL | 2673 | 2322 | 1689 | 100 |
| Overlap | 2428 | 1536 | 91 | 0 |
| RIL | 2428 | 1569 | 107 | 0 |
| DIL | 2428 | 2121 | 1470 | 101 |
Effects of sex and genotype on constitutive exon expression:
We next examined expression variation of constitutive exons for the 2724 genes with constitutive exon probes. Hybridization of constitutive probe(s) was detected for 2340 loci in both the RIL and the DIL experiments (Table 2). Similar to the overall model, in the RIL data, 67.7% of the 2340 genes examined showed significant sex-biased expression for constitutive exons, and 2.69% were significantly different among genotypes (Table 2, FDR 0.1, 0.2; supplemental Table 3). In DIL data, 88.7% of the 2340 genes showed significant sex-biased expression of constitutive exons, and 67.1% were variable among genotypes. Line-by-sex interaction was significant for 6.06% of the genes (Table 2, FDR 0.1, 0.2; supplemental Table 3).
TABLE 2.
Results from ANOVA model by gene for constitutive probes detected in the RIL and DIL data sets, with significant genes shown at FDR 5%
| Genes | Sex | Line | Line × sex | |
|---|---|---|---|---|
| Multitranscript genes with constitutive probe(s) | ||||
| RIL | 2346 | 1586 | 72 | 2 |
| DIL | 2651 | 2327 | 1851 | 142 |
| Overlap | 2340 | 1562 | 63 | 0 |
| RIL | 2340 | 1586 | 71 | 2 |
| DIL | 2340 | 2076 | 1570 | 144 |
Effects of sex and genotype on alternative exon representation:
For probes that target specific exons, we fitted a model that tests for interaction between probe and sex and between probe and genotype (model 2). A significant interaction indicates that a particular exon is differentially spliced or transcribed either in males vs. females or in different genotypes. Hybridization of 243 genes was detected in both the RIL and the DIL data sets (n = 644 and 1200 probes, RIL and DIL experiments, respectively). In the RIL data, 58% of genes showed significant sex-by-probe interactions and 3.3% of genes were significant for line-by-probe interactions (Table 3). In the DIL data, 87.2% of genes showed significant sex bias for expression of alternative exons (sex-by-probe interaction), and 3.3% of genes showed genetic variation for alternative transcript expression (line-by-probe interaction, Table 3; supplemental Table 4). The fact that so little variation is observed among the DIL crosses (n = 72) despite high levels of genetic variation for expression (Table 3) indicates that sex-biased exon preference is common while genotype-biased exon preference is relatively rare. Eighty-eight genes (36.2%) had consistent sex-by-probe interactions in the two experiments, with no significant line-by-probe or sex-by-line interactions. At these loci, the difference between the female and the male exon abundance was highly consistent between experiments (supplemental Figure 1). These genes perform a range of biological and molecular roles, including learning and memory, gametogenesis, and cell signaling; however, they also include many genes with unknown functions (supplemental Table 4).
TABLE 3.
Results from ANOVA model by gene for at least two alternative exon probes detected in the RIL and DIL data sets, with significant genes shown at FDR 5%
| Genes | Sex | Line | Line × sex | Sex × probe | Line × probe | |
|---|---|---|---|---|---|---|
| RIL | 245 | 165 | 177 | 5 | 142 | 9 |
| DIL | 363 | 309 | 342 | 72 | 283 | 10 |
| Overlap | 243 | 159 | 166 | 3 | 135a (88) | 1 |
| RIL | 243 | 165 | 175 | 5 | 142 | 9 |
| DIL | 243 | 212 | 229 | 70 | 196 | 8 |
Genes common to the RIL and DIL data sets with sex-by-probe interaction at FDR 5% when significant line-by-probe and sex-by-probe interactions at FDR 5% were removed.
There was little evidence for an interaction between sex and genotype effects. For constitutive exons, no significant interactions were observed, and for alternative exons such interactions were seen for only 2% of the genes in the RIL data (Table 3). Interestingly, even with the increased power of the DIL experiment to detect interactions with genotypic effects, few genes showed a line-by-sex interaction for constitutive exons (∼6%) while the interaction between the sex and genotype effects was detectable in 28.2% of the genes in the alternative exon model (Table 3). This apparent conflict in findings between the alternative exon model and the constitutive exon model can be explained by a possible interaction between sex and probe by genotype. That is, the magnitude of the sex effect may vary by genotype. The failure to detect the three-way interaction as statistically significant does not mean that this effect is not present, as this design is underpowered to detect this effect.
Sex-specific expression of alternative transcripts confirmed by RT–PCR:
Of the 88 multitranscript genes with sex-biased alternative transcript expression, 34 showed a clear qualitative exon preference between the two sexes, with one exon more abundant in males and another in females (supplemental Table 4, supplemental Figure 2). Seven of these loci were selected for confirmation by RT–PCR. We selected genes to represent diverse biological and molecular functions, including gametogenesis (hts), learning and memory (Mob1 and ltd), pathogen phagocytosis (garz), and protein folding (jdp). To examine potentially novel roles of sex-specific splicing, we also included 2 uncharacterized genes (CG6016 and CG4662). At 5 of these loci (hts, garz, jdp, CG6016, and CG4662), different transcripts are produced by alternative splicing. The remaining 2 genes (ltd and Mob1) produce alternative transcripts due to the use of multiple promoters. The relative abundance of alternative exons in males and females for these genes was almost perfectly correlated between the RIL and the DIL microarray experiments (supplemental Figure 3).
For each gene, primer pairs specific to two alternative exon junctions were designed (supplemental Table 1, Figure 1). Primers for the dsx male-specific and female-specific transcripts were designed and amplified in D. melanogaster to ensure that the PCR data showed the expected sex-biased transcript ratios before proceeding to the other genes (data not shown). To determine consistency across platforms, we compared RT–PCR for D. melanogaster whole bodies to the array results and found the results to be concordant for six of the seven genes tested (supplemental Figure 4, A and B). This is remarkable given that the samples compared were from flies reared almost 2 years after the initial experiment. The sex-specific exon expression for Mob1 was not verified in the strain used for RT–PCR, and this gene was not considered further.
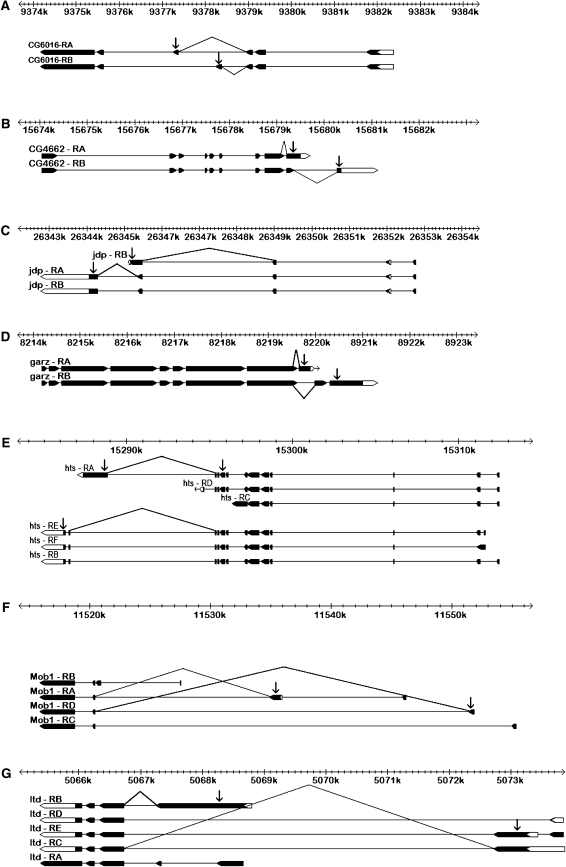
Figure 1.—
Gene structure schematic illustrating transcript targets of exon-specific microarray probes (arrows) and exon-junction RT–PCR primers (connected lines) for each of the seven nonregulatory genes investigated by RT–PCR for D. melanogaster. Exons are shown as boxes (solid boxes, coding regions; open boxes, UTRs) connected by introns (lines), with each transcript aligned against the cytogenic scale. (A) Microarray probes targeting alternatively spliced exons CG6016:7 (transcript CG6016-RA) and CG6016:4 (transcript CG6016-RB) and RT–PCR primers for transcripts CG6016-RA and CG6016-RB. (B) Microarray probes targeting alternatively spliced exons CG4662:9 (transcript CG4662-RA) and CG4662:11 (transcript CG4662-RB) and RT–PCR primers for transcripts CG4662-RA and CG4662-RB. (C) Microarray probes targeting alternatively spliced exons jdp:4 (transcripts jdp-RA and jdp-RB) and jdp:7 (transcript jdp-RC) and RT–PCR primers targeting transcripts jdp-RA and jdp-RC. (D) Microarray probes targeting alternatively spliced exons garz:11 (transcript garz-RA) and garz:9 (transcript garz-RB) and RT–PCR primers targeting transcripts garz-RA and garz-RB. (E) Microarray probes targeting exons hts:16 (transcript hts-RA) and hts:13 (transcripts hts-RB, hts-RE, and hts-RF) and RT–PCR probes targeting transcripts hts-RA and hts-BEF. (F) Microarray probes targeting exons derived from alternative initiation sites, Mob1:6 (transcript Mob1-RA) and Mob1:5 (transcript Mob1-RD) and RT–PCR primers for Mob1-RA and Mob1-RD—note that the microarray results for this gene were not verified by PCR and it was not considered further. (G) Microarray probes targeting exons derived from alternative initiation sites, ltd:7 (transcript ltd:RB), ltd:6 (transcript ltd:RC), and ltd:1 (transcript ltd:RE) and RT–PCR primers for transcript ltd-RB and transcripts ltd-RC and ltd-RE. Gene structures were adapted from FlyBase V. 5.4.
Tissue specificity of sex-specific expression of alternative transcripts:
Most sex-biased differences in transcript abundance in D. melanogaster and other organisms can be attributed to the gonads (Ellegren and Parsch 2007; Mank et al. 2008). To test the hypothesis that the sex-specific preference for particular exons may be largely ovary or testes specific, flies were dissected into three parts: heads, soma, and gonads. Expression profiles were compared between the sexes in each of these tissues as well as in intact whole bodies. For each sex, the CTs were expressed as a ratio of one exon junction to the other exon junction.
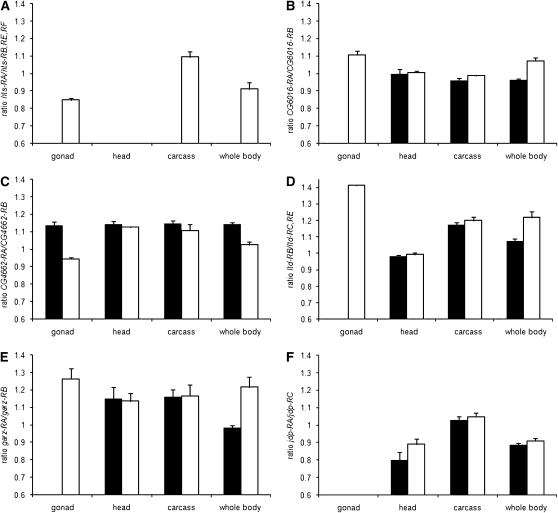
Most, but not all, sex-specific expression of alternative transcripts observed in whole adults can be attributed to ovary- and testes-specific splicing (Figure 2, A–D). At the hts locus, which has a known function in oogenesis (Yue and Spradling 1992; Whittaker et al. 1999), the RB, RE, and RF transcripts were detected in both sexes, while hts-RA was restricted to females. The latter transcript is most abundant in ovaries and was detected in some somatic tissues, excluding the head (Figure 2A). For the remaining genes, we observed highly significant differences in the ratios of alternative exons in whole bodies between the sexes (P < 0.001) (supplemental Table 5). Three genes, ltd, CG6016, and garz show extreme sex bias in the gonads, where one of the transcripts is present in the ovaries and absent in the testes (Figure 2, C–E). For CG6016, slight but significant sex-specific expression was observed in the gonadectomized carcass (P < 0.01), whereas ltd and garz were sexually monomorphic in the soma. At the CG4662 locus, the RA transcript was overrepresented in females and the RB transcript was overrepresented in males, and this bias is also explained by the gonads (Figure 2B). The jdp gene, however, was not detected in either male or female gonads, but showed significant sexual dimorphism in adult heads (P < 0.001) (Figure 2F).
Figure 2.—
Sex- and tissue-specific expression of alternative transcripts from six D. melanogaster genes that showed significant probe-by-sex interaction in the microarray data (solid bars, males; open bars, females). The RT–PCR results are analyzed as the average ratio of CT values for the two exon junctions amplified for each gene (y-axis), and error bars show the standard deviation. (A) Ratio of transcripts hts-RA and hts-RB, -RE, and -RF. (B) Ratio of transcripts CG6016-RA and CG6016-RB. (C) Ratio of transcripts CG4662-RA and CG-4662-RB. (D) Ratio of transcripts ltd-RB and ltd-RC,-RE. (E) Ratio of transcripts garz-RA and garz-RB. (F) Ratio of transcripts jdp-RA and jdp-RC.
Evolutionary conservation of sex-specific splicing:
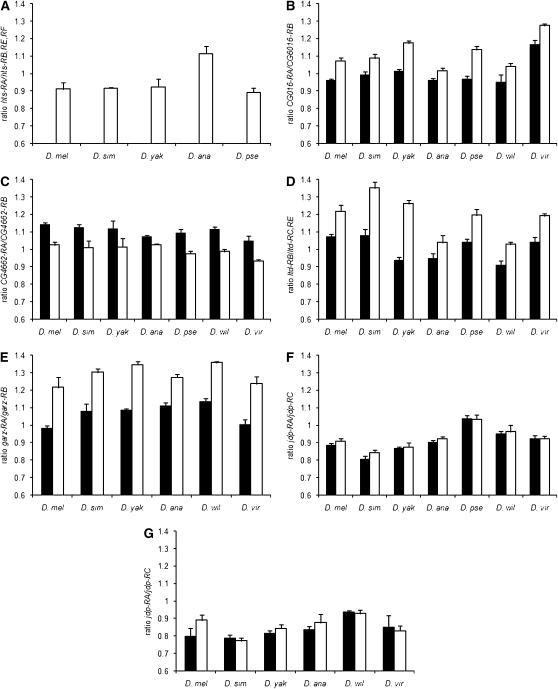
Sex-specific splicing patterns observed in D. melanogaster whole bodies were tested in six other Drosophila species: D. simulans, D. yakuba, D. ananassae, D. pseudoobscura, D. willistoni, and D. virilis (see supplemental Table 1 for primer design). These species represent both major subgenera of Drosophila and span over 60 million years of evolutionary divergence (Markow and O'Grady 2007). Female-restricted expression of hts-RA was conserved in all five species tested (Figure 3A). Transcript levels varied among species, but sexually dimorphic splicing of alternative transcripts was conserved in all seven species for CG4622, CG6016, and ltd and in six of the seven species tested for garz (Figure 3, B–E, supplemental Table 5). Sequencing of the garz PCR products in D. pseudoobscura revealed that the correct garz-RB junction was not being amplified; therefore this species was not included in the analysis of this gene.
Figure 3.—
Sex-specific expression of alternative transcripts for six genes that showed significant probe-by-sex interaction in the microarray data (solid bars, males; open bars, females) among Drosophila species. The RT–PCR results are analyzed as the average ratio of CT values for the two exon junctions amplified for each gene (y-axis), and error bars show the standard deviation. D. mel, D. melanogaster; D. sim, D. simulans; D. yak, D. yakuba; D. ana, D. anannasae; D. pse, D. pseudoobscura; D. wil, D. willistoni; D. vir, D. virilis. (A) Ratio of hts-RA and hts-RB, -RE, and -RF for female whole bodies of D. melanogaster, D. simulans, D. yakuba, D. anannasae, and D. pseudoobscura. (B) Ratio of CG6016-RA and CG6016-RB male and female whole bodies for D. melanogaster, D. simulans, D. yakuba, D. anannasae, D. pseudoobscura, D. willistoni, and D. virilis. (C) Ratio of CG4662-RA and CG4662-RB male and female whole bodies for all seven species. (D) Ratio of ltd-RB and ltd-RC and -RE for male and female whole bodies for all seven species. (E) Ratio of garz-RA and garz-RB for male and female whole bodies of all seven species. (F) Ratio of jdp-RA and jdp-RC for male and female whole bodies tested for all seven species. (G) Ratio of jdp-RA and jdp-RC for male and female heads for D. melanogaster, D. simulans, D. yakuba, D. anannasae, D. willistoni, and D. virilis.
Interestingly jdp, the only gene not expressed in the reproductive tissues, showed the greatest evolutionary variation for sex-specific splicing. Significant sex bias in the RA/RC transcript ratio was found in whole bodies of D. simulans and D. ananassae (P < 0.01), while nonsignificant bias in the same direction was seen in D. yakuba (Figure 3F). As alternative jdp transcripts are expressed sex specifically in D. melanogaster heads, we examined jdp expression in the heads of adult males and females of other species. No significant biases were observed, although D. yakuba and D. ananassae showed a trend that was similar to D. melanogaster (Figure 3G). Overall these data suggest that strong sex bias in the splicing of jdp in the heads may be restricted to D. melanogaster.
Sex-specific splicing of splicing regulators:
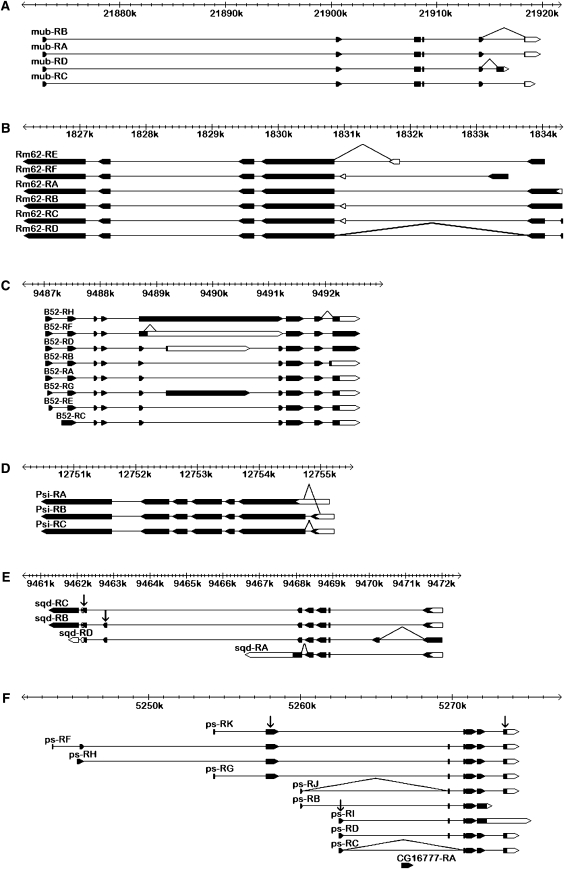
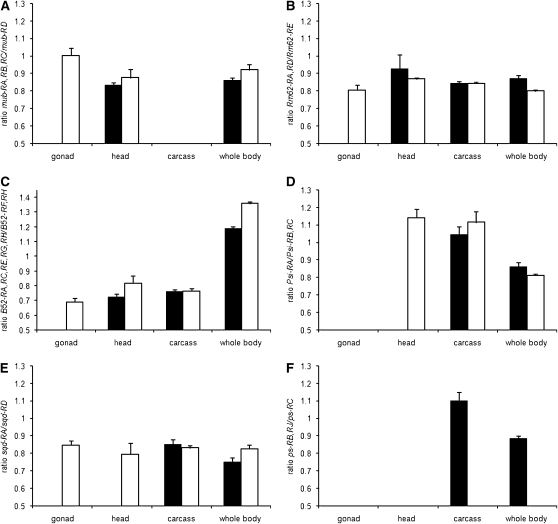
The Drosophila sex determination pathway operates as a multilevel alternative splicing cascade, suggesting that analogous cascades might play a role in other aspects of cell differentiation. We therefore examined the splicing of six known regulators of alternative splicing: mub, Rm62, Psi, B52, sqd, and ps (Park et al. 2004; Blanchette et al. 2005; Robida et al. 2007). All six genes had probes on the array and showed significant sex bias for the constitutive exons (supplemental Table 6). Due to the structure of these genes (Figure 4), only sqd and ps could be included in the analysis of alternative exons (model 2). Both showed significant sex-by-probe interactions (supplemental Table 2). We designed primers to amplify alternative exon junctions for each of the six regulators (Figure 4) and compared the relative abundance of alternative transcripts between male and female whole bodies and dissected tissues as described above (Figure 5, A–F). At the ps locus, the ps-RB and RJ transcripts were male restricted in all tissues (Figure 5F). At the mub, Rm62, Psi, B52, and sqd loci, transcripts were significantly sex biased in whole bodies (supplemental Table 7; Figure 5, A–E). For mub, Rm62, B52, and sqd, much of this sexual dimorphism is explained by the gonads (Figure 5, A–C and E). However, sqd also showed strongly sex-specific splicing in the head, though not in the rest of the soma (Figure 5E). Psi showed no expression in either male or female gonads, but was spliced sex specifically in adult heads (Figure 5D). Overall, this analysis shows that the regulators of alternative splicing are themselves spliced in complex sex- and tissue-specific patterns.
Figure 4.—
Gene structure schematic illustrating transcript targets of exon-junction RT–PCR primers (connected lines) for the six mRNA splicing regulator genes investigated by RT–PCR for D. melanogaster. Exons are shown as boxes (solid boxes, coding regions; open boxes, UTRs) connected by introns (lines), with each transcript aligned against the cytogenic scale. (A) RT–PCR primers for transcripts mub-RA,-RB and mub-RD. (B) RT–PCR primers for transcripts Rm62-RA,-RD and Rm62-RE. (C) RT–PCR primers for transcripts B52-RA,-RC,-RE,-RG,-RH and B52-RB. (D) RT–PCR primers for transcripts Psi-RA/Psi-RB,-RC. (E) RT–PCR primers for transcripts sqd-RD and sqd-RA and exon-specific microarray probes (arrows). (F) RT–PCR primers for transcripts ps-RB,-RJ and ps-RC and exon-specific microarray probes (arrows).
Figure 5.—
Sex- and tissue-specific expression of alternative transcripts from six D. melanogaster mRNA splicing regulator genes (solid bars, males; open bars, females). The RT–PCR results are analyzed as the average ratio of CT values for the two exon junctions amplified for each gene (y-axis), and error bars show the standard deviation. (A) Ratio of transcripts mub-RA,-RB,-RC and mub-RD. (B) Ratio of transcripts Rm62-RA,-RD and Rm62E-RB. (C) Ratio of transcripts B52-RA,-RC,-RE,-RG,-RH and B52-RF,RH. (D) Ratio of transcripts Psi-RA/Psi-RB,-RC. (E) Ratio of transcripts sqd-RD and sqd-RA. (F) Ratio of transcripts ps-RB,-RJ and ps-RC.
DISCUSSION
The sex determination hierarchy of Drosophila has long served as the leading paradigm for the role of alternative splicing in development (McKeown 1992). More recently, a microarray analysis based on a small number of genotypes revealed an unexpectedly high prevalence of sex-biased splicing in the Drosophila genome (McIntyre et al. 2006), suggesting that alternative splicing may play an important role in sexual differentiation beyond the canonical sex determination pathway. The splicing of several sex determination components was also found to vary across genotypes (Tarone et al. 2005). Unfortunately, the scale of previous analyses was insufficient to estimate the genomewide extent of genotype-specific splicing or the importance of sex-by-genotype interactions in splicing variation.
In this study, we examined variation in the splicing of 243 multitranscript genes among both the RIL and the DIL experiments. Sex-biased exon expression was evident for ∼80% of these genes. In contrast, while ample genetic variation was observed in transcript abundance, genotype-specific splicing and sex-by-genotype interactions were rare despite the number of genotypes examined for the DIL experiment. Sex-biased splicing of many genes was remarkably consistent across all 80 genotypes. We conclude that sex-specific splicing of these genes is a fundamental feature of Drosophila biology, rather than a rare occurrence restricted to some genotypes.
An increasing amount of evidence supports the importance of alternative splicing in regulating distinct developmental and physiological phenotypes (Ben-Dov et al. 2008). In Drosophila, alternatively spliced isoforms of the lola transcription factors play nonredundant roles in axon guidance in the embryonic central nervous system (Goeke et al. 2003) and in apoptosis in the ovary (Bass et al. 2007). The Drosophila Dscam locus has enormously complicated patterns of alternative splicing that can produce >38,000 distinct cell adhesion proteins (Schmucker et al. 2000), at least some of which have distinct binding affinities that are essential for neuron wiring specificity (Hattori et al. 2007; Matthews et al. 2007; Meijers et al. 2007; Wojtowicz et al. 2007). In the vertebrate CNS, alternatively spliced variants of the Robo3 receptor control midline crossing by spinal commissural axons, with one variant silencing and the other favoring midline repulsion mediated by the Slit ligand (Chen et al. 2008). Misregulation of alternative splicing in humans is implicated in such diseases as neurofibromatosis and myotonic dystrophy (reviewed in Ben-Dov et al. 2008), highlighting the importance of splicing regulation for normal development.
Like many eukaryotes, the Drosophila gonads are the most sexually dimorphic tissue and are the main source of sex-biased transcription (Parisi et al. 2004; Reinke et al. 2004; Cutter and Ward 2005; Mank et al. 2008). Our data suggest that sex-biased splicing also occurs predominantly in the reproductive organs. Nine of the 12 examined genes were differentially spliced in D. melanogaster gonads. Interestingly, for 8 of these 9 genes, one or more splice variants were completely restricted to the ovaries, but no strictly testis-specific alternative transcripts were observed. The prevalence of sex-biased splicing in the Drosophila gonads suggests that alternatively spliced transcripts may play distinct roles in male and female reproduction. Among the genes examined in this study, hts and sqd have well-characterized roles in oogenesis. Different isoforms of the sqd hnRNPs perform different functions in the dorsal–ventral patterning of the oocyte, including gurken mRNA localization and protein accumulation (Norvell et al. 1999). hts encodes an essential component of cytoskeletal structures that regulate germline stem cell maintenance and gamete differentiation in both sexes (Yue and Spradling 1992; Deng and Lin 1997; Wilson 2005; Lighthouse et al. 2008). Interestingly, different hts transcripts are expressed in different cell types in the ovary: the RB, RE, and RF transcripts are localized in the somatic cells of the egg chambers, while the RA transcript is germline specific and is conveyed to the maturing oocyte from the nurse cells (Whittaker et al. 1999; Chen et al. 2008). The extent to which the proteins encoded by alternatively spliced hts transcripts are functionally distinct remains to be determined. The other 7 genes that show sex-specific splicing in the gonads (ltd, garz, CG6016, and CG4662 and splicing regulators mub, Rm62, and B52) have no characterized functions in gonad development or gametogenesis. Experimental analysis of these and other candidate genes will be needed to determine the functional significance of sex-specific splicing for these genes.
However, sex-biased splicing is not limited to the gonads. Among somatic tissues, it appears to be especially prevalent in adult heads, where 3 of 12 examined genes show significant differences in splicing between males and females. Although sex-biased transcription is less extensive in somatic tissues than in the gonad, it nevertheless plays an important role in sex-specific development, particularly in the formation of neuronal circuits that control sexual behavior in both vertebrates (Isensee and Noppinger 2007; Mank et al. 2008) and insects (Goldman and Arbeitman 2007; Iatrou and Biessmann 2008). It is possible that sex-biased splicing contributes to CNS development and function, as well. For example, jdp encodes a molecular chaperon-like protein that has been shown to be phosphorylated by a cAMP-dependent kinase in the Drosophila brain (Inoue et al. 2000). cAMP signaling is involved in synaptic function and is implicated in learning and memory (Silva and Murphy 1999; Siwicki and Ladewski 2003), suggesting that sex-biased jdp transcripts could be involved in these processes.
Sex-specific splicing showed remarkably little intraspecific variation in D. melanogaster. This observation is consistent with evolutionary conservation of sex-specific splicing between Drosophila species that diverged ∼60 million years ago. All seven species compared exhibit the same sex-biased alternative exon expression as observed in the D. melanogaster gonads, suggesting that these differences are under purifying selection. In contrast, sex-specific splicing of jdp in adult heads appears to be limited to D. melanogaster. Studies in mammals and insects have shown that the levels of alternative exon conservation are typically low in comparison to constitutive exons, and the loss of splice variants is frequent (Malko et al. 2006; Nurtdinov et al. 2007). However, some alternative splicing events exhibit strong similarity among diverse eurkaryotic lineages (Irimia et al. 2007, 2008; Katyal et al. 2007), indicating that such loci are under strong selection to preserve functional protein structure (Resch et al. 2004; Kim et al. 2008). Conservation of sex-specific splicing of the sex determination genes dsx and fru in holometabolous insects is an important example, reflecting their functional importance in establishing sexual dimorphism at both the morphological and the behavioral levels (Gailey et al. 2006; Cho et al. 2007).
Perhaps the most intriguing question raised by the high prevalence of sex-specific splicing is how it is regulated in each tissue. We have shown that all six examined regulators of alternative mRNA splicing are themselves spliced sex specifically in the gonads and heads. It remains to be tested whether their splicing is controlled by Sxl, tra, and other components of the canonical sex determination pathway. The proteins encoded by mub, Rm62, Psi, B52, sqd, and ps are known to affect the splicing of anywhere between a few and several hundred target genes (Park et al. 2004; Blanchette et al. 2005). This raises the possibility that multilevel splicing cascades acting downstream of, or in parallel with, the sex determination pathway may be involved in the differentiation of multiple cell types. Genetic analysis will be necessary to identify and dissect these regulatory hierarchies.
Acknowledgments
We thank Muneo Matsuda and the Tucson Drosophila species stock center for fly strains, as well as two anonymous reviewers. M.T.S., L.M.M., S.V.N., and M.L.W. are supported by National Institutes of Health grant 5R01GM077618-02. A.K. is supported by National Science Foundation grant DEB-0548991.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Baker, B. S., 1989. Sex in flies: the splice of life. Nature 340 521–524. [DOI] [PubMed] [Google Scholar]
- Barberan-Soler, S., and A. M. Zahler, 2008. Alternative splicing regulation during C. elegans development: splicing factors as regulated targets. PLoS Genet. 4(2): e1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, B. P., K. Cullen and K. McCall, 2007. The axon guidance gene lola is required for programmed cell death in the Drosophila ovary. Dev. Biol. 304 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, L. R., E. M. Maine, P. Schedl and T. W. Cline, 1988. Sex-Lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA-binding proteins. Cell 55 1037–1046. [DOI] [PubMed] [Google Scholar]
- Bell, L. R., J. I. Horabin, P. Schedl and T. W. Cline, 1991. Positive autoregulation of Sex-Lethal by alternative splicing maintains the female determined state in Drosophila. Cell 65 229–239. [DOI] [PubMed] [Google Scholar]
- Ben-Dov, C., B. Hartmann, J. Lundgren and J. Valcarcel, 2008. Genome-wide analysis of alternative Pre-mRNA splicing. J. Biol. Chem. 283 1229–1233. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Y. Hochberg, 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57 289–300. [Google Scholar]
- Black, D. L., 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72 291–336. [DOI] [PubMed] [Google Scholar]
- Blanchette, M., R. E. Green, S. E. Brenner and D. C. Rio, 2005. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev. 19 1306–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe, B. J., 2006. Alternative splicing: new insights from global analyses. Cell 126 37–47. [DOI] [PubMed] [Google Scholar]
- Chen, Z., B. B. Gore, H. Long, L. Ma and M. Tessier-Lavigne, 2008. Alternative splicing of the Robo3 axon guidance receptor governs the midline switch from attraction to repulsion. Neuron 58 325–332. [DOI] [PubMed] [Google Scholar]
- Cho, S., Z. Y. Huang and J. Z. Zhang, 2007. Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics 177 1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, T. A., A. C. Schweitzer, T. X. Chen, M. K. Staples, G. Lu et al., 2007. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 8(4): R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, T. W., 1993. The Drosophila sex determination signal: How do flies count to 2. Trends Genet. 9 385–390. [DOI] [PubMed] [Google Scholar]
- Connallon, T., and L. L. Knowles, 2005. Intergenomic conflict revealed by patterns of sex-biased gene expression. Trends Genet. 21 495–499. [DOI] [PubMed] [Google Scholar]
- Cutter, A. D., and S. Ward, 2005. Sexual and temporal dynamics of molecular evolution in C. elegans development. Mol. Biol. Evol. 22 178–188. [DOI] [PubMed] [Google Scholar]
- Deng, W., and H. F. Lin, 1997. Spectrosomes and fusomes anchor mitotic spindles during asymmetric germ cell divisions and facilitate the formation of a polarized microtubule array for oocyte specification in Drosophila. Dev. Biol. 189 79–94. [DOI] [PubMed] [Google Scholar]
- Ellegren, H., and J. Parsch, 2007. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 8 689–698. [DOI] [PubMed] [Google Scholar]
- Erickson, J. W., and T. W. Cline, 1998. Key aspects of the primary sex determination mechanism are conserved across the genus Drosophila. Development 125 3259–3268. [DOI] [PubMed] [Google Scholar]
- Gailey, D. A., J. C. Billeter, J. H. Liu, F. Bauzon, J. B. Allendorfer et al., 2006. Functional conservation of the fruitless male sex-determination gene across 250 Myr of insect evolution. Mol. Biol. Evol. 23 633–643. [DOI] [PubMed] [Google Scholar]
- Goeke, S., E. A. Greene, P. K. Grant, M. A. Gates, D. Crowner et al., 2003. Alternative splicing of lola generates 19 transcription factors controlling axon guidance in Drosophila. Nat. Neurosci. 6 917–924. [DOI] [PubMed] [Google Scholar]
- Goldman, T. D., and M. N. Arbeitman, 2007. Genomic and functional studies of Drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS Genet. 3 2278–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B. R., 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17 100–107. [DOI] [PubMed] [Google Scholar]
- Gupta, S., D. Zink, B. Korn, M. Vingron and S. A. Haas, 2004. Genome wide identification and classification of alternative splicing based on EST data. Bioinformatics 20 2579–2585. [DOI] [PubMed] [Google Scholar]
- Hattori, D., E. Demir, H. W. Kim, E. Viragh, S. L. Zipursky et al., 2007. Dscam diversity is essential for neuronal wiring and self-recognition. Nature 449 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iatrou, K., and H. Biessmann, 2008. Sex-biased expression of odorant receptors in antennae and palps of the African malaria vector Anopheles gambiae. Insect Biochem. Mol. Biol. 38 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, H., Y. Chikaoka, M. Takahashi and T. Yoshioka, 2000. Identification of a protein phosphorylated by cAMP-dependent protein kinase in Drosophila brain. Brain Res. 875 160–163. [DOI] [PubMed] [Google Scholar]
- Irimia, M., J. L. Rukov, D. Penny and S. W. Roy, 2007. Functional and evolutionary analysis of alternatively spliced genes is consistent with an early eukaryotic origin of alternative splicing. BMC Evol. Biol. 7 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia, M., J. L. Rukov, D. Penny, J. Garcia-Fernandez, J. Vinther et al., 2008. Widespread evolutionary conservation of alternatively spliced exons in Caenorhabditis. Mol. Biol. Evol. 25 375–382. [DOI] [PubMed] [Google Scholar]
- Isensee, J., and P. R. Noppinger, 2007. Sexually dimorphic gene expression in mammalian somatic tissue. Gend. Med. 4 S75–S95. [DOI] [PubMed] [Google Scholar]
- Jin, W., R. M. Riley, R. D. Wolfinger, K. P. White, G. Passador-Gurgel et al., 2001. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat. Genet. 29 389–395. [DOI] [PubMed] [Google Scholar]
- Johnson, J. M., J. Castle, P. Garrett-Engele, Z. Y. Kan, P. M. Loerch et al., 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302 2141–2144. [DOI] [PubMed] [Google Scholar]
- Katyal, S., Z. Gao, R. Z. Liu and R. Godbout, 2007. Evolutionary conservation of alternative splicing in chicken. Cytogenet. Genome Res. 117 146–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, E., A. Magen and G. Ast, 2007. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 35 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, E., A. Goren and G. Ast, 2008. Alternative splicing: current perspectives. BioEssays 30 38–47. [DOI] [PubMed] [Google Scholar]
- Kulathinal, R. J., L. Skwarek, R. A. Morton and R. S. Singh, 2003. Rapid evolution of the sex-determining gene, transformer: structural diversity and rate heterogeneity among sibling species of Drosophila. Mol. Biol. Evol. 20 441–452. [DOI] [PubMed] [Google Scholar]
- Lande, R., 1980. Sexual dimorphism, sexual selection, and adaptation in polygenic characters. Evolution 34 292–305. [DOI] [PubMed] [Google Scholar]
- Lighthouse, D. V., M. Buszczak and A. C. Spradling, 2008. New components of the Drosophila fusome suggest it plays novel roles in signaling and transport. Dev. Biol. 317 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malko, D. B., V. J. Makeev, A. A. Mironov and M. S. Gelfand, 2006. Evolution of exon-intron structure and alternative splicing in fruit flies and malarial mosquito genomes. Genome Res. 16 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank, J. E., L. Hultin-Rosenberg, M. T. Webster and H. Ellegren, 2008. The unique genomic properties of sex-biased genes: insights from avian microarray data. BMC Genomics 9 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow, T. A., and P. M. O'Grady, 2007. Drosophila biology in the genomic age. Genetics 177 1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B. J., M. E. Kim, J. J. Flanagan, D. Hattori, J. C. Clemens et al., 2007. Dendrite self-avoidance is controlled by Dscam. Cell 129 593–604. [DOI] [PubMed] [Google Scholar]
- McIntyre, L. M., L. M. Bono, A. Genissel, R. Westerman, D. Junk et al., 2006. Sex-specific expression of alternative transcripts in Drosophila. Genome Biol. 7(8): R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown, M., 1992. Alternative messenger-RNA splicing. Annu. Rev. Cell Biol. 8 133–155. [DOI] [PubMed] [Google Scholar]
- Meijers, R., R. Puettmann-Holgado, G. Skiniotis, J. H. Liu, T. Walz et al., 2007. Structural basis of Dscam isoform specificity. Nature 449 487–491. [DOI] [PubMed] [Google Scholar]
- Nagoshi, R. N., M. McKeown, K. C. Burtis, J. M. Belote and B. S. Baker, 1988. The control of alternative splicing at genes regulating sexual-differentiation in Drosophila melanogaster. Cell 53 229–236. [DOI] [PubMed] [Google Scholar]
- Norvell, A., R. L. Kelley, K. Wehr and T. Schupbach, 1999. Specific isoforms of Squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes Dev. 13 864–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurtdinov, R. N., A. D. Neverov, A. V. Favorov, A. A. Mironov and M. S. Gelfand, 2007. Conserved and species-specific alternative splicing in mammalian genomes. BMC Evol. Biol. 7 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin, S. V., P. D. Keightley, E. G. Pasyukova and E. A. Morozova, 1998. Mapping quantitative trait loci affecting sternopleural bristle number in Drosophila melanogaster using changes of marker allele frequencies in divergently selected lines. Genet. Res. 72 79–91. [DOI] [PubMed] [Google Scholar]
- Pan, Q., O. Shai, C. Misquitta, W. Zhang, A. L. Saltzman et al., 2004. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell 16 929–941. [DOI] [PubMed] [Google Scholar]
- Parisi, M., R. Nuttall, D. Naiman, G. Bouffard, J. Malley et al., 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299 697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M., R. Nuttall, P. Edwards, J. Minor, D. Naiman et al., 2004. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 5(6): R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. W., K. Parisky, A. M. Celotto, R. A. Reenan and B. R. Graveley, 2004. Identification of alternative splicing regulators by RNA interference in Drosphila. Proc. Natl. Acad. Sci. USA 101 15974–15979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalva, L. O. F., and L. Sanchez, 2003. RNA binding protein sex-lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiol. Mol. Biol. Rev. 67 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomiankowski, A., R. Nothiger and A. Wilkins, 2004. The evolution of the Drosophila sex-determination pathway. Genetics 166 1761–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz, J. M., C. I. Castillo-Davis, C. D. Meiklejohn and D. L. Hartl, 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300 1742–1745. [DOI] [PubMed] [Google Scholar]
- Reinke, V., H. E. Smith, J. Nance, J. Wang, C. Van Doren et al., 2000. A global profile of germline gene expression in C. elegans. Mol. Cell 6 605–616. [DOI] [PubMed] [Google Scholar]
- Reinke, V., I. S. Gil, S. Ward and K. Kazmer, 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131 311–323. [DOI] [PubMed] [Google Scholar]
- Resch, A., Y. Xing, A. Alekseyenko, B. Modrek and C. Lee, 2004. Evidence for a subpopulation of conserved alternative splicing events under selection pressure for protein reading frame preservation. Nucleic Acids Res. 32 1261–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robida, M. D., A. Rahn and R. Singh, 2007. Genome-wide identification of alternatively spliced mRNA targets of specific RNA-binding proteins. PLoS ONE 6 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker, D., J. C. Clemens, H. Shu, C. A. Worby, J. Xiao et al., 2000. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101 671–684. [DOI] [PubMed] [Google Scholar]
- Silva, A. J., and G. G. Murphy, 1999. cAMP and memory: a seminal lesson from Drosophila and Aplysia. Brain Res. Bull. 50 441–442. [DOI] [PubMed] [Google Scholar]
- Siwicki, K. K., and L. Ladewski, 2003. Associative learning and memory in Drosophila: beyond olfactory conditioning. Behav. Processes 64 225–238. [DOI] [PubMed] [Google Scholar]
- Stolc, V., Z. Gauhar, C. Mason, G. Halasz, M. F. van Batenburg et al., 2004. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science 306 655–660. [DOI] [PubMed] [Google Scholar]
- Tarone, A. M., Y. M. Nasser and S. V. Nuzhdin, 2005. Genetic variation for expression of the sex determination pathway genes in Drosophila melanogaster. Genet. Res. 86 31–40. [DOI] [PubMed] [Google Scholar]
- Thoemke, K., W. S. Yi, J. M. Ross, S. Kim, V. Reinke et al., 2005. Genome-wide analysis of sex-enriched gene expression during C. elegans larval development. Dev. Biol. 284 500–508. [DOI] [PubMed] [Google Scholar]
- Wayne, M. L., M. Telonis-Scott, L. M. Bono, L. Harshman, A. Kopp et al., 2007. Simpler mode of inheritance of transcriptional variation in male Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 104 18577–18582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker, K. L., D. L. Ding, W. W. Fisher and H. D. Lipshitz, 1999. Different 3′ untranslated regions target alternatively processed hu-li tai shao (hts) transcripts to distinct cytoplasmic locations during Drosophila oogenesis. J. Cell Sci. 112 3385–3398. [DOI] [PubMed] [Google Scholar]
- Wilson, P. G., 2005. Centrosome inheritance in the male germ line of Drosophila requires hu-li tai-shao function. Cell Biol. Int. 29 360–369. [DOI] [PubMed] [Google Scholar]
- Wilson, R. J., J. L. Goodman, V. B. Strelets and FlyBase Consortium, 2008. FlyBase: integration and improvements to query tools. Nucleic Acids Res. 36 D588–D593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz, W. M., W. Wu, I. Andre, B. Qian, D. Baker et al., 2007. A vast repertoire of Dscam binding specificities arises from modular interactions of variable ig domains. Cell 130 1134–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. P., and S. V. Nuzhdin, 2003. Fitness costs of Doc expression are insufficient to stabilize its copy number in Drosophila melanogaster. Mol. Biol. Evol. 20 800–804. [DOI] [PubMed] [Google Scholar]
- Yang, X., E. E. Schadt, S. Wang, H. Wang, A. P. Arnold et al., 2006. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 16 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, L., and A. C. Spradling, 1992. hu-li-tai-shao, a gene required for ring canal formation during Drosophila oogenesis, encodes a homolog of adducin. Genes Dev. 6 2443–2454. [DOI] [PubMed] [Google Scholar]