Abstract
In this work we addressed the role of ubiquitination in the function of the nascent polypeptide-associated complex (NAC), named EGD in the yeast Saccharomyces cerevisiae. To this end, we first identified the lysines residues required for ubiquitination of EGD/NAC. While simultaneous mutation of many lysines in the α-subunit of NAC (Egd2p) was required to abolish its ubiquitination, for the β-subunit of NAC (Egd1p), mutation of K29 and K30 was sufficient. We determined that the ubiquitination of the two EGD subunits was coordinated, occurring during growth first on Egd1p and then on Egd2p. Egd2p was ubiquitinated earlier during growth if Egd1p could not be ubiquitinated. The use of mutants revealed the importance of EGD ubiqutination for its ribosome association and stability. Finally, our study demonstrated an interaction of EGD/NAC with the proteasome and revealed the importance of the Not4p E3 ligase, responsible for EGD/NAC ubiquitination, in this association.
WHEN emerging from the ribosome, newly synthesized polypeptide chains are immediately bound by molecular chaperones that assist in subsequent folding or by specifically targeting factors that help nascent chains to reach their proper cellular localization (Bukau et al. 2000; Hartl and Hayer-Hartl 2002). In yeast and metazoans, the Hsp70-type chaperone, Ssz, and its DnaJ co-chaperone, Zuotin, form a ribosome-associated complex (RAC) (Gautschi et al. 2001; Hundley et al. 2005; Otto et al. 2005). In yeast, RAC interacts with another ribosome-bound Hsp70 homolog, Ssb (Gautschi et al. 2001; Hundley et al. 2002), where only Ssb is in direct association with nascent chains. In addition to the Hsp70-RAC machinery, there is a protein complex, the nascent polypeptide-associated complex (NAC), which associates with ribosomes and nascent chains in an apparent 1:1 stoichiometry (Wiedmann et al. 1994; Funfschilling and Rospert 1999; Rospert et al. 2002).
NAC is a highly conserved heterodimeric complex composed of two subunits (α and β) that are both in direct contact with nascent polypeptide chains (Wiedmann et al. 1994); however, βNAC alone is responsible for binding to the ribosome (Beatrix et al. 2000). The yeast genome encodes three known NAC homologs: a single α-subunit (encoded by EGD2) and two β-subunits (encoded by EGD1 and BTT1), where Btt1p is expressed at 100 times lower levels than Egd1p (George et al. 1998; Reimann et al. 1999). All NAC homologs contain a NAC domain, responsible for dimerization. Both β-subunits can form heterodimeric complexes with αNAC. Egd1p can also form homodimers (Panasenko et al. 2006), and probably so can Egd2p since archaeal NAC is an αNAC homodimer (Spreter et al. 2005). Although NAC is highly conserved and present in archaea, yeast, and mammalian cells, our knowledge of its function in vivo is still far from complete.
It has been shown that NAC associates with the ribosome through binding of the ribosomal protein Rpl25p, near the site where newly synthesized polypeptide chains emerge (Wegrzyn et al. 2006). These data, together with the observation that NAC crosslinks to short nascent polypeptides (Wiedmann et al. 1994), have led to the speculation that NAC might play a role in the folding of newly synthesized proteins, protecting them from interaction with inappropriate cytosolic factors. It was proposed that cycles of binding and releasing NAC would expose the polypeptide to the cytosol in quantal units, rather than amino acid by amino acid. NAC would thus contribute to fidelity in cotranslational processes such as targeting and folding (Wang et al. 1995). There has also been evidence that NAC directly interacts with the signal recognition particle and is involved in correct translocation of proteins to the endoplasmic reticulum by regulating the accessibility of the translocation pore and by preventing the mistargeting of nonsecretory proteins (Lauring et al. 1995; Moller et al. 1998). In addition, a regulatory role for NAC in the import of proteins into mitochondria was proposed (George et al. 1998; Funfschilling and Rospert 1999); however, direct evidence to support this hypothesis is still lacking. Finally, NAC has also been associated with transcription regulation (Zheng et al. 1987, 1990; Quelo et al. 2002, 2005; Akhouayri et al. 2005) and with human cell differentiation (Lopez et al. 2005), mostly in situations of unequal expression of either NAC subunit, suggesting individual functions of the α- and β-subunits.
The biological importance of NAC is highlighted by the embryonic lethality of NAC mutants in mice (Deng and Behringer 1995), nematodes (Bloss et al. 2003), and fruit flies (Markesich et al. 2000). In contrast, deletion of EGD/NAC in yeast (referred to as EGD from here on) is not lethal and leads to only insignificant growth defects at high temperature (Reimann et al. 1999). αNAC contains a ubiquitin-associated (UBA) domain, which is found in several proteins involved in the ubiquitin–proteasome pathway for protein degradation. The UBA domain is structurally distinct from the NAC domain (Spreter et al. 2005) and it is not necessary for heterodimer formation, but it is required for stability of EGD (Panasenko et al. 2006). Recently, we found that the Not4p E3 ligase, a component of the nine-subunit evolutionarily conserved Ccr4-Not complex, was responsible for regulated ubiquitination of EGD in yeast and influenced its cellular localization (Panasenko et al. 2006). However, the exact role of EGD ubiquitination in vivo remains unknown.
In this work, we undertook the identification of the ubiquitinated residues in EGD to investigate the role of ubiquitination for this chaperone. We determined that the ribosome association and stability of Egd1p required its own ubiquitination, particularly in the absence of Egd2p. In addition, we found that Egd2p binds to the proteasome, a 2.5-MDa protease present in all eukaryotes, which degrades proteins conjugated to ubiquitin. The proteasome can be subdivided into two major subcomplexes: (1) the 20S core particle (CP), a hollow cylinder that consists of a stack of four rings, two outer rings with seven different α-type subunits, and two inner rings with seven different β-type subunits each and (2) two additional 19S regulatory particles (RP; also called PA700 in mammals) attached to the ends of the 20S cylinder to build up the 26S proteasome holoenzyme (Peters et al. 1994). In particular, we found that Egd2p associates with the CP, containing the proteolytically active sites of the proteaseome, in a Not4p E3 ligase-dependent manner. This result suggests a role for EGD ubiquitination in its association with the proteasome. Our finding of the importance of EGD Not4p-dependent ubiquitination in its association with the ribosome at the site of nascent chain emergence, on the one hand, and with the proteasome, on the other hand, is provocative. Indeed, ubiquitination is likely to play a role in the targeting of inappropriately folded nascent polypeptides to the proteasome, indicating that the Not4p E3 ligase and EGD are possible candidates for playing a role in this process.
MATERIALS AND METHODS
Media and strains:
All media were standard. The strains used in this work derive from MY1 or from the Euroscarf strain SC0000 (Table 1). Single-step deletions and/or tagging of genes were performed by PCR according to Longtine et al. (1998).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| MY1 | MATaura3-52 trp1-1 leu2∷PET56 gcn4Δ | Collart and Struhl (1994) |
| MY1719 | Isogenic to MY1 except MATα not5∷LEU2 | Lenssen et al. (2005) |
| MY3595 | Isogenic to MY1 except MATα not4∷KanMX4 | Lenssen et al. (2007) |
| MY3609 | Isogenic to MY1 except egd2∷KanMX4 | Panasenko et al. (2006) |
| MY3610 | Isogenic to MY1 except egd1∷KanMX4 | Panasenko et al. (2006) |
| MY3622 | Isogenic to MY1 except not4∷LEU2 egd1∷KanMX4 | This work |
| MY3624 | Isogenic to MY1 except not4∷LEU2 egd1∷EGD1-HA3-KanMX4 | Panasenko et al. (2006) |
| MY3894 | Isogenic to MY1 except egd1∷EGD1-HA3-KanMX4 ubc4∷TRP1 | This work |
| MY3895 | Isogenic to MY1 except egd1∷EGD1-HA3-KanMX4 ubc5∷TRP1 | This work |
| MY3897 | Isogenic to MY1 except not4∷LEU2 egd1∷EGD1-HA3-KanMX4 ubc5∷TRP1 | This work |
| MY4272 | Isogenic to MY1 except ubc5∷TRP1 | This work |
| MY4519 | Isogenic to MY1 except not4∷KanMX4:NOT4-URA3 egd1∷EGD1-HA3-KanMX4 | Panasenko et al. (2006) |
| MY4644 | Isogenic to MY1 except egd1∷NatMX4 egd2∷KanMX4 | This work |
| MY4863 | Isogenic to MY1 except ubc4∷NatMX4 | This work |
| MY5125 | Isogenic to MY1 except not4∷LEU2 egd1∷EGD1-HA3-KanMX4 ubc4∷TRP1 | This work |
| MY5561 | MATaade2 arg4 leu2-3,112 trp1-289 ura3-52 rpn1∷RPN1-TapTag-URA3 | Gavin et al. (2006) |
| MY5693 | MATarpn1∷RPN1-TapTag-URA3 not4∷KanMX4 | 3595 × 5561 |
| MY5699 | MATarpn1∷RPN1-TapTag-URA3 | 1719 × 5561 |
DNA constructs:
Plasmids expressing HA-tagged Egd1p (pADH1-EGD1-HA) and Egd2p (pADH1-EGD2-HA) under the control of the ADH1 promoter were made by cloning EGD1 or EGD2 sequences with an HA tag, amplified by PCR, into a pRS414-derived plasmid containing the ADH1 promoter (pMAC392; Panasenko et al. 2006) between the BamHI and XhoI sites, leading to pMAC594 and pMAC596. Mutagenesis of Egd1p and Egd2p was performed on the basis of pMAC594 and pMAC596 with the standard Promega protocol, leading to plasmids pMAC595 (Egd1K10,13Rp-HA), pMAC768 (Egd1K29,30Rp-HA), pMAC604 (Egd1K10,13,29,30Rp-HA), pMAC629 (Egd1K10,13,29,30,43,50,66Rp-HA), pMAC624 (Egd1K43,50,66Rp-HA), pMAC625 (Egd1K138,139Rp-HA), pMAC636 (Egd1R24A,R25A,K26Rp-HA), pMAC599 (Egd2K18,19,26Rp-HA), pMAC626 (Egd2K18,19,26,42,52,70Rp-HA), pMAC628 (Egd2K42,52,70Rp-HA), and pMAC600 (Egd2K159Rp-HA). The marker gene in pMAC768 was changed from URA3 to TRP1 by homologous recombination in yeast, leading to pMAC770.
Plasmids expressing Myc6-tagged Ubc4p and Ubc5p were made by cloning UBC4 or UBC5 sequences, amplified by PCR, into pGREG516 (Jansen et al. 2005) between the SalI sites, by homologous recombination in yeast. The GAL1 promoter in these plasmids was changed to the UBC4 or UBC5 promoters amplified by PCR and cloned between the AscI and NotI sites by homologous recombination in yeast, leading to pMAC640 (pUBC4-Myc6-UBC4) and pMAC648 (pUBC5-Myc6-UBC5), respectively. The sequences of all plasmids were verified.
Co-immunoprecipitation:
A total of 100 OD units of cells grown to an OD600 of 0.8 were broken with 1 ml of glass beads in 0.8 ml of an immunoprecipitation (IP) buffer (40 mm HEPES–KOH, pH 7.5, 150 mm K-acetate, 40 mm KCl, 1 mm EDTA, 20% glycerol, 1 mm PMSF) during 15 min at 4°. The supernatant was clarified by centrifugation for 20 min at 14,000 × g at 4° and the total protein concentration in the supernatant was measured by the Bradford assay. To analyze the protein levels in the lysates, we loaded on SDS–PAGE gels 12 μg of total protein extract (TE). For immunoprecipitation, 0.4 ml of the lysates containing 2.5 mg of total protein was mixed with 100 μl of 20% Protein A-sepharose and 1 μl of anti-HA antibodies (IP anti-HA) for 10 hr at 4°. For control (IP control), we used the same mixture but without antibodies. After incubation, beads were washed three times with 1 ml of IP buffer and boiled with 50 μl of two times concentrated SDS–sample buffer, 15 μl of which was analyzed by SDS–PAGE. The efficiency of immunoprecipitation was analyzed by Western blot with antibodies against HA. Co-immunoprecipitated proteins were analyzed by Western blot with antibodies against Rpl25p and Egd2p.
Stability assay and in vivo ubiquitination assay:
These assays were performed as described earlier (Panasenko et al. 2006). Equal loading of the gels in these experiments was verified by ponceau S staining of all membranes prior to Western blotting.
Modeling:
To model NAC–ribosome interaction, we use the described structure of the trigger factor (TF) from Deinococcus radiodurans interacting with the ribosome, particularly with Rpl23p (Schlunzen et al. 2005). To build a homology model of the interaction of Egd1p with Rpl25p, the ribosome-bound state of the TF loop (TF-BD, PDB:2d3o) was used as a template in Swiss-PDB-Viewer (Guex and Peitsch 1997). This modeling is supported mainly by a multiple alignment of numerous TF and Egd1p homologous sequences that shows a strong conservation of several residues in the loop region. The three-dimensional structure of the ribosomal proteins interacting with TF (Rpl23p, Rpl24p, and Rpl29p) was superimposed to the low-resolution, three-dimensional structure of their eukaryotic homologs (Rpl25p, Rpl26Ap, and Rpl34p, respectively) in an unbound state (PDB:1s1i) (Spahn et al. 2004).
The model of Egd1p–Egd2p interaction was performed with modeler8 v2 (Sali and Blundell 1993), using as a template the structure proposed for archaeal NAC (aeNAC, PDB:1tr8) as done previously (Spreter et al. 2005). Reconstruction of the loops and energy minimization were done with Swiss-PDB-Viewer. ANOLEA profiles were computed for each chain as a validation indicator for the model (Melo et al. 1997). The secondary structures of Egd1p and TF homologs were predicted using PSIPRED v2.6 (Jones 1999; McGuffin et al. 2000).
A protein–protein rigid body docking experiment between Rpl25p-Rpl26Ap ribosomal proteins and ubiquitin was done with Hex v4.5 (Ritchie and Kemp 2000). The search was performed using full rotation of molecules and was based on both shape and electrostatics.
Proteasome purification:
Proteasomes were purified from yeast strains expressing the RP subunit Rpn1p with a tobacco etch virus (TEV)-ProA tag (Tap-tag) at the C terminus as described in Leggett et al. (2005) with some modifications as outlined below, but all of the buffers were the same. Briefly, cells from 10 liters of culture, grown to stationary phase (OD600 of 12), were harvested, washed with buffer 1, resuspended in a twofold volume of the same buffer containing 10% glycerol, and disrupted with glass beads. Lysates were clarified by centrifugation at 43,000 × g for 45 min and frozen at −80°. A total of 100 ml of lysates, containing 2 g of total protein, were incubated with immunoglobulin–sepharose (IgG–sepharose) for 1.5 hr at 4° and the resin was washed with 50 bed volumes (BV) of buffer 2. We divided the resin into two parts: one was used for the purification of the holoenzyme and the other for the purification of RP and CP. The holoenzyme was eluted by equilibrating the IgG resin with buffer 5 and then incubating with 1.5 BV of buffer 5, containing 100 units/ml of His6-TEV protease (Invitrogen) at 30° for 1.5 hr. TEV protease was subsequently removed from the eluate by incubation with Ni-NTA resin (Qiagen) at 4° for 15 min. To purify the CP, we incubated the second part of the IgG resin with 5 BV of buffer 3 for 1 hr at 4° and collected the flow through. To purify the RP, resin was washed with 50 BV of buffer 3, equilibrated with buffer 5, and incubated with 1.5 BV of buffer 5, containing 100 units/ml of His6-TEV protease at 30° for 1.5 hr. TEV protease was removed with Ni-NTA resin.
Antibodies:
Antibodies against Rpl25p were kindly provided by Elke Deuerling. Production of antibodies against Egd1p and Egd2p was described previously (Panasenko et al. 2006). Antibodies against Rpt1p, Rpt2p, and α1, -2, -3, -5, -6, and -7 were purchased from Biomol.
RESULTS
Ubc4p and Ubc5p play different roles in Egd1p ubiquitination:
Ubiquitination of the EGD complex in vivo depends upon Not4p (Panasenko et al. 2006), and Not4p can interact in the two-hybrid assay with two yeast E2-conjugating enzymes, Ubc4p and Ubc5p (Albert et al. 2002; Panasenko et al. 2006). In vitro the EGD complex can be ubiquitinated by Ubc4p and Not4p (Panasenko et al. 2006), so we investigated the role of Ubc4p and Ubc5p in ubiquitination of the EGD complex in vivo using wild-type or not4Δ strains expressing HA-tagged Egd1p and deleted (or did not delete) UBC4 or UBC5. As previously published (Panasenko et al. 2006), ubiquitinated forms of Egd1-HA were detectable in wild-type cells and increased as glucose was depleted from the growth medium (Figure 1A). Deletion of NOT4 led to reduced Egd1p ubiquitination, as expected, whereas deletion of UBC4 did not affect Egd1p ubiquitination, which remained mostly Not4p dependent. Surprisingly, Egd1p ubiquitination strongly increased in ubc5Δ cells, although it remained Not4 dependent. Unfortunately, we could not measure Egd1p ubiquitination in the double ubc4Δ ubc5Δ mutant because the double deletion was not viable in our genetic background.
Figure 1.—
Different roles of Ubc4p and Ubc5p in Egd1p ubiquitination. (A) Total protein extracts were prepared from wild-type (WT), ubc4Δ, ubc5Δ, not4Δ, ubc4Δ not4Δ, and ubc5Δ not4Δ cells collected from exponential phase to glucose depletion and expressing Egd1p-HA3 and His6-ubiquitin. TEs and proteins eluted from a nickel column (Ni-eluate) were analyzed by Western blot with antibodies against the epitope tag. The molecular weights of protein markers are indicated on the left. (B) ubc4Δ or ubc5Δ strains expressing Myc6-Ubc4p or Myc6-Ubc5p were harvested after the indicated times in medium with cycloheximide (CHX, 50 μg/ml). The levels of the tagged proteins (indicated on the left) were revealed by rapid alkaline lysis and Western blot using antibodies against the tag (left), and equal protein loading was verified by ponceau S staining (right). (C) Total protein extracts prepared from ubc4Δ or ubc5Δ cells expressing Myc6-Ubc4p or Myc6-Ubc5p and His6-ubiquitin were collected from exponential phase to glucose depletion and analyzed as described in A.
Previous work has suggested that Ubc4p and Ubc5p are functionally redundant E2 enzymes (Chuang and Madura 2005), yet our results now show that Ubc4p and Ubc5p are not functionally identical with regard to Egd1p ubiquitination. In addition, it was reported that Ubc4p is expressed at much higher levels than Ubc5p and is the E2 enzyme primarily detected in actively growing cells (Chuang and Madura 2005). However, we observed that deletion of UBC5 had a more dramatic effect on Egd1p ubiquitination than did the deletion of UBC4. Both proteins were expressed at similar levels at the early stage of growth at high glucose concentration in the medium and 18 hr later when glucose had been depleted (Figure 1B). Interestingly, Ubp5p was unstable, with a half-life of <30 min, whereas Ubc4p was much more stable (Figure 1B). Thus, the differences in the cellular levels of Ubc4p and Ubc5p described in previous studies might have stemmed from the difference in their stabilities rather than from differences in their expression. A slower migrating form of Ubc4p, and to a lesser extent of Ubc5p, was visible in our experiments (Figure 1B, left) and compatible with ubiquitination of these E2 enzymes. Indeed, using the same assay described above for Egd1p and Egd2p, we observed that both Ubc4p and Ubc5p were ubiquitinated (Figure 1C). Ubc4p was more extensively ubiquitinated, yet more stable, than Ubc5p, suggesting that the observed ubiquitination was not associated with destabilization for these E2 enzymes.
Taken together, these results show that the Ubc4p and Ubc5p E2 enzymes are expressed at similar levels but have different stabilities, are both ubiquitinated, and play different roles in EGD complex ubiquitination.
Ubiquitination of the two EGD subunits is tightly coordinated:
To investigate EGD ubiquitination further, we tried to identify on which lysine residues the ubiquitination occurs. There are 18 lysine residues in Egd1p and 16 in Egd2p, all of which could potentially be ubiquitinated (Figure 2A). We mutated to arginine the most conserved lysines in Egd1p and Egd2p and performed a ubiquitination assay.
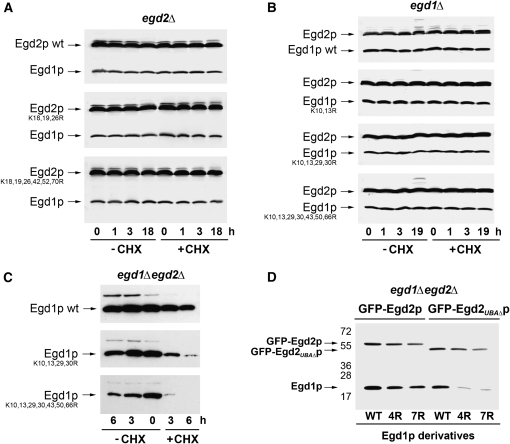
Figure 2.—
Ubiquitination of Egd1p and Egd2p is linked. (A) Primary structure of Egd1p and Egd2p. The most conserved lysine residues are marked in boldface. NAC and UBA domains are underlined with one or two lines, respectively. The mapped ribosome-binding site in the structure of Egd1p is indicated by italics. (B) Total protein extracts were prepared from egd1Δ cells collected from exponential phase to glucose depletion and expressing from episomes His6-ubiquitin and HA-tagged Egd1p, either wild type or carrying the indicated mutations, and analyzed as in Figure 1A. Membranes were analyzed with antibodies against HA (top) or, after stripping, with antibodies against Egd2p (bottom). Molecular weights of protein markers are indicated on the left. (C) Total protein extracts were prepared from egd2Δ cells expressing from episomes His6-ubiquitin and HA-tagged Egd2p, either wild type or carrying the indicated mutations, and analyzed as described in Figure 1A. The signal in C for Egd2p is stronger than in B because different antibodies were used (HA vs. Egd2p). (D) Total protein extracts were prepared from egd1Δ (lanes 1 and 2) or egd1Δ not4Δ (lanes 3) cells expressing His6-ubiquitin and HA-tagged Egd1p, either wild type (lanes 1 and 3) or K29,30R (lane 2). Cells were grown and collected as described in Figure 1A. Molecular weights of protein markers are indicated on the left.
In the case of Egd1p (Figure 2B, top), some mutations, such as K43,50,66R (lane 6, right) or K138,139R (lane 7, right) did not affect its ubiquitination (compare to lane 1, right), whereas mutation of all seven lysine residues, K10,13,29,30,43,50,66R (lane 5, right), led to a striking reduction of Egd1p ubiquitination at both the exponential and post-diauxic phases of growth. K10,13,29,30R and K29,30R had the same reducing effect (lanes 3 and 4, left and right), whereas mutations K10,13R had only a minor impact on Egd1p ubiquitination (lane 2, right). All derivatives were equally expressed (left). These results suggest that ubiquitination of Egd1p might occur on lysines 29 and/or 30.
In the case of Egd2p, only upon mutation of a large number of lysine residues, such as in the mutant K18,19,26,42,52,70R, was there any measurable reduction in Egd2p ubiquitination (Figure 2C, lane 4). These results suggest that Egd2p might be carrying ubiquitin entities on several different lysine residues, rather than a polyubiquitin chain on one given lysine, but it is also possible that ubiquitination can occur on different lysines when the appropriate one is mutated.
During exponential growth, Egd1p, but not Egd2p, was ubiquitinated (compare lane 1 in Figure 2, B and C), whereas both Egd1p and Egd2p were ubiquitinated after the diauxic shift, most likely in response to depletion of several nutrients, including glucose (data not shown). According to these observations, one can imagine that, in the EGD complex, Egd1p is ubiquitinated first and Egd2p ubiquitination occurs later. Thus, it was interesting to investigate whether there was any correlation between ubiquitination of the two subunits. We followed Egd2p ubiquitination in egd1Δ cells expressing wild-type or non-ubiquitinated forms of Egd1p (Figure 2B, bottom). While Egd2p was moderately ubiquitinated in cells expressing wild-type Egd1p—and this only after the diauxic shift (lane 1, left and right)—its ubiquitination was strikingly increased in cells growing exponentially and after the diauxic shift, when they expressed a mutant form of Egd1p that was not ubiquitinated (K29,30R, K10,13,29,30R or K10,13,29,30,43,50,66R) (lanes 3–5). Thus, Egd1p is the major subunit of the EGD complex that is ubiquitinated, but if it cannot be ubiquitinated, Egd2p becomes more extensively ubiquitinated.
As previously reported (Panasenko et al. 2006), ubiquitination of endogenous Egd1p is decreased in not4Δ cells (see Figure 1A). Deletion of Not4p similarly abolished ubiquitination of Egd1p expressed from a plasmid, and this to the same extent as mutation K29,30R in Egd1p (Figure 2D: compare lanes 2 and 3 to lane 1).
Stability of Egd1p requires its own ubiquitination and Egd2p:
Our results indicate that ubiquitination of the two subunits of the EGD complex is connected, and this raises the question of the role of this ubiquitination. One possibility is that it mediates destabilization of the EGD complex. However, the stable levels of both subunits were constant during 18 hr of cell growth, with or without cycloheximide, whether the derivatives were ubiquitinated or not (Figure 3, A and B). These observations suggest that the EGD subunits are generally very stable and that their ubiquitination does not mediate their degradation, although one cannot exclude the possibility that only a small proportion of the EGD subunits is ubiquitinated and turned over. Furthermore, the stability of the mutants also suggests that they are probably not misfolded.
Figure 3.—
Stability of Egd1p is dependent upon its own ubiquitination and Egd2p. (A) egd2Δ cells expressing HA-tagged Egd2p, either wild type or carrying the indicated mutations, were treated and analyzed as in Figure 1B, with antibodies against HA or against Egd1p to reveal endogenous Egd1p. (B) egd1Δ cells expressing HA-tagged Egd1p, either wild type or carrying the indicated mutations, were treated and analyzed as in Figure 1B, with antibodies against HA or against Egd2p to reveal endogenous Egd2p. (C) egd1Δ egd2Δ cells expressing HA-tagged Egd1p, either wild type or carrying the indicated mutations, were analyzed as in Figure 1B. (D) egd1Δ egd2Δ cells expressing GFP-Egd2p or GFP-Egd2ΔUBAp and HA-tagged Egd1p, either wild type (WT), K10,13,29,30R (4R), or K10,13,29,30,43,50,66R (7R) were grown to an OD600 of 0.8 and collected after 6 hr when the drop of Egd1p in cells totally lacking Egd2p was maximal (C). Total extracts were analyzed by Western blot for the presence of the Egd1p and Egd2p derivatives with antibodies against the HA or GFP tags.
Previously, we determined that the presence of Egd2p was required for Egd1p stability (Panasenko et al. 2006), and we thus considered that ubiquitination of Egd1p might be required to degrade Egd1p in the absence of Egd2p. To test this, we compared the stability of wild-type or mutant Egd1p in cells lacking Egd2p. As previously observed, wild-type Egd1p was slightly unstable in the absence of Egd2p with a half life of ∼3 hr (Figure 3C, top). Surprisingly, non-ubiquitinated Egd1p derivatives were much less stable than wild-type Egd1p in cells lacking Egd2p (Figure 3C: compare the two bottom panels to the top panel). Furthermore, we noted that, in egd2Δ cells, the levels of non-ubiquitinated Egd1p derivatives decreased during the growth of cells, even in the absence of cycloheximide, at the time of growth when ubiquitination of wild-type Egd1p usually increases. Thus, our results indicate that, in contrast to our initial hypothesis, in egd2Δ cells Egd1p ubiquitination correlates with its stability.
As mentioned above, Egd2p has a UBA domain that is important for Egd2p's own stable expression (Panasenko et al. 2006). To analyze if this UBA domain may influence the stability of Egd1p, we used plasmids expressing N-terminally GFP-tagged Egd2p with and without a UBA domain. Indeed, we observed that the presence of a GFP entity at the N-terminal end of Egd2p was able to stabilize UBA-less Egd2p (Figure 3D) such that, by using fusion proteins, we could compare expressed Egd2p and UBA-less Egd2p to define the role of the UBA domain. We measured wild-type and mutant Egd1p levels in cells expressing GFP-Egd2p or GFP-Egd2ΔUBAp. Wild-type Egd1p was expressed at slightly lower levels in cells expressing UBA-less Egd2p compared to full-length Egd2p. The level of Egd1p mutants was not significantly reduced compared to wild-type Egd1p in cells expressing full-length Egd2p. In contrast, in cells expressing UBA-less Egd2p, the level of Egd1K10,13,29,30Rp and Egd1K10,13,29,30,43,50,66Rp was dramatically reduced compared to wild-type Egd1p or compared to the level of these mutants in cells expressing full-length Egd2p [Figure 3D: compare 4R and 7R lanes to wild-type (WT) lanes]. Interestingly, UBA-less Egd2p was also less ubiquitinated than full-length Egd2p (supplemental Figure 1).
Thus, the UBA domain of Egd2p is required not only for stability of Egd2p itself as previously observed, but also for its own ubiquitination and for the stability of non-ubiquitinated Egd1p.
Ribosome association of the EGD complex is dependent upon its ubiquitination, Not4p, and Egd2p:
We showed above that Egd1p ubiquitination requires its lysines K29 and K30 and is necessary for Egd1p stability in the absence of Egd2p, but the question of the relevance of this ubiquitination for the cellular function of the EGD complex remains. One clear function attributed to the EGD complex is its association with the ribosome, so we investigated how ubiquitination of the EGD complex might affect its ribosome binding. Indeed, K29 and K30, required for Egd1p ubiquitination as shown above, are part of the ribosome-binding site of Egd1p (RRK(x)nK29K30), and the mutation K29,30A has been reported to reduce the ribosomal association of Egd1p (Wegrzyn et al. 2006). To thus analyze the role of ubiquitination for ribosome association of Egd1p, we immunoprecipitated wild-type and mutant Egd1p. Egd1p was expressed at roughly the same level in all strains during early exponential growth and was immunoprecipitated with similar efficiency (Figure 4A, left). The ribosomal protein Rpl25p, with which the EGD complex interacts (Wegrzyn et al. 2006), was expressed at similar levels in all strains and was co-immunoprecipitated with wild-type or mutant Egd1p in cells expressing Egd2p (Figure 4A, middle), although maybe with slightly less efficiency in the case of the non-ubiquitinated derivatives of Egd1p (Figure 4A, middle). In egd2Δ cells, Rpl25p was co-immunoprecipitated with wild-type Egd1p, albeit less efficiently than in the presence of Egd2p (Figure 4A: compare each lane 1 of the middle). In contrast, only traces of Rpl25p were co-immunoprecipitated with Egd1K10,13,29,30Rp and Egd1K10,13,29,30,43,50,66Rp. The interaction of Egd2p was similar with wild-type or mutant forms of Egd1p (Figure 4A, right), indicating that ubiquitination of Egd1p is not essential for heterodimer formation and, also again, that the Egd1p mutants are appropriately folded.
Figure 4.—
Ribosome association of the EGD complex is dependent upon its own ubiquitination and Egd2p. (A) Egd1p derivatives were immunoprecipitated from total protein extracts of egd1Δ or egd1Δ egd2Δ cells expressing tagged Egd1p, either wild type (WT, lanes 1), K10,13,29,30R (4R, lane 2, or K10,13,29,30,43,50,66R (7R, lane 3), with antibodies against the HA tag. TE and immunoprecipitates were analyzed by Western blot for the presence of tagged Egd1p (left), Rpl25p (middle), and Egd2p (right) with the antibodies indicated below the panels. (B) Egd1p-HA was immunoprecipitated from egd1Δ (lane 1) or egd1Δ not4Δ (lane 2) cells. TE and immunoprecipitates were analyzed for the presence of Egd1p-HA (left) or Rpl25p (right). (C) Total protein extracts were prepared from egd1Δ egd2Δ cells expressing His6-ubiquitin, Egd1K29,30Rp-HA, and wild-type or mutant Egd2p-HA as indicated. TE and proteins eluted from a nickel column were analyzed by Western blot with antibodies against Egd2p. The molecular weights of protein markers are indicated on the left. (D) The EGD complex was immunoprecipitated from total protein extracts collected from egd1Δ egd2Δ cells expressing Egd1K29,30Rp-HA and wild-type or mutant Egd2p-HA. TE and immunoprecipitates were analyzed with antibodies against HA or Rpl25p as indicated on the left. (E) Total protein extracts were prepared from egd1Δ cells expressing His6-ubiquitin and HA-tagged Egd1p, either wild type (Egd1WTp) or mutated in its ribosome-binding domain (Egd1RRK/AAAp). The ubiquitination assay was performed as in Figure 1A. (F) egd1Δ egd2Δ or egd1Δ egd2Δ not4Δ cells expressing HA-tagged Egd1RRK/AAAp were analyzed as in Figure 1B for the presence of the mutant Egd1p.
Our result indicates that ribosome association of Egd1p is dependent upon its own ubiquitination and Egd2p. As shown above, Egd1p ubiquitination depends upon Not4p (see Figure 1A and Figure 2D), so we analyzed how deletion of Not4p might influence the ribosome binding of Egd1p. Egd1p was expressed at the same level in wild-type and not4Δ strains and was immunoprecipitated with similar efficiency (Figure 4B, left). Rpl25p was also expressed at similar levels in both strains and was co-immunoprecipitated with Egd1p from wild-type cell extracts, but not from cell extracts lacking Not4p (Figure 4B, right). Thus, deletion of Not4p, which reduces ubiquitination of both Egd1p and Egd2p (Panasenko et al. 2006), disrupts Egd1p ribosome association even in the presence of Egd2p.
Loss of Egd1p ubiquitination results in reduced association of Egd1p with its ribosomal partner Rpl25p in the absence but not in the presence of Egd2p. Deletion of NOT4 reduces Egd1p ribosomal association even in the presence of Egd2p, but we know that in this case ubiquitination of both Egd1p and Egd2p is reduced. These results suggest that increased ubiquitination of Egd2p might compensate for reduced Egd1p ubiquitination and support association of the EGD complex with ribosome in the first situation. To test this, we studied Rpl25p binding of a non-ubiquitinated mutant of Egd1p (K29,30R) in cells expressing a mutant of Egd2p (K18,19,26,42,52,70) that is much less ubiquitinated than wild-type Egd2p after glucose depletion (see Figure 2C). First, we found that, indeed, this mutant Egd2p was much less ubiquitinated than wild-type Egd2p in cells expressing a non-ubiquitinated mutant of Egd1p (Figure 4C). Second, immunoprecipitation of Egd1K29,30Rp from cells expressing wild-type or mutant Egd2p was equally efficient (Figure 4D, top), but co-immunoprecipitation of Rpl25p was significantly decreased from cells expressing the K18,19,26,42,52,70R mutant of Egd2p compared to wild-type Egd2p (Figure 4D, bottom). These results suggest that increased ubiquitination of Egd2p indeed can compensate for reduced Egd1p ubiquitination to support EGD ribosome association.
Our experiments show that, in the absence of Egd2p, reduction of Egd1p ubiquitination leads to instability and loss of ribosome binding. It is unclear whether instability might be a consequence of ribosome dissociation or vice versa. To solve this issue, we created an Egd1p mutant in which the conserved ribosome-binding motif R24R25K26 was replaced by alanines (Egd1RRK/AAAp). This mutant does not associate with the ribosome (Wegrzyn et al. 2006) and contains one mutated lysine residue, so we first tested whether it could be ubiquitinated. Indeed, ubiquitination of Egd1RRK/AAAp was similar to ubiquitination of wild-type Egd1p (Figure 4E). This mutant was also as stable as wild-type Egd1p in cells lacking Egd2p (Figure 4F, top: compare to Figure 3C, top), indicating that loss of ribosome association per se does not lead to Egd1p instability. Interestingly, the deletion of NOT4, which leads to reduced Egd1p ubiquitination (Figure 1A and Figure 2D; Panasenko et al. 2006), led to reduced stability of Egd1RRK/AAAp in not4Δ egd2Δ cells (Figure 4F). Thus, Egd2p and ubiquitination, but not ribosome association, are required for Egd1p stability.
Modeling EGD heterodimer interaction with Rpl25p is compatible with ubiquitination at K29 and/or K30 of Egd1p and possible docking of ubiquitin to the ribosome:
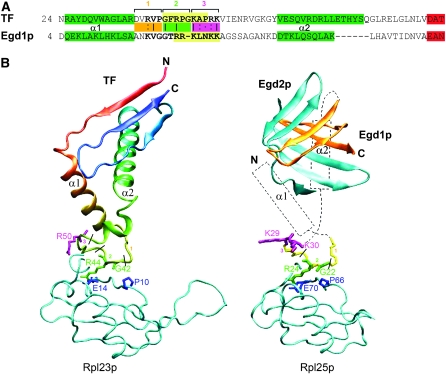
Our finding that ubiquitination of Egd1p was important for its stability and ribosome association was unexpected and led us to investigate the available structural information on the EGD complex, on the one hand, and, on the other hand, on the ribosome binding of a prokaryotic chaperone, TF, which associates with the ribosomal subunit Rpl23p, the homolog of eukaryotic Rpl25p with which Egd1p interacts (Schlunzen et al. 2005; Wegrzyn et al. 2006). The structures of Rpl23p and Rpl25p present well-conserved folds (Spahn et al. 2004), and the region of Rpl23p interacting with TF (L/IxxPxxS/TExA) is also conserved in all kingdoms of life (Wegrzyn et al. 2006). From this observation, one can propose that the ribosome-binding site might be similar between prokaryotic TF and eukaryotic EGD/NAC. The ribosome-binding domain of TF has a helix-loop-helix fold and interacts with Rpl23p via this loop (Kramer et al. 2002). A similar helix-loop-helix region was found in the predicted secondary structure of Egd1p, whose ribosome-binding loop contains three segments corresponding to the same segments of TF (Figure 5). Despite the reported absence of similarity between TF and Egd1p, we found 38% of sequence identity between the ribosome interacting loop of the D. radiodurans TF and the corresponding region of yeast Egd1p (Figure 5). The loop includes both the RRK(x)nKK and GFRxGxxP motifs conserved among eukaryotic and bacterial species, respectively (Kramer et al. 2002; Wegrzyn et al. 2006). In the multiple alignment, G42 and R44 in TF, corresponding to G22 and R24 in Egd1p, are fully conserved. R44 of TF interacts with E14 of Rpl23p (which corresponds to E70 of Rpl25p) and with 23S rRNA (Schlunzen et al. 2005). To allow this tripartite interaction, E14 of Rpl23p and R44 of TF have to be ideally placed and oriented, suggesting the unique bound conformation for the ribosome-binding loop of TF. We found that, in spite of the divergence among prokaryotic sequences, predicted secondary structures for the helix-loop-helix region are well conserved. Finally, we noted a strong fold conservation between Rpl23p in prokaryotes and Rpl25p in eukaryotes, in particular in the region interacting with TF or βNAC/Egd1p (Figure 5). Taking into account all of these observations, we can hypothesize that the main requirement for the binding of TF or Egd1p to the ribosome is the ability of the loop to adopt specifically this bound conformation. Thus, we used the ribosome-binding domain of TF as a template and proposed a model for the interaction of the yeast EGD complex with the ribosome.
Figure 5.—
Comparison of the interaction of bacterial TF and yeast EGD with the ribosome. (A) Sequence alignments of D. radiodurans TF and S. cerevisiae Egd1p was derived from a multiple alignment that confirms the high conservation rate of several residues in this region over homologous sequences of TF and βNAC/Egd1p. The three parts of the ribosome-binding loop are indicated by numbers and orange, green, and purple, respectively. Identical, equivalent, and similar residues are shown with “|” (vertical line), “:” (colon), and “.” (period), respectively. Pale yellow indicates the conserved motifs known from the literature (Kramer et al. 2002; Wegrzyn et al. 2006). The modeled regions are indicated in boldface type. Predicted α-helices and β-strands are in green and red, respectively. (B) TF is displayed colored from red to blue according to sequence position. The three parts of the ribosome-binding loop are indicated by numbers and orange, green and purple, respectively, and conserved residues interacting with D. radiodurans Rpl23p (G42, R44) or S. cerevisiae Rpl25p (G22, R24) are in green. Conserved P10 and E14 in Rpl23p and the corresponding P66 and E70 in Rpl25p are in blue. Egd1p and Egd2p are in orange and blue, respectively. K29 of Egd1p, the corresponding R50 of TF, and K30 of Egd1p, pointing to the surface of the ribosome, are in purple. From modeling of Egd1p and Egd2p, we can propose that EGD complex architecture is similar to the TF β-sheet structure although the exact position of α-helices (α1 and α2), indicated by dotted line in Egd1p, and the precise orientation of the Egd1p/Egd2p-interacting domains cannot be determined.
According to our model, lysines K29 and K30 of Egd1p are the most exposed residues of the loop and could be post-translationally modified (Figure 5). This observation is in good agreement with our data that one or both of these lysines are ubiquitinated. Furthermore, the proximity of K29 and K30 of Egd1p to the two ribosomal proteins Rpl25p and Rpl26Ap (not shown) in our model allows us to suppose that these ribosomal proteins might interact with ubiquitinated Egd1p and stabilize the interaction of the EGD complex with the ribosome. Protein–protein docking experiments of ubiquitin (ligand) against Rpl25p-Rpl26Ap (receptor) produces a collection of solutions, all of which indicate binding of ubiquitin to Rpl25p. In one of the best solutions in terms of electrostatics and shape (total interaction energy of −335 kJ/mol against −377 kJ/mol for the first solution), ubiquitin presents its C terminus to K29 and K30. These findings are in good agreement with our finding that ubiquitination of Egd1p at these residues supports its ribosome association.
Egd2p interacts with proteasome:
Recently, it has been shown that the Ccr4-Not complex is associated with the proteasome (Laribee et al. 2007), and previous studies have described proteasome association of the ubiquitin-conjugating enzymes Ubc4p and Ubc5p in the presence of translationally damaged proteins (Chuang and Madura 2005). Taking into account that the EGD complex is thought to be a chaperone protecting newly synthesized polypeptides as they emerge from the ribosome, and that it forms a tertiary complex with Not4p and Ubc4p/Ubc5p (Panasenko et al. 2006), we investigated whether the EGD complex might interact with the proteasome. Using Tap-tagged Rpn1p, we purified different subcomplexes of the proteasome: holoenzyme, RP, and CP. In the purified holoenzymes from both strains, we found all of the analyzed proteasomal subunits, while in the subcomplexes we found only the subunits specific for these particles, namely for the regulatory subunits Rpt1p and Rpt2p and for the core particles α1, -2, -3, -5, -6, and -7 (Figure 6). Egd2p copurified with the holoenzyme from wild-type but not from not4Δ cells, while the level of Egd2p was the same in total extracts from both strains (Figure 6, bottom). We observed an enrichment of Egd2p in the purification of the CP, and again, the levels of copurifying Egd2p were higher from wild-type than from not4Δ cells. We conclude that Egd2p can interact with the proteasome, mostly with the CP, and that the deletion of NOT4 reduces this interaction.
Figure 6.—
Egd2p interacts with the proteasome. Proteasome holoenzyme (holo), RP, and CP were purified from wild-type or not4Δ cell extracts by tandem affinity purification through the Rpn1p subunit. The presence of the indicated proteasome subunits was analyzed in TEs and in the different proteasome fractions by Western blot using antibodies against Rpt1p and Rpt2p or the α-subunits. α1, -2, -3, -5, -6, and 7 levels in TE were detectable only upon longer exposure times and were the same in wild type and not4Δ (data not shown). Copurification of Egd2p was analyzed by probing the blots with antibodies against Egd2p.
DISCUSSION
Characterization of EGD ubiquitination:
Recently, we determined that the Not4p E3 ligase, a subunit of the evolutionary conserved Ccr4-Not complex, could ubiquitinate the EGD complex, called NAC in higher eukaryotes, in a regulated manner (Panasenko et al. 2006). However, the role of EGD ubiquitination in vivo is unknown. In this work, we set out to investigate the function of this ubiquitination by first creating and analyzing a series of Egd1p and Egd2p mutants, where the most conserved lysine residues were changed to arginine. For Egd1p, mutation of lysines 29 and 30 to arginine was sufficient to prevent ubiquitination, suggesting that ubiquitination of Egd1p occurs mainly at lysine residues 29 and/or 30 and might involve the addition of more than one ubiquitin residue at these sites. For Egd2p, ubiquitination was reduced only when a large number of lysine residues were simultaneously mutated to arginine, such as lysines 18, 19, 26, 42, 52, and 70. These results may indicate that in Egd2p multiple sites can be monoubiquitinated.
Interestingly, we observed that ubiquitination of the EGD subunits is linked. It seems that Egd1p is the main ubiquitinated subunit, but that Egd2p can be ubiquitinated either when Egd1p is unable to be ubiquitinated or in response to an environmental change.
We made the surprising observation that deletion of Ubc5p, but not deletion of Ubc4p, both E2 partners of the Not4 E3 ligase, leads to an increase in Egd1p ubiquitination, and at the same time that Ubc5p, but not Ubc4p, is an unstable protein. It could be that Ubc5p is more readily recruited to ubiquitinate Egd1p with Not4p, but is not the most efficient E2 for Egd1p ubiquitination. In its absence, Not4p would then work with another E2, possibly Ubc4p, more efficiently in Egd1p ubiquitination. In this regard, it is interesting to note that we observed an increase in Ubc4p levels in cells lacking Ubc5p (data not shown). Thus, a reduction of de novo Ubc5p synthesis is one way to obtain more efficient Egd1p ubiquitination, for example, in response to environmental clues.
The UBA domain of Egd2p is important for EGD complex stability:
The structure of aeNAC has revealed that it is a homodimer that associates through the NAC domains, indicating that eukaryotic α- and βNAC subunits are likely to dimerize via their NAC domains. This idea was supported by homology modeling using the homodimeric NAC domain of aeNAC as a template for the yeast and human heterodimeric NAC domains as shown in previous work (Spreter et al. 2005) and supported in our own study. Egd2p is stable in yeast cells from which Egd1p has been depleted (Panasenko et al. 2006), but is no longer associated with ribosomes (Reimann et al. 1999). In contrast, Egd1p is unstable in yeast in the absence of Egd2p, even though it can form homodimers in vivo (Panasenko et al. 2006). An obvious question is, since Egd1p can exist in the form of homodimers, why is it unstable in the absence of Egd2p? In other words, what is specific in the structure of Egd2p that stabilizes its partner? Egd2p contains a UBA domain, and this has interesting implications. UBA domains are found in very diverse proteins involved in protein degradation, cell cycle control, and DNA repair and have been shown to bind ubiquitin or polyubiquitin. NMR studies have revealed that UBA domains interact via a hydrophobic patch within ubiquitin (Mueller et al. 2004). Such a solvent-exposed hydrophobic patch is also present on the aeNAC UBA domain, and the hydrophobic residues are conserved in all NAC homologs (Spreter et al. 2005). In the absence of its UBA domain, Egd2p is very unstable (Panasenko et al. 2006), although the truncated protein is also expected to be able to homodimerize because the UBA domain is structurally separate from the NAC domain (Spreter et al. 2005). This protein can be stabilized if it is fused to an N-terminal tag such as GFP or B42-HA (Panasenko et al. 2006). Nevertheless, even if yeast cells express stable UBA-less Egd2p, Egd1p, particularly non-ubiquitinated Egd1p, is unstable. This observation suggests that the UBA domain of Egd2p is involved in Egd1p stabilization directly, and not only via stabilization of Egd2p itself.
Recently, it was found that in the fission yeast Schizosaccharomyces pombe αNAC did not interact with monoubiquitin or with K48-linked ubiquitin chains in vitro (Andersen et al. 2007). The same results were obtained earlier for the baker's yeast Egd2p UBA domain (Raasi et al. 2005). All these data were obtained in vitro, so it remains possible that the UBA domain of Egd2p can interact with ubiquitinated Egd1p or maybe with ubiquitinated Egd2p itself, within the EGD complex, whose structure might favor such an interaction. Heterodimerization of Egd1p and Egd2p might subsequently protect the complex from degradation. The surprise was the finding that the UBA domain of Egd2p is particularly important for Egd1p stability if Egd1p is not ubiquitinated. However, we observed that, in this case, ubiquitination of Egd2p increases, but much less so in the absence of the UBA domain. It is possible that the Egd2p UBA domain can bind either ubiquitinated Egd1p or Egd2p (if and when Egd1p is not ubiquitinated) to stabilize ubiquitination and thus the EGD complex.
The demonstration of the importance of the Egd2p UBA domain for the stability of the EGD complex is exciting. Indeed, while a number of ubiquitin-binding domains have now been described, there are still few examples of the role of these domains in vivo. The UBL/UBA domain protein Rad23p contributes to shuttling polyubiquitinated substrates to the proteasome, and the Rad23p UBA domains might compete with the proteasome for these substrates, thereby inhibiting ubiquitin-mediated degradation and stabilizing proteins (Raasi and Pickart 2003). Such a protective role for UBA domains has been shown for several other yeast proteins, namely Dsk2p, Ddi1p, and Ede1p (Heessen et al. 2005). Our work is clearly in line with these published results, except that it additionally suggests that the Egd2p UBA domain is required not only for Egd2p stability, but also for the stability of its interacting partner, Egd1p.
Ubiquitination and Egd2p support Egd1p association with the ribosome:
EGD was originally characterized as the first ribosome-associated protein to contact the emerging, not yet properly folded, polypeptide chains and to contribute to fidelity in cotranslational processes such as targeting and folding (Wiedmann et al. 1994). Only the Egd1p subunit contacts the ribosome directly, while both subunits of EGD interact with newly synthesized polypeptide chains (Wiedmann et al. 1994; Beatrix et al. 2000). In this work, we found that efficient Egd1p ribosome association is dependent upon Not4p, Egd2p, and its own ubiquitination. These results were considered in light of a modeling of the EGD heterodimer interaction with ribosomal protein Rpl25p. According to our model, the N terminus of Egd1p is in direct contact with Rpl25p in agreement with earlier data (Beatrix et al. 2000). However, Egd2p contributes with Egd1p to form a conformation suitable for ribosome binding, which might be similar to the described association of prokaryotic TF with the ribosome (Kramer et al. 2002). Our experimental data revealing that Rpl25p is co-immunoprecipitated with non-ubiquitinated Egd1p less efficiently in the absence of Egd2p are in good agreement with this model. Concerning the role of ubiquitination, our model suggests that lysines K29 and K30 are the most exposed residues of the ribosome-binding loop of Egd1p, and therefore they could indeed be ubiquitinated. Moreover, interactions between Rpl25p and ubiquitin are likely to occur if ubiquitin is C-terminally linked to either K29 or K30 and might stabilize the interaction of the EGD complex with the ribosome.
Obviously, our results are intriguing in light of the knowledge that EGD can bind Rpl25p in vitro in the absence of any ubiquitination (Wegrzyn et al. 2006), and, furthermore, that in cell extracts only a small portion of Egd1p seems to be ubiquitinated, yet binding of EGD to the ribosome has been reported to be stoichiometric. While we have no definitive explanations to answer these questions, it is likely that, in vivo, binding to the ribosome is more complex and competitive than in the experiments performed in vitro, where only the two binding partners were put together, and thus it is not unreasonable to suggest that additional mechanisms contribute to binding in vivo. Concerning the efficient binding to ribosomes in vivo and the relatively small percentage of visible ubiquitinated Egd1p in total extracts, one can imagine that the contribution of ubiquitination is transient upon initial binding yet is not needed subsequently. But it is also likely that Egd1p in vivo is ubiquitinated to higher extents than visible in total extracts and simply becomes rapidly deubiquitinated upon cell lysis. Indeed, it is generally the case that ubiquitinated proteins are modified only to a reduced degree when analyzed in cell extracts. We observed that although EGD ubiquitination increases as cells grow from exponential phase to saturation, no difference in ribosome association could be measured (our unpublished results), revealing, not surprisingly, that ubiquitination is not the only determining factor for the level of EGD ribosome association. In this context, we found that the mutation of lysines 10, 13, 29, and 30 to positively charged arginine had only a weak effect on Egd1p ribosome association when Egd2p was still present, while former work (Wegrzyn et al. 2006) showed that the mutation K29,30A disrupted ribosome association. This leads us to suggest that, in addition, a positive charge in this region of Egd1p might contribute to ribosome binding.
In any event, taken together, our results lead us to suppose that there are two factors important for interaction of EGD with the ribosome: ubiquitination of the ribosome-binding loop of Egd1p and Egd2p. The role of Egd2p may be related to its capacity to be ubiquitinated if Egd1p is not, since the ribosome association of Egd1p is disrupted in cells expressing mutants of Egd1p and Egd2p that both have reduced ubiquitination or in cells lacking Not4p where ubiquitination of both Egd1p and Egd2p is lost.
The importance of ubiquitination for EGD ribosome association is interesting. Indeed, originally linked to protein degradation by the proteasome, it is now clear that, in cells, ubiquitination serves as a reversible modification, similarly to phosphorylation, by the concerted action of enzymes that place and remove the modification. Many functions have been attributed to ubiquitination. Indeed, ubiquitin is a particularly versatile modification because of its ability to modify proteins in a monomeric form or to be conjugated to preceding ubiquitin moieties and thus form many types of ubiquitin chains, thereby creating a great variety of molecular signals in the cell (for review, see Ikeda and Dikic 2008). A large number of ubiquitin-binding domains have been identified, some of which bind preferentially to ubiquitin chains with a linkage specificity, whereas others are more promiscuous. The binding affinity between ubiquitin-binding domains and ubiquitin is generally relatively low for a physiological interaction, suggesting that physiological interactions between ubiquitinated substrates and ubiquitin-binding proteins are likely to be mediated by multivalent interactions. This might be the case for the interaction revealed in this study since EGD is able to bind Rpl25p in vitro in the absence of any ubiquitination, which becomes relevant in vivo. Understanding exactly how ubiquitination of both Egd1p and Egd2p contribute to this binding in vivo is certainly an exciting challenge for future studies.
A function for EGD linking the ribosome and the proteasome:
Our study also revealed an interaction between Egd2p and the proteasome. This finding can be considered in light of the demonstrated cotranslational ubiquitination (Schubert et al. 2000; Turner and Varshavsky 2000), which implies the existence of a ribosome-associated ubiquitin ligase. Ubiquitination of EGD maybe connected in some way to ubiquitination of newly synthesized polypeptide chains on the ribosome or to the transport of these chains to the proteasome. In this context, first, Not4p, which is an E3 ubiquitin ligase that interacts with EGD and can ubiquitinate it in vitro, has been shown to interact with the proteasome (Laribee et al. 2007). Second, Ubc4p, which works as an E2 enzyme together with Not4p to ubiquitinate EGD, has also been shown to associate with the proteasome in response to translationally damaged proteins (Chuang and Madura 2005). Thus one can imagine a possible new function of EGD, which involves connecting events occurring at the ribosome with the proteasome. Maybe Egd2p can be recruited to the proteasome via its UBA domain, as in the case of Rad23p (Raasi and Pickart 2003). Interestingly also, the efficient interaction of Egd2p with the proteasome required Not4p and probably therefore the Ccr4-Not complex. Clearly understanding the coordination of protein synthesis with protein degradation, and the role of the EGD and Ccr4-Not complexes in this coordination, opens intriguing new areas of investigation in the future.
Acknowledgments
We thank Bruno André for the plasmid-expressing His6-ubiquitin and Elke Deuerling for antibodies against Rpl25p. We thank Olivier Michielin for a critical reading of the manuscript. This work was supported by grant 3100AO-100793 from the National Science Foundation to M.A.C.
References
- Akhouayri, O., I. Quelo and R. St-Arnaud, 2005. Sequence-specific DNA binding by the alphaNAC coactivator is required for potentiation of c-Jun-dependent transcription of the osteocalcin gene. Mol. Cell. Biol. 25 3452–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert, T. K., H. Hanzawa, Y. I. Legtenberg, M. J. de Ruwe, F. A. van den Heuvel et al., 2002. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 21 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, K. M., C. A. Semple and R. Hartmann-Petersen, 2007. Characterisation of the nascent polypeptide-associated complex in fission yeast. Mol. Biol. Rep. 34 275–281. [DOI] [PubMed] [Google Scholar]
- Beatrix, B., H. Sakai and M. Wiedmann, 2000. The alpha and beta subunit of the nascent polypeptide-associated complex have distinct functions. J. Biol. Chem. 275 37838–37845. [DOI] [PubMed] [Google Scholar]
- Bloss, T. A., E. S. Witze and J. H. Rothman, 2003. Suppression of CED-3-independent apoptosis by mitochondrial betaNAC in Caenorhabditis elegans. Nature 424 1066–1071. [DOI] [PubMed] [Google Scholar]
- Bukau, B., E. Deuerling, C. Pfund and E. A. Craig, 2000. Getting newly synthesized proteins into shape. Cell 101 119–122. [DOI] [PubMed] [Google Scholar]
- Chuang, S. M., and K. Madura, 2005. Saccharomyces cerevisiae Ub-conjugating enzyme Ubc4 binds the proteasome in the presence of translationally damaged proteins. Genetics 171 1477–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart, M. A., and K. Struhl, 1994. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 8 525–537. [DOI] [PubMed] [Google Scholar]
- Deng, J. M., and R. R. Behringer, 1995. An insertional mutation in the BTF3 transcription factor gene leads to an early postimplantation lethality in mice. Transgenic Res. 4 264–269. [DOI] [PubMed] [Google Scholar]
- Funfschilling, U., and S. Rospert, 1999. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell 10 3289–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi, M., H. Lilie, U. Funfschilling, A. Mun, S. Ross et al., 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. USA 98 3762–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin, A. C., P. Aloy, P. Grandi, R. Krause, M. Boesche et al., 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440 631–636. [DOI] [PubMed] [Google Scholar]
- George, R., T. Beddoe, K. Landl and T. Lithgow, 1998. The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc. Natl. Acad. Sci. USA 95 2296–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex, N., and M. C. Peitsch, 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hartl, F. U., and M. Hayer-Hartl, 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295 1852–1858. [DOI] [PubMed] [Google Scholar]
- Heessen, S., M. G. Masucci and N. P. Dantuma, 2005. The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Mol. Cell 18 225–235. [DOI] [PubMed] [Google Scholar]
- Hundley, H., H. Eisenman, W. Walter, T. Evans, Y. Hotokezaka et al., 2002. The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc. Natl. Acad. Sci. USA 99 4203–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley, H. A., W. Walter, S. Bairstow and E. A. Craig, 2005. Human Mpp11 J protein: ribosome-tethered molecular chaperones are ubiquitous. Science 308 1032–1034. [DOI] [PubMed] [Google Scholar]
- Ikeda, F., and I. Dikic, 2008. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 9 536–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, G., C. Wu, B. Schade, D. Y. Thomas and M. Whiteway, 2005. Drag&Drop cloning in yeast. Gene 344 43–51. [DOI] [PubMed] [Google Scholar]
- Jones, D. T., 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292 195–202. [DOI] [PubMed] [Google Scholar]
- Kramer, G., T. Rauch, W. Rist, S. Vorderwulbecke, H. Patzelt et al., 2002. L23 protein functions as a chaperone docking site on the ribosome. Nature 419 171–174. [DOI] [PubMed] [Google Scholar]
- Laribee, R. N., Y. Shibata, D. P. Mersman, S. R. Collins, P. Kemmeren et al., 2007. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proc. Natl. Acad. Sci. USA 104 5836–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring, B., H. Sakai, G. Kreibich and M. Wiedmann, 1995. Nascent polypeptide-associated complex protein prevents mistargeting of nascent chains to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 92 5411–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett, D. S., M. H. Glickman and D. Finley, 2005. Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol. Biol. 301 57–70. [DOI] [PubMed] [Google Scholar]
- Lenssen, E., N. James, I. Pedruzzi, F. Dubouloz, E. Cameroni et al., 2005. The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation—via a newly identified Glc7/Bud14 type I protein phosphatase module—and TFIID promoter distribution. Mol. Cell. Biol. 25 488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenssen, E., N. Azzouz, A. Michel, E. Landrieux and M. A. Collart, 2007. The Ccr4-not complex regulates Skn7 through Srb10 kinase. Eukaryot. Cell 6 2251–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Lopez, S., L. Stuhl, S. Fichelson, A. Dubart-Kupperschmitt, R. St. Arnaud et al., 2005. NACA is a positive regulator of human erythroid-cell differentiation. J. Cell Sci. 118 1595–1605. [DOI] [PubMed] [Google Scholar]
- Markesich, D. C., K. M. Gajewski, M. E. Nazimiec and K. Beckingham, 2000. bicaudal encodes the Drosophila beta NAC homolog, a component of the ribosomal translational machinery*. Development 127 559–572. [DOI] [PubMed] [Google Scholar]
- McGuffin, L. J., K. Bryson and D. T. Jones, 2000. The PSIPRED protein structure prediction server. Bioinformatics 16 404–405. [DOI] [PubMed] [Google Scholar]
- Melo, D. R., J. L. Lipsztein, C. A. Oliveira, D. L. Lundgren, B. A. Muggenburg et al., 1997. A biokinetic model for 137Cs. Health Phys. 73 320–332. [DOI] [PubMed] [Google Scholar]
- Moller, I., B. Beatrix, G. Kreibich, H. Sakai, B. Lauring et al., 1998. Unregulated exposure of the ribosomal M-site caused by NAC depletion results in delivery of non-secretory polypeptides to the Sec61 complex. FEBS Lett. 441 1–5. [DOI] [PubMed] [Google Scholar]
- Mueller, T. D., M. Kamionka and J. Feigon, 2004. Specificity of the interaction between ubiquitin-associated domains and ubiquitin. J. Biol. Chem. 279 11926–11936. [DOI] [PubMed] [Google Scholar]
- Otto, H., C. Conz, P. Maier, T. Wolfle, C. K. Suzuki et al., 2005. The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc. Natl. Acad. Sci. USA 102 10064–10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasenko, O., E. Landrieux, M. Feuermann, A. Finka, N. Paquet et al., 2006. The yeast Ccr4-Not complex controls ubiquitination of the nascent-associated polypeptide (NAC-EGD) complex. J. Biol. Chem. 281 31389–31398. [DOI] [PubMed] [Google Scholar]
- Peters, J. M., W. W. Franke and J. A. Kleinschmidt, 1994. Distinct 19 S and 20 S subcomplexes of the 26 S proteasome and their distribution in the nucleus and the cytoplasm. J. Biol. Chem. 269 7709–7718. [PubMed] [Google Scholar]
- Quelo, I., M. Hurtubise and R. St-Arnaud, 2002. alphaNAC requires an interaction with c-Jun to exert its transcriptional coactivation. Gene Expr. 10 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quelo, I., C. Gauthier and R. St-Arnaud, 2005. Casein kinase II phosphorylation regulates alphaNAC subcellular localization and transcriptional coactivating activity. Gene Expr. 12 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raasi, S., and C. M. Pickart, 2003. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J. Biol. Chem. 278 8951–8959. [DOI] [PubMed] [Google Scholar]
- Raasi, S., R. Varadan, D. Fushman and C. M. Pickart, 2005. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct. Mol. Biol. 12 708–714. [DOI] [PubMed] [Google Scholar]
- Reimann, B., J. Bradsher, J. Franke, E. Hartmann, M. Wiedmann et al., 1999. Initial characterization of the nascent polypeptide-associated complex in yeast. Yeast 15 397–407. [DOI] [PubMed] [Google Scholar]
- Ritchie, D. W., and G. J. Kemp, 2000. Protein docking using spherical polar Fourier correlations. Proteins 39 178–194. [PubMed] [Google Scholar]
- Rospert, S., Y. Dubaquie and M. Gautschi, 2002. Nascent-polypeptide-associated complex. Cell. Mol. Life Sci. 59 1632–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali, A., and T. L. Blundell, 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234 779–815. [DOI] [PubMed] [Google Scholar]
- Schlunzen, F., D. N. Wilson, P. Tian, J. M. Harms, S. J. McInnes et al., 2005. The binding mode of the trigger factor on the ribosome: implications for protein folding and SRP interaction. Structure 13 1685–1694. [DOI] [PubMed] [Google Scholar]
- Schubert, U., L. C. Anton, J. Gibbs, C. C. Norbury, J. W. Yewdell et al., 2000. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404 770–774. [DOI] [PubMed] [Google Scholar]
- Spahn, C. M., E. Jan, A. Mulder, R. A. Grassucci, P. Sarnow et al., 2004. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell 118 465–475. [DOI] [PubMed] [Google Scholar]
- Spreter, T., M. Pech and B. Beatrix, 2005. The crystal structure of archaeal nascent polypeptide-associated complex (NAC) reveals a unique fold and the presence of a ubiquitin-associated domain. J. Biol. Chem. 280 15849–15854. [DOI] [PubMed] [Google Scholar]
- Turner, G. C., and A. Varshavsky, 2000. Detecting and measuring cotranslational protein degradation in vivo. Science 289 2117–2120. [DOI] [PubMed] [Google Scholar]
- Wang, S., H. Sakai and M. Wiedmann, 1995. NAC covers ribosome-associated nascent chains thereby forming a protective environment for regions of nascent chains just emerging from the peptidyl transferase center. J. Cell Biol. 130 519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn, R. D., D. Hofmann, F. Merz, R. Nikolay, T. Rauch et al., 2006. A conserved motif is prerequisite for the interaction of NAC with ribosomal protein L23 and nascent chains. J. Biol. Chem. 281 2847–2857. [DOI] [PubMed] [Google Scholar]
- Wiedmann, B., H. Sakai, T. A. Davis and M. Wiedmann, 1994. A protein complex required for signal-sequence-specific sorting and translocation. Nature 370 434–440. [DOI] [PubMed] [Google Scholar]
- Zheng, X. M., V. Moncollin, J. M. Egly and P. Chambon, 1987. A general transcription factor forms a stable complex with RNA polymerase B (II). Cell 50 361–368. [DOI] [PubMed] [Google Scholar]
- Zheng, X. M., D. Black, P. Chambon and J. M. Egly, 1990. Sequencing and expression of complementary DNA for the general transcription factor BTF3. Nature 344 556–559. [DOI] [PubMed] [Google Scholar]