Abstract
Cell–cell fusion is essential for a variety of developmental steps in many eukaryotic organisms, during both fertilization and vegetative cell growth. Although the molecular mechanisms associated with intracellular membrane fusion are well characterized, the molecular mechanisms of plasma membrane merger between cells are poorly understood. In the filamentous fungus Neurospora crassa, cell fusion events occur during both vegetative and sexual stages of its life cycle, thus making it an attractive model for studying the molecular basis of cell fusion during vegetative growth vs. sexual reproduction. In the unicellular yeast Saccharomyces cerevisiae, one of the few proteins implicated in plasma membrane merger during mating is Prm1p; prm1Δ mutants show an ∼50% reduction in mating cell fusion. Here we report on the role of the PRM1 homolog in N. crassa. N. crassa strains with deletions of a Prm1-like gene (Prm1) showed an ∼50% reduction in both vegetative and sexual cell fusion events, suggesting that PRM1 is part of the general cell fusion machinery. However, unlike S. cerevisiae, N. crassa strains carrying a Prm1 deletion exhibited complete sterility as either a male or female mating partner, a phenotype that was not complemented in a heterokaryon with wild type (WT). Crosses with ΔPrm1 strains were blocked early in sexual development, well before development of ascogenous hyphae. The ΔPrm1 sexual defect in N. crassa was not suppressed by mutations in Sad-1, which is required for meiotic silencing of unpaired DNA (MSUD). However, mutations in Sad-1 increased the number of progeny obtained in crosses with a ΔPrm1 (Prm1-gfp) complemented strain. These data indicate multiple roles for PRM1 during sexual development.
CELL–cell fusion is an essential part of the development of most eukaryotic organisms. Cell fusion events are required during sexual development in eukaryotic species, including fusion between egg and sperm, mating between two uninucleate yeast cells of different mating type, and fusion between pollen and female gametophytes in plant cells (Dresselhaus 2006; Chen et al. 2007). Cell fusion is also important in developmental processes during vegetative growth (Chen et al. 2007), such as fusion between myoblasts during muscle development, the formation of multinucleate osteoclasts involved in bone readsorption, epidermal and vulva development in Caenorhabditis elegans, the formation of multinucleate plasmodia in the slime mold Plasmodium polycephalum, and the formation of the mycelial network characteristic of filamentous fungi. Despite the diverse role of cell–cell fusion in different species, fusion events require similar cellular processes. These include competency and recognition of a fusion partner, adhesion of fusing cells, and plasma membrane merger. Of these, the molecular mechanisms associated with plasma membrane merger are the least understood.
The filamentous fungus Neurospora crassa provides an excellent model system to study cell fusion mechanisms. Cell fusion occurs during vegetative growth, mating, and sexual development, allowing a comparison of the molecular machinery associated with both vegetative and sexual cell fusion events. The organism is haploid, genetically tractable, has a completed and well-annotated genome sequence, and approximately two-thirds of a full genome deletion strain set is available (Davis 2000; Galagan et al. 2003; Dunlap et al. 2007). During vegetative growth in N. crassa, germinating spores (conidia) of identical genotype show mutual attraction followed by cell (germling) fusion (Köhler 1930; Roca et al. 2005), thus forming a meshwork-like multinucleate syncytium, which develops into a mycelial colony. Mature colonies also undergo fusion between hyphal branches within the interior of the colony (hyphal fusion) (Buller 1933; Hickey et al. 2002; Glass et al. 2004) or even between separate colonies of the same or different genotype (Glass and Dementhon 2006).
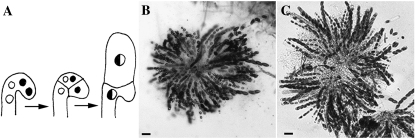
During sexual development in N. crassa, specialized hyphae (trichogynes) emerge from prefruiting bodies (protoperithecia) and recognize pheromones secreted by spores of the opposite mating type (Raju 1980; Bistis 1981; Kim and Borkovich 2004, 2006). Cell fusion between trichogynes and a male cell of the opposite mating type (microconidia, conidia, or hyphae) is followed by migration of the male nucleus through the trichogyne and into the protoperithecium. Approximately 4 days after fertilization, fertile ascogenous hyphae are observed (Raju 1980). Within the ascogenous hyphae, two nuclei of opposite mating type migrate into a hook-shaped crozier; a cell fusion event then occurs between the first and third cells of the crozier during ascus development (see Figure 10A). Asci develop from the second cell of the crozier, in which nuclei of opposite mating type undergo karyogamy and meiosis. Thus, crozier fusion represents a fourth cell fusion event that occurs during the life cycle of N. crassa.
Figure 10.—
(A) Cartoon showing crozier formation and karyogamy in N. crassa. Karyogamy occurs in the penultimate cell, which becomes the ascus mother cell. Cell fusion occurs between the first and third compartments of the crozier within a similar time frame to karyogamy in the penultimate cell. Adapted from Raju (1980). (B) Rosette of asci in a WT × WT cross (FGSC 988 × FGSC 2489). (C) Rosette of asci from a Sad-1 × ΔPrm1 (Prm1-gfp) cross [his-3 Sad-1 (RIP141); mep a (R16-20) × his-3∷ccg1-Prm1-gfp; ΔPrm1∷hph; Δmus-51∷bar A (A24)]. Note that the number of asci formed in the Sad-1 × ΔPrm1 (Prm1-gfp) cross is comparable to a WT cross. Bars, 50 μm.
One of the most intensively studied cell fusion pathways is that of mating cell fusion in the budding yeast Saccharomyces cerevisiae. Although mechanisms mediating cell communication, signal transduction, and polarization during mating are very well understood in S. cerevisiae, only a few proteins have been identified to be involved in membrane merger. These include Fus1p, Fus2p, Fig1p, and Prm1p. Prm1p and Fig1p are implicated in formation of the fusion pore, while Fus1p and Fus2p localize to the expanding fusion pore (Elion et al. 1995; Erdman et al. 1998; Heiman and Walter 2000; Proszynski et al. 2006; Heiman et al. 2007; Paterson et al. 2008). Analysis of the conservation of genes involved in mating cell fusion in S. cerevisiae and N. crassa reveal that most of the components of the signal transduction/polarization pathway are highly conserved (Glass et al. 2004). In fact, mutations in N. crassa genes encoding the orthologs of the S. cerevisiae pheromone response MAP kinase pathway (STE11, STE7, and FUS3) result in strains that are defective in germling and hyphal fusion (Pandey et al. 2004; Maerz et al. 2008). So far, homologs of S. cerevisiae genes involved in membrane merger, including FUS1, FUS2, and FIG1 have not been identified in the N. crassa genome (Glass et al. 2004). However, a putative N. crassa ortholog of PRM1 is present.
In S. cerevisiae, the PRM1 locus encodes a multispan transmembrane protein which is exclusively expressed in the presence of mating pheromone (Heiman and Walter 2000). Approximately 50% of prm1Δ mating pairs do not fuse; they arrest at a stage after cell wall degradation as prezygotes, indicating defects in plasma membrane merger. In addition, ∼20–40% of these nonfusing pairs lyse (Jin et al. 2004; Aguilar et al. 2007). prm1Δ mating pairs that are able to complete mating undergo normal karyogamy, meiosis, and ascosporogenesis. Analysis of the subcellular localization of Prm1p using green fluorescent protein (GFP) fusion constructs revealed that Prm1p localizes to the endoplasmic reticulum before concentrating at shmoo tips; Prm1p also accumulates at the point of cell contact in fusion pairs (Heiman and Walter 2000; Jin et al. 2008). Although Prm1p has been implicated in either formation or stabilization of the fusion pore during mating in S. cerevisiae, much of its role in this process remains obscure.
To determine whether the fusion functions of PRM1 homologs are conserved among fungi and whether Prm1p plays a general role in cell fusion events, we analyzed the role of the PRM1 homolog in N. crassa during vegetative fusion events, including germling and hyphal fusion, as well as its role during fertilization and sexual development. We found that N. crassa strains containing a deletion of the Prm1-like gene (Prm1) exhibited a cell fusion defect during vegetative fusion as well as during mating cell fusion, comparable to that of the S. cerevisiae prm1Δ mutant. However, additional sexual defects were observed in the N. crassa ΔPrm1 mutants. Surprisingly, although mating cell fusion was only reduced by ∼50%, crosses between wild type (WT) and ΔPrm1 were completely sterile, even when ΔPrm1 strains were used as a male. The dominant male and female sterility phenotype of ΔPrm1 mutants in N. crassa was not suppressed by mutations in the meiotic gene silencing pathway (MSUD). Our analysis revealed functions for PRM1 during sexual development that occur postfertilization, indicating either novel functions for PRM1 or a heretofore uncharacterized cell fusion event(s) required early in sexual development in N. crassa.
MATERIALS AND METHODS
N. crassa strains and growth media:
Strains used in this study are listed in Table 1. Strains were grown on Vogel's minimal medium (Vogel 1956). For strains carrying auxotrophic markers, required supplements were added to the medium. Crosses were performed on Westergaard's medium (Westergaard and Mitchell 1947). If strains carrying auxotrophic markers were used as females, a heterokaryon between the helper strain Fungal Genetics Stock Center (FGSC) 4564 (Table 1) and the auxotrophic strain was used in crosses (Perkins 1984). To test the mating type of strains, they were crossed with mating type tester strains fl a (FGSC 4347) and fl A (FGSC 4317) (Table 1).
TABLE 1.
Strains used in this study
| Strain | Genotype | Origin |
|---|---|---|
| A7 | his-3∷ccg1-gfp; ΔPrm1∷hph; Δmus-51∷bar A | This study |
| A8 | his-3∷ccg1-dsRed; ΔPrm1∷hph; Δmus-51∷bar A | This study |
| A9 | his-3∷ccg1-gfp A | This study |
| A10 | his-3∷ccg1-dsRed A | This study |
| A20 | his-3∷ccg1-dsRed; Δmus-51∷bar A | This study |
| A21 | his-3∷ccg1-HI-gfp; ΔPrm1∷hph; Δmus-51∷bar A | This study |
| A24 | his-3∷ccg1-Prm1-gfp; ΔPrm1∷hph; Δmus-51∷bar A | This study |
| A26 | his-3∷ccg1-gfp-Prm1; ΔPrm1∷hph; Δmus-51∷bar A | This study |
| A29 | ΔPrm1∷hph a | This study |
| A32 | ΔPrm1∷hph A | This study |
| FGSC 988 | Oak Ridge WT a | FGSC |
| FGSC 2489 | Oak Ridge WT A | FGSC |
| FGSC 4317 | fl A | FGSC |
| FGSC 4347 | fl a | FGSC |
| FGSC 4564 | ad-3B cyh-1 am1 | FGSC |
| FGSC 9717 | his-3; Δmus-51∷bar A | Colot et al. (2006) |
| R11-03 | his-3∷ccg1-H1-gfp A | Gift from David Jacobson |
| R16-19 | his-3 Sad-1 (RIP78); mep A | Gift from Patrick Shiu |
| R16-20 | his-3 Sad-1 (RIP141); mep a | Gift from Patrick Shiu |
| R16-51 | his-3; ΔPrm1∷hph; Δmus-51∷bar A | This study |
| R16-53 | his-3; ΔPrm1∷hph; Δmus-51∷bar A | This study |
| R21-04 | his-3∷Prm1-gfp A | This study |
Construction of N. crassa ΔPrm1 mutant:
N. crassa ΔPrm1 mutants were constructed using the “Neurospora Knockout Strain Kit” from the FGSC (as described in http://www.dartmouth.edu/∼neurosporagenome/) (Colot et al. 2006). The transformation cassette was assembled using yeast recombinational cloning (Colot et al. 2006); primer sequence information was provided by the Neurospora Functional Genomics Project (http://www.dartmouth.edu/∼neurosporagenome/). N. crassa strain FGSC 9717 was transformed with the replacement cassette by electroporation of macroconidia (Margolin et al. 1997; R. L. Metzenberg and K. Black, personal communication). The obtained transformants were tested for homologous single-copy integration of the replacement cassette by using Southern blot hybridization analysis. Homokaryotic ΔPrm1 strains were obtained from primary transformants through single spore isolation. Macroconidia were spread on Brockman de Serres medium (Brockman and de Serres 1963) containing 200 μg/ml hygromycin. After an overnight incubation at 30° colonies derived from single spores were transferred to slant tubes containing minimal medium (MM) with 200 μg/ml hygromycin. The described purification was repeated at least three times for each transformant. The homokaryotic state of the final isolates was confirmed by using PCR analysis using Prm1- and hygB-specific primers.
Deconvolution microscopy:
Growth media for microscopy was either Vogel's minimal media (Vogel 1956) or minimal medium containing 1.2% sodium acetate as the sole carbon source. Germling samples were grown for 3–4 hr at 30° until points of fusion could be observed. Hyphal samples were grown overnight at 30° and squares of agar were cut from the edge of the colony where hyphal density was low and fusion between hyphae was observed. All samples were observed on an inverted Olympus IX70 deconvolution microscope using an Olympus 100× PlanApo oil immersion objective and using the DeltaVision Spectris system. Images were captured with a Photometrics CH350 liquid-cooled camera and taken in stacks of 20–30 with exposures from 0.5 to 2 sec, depending on GFP intensity. All image stacks selected for deconvolution showed normal pixel intensity distributions with a difference between minimal and maximal intensity of at least 800. Image stacks were deconvolved using Huygens deconvolution software, Scientific Volume Imaging (SVI), in classic mode with up to 100 possible iterations.
Electron microscopy:
For electron microscopy, 300 μl of ΔPrm1 conidia (A7 + A8; Table 1) or 300 μl of WT conidia (A9 + A10; Table 1) at a concentration of 5 × 107 cells/ml were spread on minimal media plates and allowed to undergo germling fusion at 30° for 3–4 hr. EM fix (1% glutaraldehyde, 0.2% paraformaldehyde, 0.04 m KPO4, pH 7) was added to the plates for 5 min. Cells were subsequently scraped off the plates and incubated in EM fix on ice for 50 min. Cells were washed twice with 0.9% NaCl, once with water, and once with 2% KMnO4 for 5 min each. Cells were subsequently incubated in 2% KMnO4 for 45 min at room temperature. Samples were then dehydrated through graded ethanol series in a Lecia automatic freeze substitution unit. Samples were incubated for 30 min at each ethanol concentration from 50 to 100%. Temperature was decreased by 10° each cycle until a minimum of −50°. Samples were subsequently infiltrated with epon/araldite resin. Resin was added at the following concentrations for 2 hr each: 3:1, 1:1, and 1:3 (acetone:resin). Pure resin was then added and incubated overnight. The following day, fresh pure resin was added along with accelerant and incubated for 4 hr. Samples were embedded in molds for 72 hr. Sections of 60–70 nm were cut on a Leica microtome and stained with 2% methanol uranyl acetate and lead citrate. Samples were imaged under an FEI Tecnai 12 transmission electron microscope at 100 kV.
Crozier staining:
Crozier fixation was performed according to Barry and Perkins (http://www.fgsc.net/neurosporaprotocols/How%20to%20prepare%20aceto-orcein%20squashes,%20especially%20for%20pachytene%20chro.pdf). Samples were grown on Westergaards media (Westergaard and Mitchell 1947) for 7 days, to induce the formation of female reproductive structures (protoperithecia). These plates were subsequently inoculated with 300 μl of conidial suspension from strains of the opposite mating type, to induce perithecial development. Small strips of agar containing perithecia were cut out from the plates at 3, 4, and 7 days postfertilization. These strips were fixed in a solution of EtOH 100%:glacial acetic acid:lactic acid 85% (6:1:1). After fixation, individual perithecia were removed from strips and their contents extruded by pressure with two syringe needles into a drop of DAPI stain (0.5 μg/ml). Perithecial casings were discarded and a coverslip was placed over the DAPI drop. Cells were observed on a Zeiss AxioImager M1 fluorescence microscope with a 40× EC Plan-Neofluor oil immersion objective. Images were taken with a Qimaging 5Mpix MicroPublisher camera.
Fusion assays:
Strains expressing either cytoplasmic GFP or dsRED (see above) were grown on Vogel's minimal media (Vogel 1956) slant tubes for 4–6 days or until significant conidiation occurred. Conidia were harvested by vortexing slant tubes with 2 ml ddH2O. The conidial suspension was filtered by pouring over cheese cloth to remove hyphal fragments. Conidia were diluted to a concentration of ∼2 × 106 conidia/ml. For each sample, 500 μl of spore suspension of both GFP- and dsRED-expressing strains were mixed, and 300 μl of this final mixture were spread on a minimal media plate. The plates were incubated for 3–4 hr at 30°. Squares of 1 cm were excised and observed with a Zeiss Axioskop 2 using a 40× Plan-Neofluor oil immersion objective. Fusion events were counted for all germling pairs that expressed fluorescence in both red and green channels.
Trichogyne fusion assays were performed as described in Fleißner et al. (2005).
RESULTS
Construction and phenotype of a N. crassa ΔPrm1 mutant:
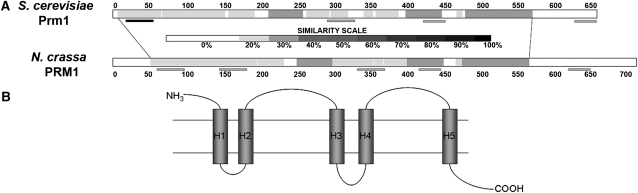
We identified gene NCU09337.3 as a potential N. crassa homolog of PRM1 of S. cerevisiae. The N. crassa Prm1-like gene lacks introns and is predicted to encode a protein of 764 amino acids (aa) in length. The N. crassa PRM1 and S. cerevisiae Prm1p proteins showed 26% identity (E-value = 2e-129). Kyte-Doolittle hydrophobicity analysis showed five hydrophobic regions in the N. crassa PRM1 protein. Further analysis on PRM1 using the transmembrane prediction software TMHMM, TMpred, and SOSUI all supported the presence of five transmembrane helixes in similar locations as well as placement of the N terminus on the outside of the cell (Figure 1B). Prm1p of S. cerevisiae encodes a 661-aa protein; the original publication on Prm1p predicted five TM domains (Heiman and Walter 2000). However, by using TMHMM, we only identified four transmembrane domains in S. cerevisiae Prm1p, one of which may function as a signal peptide, an identical prediction to that provided by the Saccharomyces Genome Database (http://db.yeastgenome.org/cgi-bin/protein/domainPage.pl?dbid=S000005223; Figure 1A). An alignment using ClustalW of the N. crassa PRM1 protein and S. cerevisiae Prm1p showed that four of the five predicted TM domains are conserved. One TM domain in N. crassa PRM1 between amino acids 149–171 is not predicted in Prm1p in S. cerevisiae. This predicted TM domain is conserved among predicted PRM1 homologs in filamentous ascomycete genomes, which also contain five predicted TM domains in similar positions (data not shown). The predicted signal peptide region identified in S. cerevisiae Prm1p is not at the N terminus of the N. crassa PRM1 protein (nor in the predicted PRM1 homologs in other filamentous ascomycete genomes). We therefore evaluated both the start site for Prm1 and whether a cryptic intron might occur in the 5′ region; RT-PCR data confirmed the predicted ORF for Prm1 (Figure 1; data not shown).
Figure 1.—
(A) Comparative similarity between Saccharomyces cerevisiae Prm1p and Neurospora crassa PRM1. Regions of higher similarity are shown as darker on the scale. Shaded bars at the bottom of each sequence represent predicted transmembrane domains, while the solid bar indicates the predicted signal peptide in Prm1p. For N. crassa PRM1, transmembrane domains are represented as H1–H5 in B. (B) Depicts the predicted topology of the five predicted transmembrane helixes in N. crassa PRM1. PRM1 has a predicted extracellular N-terminal region and intracellular C-terminal region.
We predicted that a Prm1 mutant in N. crassa may be affected in cell fusion processes, either during germling/hyphal fusion or during mating. To determine the phenotype of a N. crassa Prm1 deletion mutant, we replaced the entire Prm1 coding sequence with a gene encoding hygromycin phosphotransferase (hph) (see materials and methods). Transformants were tested for homologous integration of the knockout cassette as well as for the absence of additional heterologous integrated fragments using Southern blot hybridization analysis (data not shown). Two homokaryotic ΔPrm1 isolates (R16-51 and R16-53; Table 1) were purified from primary heterokaryotic transformants by single spore isolation.
Macroscopically, all of the ΔPrm1 strains (Table 1) growing on MM slant tubes were indistinguishable from wild-type isolates (FGSC 2489 and FGSC 988) (data not shown). The average growth rate as well as the development of aerial hyphae of the ΔPrm1 mutants was also comparable to wild type. The wild-type strains FGSC 2489 and FGSC 988 grew 8.2 (±0.4) cm and 8.3 (±0.4) cm per day, while a ΔPrm1∷hph a strain (A29; Table 1) grew 8.3 (±0.6) cm and a ΔPrm1∷hph A strain (A32; Table 1) grew 8.1 (±0.9) cm per day. Aerial hyphal growth was 1.0 (± 0.4) cm per day in FGSC 2489, 0.9 (± 0.5) cm in FGSC 988, 1.1 (±0.4) cm in A29, and 1.2 (±0.5) cm in A32. The ΔPrm1 strains A29 and A32 produced 2.0 (± 0.8) × 108 and 1.9 (± 0.2) × 108 asexual spores (conidia) per culture tube, respectively, slightly more than the wild-type strains FGSC 988 and FGSC 2489 [1.0 (± 0.2) × 108 and 1.5 (± 0.3) × 108, respectively]. Together these data indicate that ΔPrm1 mutants exhibited no general growth defects.
In S. cerevisiae, Prm1p contributes to plasma membrane fusion during mating of a- and α-cells, such that homozygous prm1Δ crosses show a ∼50% reduction in mating cell fusion (Heiman and Walter 2000). In S. cerevisiae, mating is the only cell fusion event associated with the life cycle. In contrast, N. crassa undergoes cell fusion during vegetative growth, both between conidial germlings (Roca et al. 2005) and between mature hyphae within a colony (Buller 1933; Hickey et al. 2002). First, we evaluated germination kinetics and chemotropic interactions between ΔPrm1 germlings as compared to WT germlings. The conidial germination rate of wild type (FGSC 2489) vs. the ΔPrm1 (A32) mutant was comparable (WT: 84.9 ± 1.7% vs. ΔPrm1: 88.6 ± 2.2%). Also similar to wild-type germlings, ΔPrm1 germlings exhibited normal chemotropic attraction and directed growth (WT: 71.0 ± 10.7% vs. ΔPrm1: 62.4 ± 4.4%), resulting in normal appearing cell–cell contacts associated with germling fusion (Figure 2, A and B). These data indicate that in N. crassa Prm1 is dispensable for cell–cell communication and directed growth leading to cell–cell contact during germling fusion.
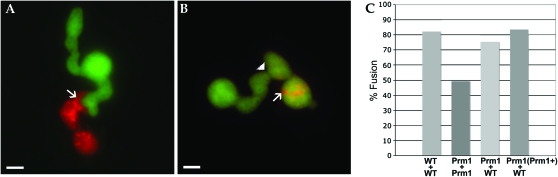
Figure 2.—
(A) A his-3∷gfp; ΔPrm1∷hph (A7) and his-3∷dsRed; ΔPrm1∷hph (A8) germling pair that has remained unfused after 4 hr of interaction. A small membrane invagination can be seen protruding from the GFP-expressing cell into the dsRED-expressing cell (white arrow). (B) A his-3∷gfp; ΔPrm1∷hph (A7) and his-3∷dsRed; ΔPrm1∷hph (A8) germling pair that has undergone fusion (arrowhead) showing mixing of the green cytoplasm of the GFP-expressing cell and red cytoplasm of the dsRED-expressing cell (dsRed accumulates in vacuoles), white arrow; not all fusion events show equal sharing of cytoplasm (Hickey et al. 2002). (C) Graph showing the percentages of germling fusion between two wild-type strains (WT + WT) [his-3∷gfp (A9) + his-3∷dsRed (A10)], between two ΔPrm1 strains (Prm1 + Prm1) [his-3∷gfp; ΔPrm1∷hph (A7) + his-3∷dsRed; ΔPrm1∷hph (A8)], between a ΔPrm1 strain plus a WT strain (Prm1 + WT) [his-3∷gfp; ΔPrm1∷hph (A7) + his-3∷dsRed (A10)] and between a ΔPrm1 complemented strain and WT [Prm1 (Prm1+) + WT] [his-3∷Prm1-gfp; ΔPrm1∷hph (A24) + his-3∷gfp (A9)]. In the A24 strain, fluorescence of the PRM1-GFP was dim and localized to vacuoles as compared to bright cytoplasmic GFP fluorescence in the his-3∷gfp strain; strains were distinguished by both GFP localization and fluorescence intensity. For assessing fusion frequency, only pairs were evaluated that showed this differential fluorescence intensity and localization. Bars, 1 μm.
We next evaluated whether the N. crassa ΔPrm1 mutant showed a cell fusion defect during vegetative growth by comparing the frequency of germling fusion between wild-type conidia vs. between ΔPrm1 conidia. In N. crassa, contact between chemotropic germlings is followed by cell wall breakdown and plasma membrane fusion, which are difficult to unambiguously evaluate using light microscopy. Thus, to obtain quantitative data for cell fusion events in wild type and the ΔPrm1 mutants, we constructed one set of strains (wild type and ΔPrm1, A9 and A7, respectively; Table 1) that expressed cytoplasmic GFP under the regulation of the ccg-1 promoter (McNally and Free 1988; Loros et al. 1989) and a second set of strains (wild type and ΔPrm1, A10 and A8, respectively; Table 1) that expressed cytoplasmic dsRED under the regulation of the ccg-1 promoter.
Conidia from wild-type strains A9 and A10 (Pccg1-gfp and Pccg1-dsRed, respectively) were mixed in an equal ratio and ∼106 conidia were plated onto minimal medium. After 3–4 hr, wild-type fusion pairs consisting of a green and a red fluorescent germling were analyzed. If germlings had undergone fusion, red and green fluorescence could be detected in both cells of the fusion pair. If the pair remained unfused, one germling remained red and the other remained green. We evaluated 107 wild-type pairs (A9/A10) for germling fusion; 82% (88/107) of the wild-type pairs fused after 4 hr of incubation (Figure 2C). In contrast to WT, fusion between ΔPrm1 (Pccg1-gfp; A7) and ΔPrm1 (Pccg1-dsRed; A8) germlings was reduced to 49% (62/126) (Figure 2, A–C). The percentage of germlings fused in the ΔPrm1 (Pccg1-gfp) + ΔPrm1 (Pccg1-dsRed) sample was not significantly increased by extended incubation times (data not shown). In pairings between WT (Pccg1-dsRed) and ΔPrm1 (Pccg1-gfp) (A10/A7; Table 1) germlings, a reduction in fusion frequency was also observed (75%; 83/110) (Figure 2C), suggesting that PRM1 function is required in both germlings of a fusion pair. In S. cerevisiae strains, a slight reduction in the frequency of successful fusion events was also observed in heterozygous matings between WT and prm1 strains (Heiman and Walter 2000). Thus, similar to what was observed in S. cerevisiae, PRM1 contributes to, but is not essential for vegetative germling fusion in N. crassa.
ΔPrm1 mutants are blocked at membrane merger:
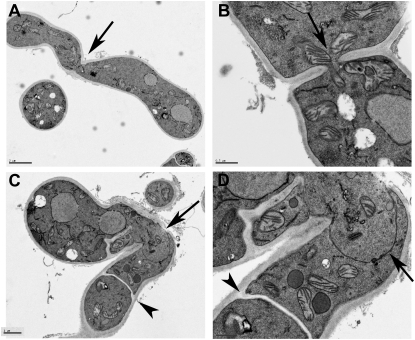
Similar to what was observed in prm1Δ × prm1Δ crosses in S. cerevisiae, we often observed internal invaginations at the fusion site between ΔPrm1 (Pccg1-gfp; A7) and ΔPrm1 (Pccg1-dsRed; A8) germling pairs (Figure 2A; white arrow). These observations suggested that, similar to S. cerevisiae, N. crassa ΔPrm1 mutants are blocked in plasma membrane merger during germling fusion. We therefore compared germling fusion between two WT strains (A9 + A10; his-3∷Pccg-1-gfp A + his-3∷Pccg1-dsRed A, respectively) vs. two ΔPrm1 strains (A7 + A8) by thin section electron microscopy (see materials and methods). As shown in Figure 3B, cell wall dissolution at the point of contact between WT + WT germling fusion pairs was also associated with plasma membrane merger. In no case was naked plasma membrane observed at the fusion point between two WT germling pairs. By contrast, we observed juxtaposed plasma membranes in ΔPrm1 + ΔPrm1 germling fusion pairs in the absence of cell wall material (Figure 3, C and D; arrow), a phenotype remarkably similar to that observed via EM studies of prm1Δ × prm1Δ crosses in S. cerevisiae (Heiman and Walter 2000). These data indicate that ΔPrm1 + ΔPrm1 germling fusion pairs are able to undergo cell wall breakdown, but are often blocked in the subsequent step of plasma membrane merger.
Figure 3.—
(A) Transmission electron micrograph (TEM) of a WT germling pair (A9 + A10; his-3∷Pccg1-gfp A + his-3∷Pccg1-dsRed A) showing a complete fusion event. Arrow shows fusion point. Bar, 2 μm. (B) TEM of pore opening at fusion point between an A9 and A10 conidium. Arrow shows fusion pore with mitochondrion traveling through the pore. Bar, 0.5 μm. (C) TEM of fusion event between ΔPrm1 (Pccg1-gfp; A7) and ΔPrm1 (Pccg1-dsRed; A8) germlings. Arrowhead shows a septum; septation during conidial germination is often associated with germling fusion. Arrow indicates fusion point between two germlings. Bar, 1 μm. (D) Enlargement of fusion area shown in C. Arrowhead shows septum in a conidial germling, while the arrow shows juxtaposed plasma membranes that have remained between the germlings, even after cell wall degradation had occurred at the fusion point. Bar, 0.5 μm.
GFP-tagged PRM1 localizes to the plasma membrane and fusion regions in hyphae:
PRM1 is predicted to be an integral membrane protein. Since mutations in Prm1 reduced germling fusion frequency, we predicted that PRM1 would localize to the point of fusion. In S. cerevisiae, Prm1p-GFP shows endomembrane localization; upon treatment of α-factor, Prm1p showed localization to the shmoo tip and points of cell fusion between a- and α-cells (Heiman and Walter 2000). To test this hypothesis, we constructed a 3′ (R21-04; Table 1) GFP-tagged allele of Prm1 under its native promoter, and both 5′ and 3′ GFP-tagged alleles of Prm1 under the regulation of the ccg-1 promoter (A24/A26). Pccg1 has been routinely used as a heterologous promoter in N. crassa, especially for expression of fluorescently labeled proteins (Freitag et al. 2004). GFP-tagged Prm1 alleles were targeted to the his-3 locus in a ΔPrm1 his-3 strain (R16-51). Full complementation of germling fusion defects was observed with the ΔPrm1 (his-3∷Prm1-gfp) (A24) strain (Figure 2C).
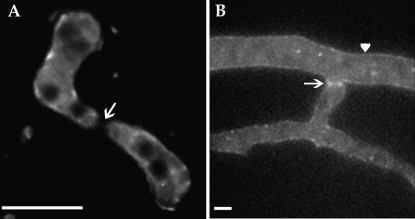
GFP localization during germling fusion was evaluated in all strains bearing Prm1-gfp-tagged alleles using live cell imaging and deconvolution microscopy (see materials and methods). The native promoter Prm1-gfp construct (R21-04; Table 1) displayed extremely low fluorescence intensity in all cell types (data not shown). ΔPrm1 germlings bearing the Pccg1 Prm1-gfp construct (A24; Table 1) revealed only dim cytoplasmic, membrane, and punctate fluorescence; GFP fluorescence was never detected at germling fusion points (Figure 4A, arrow). We therefore evaluated PRM1-GFP fluorescence in vegetative hyphae by inoculating A24 onto MM containing 1.2% sodium acetate as the sole carbon source (which increases expression from the ccg-1 promoter in vegetative hyphae; ccg-1 is regulated by carbon source) (McNally and Free 1988). Low but detectable PRM1-GFP fluorescence was observed within hyphae in punctate bodies and also at the plasma membrane. A higher intensity of GFP fluorescence was detected at points of fusion (Figure 4B, arrow). Thus, the localization of PRM1 in fusion hyphae of N. crassa is consistent both with its prediction as a plasma membrane protein and with its predicted role in membrane fusion.
Figure 4.—
(A) Deconvolution microscopy of germling fusion and localization of PRM1-GFP in ΔPrm1 germling pairs [his-3∷Prm1-gfp; ΔPrm1∷hph (A24)]. PRM1-GFP showed vacuolar/endomembrane and some plasma membrane localization, but lacked localization to the fusion tips (arrow). (B) Strain A24 grown on MM agar containing sodium acetate as the sole carbon source for 4 hr. Examination of hyphae by deconvolution microscopy showed membrane localized GFP fluorescence (arrowhead) with higher intensity GFP fluorescence at points of hyphal fusion (arrow). Bars, 5 μm.
ΔPrm1 mutants show a comparable reduction in trichogyne–conidium fusion during fertilization as during germling fusion:
Many fusion mutants in N. crassa are defective in the production of prefruiting bodies (protoperithecia) that function as female reproductive structures (Pandey et al. 2004; Maerz et al. 2008), suggesting that hyphal fusion contributes to the formation of these structures. We therefore evaluated whether the ΔPrm1 mutants form normal protoperithecia. Strain A29 (ΔPrm1∷hph a) was inoculated onto crossing medium and after 7 days, the ΔPrm1 mutant strain formed protoperithecia that were indistinguishable in structure and size from those of a wild-type strain (FGSC 988). However, the number of protoperithecia produced by the ΔPrm1 mutant was reduced (18 ± 12.6 protoperithecia/cm2) as compared to a wild-type strain of identical mating type (FGSC 988; 34 ± 15.3 protoperithecia/cm2).
In N. crassa, germling fusion occurs between conidia that are genetically identical to each other. In contrast, during fertilization, fusion requires cells to be of opposite mating type. In N. crassa, as in many other heterothallic filamentous ascomycete species, fertilization occurs between a specialized hypha (trichogyne) of one mating type that emanates from the protoperithecium and a male cell of the opposite mating type, which may be a conidium, a microconidium, or even a hypha (Davis 2000). As a result of the fertilization event, the male nucleus migrates through the trichogyne and into the ascogonium within the developing fruiting body (perithecium); karyogamy and meiosis occur in the dikaryotic crozier ∼4 days postfertilization (Raju 1980). In filamentous ascomycete species, the cell biological events associated with the interval between fertilization and formation of the dikaryotic crozier are not well described, primarily due to the difficulty in differentiating the ascogonium/dikaryotic hyphae from maternal tissue.
Since PRM1 is involved in vegetative cell fusion, we hypothesized that cell fusion between a trichogyne and a conidium during fertilization might be affected in the ΔPrm1 mutants. Such a result would indicate that PRM1 is required for both vegetative and sexual fusion, and thus may be an integral part of the fusion machinery. To test this hypothesis, we performed trichogyne fusion assays. Trichogynes have the capacity to sense a spore or a hypha of the opposite mating type via pheromone-receptor signaling, resulting in chemotropic growth toward and subsequent fusion with the mating partner (Bistis 1981; Kim and Borkovich 2004, 2006). In our trichogyne-fusion assay, microconidia containing a single H1-GFP-labeled nucleus were used as the male crossing partner (see materials and methods). Successful fusion between a microconidium and trichogyne of opposite mating type can be detected as a disappearance of the H1-GFP nucleus from the male cell as a consequence of its migration through the trichogyne and into the protoperithecium (Fleißner et al. 2005).
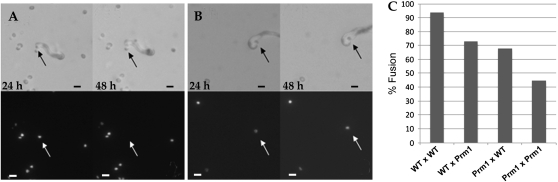
We evaluated >50 trichogyne/microconidium interactions for WT crosses, ΔPrm1 homozygous crosses, and crosses between ΔPrm1 and WT, where ΔPrm1 functioned as either a female or a male. In the WT × WT control crosses (FGSC 988 × R11-03, cross descriptions are always “female × male”), 94% of the trichogyne–microconidium pairs fused (Figure 5, A and C). By contrast, the ΔPrm1 homozygous cross (A29 × A21) showed the large reduction in successful sexual fusions, with only 45% of trichogyne–microconidium pairs undergoing successful fusion events (Figure 5, B and C). In a cross using WT as a female and ΔPrm1 as a male (FGSC 988 × A21), we observed that 73% of the trichogyne–microconidium pairs fused (Figure 5A). When ΔPrm1 was used as the female and crossed with WT (A29 × R11-03), 68% of the trichogyne–microconidium pairs fused; an example of a nonfusion pair is shown in Figure 5B. Thus, similar to defects in cell fusion observed between germling fusion pairs, mutations in Prm1 decrease, but do not abolish, cell fusion during fertilization. The defect in sexual fusion in homozygous ΔPrm1 crosses (45%) is comparable to the fusion defect observed between ΔPrm1 germling pairs (49%). This result is consistent with the hypothesis that PRM1 is a part of the general membrane fusion machinery in N. crassa, without specificity for developmental stage.
Figure 5.—
Trichogyne–conidium fusion assays in wild type and ΔPrm1 mutants. (A) Fertilization between a WT trichogyne (FGSC 988) and a microconidium from strain A21 (his-3∷HI-gfp; ΔPrm1∷hph A) showing transfer of a ΔPrm1 nucleus into a WT trichogyne between 24 and 48 hr. The WT trichogyne can be seen curling around the ΔPrm1 microconidium in the DIC image (solid arrows). (Bottom) GFP fluorescence. The ΔPrm1 nucleus (open arrows) is apparent at 24 hr, but is no longer present after 48 hr. (B) Lack of trichogyne–conidium fusion between an A29 (his-ΔPrm1∷hph a) trichogyne and an WT H1-GFP-tagged microconidium [his-3∷H1-gfp A (R11-03)]. The ΔPrm1 trichogyne can be seen in the DIC image (solid arrows), which has curled around the WT microcondium (top). However, even after 48 hr the H1-GFP-labeled WT nucleus has not been transferred to the ΔPrm1 trichogyne (bottom), indicating a defect in cell fusion during fertilization. (C) Graph showing percentage of trichogyne–conidium fusion events between two WT strains [FGSC 988 a × his-3∷H1-gfp A (R11-03)], vs. between WT and a ΔPrm1 strain [FGSC 988 a × his-3∷HI-gfp; ΔPrm1∷hph A (A21)], a ΔPrm1 strain and WT [ΔPrm1∷hph a (A29) × his-3∷H1-gfp A (R11-03)] and trichogyne–conidium fertilization between two ΔPrm1 strains [ΔPrm1∷hph a (A29) × his-3∷HI-gfp; ΔPrm1∷hph A (A21)]. The reduction in successful trichogyne–conidium fusion events between ΔPrm1 pairs was similar to the reduction observed in germling fusion events (∼50%) (Figure 2). Bars, 10 μm.
Mutations in Prm1 completely block sexual reproduction postfertilization:
A common strategy of purifying homokaryotic strains in N. crassa consists of taking heterokaryotic transformants through a sexual cross. Homokaryotic ascospores are produced following meiosis, thus allowing purification of a gene deletion strain via selection for their selectable marker (such as hygromycin resistance). When the primary heterokaryotic ΔPrm1 deletion transformants were crossed with a wild-type strain (FGSC 988), hygromycin-resistant ascospore progeny were not obtained, indicating that either ΔPrm1 nuclei were unable to pass through a cross or ΔPrm1 ascospores were unable to germinate. To resolve this question, a homokaryotic Prm1 strain (R16-51, purified by macroconidum isolation, see materials and methods; Table 1) was crossed with wild type (FGSC 988). Surprisingly, no ascospore progeny were produced. As shown above, fertilization was observed in 73% of the trichogyne–conidium interactions in a WT × ΔPrm1 cross. These data suggest that PRM1 performs an essential function during sexual reproduction that is required postfertilization.
Many N. crassa mutants that show defects in female fertility and protoperithecial development can be rescued in a heterokaryon with the am1 mutant (FGSC 4564, the helper strain) (Perkins 1984) when such heterokaryons are used as a female in a cross. However, the am1 nuclei cannot participate in karyogamy, meiosis, or the formation of ascospores because the mating-type locus in the am1 strains is inactive (Griffiths and Delange 1978; Staben and Yanofsky 1990). To test if the am1 helper strain could complement the female or male sterility of the ΔPrm1 mutant, a cross between a ΔPrm1 + am1 heterokaryon and WT [(R16-51 + FGSC 4564) × FGSC 988] and a second cross of WT crossed to the ΔPrm1 + am1 heterokaryon FGSC 988 × (R16-51 × FGSC 4564)] were analyzed. Both combinations failed to produce any ascospore progeny (data not shown). However, perithecia were slightly larger and darker in the (ΔPrm1 + am1) × WT cross as compared to the ΔPrm1 × WT cross. These data indicate that PRM1 function is required when nuclei of opposite mating type are compartmentalized during ascogenous hyphae development in the perithecium, a stage where complementation with a wild-type copy of Prm1 in a am1 mating-type mutant does not occur (Perkins 1984; Raju 1992).
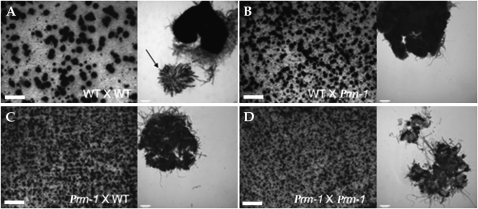
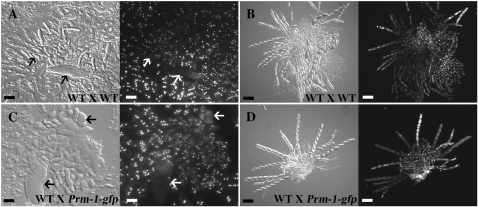
To evaluate where the block in postfertilization occurs in the ΔPrm1 mutant, the following crosses were made and their development was followed and compared (strain used as female is listed first): WT × WT (FGSC 988 × FGSC 2489 and FGSC 2489 × FGSC 988), WT × ΔPrm1 (FGSC 988 × A32 and FGSC 2489 × A29), ΔPrm1 × WT (A29 × FGSC 2489 and A32 × FGSC 988) and ΔPrm1 × ΔPrm1 (A29 × A32 and A32 × A29). After 6 days, perithecia in the WT × WT crosses were large, black, and pear shaped, with a pronounced beak (Figure 6A). In contrast, perithecia in the WT × ΔPrm1 crosses were smaller, lighter colored, and lacked beaks, indicating that their development was arrested early (Figure 6B). When used as a female, perithecia in the ΔPrm1 × WT cross were even smaller and lighter (Figure 6C). In homozygous ΔPrm1 × ΔPrm1 crosses, protoperithecia enlarged only slightly after fertilization and showed only a slight browning of perithecial walls (Figure 6D). While WT × WT perithecia typically enclosed rosettes of asci containing eight ascospores each (Figure 6A, right, arrow), perithecia of all crosses with at least one ΔPrm1 partner were completely barren (Figure 6, B–D). The fact that some protoperithecial development occurred in the ΔPrm1 crosses is consistent with the trichogyne–conidium fusion data and indicates that the block in sexual development in ΔPrm1 crosses occurs postfertilization.
Figure 6.—
ΔPrm1 mutants are male and female sterile in crosses with a wild-type strain. (A) A cross between two WT strains (FGSC 988 × FGSC 2489) shows perithecial development (left) and a rosette of asci with ascospore progeny 7 days postfertilization (right; solid arrow). (B) A cross between WT (FGSC 988) as a female and A32 (ΔPrm1∷hph A) as a male, showed only immature black perithecia (left); asci and ascospore progeny were not observed (right). (C) When ΔPrm1 was used as a female (ΔPrm1∷hph A) and crossed to WT (FGSC 988), perithecial development was blocked at an earlier stage of development (left) and no asci or ascospore progeny were produced (right). (D) Perithecial development in homozygous crosses between two ΔPrm1 strains [ΔPrm1∷hph A × ΔPrm1∷hph a (A29 × A32)] was blocked very early (left), and as with the heterozygous crosses, no asci or ascospore progeny were observed (right). Large bars, 1 mm; small bars, 50 μm.
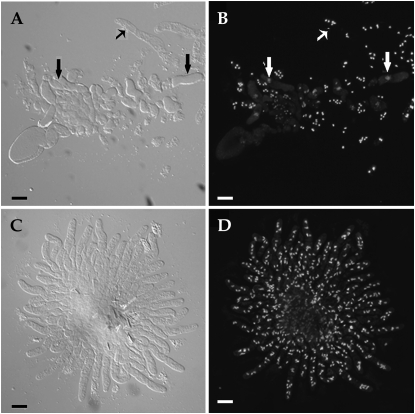
In addition to mating, cell fusion within the crozier is associated with development of asci in N. crassa and other filamentous ascomycete species (Raju 1980, 1992; Read and Beckett 1996). Karyogamy and meiosis occur in the penultimate cell of the crozier, while the first and third cells of the crozier undergo both a cell and nuclear fusion event; this compartment subtends the developing ascus (see Figure 10A). We therefore evaluated whether crozier formation was blocked in the ΔPrm1 crosses by specifically assaying for ascogenous hyphae or crozier formation by staining with DAPI to visualize nuclei (see materials and methods). In WT crosses (FGSC 988 × FGSC 2489) 4 days postfertilization, numerous crozier cells containing diploid nuclei were observed embedded in sterile hyphae called paraphyses (Figure 7, A and B). In contrast, in WT × ΔPrm1 crosses (FGSC 988 × A32), only paraphyses were observed; no crozier cells were detected (Figure 7, C and D). Furthermore, ΔPrm1 × WT and ΔPrm1 homozygous crosses produced only empty perithecia; not even paraphyses were observed. These data indicate that the loss of ΔPrm1 in at least one of the crossing partners arrests perithecial development prior to crozier formation and that functional Prm1 in the female is required to reach the developmental stage of paraphyses formation.
Figure 7.—
Crosses with ΔPrm1 as a male or female failed to produce croziers during ascospore development. (A) DIC image of a perithecial squash from a WT cross (FGSC 2489 × FGSC 988) showing developing asci/croziers (thick solid arrows) as well as paraphyses (thin solid arrow). (B) DAPI-stained nuclei from A. Nuclei in paraphyses stain well with DAPI (thin open arrow), while thick open arrows show diploid nucleus in developing asci. (C) Perithecial squash from a FGSC 988 (OR a) × A32 (ΔPrm1∷hph A) cross. Note the lack of croziers and developing asci. (D) DAPI-stained exudates from C. All nuclei show characteristic staining pattern for paraphyses. No croziers or developing asci were observed. Bars, 10 μm.
The ΔPrm1 sexual phenotype is not caused by meiotic silencing of unpaired DNA:
Since the absence of Prm1 in only one of the mating partners is sufficient to cause complete sterility, the mutation can be classified as “ascus dominant” (thus Prm1 for the locus designation). In many cases, ascus dominance in N. crassa is caused by a gene silencing mechanism (MSUD) (Shiu et al. 2001; Nakayashiki 2005; Bardiya et al. 2008), which leads to silencing of unpaired DNA segments during meiosis. If a knockout mutant of a gene essential for meiosis or the subsequent development of ascospores is crossed with a wild-type strain, the wild-type copy remains unpaired during meiosis (since the respective copy is deleted in the mutant) and is silenced by MSUD (Shiu et al. 2001). As a consequence no gene product is made and the cross will produce none or defective ascospores (Aramayo and Metzenberg 1996; Shiu et al. 2001; Shiu and Metzenberg 2002).
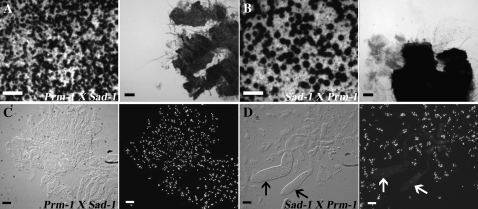
In N. crassa, mutations in Sad-1 in one of the mating partners suppresses ascus dominant mutations, including round spore (Rsp) and banana (Ban) (Shiu et al. 2001). Sad-1 encodes an RNA-directed RNA polymerase (Shiu et al. 2001) required for MSUD. To test if the observed ascus-dominant phenotype of the ΔPrm1 mutant was caused by MSUD, we crossed the ΔPrm1 strains (A32 and A29) with Sad-1 strains (R16-20 or R16-19) in which MSUD is suppressed (Shiu et al. 2001). Crosses between the ΔPrm1 and Sad-1 strains in all combinations were completely sterile (Figure 8, A and B), although perithecia resulting from these crosses were both larger and darker than perithecia in equivalent ΔPrm1 crosses with WT strains.
Figure 8.—
ΔPrm1 crosses with a Sad-1 strain, which suppresses meiotic silencing of unpaired DNA (MSUD) (Shiu et al. 2001) fail to rescue asci and ascospore development. (A) Only small perithecia (left) are observed in crosses where ΔPrm1 is used as a female (R16-51) and crossed to Sad-1 (R16-20) as a male and which show no rosettes (right). (B) When the Sad-1 strain is used as a female, crosses between Sad-1 (R16-20) and ΔPrm1 (R16-51) show more developed perithecia (left), but which are also sterile and lack asci (right). (C) The ΔPrm1 (R16-51) × Sad-1 (R16-20) crosses form paraphyses, but lack croziers and asci. (Left) DIC. (Right) DAPI-stained nuclei of image in left. (D) When the Sad-1 is used as a female, the Sad-1 (R16-20) × ΔPrm1 (R16-51) crosses showed a few abnormally shaped asci (left, DIC; solid arrows), even though ascospores are never delimited. (Right) DAPI-stained nuclei of image in left. Open arrows show abnormal asci shown in left. (A and B) Large bars, 1 mm; small bars, 50 μm. (C and D) Bars, 10 μm.
To elucidate the developmental stage at which these crosses arrest, perithecia were fixed, squashed, and stained with DAPI 3, 4, and 7 days postfertilization. When ΔPrm1 was used as a female in a cross with Sad-1, perithecia only produced paraphyses; asci and croziers were not observed (Figure 8C). However, when the Sad-1 mutant was used as a female, we observed some crozier development and abnormal asci (Figure 8D). The asci-like structures that formed appeared to be arrested in prophase. However, none of these croziers developed asci-containing ascospores, even when the cross was extended up to 21 days. These data indicate that mutations in Sad-1 partially alleviate ΔPrm1 defects during sexual reproduction when used as a female, although apparently not at the time point when SAD1 is predicted to function, which occurs after karyogamy (Shiu et al. 2001; Shiu and Metzenberg 2002; Bardiya et al. 2008).
ΔPrm1 (Prm1-gfp) strains show full complementation of female/male sterility in a cross with Sad-1:
Prm1 deletion strains that contained a Pccg1 Prm1-gfp construct integrated at the his-3 locus (A24; Table 1) showed complementation of germling fusion (Figure 2). To evaluate whether the (his-3∷Prm1-gfp) construct also complemented the sexual defect of the ΔPrm1 mutant, we performed crosses between a ΔPrm1 (his-3∷Prm1-gfp) strain and WT (A24 × FGSC 988 and FGSC 988 × A24, respectively). In contrast to crosses with ΔPrm1 strains, crosses with ΔPrm1 (Prm1-gfp) strains produced some viable ascospore progeny (Figure 9D); both ΔPrm1 (Prm1-gfp) and ΔPrm1 progeny were recovered. However, the number of asci and ascospore progeny produced from these crosses was significantly less than that produced by a WT cross (Figure 9B). The segregation of markers was normal, indicating that the low number of progeny recovered was not due to segregation defects or ascospore lethality caused by the ΔPrm1 mutation. These data suggested either that the Prm1-gfp allele was only partially functional or that MSUD might be playing a role in silencing Prm1 during crozier and ascus formation.
Figure 9.—
Crosses between a complemented ΔPrm1 strain and WT form some ascospore progeny, but crosses are mostly blocked during early ascus development. (A) Developing croziers and asci in early prophase (arrows) from a WT × WT cross (FGSC 988 × FGSC 2489) 4 days postfertilization. (Left) DIC. (Right) DAPI-stained nuclei of image in left. (B) WT × WT cross (FGSC 988 × FGSC 2489) 7 days postfertilization. Note numerous asci with developing ascospores. (Left) DIC. (Right) Fluorescent image of DAPI-stained nuclei. (C) Developing croziers and asci from a cross between WT (FGSC 988 a) and a ΔPrm1 complemented strain [his-3∷Pccg1-Prm1-gfp; ΔPrm1∷hph; Δmus-51∷bar A (A24)]. Note that this cross is indistinguishable from a WT × WT cross at a similar time point. (Left) DIC. (Right) Fluorescent image of DAPI-stained nuclei. (D) Cross between FGSC 988 × A24 7 days postfertilization showing only a few asci; most of the croziers and developing asci have degenerated. (Left) DIC. (Right) Fluorescent image of DAPI-stained nuclei. (A and C) Bars, 10 μm. (B and D) Bars, 50 μm.
To differentiate these possibilities, crosses were performed between the Sad-1 mutant (R16-20) and the ΔPrm1 (Prm1-gfp) complemented strain (A24). These crosses displayed wild-type levels of asci and ascospore progeny that were indistinguishable from WT crosses (Figure 10, B and C). Further, segregation of markers was not affected, as hygromycin-resistant progeny were easily recovered (ΔPrm1 marker), with some progeny displaying GFP fluorescence (Prm1-gfp). Because mutations in Sad-1 suppressed the ascospore progeny defect associated with the WT × ΔPrm1 (his-3∷Prm1-gfp) crosses, we hypothesized that silencing of Prm1 in both wild type and the ΔPrm1 (his-3∷Prm1-gfp) nuclei due to MSUD (the Prm1 alleles are unpaired in this cross) might result in defects in cell fusion of the crozier (Figure 10A) and thus disrupt ascus development. To test this hypothesis more directly, we focused on crozier and asci development in the WT × A24 [ΔPrm1 (his-3∷Prm1-gfp)] cross as compared to a WT × WT cross. As shown in Figure 9, A and C, the development of croziers and young asci was indistinguishable between a WT × WT cross and the WT × A24 cross up to 4 days postfertilization. Subsequently, asci development in the WT × A24 cross was mostly arrested, resulting in the degeneration of most asci/croziers. The timing of the block in crozier and asci development in the WT × A24 crosses suggested that a block in crozier cell fusion was responsible; we were unable to obtain quantitative data on the frequency of crozier fusion in these crosses due to the difficulty in working with croziers/ascogenous hyphae. In summary, these data suggest that, similar to mating, germling and hyphal fusion, PRM1 is also involved in cell fusion events associated with crozier/ascus formation in N. crassa.
DISCUSSION
PRM1 is part of the general fusion machinery in N. crassa:
Four cell fusion events have been reported in the life cycle of N. crassa: germling and hyphal fusion during vegetative growth and fertilization and crozier fusion during sexual reproduction. Our data implicate Prm1 function in all of these cell fusion events. In addition, our analysis of the N. crassa ΔPrm1 mutant also suggests that cell fusion may be associated with events that occur postfertilization, but prior to crozier cell fusion during sexual development. During germling fusion, the defect observed in the ΔPrm1 mutant in N. crassa is remarkably similar to that of the defect in mating cell fusion in S. cerevisiae prm1Δ, with an ∼50% reduction in cell fusion in both species. In addition, membrane blebs and juxtaposed plasma membranes are observed both in S. cerevisiae prm1Δ mating cells (Heiman and Walter 2000; Jin et al. 2004) and in N. crassa ΔPrm1 germlings (Figures 2 and 3). In S. cerevisiae, contact-dependent cell lysis is often observed in prm1Δ mutants that fail to undergo mating cell fusion (Jin et al. 2004). After extended incubation, germling pairs of N. crassa ΔPrm1 mutants that fail to undergo fusion are often highly vacuolated (data not shown), a phenotype reminiscent of the contact-dependent cell lysis of S. cerevisiae prm1Δ mutants (Jin et al. 2004).
In S. cerevisiae, Prm1p showed a localization pattern consistent with the endoplasmic reticulum (Heiman and Walter 2000), in addition to localization at shmoo tips and mating cell fusion points. We did not detect PRM1-GFP accumulation at the point of cell contact during germling fusion in N. crassa, although we observed GFP fluorescence in inner membranous structures, such as vacuoles and ER-like compartments. This discrepancy between the proposed direct function of PRM1 at the fusion point and its detected localization, could be explained by transient recruitment to the point of cell contact. In S. cerevisiae, certain alleles of PRM1 fused with GFP fully complement the prm1Δ mating defect, but fluorescence was not detected at the point of cell contact (A. Engel, personal communication). While the amount of PRM1-GFP at the fusion point could be below the level of detection, a possible function remote from the point of cell contact cannot be excluded. In contrast to localization in germlings, we detected GFP fluorescence at both the plasma membrane and fusion points of hyphae in ΔPrm1 strains carrying Prm1-gfp under the regulation of the ccg-1 promoter. Taken together, these observations strongly suggest that the molecular function of Prm1 and its role in membrane merger is conserved among ascomycete species.
In S. cerevisiae, the expression of PRM1 is induced by treatment with pheromone (Heiman and Walter 2000). The promoter of PRM1 has multiple sites for binding of the transcription factor Ste12p, which directly regulates genes involved in shmoo formation and mating cell fusion (Zeitlinger et al. 2003). Interestingly, mutations in the ortholog of STE12 in N. crassa, called pp-1, also show defects in germling/hyphal fusion (Li et al. 2005) (A. Fleißner and N. L. Glass, unpublished data), as do strains containing mutations in orthologs of the pheromone response MAPK pathway (mak-2, nrc-1, and mek-2) (Pandey et al. 2004; Maerz et al. 2008). However, N. crassa pheromone mutants, although infertile as a male, undergo normal germling and hyphal fusion (Kim and Borkovich 2006). It is unclear what the self-signaling ligand involved in germling and hyphal fusion is in N. crassa, but it presumably functions via activation of a MAPK pathway and PP1 (Glass et al. 2004; Fleißner et al. 2008); it is possible that Prm1 may be a direct target of PP1 in N. crassa.
Prm1 function is required for both germling/hyphal fusion and trichogyne–conidium fusion, with a remarkably consistent reduction in fusion frequencies in these two processes in ΔPrm1 mutants. However, not all N. crassa fusion mutants are affected in both germling/hyphal fusion and trichogyne–conidium fusion. For example, mutations in so, which encodes a filamentous ascomycete-specific protein, completely abolish germling and hyphal fusion in N. crassa, but trichogyne–conidium fusion is unaffected (Fleißner et al. 2005); so mutants show no chemotropic interactions during germling fusion (Roca et al. 2005). These data indicate that N. crassa uses different molecular mechanisms for cell–cell communication and chemotropic growth that precede cell fusion during germling/hyphal fusion vs. mating during sexual reproduction. We hypothesize that once cells are in physical contact, either during germling/hyphal fusion or during mating, the general fusion machinery is activated leading to cell wall breakdown and plasma membrane merger, a process involving PRM1. The identification of additional fusion factors and the examination of their role during both vegetative and sexual cell fusion events in N. crassa will allow us to further test this hypothesis.
Prm1 is essential for male and female fertility in both mating types:
S. cerevisiae prm1Δ mutants exhibit ∼50% reduction in mating cell fusion (Heiman and Walter 2000; Aguilar et al. 2007; Heiman et al. 2007; Jin et al. 2008), but in prm1Δ pairs that undergo mating cell fusion, karyogamy and sporulation are apparently normal. Surprisingly, the N. crassa ΔPrm1 mutants showed complete and dominant sterility when functioning as either a male or a female in a cross. The only mutants reported in N. crassa that show male and female sterility and are dominant are mating-type mutants (Griffiths and Delange 1978; Griffiths 1982; Glass et al. 1988; Saupe et al. 1996); mat A-1 and mat a-1 mating-type mutants in N. crassa show normal vegetative fusion. The complete sterility observed in N. crassa ΔPrm1 mutants is not caused by a defect in plasma membrane merger between the mating partners, because we observed up to ∼45% successful trichogyne–conidium fusion events in homozygous ΔPrm1 crosses and observed GFP-labeled nuclei migrating into the trichogyne in these crosses. These observations indicate an essential role for PRM1 during sexual reproduction after fertilization. In filamentous ascomycete fungi, the events between fertilization and crozier development, which spans an ∼4-day interval in N. crassa, remain obscure, primarily due to the difficulty in differentiating ascogenous from maternal tissue within the developing perithecium. We predict that cell fusion events are associated with development of the ascogonium/ascogenous hyphae and that this process requires PRM1. Alternatively, PRM1 may have a novel function during this stage of the sexual cycle in N. crassa. Experiments designed to identify suppressors of Prm1 may help to identify additional genes involved in the process and allow a more thorough examination of this enigmatic interval in sexual reproduction in filamentous ascomycete fungi.
The ascus dominant phenotype of the ΔPrm1 mutation is not caused by meiotic gene silencing of unpaired DNA:
In many cases, ascus dominant phenotypes in N. crassa are due to MSUD (Aramayo and Metzenberg 1996; Shiu et al. 2001; Shiu and Metzenberg 2002). In the ascus, MSUD occurs after karyogamy and silences all DNA sequences that remain unpaired. If the silenced segment carries a gene essential for meiosis or ascospore development, the respective crosses will be barren. For example, a cross involving partner nuclei that contain unpaired but functional β-tubulin genes arrested in meiosis before metaphase, due to the silencing of all β-tubulin genes in the diploid nucleus (Shiu et al. 2001). Two of our observations indicate that the dominant phenotype of the ΔPrm1 mutants is not caused by MSUD. First, mutations in Sad-1 (which is itself dominant) do not suppress the infertility of ΔPrm1 crosses. Second, examination of perithecia and ascogenous/paraphysoidal tissue in ΔPrm1 crosses showed that they were blocked well before karyogamy, when MSUD functions.
In contrast, ΔPrm1 strains carrying a Prm1-gfp allele at the his-3 locus are fertile in crosses with WT. In these crosses, the Prm1 genes remain unpaired and are thus subject to MSUD at karyogamy. Examination of ascogenous hyphae/paraphysoidal tissue in these crosses showed that development of WT × ΔPrm1 (his-3∷Prm1-gfp) crosses were indistinguishable from WT × WT crosses ∼4 days postfertilization, an interval prior to ascus and ascospore development. However, the WT × ΔPrm1 (his-3∷Prm1-gfp) crosses produced significantly fewer ascospores than a wild-type cross, a phenotype that was completely suppressed by mutations in Sad-1. These data implicate MSUD and silencing of the Prm1 alleles in the crozier, which results in a reduction in the number of asci and ascospore progeny in the WT × ΔPrm1 (his-3∷Prm1-gfp) crosses. Crozier fusion occurs at a similar time to karyogamy in N. crassa (Raju 1980) (Figure 10), suggesting that silencing of all Prm1 copies via MSUD disrupts the crozier fusion process. Interestingly, the number of asci from the WT × ΔPrm1 (his-3∷Prm1-gfp) crosses was reduced by approximately half (Figure 9D), which is a similar value to the reduction in germling and trichogyne–conidium fusion events in Prm1 mutants.
PRM1 also functions during perithecial development in N. crassa:
Our data show that PRM1 must have at least three different functions during sexual development: trichogyne–conidium fusion, crozier fusion, and at a point during sexual development between these two characterized fusion events, as evidenced by the sterility of the ΔPrm1 crosses. In addition to these stages, our experimental data also suggest some female-specific functions for PRM1. If the ΔPrm1 mutant is used as a male, perithecia show some enlargement and darkening and even develop sterile paraphyses (Figure 7, C and D). However, when a ΔPrm1 strain is used as a female, perithecia are smaller, lighter, and devoid of paraphyses (Figure 6C), while homozygous ΔPrm1 crosses showed the least perithecial development (Figure 6D). This early developmental arrest of ΔPrm1 perithecia was complemented if the female was a heterokaryon between the ΔPrm1 mutant and the helper strain (FGSC 4564). It is possible that cell fusion also plays a role in the formation/development of maternal tissue in the developing perithecium of N. crassa, a defect that would be complemented in a heterokaryon with the sterile helper strain, am1. Further analysis of both wild type and fusion mutants during development of female reproductive structures both prior to and after fertilization will help to elucidate the answer to this question.
Acknowledgments
We thank Steve Ruzin and Denise Schichnes in the College of Natural Resources Biological Imaging Facility for expert advice and training on the Delta Vision microscope. We thank Alex Engel for discussion and Abby Leeder, Charles Hall, and Anna Simonin for their critical reading of the manuscript. We thank Ulrike Brandt for excellent technical assistance in some of the experiments. We thank Patrick Shiu and Dave Jacobson for the gift of strains. We acknowledge Reena Zalpuri and Kent McDonald of the University of California Berkeley Electron Microscope Laboratory for their assistance in the TEM images. The work in this study was funded by a grant to N.L.G. from the National Science Foundation (MCB-0517660).
References
- Aguilar, P. S., A. Engel and P. Walter, 2007. The plasma membrane proteins Prm1 and Fig1 ascertain fidelity of membrane fusion during yeast mating. Mol. Biol. Cell 18 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramayo, R., and R. L. Metzenberg, 1996. Meiotic transvection in fungi. Cell 86 103–113. [DOI] [PubMed] [Google Scholar]
- Bardiya, N., W. G. Alexander, T. D. Perdue, E. G. Barry, R. L. Metzenberg et al., 2008. Characterization of interactions between and among components of the meiotic silencing by unpaired DNA machinery in Neurospora crassa using bimolecular fluorescence complementation. Genetics 178 593–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bistis, G. N., 1981. Chemotropic interactions between trichogynes and conidia of opposite mating-type in Neurospora crassa. Mycologia 73 959–975. [Google Scholar]
- Brockman, H. E., and F. J. de Serres, 1963. “Sorbose toxicity” in Neurospora. Am. J. Bot. 50 709–714. [Google Scholar]
- Buller, A. H. R., 1933. Researches on Fungi, Vol. 5. Longman, London.
- Chen, E. H., E. Grote, W. Mohler and A. Vignery, 2007. Cell-cell fusion. FEBS Lett. 581 2181–2193. [DOI] [PubMed] [Google Scholar]
- Colot, H. V., G. Park, G. E. Turner, C. Ringelberg, C. M. Crew et al., 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, R. H., 2000. Neurospora: Contributions of a Model Organism. Oxford University Press, New York.
- Dresselhaus, T., 2006. Cell-cell communication during double fertilization. Curr. Opin. Plant Biol. 9 41–47. [DOI] [PubMed] [Google Scholar]
- Dunlap, J. C., K. A. Borkovich, M. R. Henn, G. E. Turner, M. S. Sachs et al., 2007. Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv. Genet. 57 49–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion, E. A., J. Trueheart and G. R. Fink, 1995. Fus2 localizes near the site of cell fusion and is required for both cell fusion and nuclear alignment during zygote formation. J. Cell Biol. 130 1283–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman, S., L. Lin, M. Malczynski and M. Snyder, 1998. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 140 461–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleißner, A., S. Sarkar, D. J. Jacobson, M. G. Roca, N. D. Read et al., 2005. The so locus is required for vegetative cell fusion and postfertilization events in Neurospora crassa. Eukaryot. Cell 4 920–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleißner, A., A. R. Simonin and N. L. Glass, 2008. Cell fusion in the filamentous fungus, Neurospora crassa. Methods Mol. Biol. 475 21–38. [DOI] [PubMed] [Google Scholar]
- Freitag, M., P. C. Hickey, N. B. Raju, E. U. Selker and N. D. Read, 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41 897–910. [DOI] [PubMed] [Google Scholar]
- Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read et al., 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422 859–868. [DOI] [PubMed] [Google Scholar]
- Glass, N. L., and K. Dementhon, 2006. Non-self recognition and programmed cell death in filamentous fungi. Curr. Opin. Microbiol. 9 553–558. [DOI] [PubMed] [Google Scholar]
- Glass, N. L., S. J. Vollmer, C. Staben, R. L. Metzenberg and C. Yanofsky, 1988. DNAs of the two mating type alleles of Neurospora crassa are highly dissimilar. Science 241 570–573. [DOI] [PubMed] [Google Scholar]
- Glass, N. L., C. Rasmussen, M. G. Roca and N. D. Read, 2004. Hyphal homing, fusion and mycelial interconnectedness. Trends Microbiol. 12 135–141. [DOI] [PubMed] [Google Scholar]
- Griffiths, A. J. F., 1982. Null mutants of the A and a mating-type alleles of Neurospora crassa. Can. J. Genet. Cytol. 24 167–176. [Google Scholar]
- Griffiths, A. J. F., and A. M. Delange, 1978. Mutations of the a mating-type gene in Neurospora crassa. Genetics 88 239–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman, M. G., A. Engel and P. Walter, 2007. The Golgi-resident protease Kex2 acts in conjunction with Prm1 to facilitate cell fusion during yeast mating. J. Cell Biol. 176 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman, M. G., and P. Walter, 2000. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 151 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey, P. C., D. Jacobson, N. D. Read and N. Louise Glass, 2002. Live-cell imaging of vegetative hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 37 109–119. [DOI] [PubMed] [Google Scholar]
- Jin, H., C. Carlile, S. Nolan and E. Grote, 2004. Prm1 prevents contact-dependent lysis of yeast mating pairs. Eukaryot. Cell 3 1664–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., J. M. McCaffery and E. Grote, 2008. Ergosterol promotes pheromone signaling and plasma membrane fusion in mating yeast. J. Cell Biol. 180 813–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H., and K. A. Borkovich, 2004. A pheromone receptor gene, pre-1, is essential for mating type-specific directional growth and fusion of trichogynes and female fertility in Neurospora crassa. Mol. Microbiol. 52 1781–1798. [DOI] [PubMed] [Google Scholar]
- Kim, H., and K. A. Borkovich, 2006. Pheromones are essential for male fertility and sufficient to direct chemotropic polarized growth of trichogynes during mating in Neurospora crassa. Eukaryot. Cell 5 544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, E., 1930. Zur Kenntnis der vegetativen Anastomosen der Pilze (II. Mitteilung). Planta 10 495–522. [Google Scholar]
- Li, D., P. Bobrowicz, H. H. Wilkinson and D. J. Ebbole, 2005. A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa. Genetics 170 1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loros, J. J., S. A. Denome and J. C. Dunlap, 1989. Molecular cloning of genes under control of the circadian clock in Neurospora. Science 243 385–388. [DOI] [PubMed] [Google Scholar]
- Maerz, S., C. Ziv, N. Vogt, K. Helmstaedt, N. Cohen et al., 2008. The Ndr kinase COT1, and the MAP kinases MAK1 and MAK2 genetically interact to regulate filamentous growth, hyphal fusion, and sexual development in Neurospora crassa. Genetics 179 1313–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin, B. S., M. Freitag and E. U. Selker, 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newslett. 44 34–36. [Google Scholar]
- McNally, M. T., and S. J. Free, 1988. Isolation and characterization of a Neurospora glucose-repressible gene. Curr. Genet. 14 545–551. [DOI] [PubMed] [Google Scholar]
- Nakayashiki, H., 2005. RNA silencing in fungi: mechanisms and applications. FEBS Lett. 579 5950–5957. [DOI] [PubMed] [Google Scholar]
- Pandey, A., M. G. Roca, N. D. Read and N. L. Glass, 2004. Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot. Cell 3 348–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, J. M., C. A. Ydenberg and M. D. Rose, 2008. Dynamic localization of yeast Fus2p to an expanding ring at the cell fusion junction during mating. J. Cell Biol. 181 697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, D. D., 1984. Advantages of using the inactive-mating-type am1 strain as a helper component in heterokaryons. Fungal Genet. Newsl. 31 41–42. [Google Scholar]
- Proszynski, T. J., R. Klemm, M. Bagnat, K. Gaus and K. Simons, 2006. Plasma membrane polarization during mating in yeast cells. J. Cell Biol. 173 861–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju, N. B., 1980. Meiosis and ascospore genesis in Neurospora. Eur. J. Cell Biol. 23 208–223. [PubMed] [Google Scholar]
- Raju, N. B., 1992. Genetic control of the sexual cycle in Neurospora. Mycol. Res. 96 241–262. [Google Scholar]
- Read, N. D., and A. Beckett, 1996. Ascus and ascospore morphogenesis. Mycol. Res. 100 1281–1314. [Google Scholar]
- Roca, M. G., J. Arlt, C. E. Jeffree and N. D. Read, 2005. Cell biology of conidial anastomosis tubes in Neurospora crassa. Eukaryot. Cell 4 911–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe, S., L. Stenberg, K. T. Shiu, A. J. Griffiths and N. L. Glass, 1996. The molecular nature of mutations in the mt A-1 gene of the Neurospora crassa A idiomorph and their relation to mating-type function. Mol. Gen. Genet. 250 115–122. [DOI] [PubMed] [Google Scholar]
- Shiu, P. K. T., and R. L. Metzenberg, 2002. Meiotic silencing by unpaired DNA: properties, regulation, and suppression. Genetics 161 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, P. K. T., N. B. Raju, D. Zickler and R. L. Metzenberg, 2001. Meiotic silencing by unpaired DNA. Cell 107 905–916. [DOI] [PubMed] [Google Scholar]
- Staben, C., and C. Yanofsky, 1990. Neurospora crassa a mating-type region. Proc. Natl. Acad. Sci. USA 87 4917–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, H. J., 1956. A convenient growth medium for Neurospora. Microbiol. Genet. Bull. 13 42–46. [Google Scholar]
- Westergaard, M., and H. K. Mitchell, 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34 573–577. [Google Scholar]
- Zeitlinger, J., I. Simon, C. T. Harbison, N. M. Hannett, T. L. Volkert et al., 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113 395–404. [DOI] [PubMed] [Google Scholar]