Abstract
Changes in host specialization contribute to the diversification of phytophagous insects. When shifting to a new host, insects evolve new physiological, morphological, and behavioral adaptations. Our understanding of the genetic changes responsible for these adaptations is limited. For instance, we do not know how often host shifts involve gain-of-function vs. loss-of-function alleles. Recent work suggests that some genes involved in odor recognition are lost in specialists. Here we show that genes involved in detoxification and metabolism, as well as those affecting olfaction, have reduced gene expression in Drosophila sechellia—a specialist on the fruit of Morinda citrifolia. We screened for genes that differ in expression between D. sechellia and its generalist sister species, D. simulans. We also screened for genes that are differentially expressed in D. sechellia when these flies chose their preferred host vs. when they were forced onto other food. D. sechellia increases expression of genes involved with oogenesis and fatty acid metabolism when on its host. The majority of differentially expressed genes, however, appear downregulated in D. sechellia. For several functionally related genes, this decrease in expression is associated with apparent loss-of-function alleles. For example, the D. sechellia allele of Odorant binding protein 56e (Obp56e) harbors a premature stop codon. We show that knockdown of Obp56e activity significantly reduces the avoidance response of D. melanogaster toward M. citrifolia. We argue that apparent loss-of-function alleles like Obp56e potentially contributed to the initial adaptation of D. sechellia to its host. Our results suggest that a subset of genes reduce or lose function as a consequence of host specialization, which may explain why, in general, specialist insects tend to shift to chemically similar hosts.
HALF of known insect species feed primarily on plants (Jolivet 1992; Bernays and Chapman 1994), with 90% of these phytophagous insects specializing on one or a few host plant families (Bernays and Graham 1988; Jolivet 1992; Bernays and Chapman 1994). Specialists often evolve host-specific adaptations such as resistance to plant secondary compounds, changes in morphology, and new preference behaviors. An understanding of the genetic basis of traits such as these is critical to knowing how host specialization evolves (Jaenike 1987; Via 1990; Futuyma 1991; Jaenike and Holt 1991; Hawthorne and Via 2001).
Recent work has uncovered genes and genetic regions affecting host specialization (Via 1990; Sheck and Gould 1993, 1995; Cleland et al. 1996; Jones 1998; Hawthorne and Via 2001; Carsten et al. 2005; Dambroski et al. 2005; Jones 2005; Nylin et al. 2005; Matsuo et al. 2007; Kopp et al. 2008). For example, in several cactophillic Drosophila species cytochrome P450s have been implicated in detoxification of host secondary compounds (Frank and Fogleman 1992; Barker et al. 1994; Danielson and Fogleman 1997; Danielson et al. 1997; Fogleman et al. 1997, 1998; Matzkin et al. 2006). Most of these genetic studies, however, concentrated on one or a few traits. Moreover, these earlier trait-specific studies could not distinguish between genetic changes that were a consequence of host specialization vs. those that directly contributed to the host specialization per se (Bono et al. 2008; Matzkin 2008; but see Sucena and Stern 2000; Jones 2004; Orgogozo et al. 2006; McBride 2007; McGregor et al. 2007).
On its native islands, the Seychelles, Drosophila sechellia almost exclusively uses the fruit of Morinda citrifolia (Morinda), a plant common around the Indian Ocean and Polynesia (Louis and David 1986; Jones 2005). D. sechellia has evolved strong preference for and resistance to the toxins in Morinda (Louis and David 1986; R'Kha et al. 1991; Jones 1998, 2004, 2005). D. simulans, on the other hand, is a human commensal that originally arose in eastern Africa (Lachaise and Silvain 2004). Several compounds found in Morinda fruit are toxic to most Drosophila species, including D. simulans. As a result, most fruit flies avoid this plant. In contrast, D. sechellia responds positively to olfactory cues from Morinda; when female D. sechellia detect Morinda they increase egg production and ovipositioning (R'Kha et al. 1991; Jones 2004). Field experiments suggest that D. sechellia can detect Morinda at distances up to 50 m (R'Kha et al. 1991).
Several recent studies investigated how D. sechellia perceives the odor of Morinda differently from D. simulans (Dekker et al. 2006; Jones 2007; Matsuo et al. 2007; Kopp et al. 2008). Odor perception in flies occurs via antennae, although the maxillary palps and tarsi also play important roles (reviewed in Hallem et al. 2006). Odorants pass through cuticular pores in sensilla located on the antennae. These odorants are then bound by odorant binding proteins (Obps) and delivered to odorant receptors (Ors) on the surface of the insect odorant receptor neurons. These neurons converge to spatially invariant antennal lobe glomeruli. From these glomeruli, neurons project into the mushroom body where higher-order processing is believed to occur. Dekker et al. (2006) recently suggested that D. sechellia differs from D. simulans in the numbers and types of sensilla, which may alter the distribution of Ors in D. sechellia. These differences in the distribution of Ors may change the perception of odors from Morinda and may result in the behavioral differences between D. sechellia and D. simulans. Indeed, genetic ablation of antennae or sensilla changes the response of D. melanogaster to odors from Morinda (Jones 2007). Congruent with these observations, a molecular evolution study has shown that several Ors and Grs appear to have become pseudogenes in D. sechellia (McBride 2007). Loss of functional copies of these genes may also affect the perception of odors from Morinda. In contrast to the Dekker et al. and McBride results, Matsuo et al. (2007) recently suggested that a change in Obp expression in the tarsi of D. sechellia is key to the difference in oviposition-site preference between D. sechellia and other Drosophila. The chemosensory system is clearly important to host preference in D. sechellia. However, the exact role of the chemosensory system played in the evolution of D. sechellia's host preference is ambiguous as it is not known if changes in chemosensory system alone were sufficient for the host shift.
From earlier studies, we expect that the evolution of host specialization in D. sechellia involved changes in how it copes with secondary compounds found in Morinda, how it perceives the odor of Morinda, and how it behaviorally responds to its host. Our goal is to find genes responsible for these differences. To this end, we identified (i) genes in D. sechellia or D. simulans whose expression changed when flies were given a choice between medium with and without the major organic compounds from Morinda and (ii) genes whose structure and expression fundamentally differed between these two fly species. Our data show that the expression of genes involved in metabolism, olfaction, and female reproduction do indeed vary in a species- and treatment-specific manner. Surprisingly, however, we also note that in many instances reduced gene expression in D. sechellia is associated with the fixation of loss-of-function alleles. For example, the D. sechellia allele of Odorant binding protein 56e (Obp56e) harbors a premature stop codon. We show that knockdown of Obp56e activity significantly reduces the avoidance response of D. melanogaster toward M. citrifolia. The acquisition and fixation of these alleles must have been rapid as D. sechellia and D. simulans diverged <0.5 MYA (Kliman et al. 2000) and likely have lasting implications for the ability of D. sechellia to use Morinda vs. other hosts.
MATERIALS AND METHODS
Fly stocks:
Except where noted, all stocks were reared on agar–yeast–cornmeal medium at room temperature. D. sechellia line 1 [“Robertson” collected from Seychelles in 1981 by Tsacas and Bächli (Tsacas and Bächli 1981)], D. sechellia Syn A (a wild-type non-isofemale line; courtesy of J. Coyne), and D. simulans sim6 (an isofemale line from Winters, CA, courtesy of D. Begun) were used for most comparisons. J. Coyne also provided the D. sechellia wild-type lines SS77 25X and sy001. We also obtained D. simulans w501 and D. simulans Islamorada from the Drosophila Species Stock Center in Tucson, AZ. The D. melanogaster Oregon-R strain was obtained from the Drosophila Stock Center in Bloomington, IN. RNAi stocks were from the Vienna Drosophila Resource Center (Dietzl et al. 2007).
For the microarrays, two wild-type isofemale lines were used: D. sechellia line 1 and D. simulans sim6. Both were reared at 25° on agar–yeast–cornmeal medium with constant humidity in an environmental chamber, unless noted otherwise.
Preference assay—oviposition:
Following Jones (2004), oviposition-site preference was scored by presenting inseminated, ovipositing females with a choice of oviposition substrates, one with octanoic acid and one without. Media were prepared using Drosophila Instant Medium (Carolina Biological Supply, Burlington, NC). The toxic medium was 0.07% octanoic acid by weight (Sigma, St. Louis). This dose does not kill all susceptible flies (Jones 2001).
Each female was placed in a chamber with the two types of medium. She was allowed to oviposit for 2 days, after which the number of eggs laid on each type of medium was counted. The female was then shifted to a fresh pair of Morinda and control media. After 2 more days, her preference was scored again. All assays were conducted in a constant-temperature room at 20° with relative humidity between 50 and 70%.
Egg counts were converted to a preference index (PI) by the following formula:
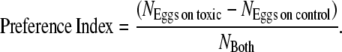 |
Positive values indicate preference for Morinda media, whereas negative values indicate avoidance of Morinda media. Unless otherwise noted, data were pooled across both days.
Preference assay—“choice–no choice”:
High-throughput assays of preference were performed in test chambers (2-liter glass beakers; Fisher Scientific, Pittsburgh) containing two standard fly bottles (Genesee Scientific, San Diego): either one bottle of control medium and one bottle of Morinda medium (above) or two bottles of control medium. Control medium was 44 ml of water with 8.5 g of Carolina 4-24 instant medium (Carolina Biological Supply). Morinda toxin medium was made by combining 44 ml of water with 8.5 g of Carolina 4-24 instant medium with 90 μl octanoic acid and 30 μl of hexanoic acid (Arcos Organics, Morris Plains, NJ). The combination was gently agitated to ensure even distribution of the hydrophobic octanoic and hexanoic acids. Morinda fruit has a 3:1 ratio of octanoic to hexanoic acid (Legal et al. 1994). The concentration of Morinda toxins in the medium was low relative to what is typically observed in nature, but not outside the normal range. This concentration was necessary to minimize mortality in D. simulans.
Approximately 90, 1-day-old females were collected and allowed to mate ad libitum with males of their own species for 3 days. Females were then separated and allowed to recover for 1 day. They were then lightly sedated and placed in test chambers, which were then placed in an environmental chamber with constant humidity and temperature (65%, 25°). They were allowed to roam the test chamber freely and to choose media to oviposit on. After a day, 80–90% of the live flies would have settled on one medium or another. Under these conditions typically 82% of the D. sechellia chose the Morinda toxin medium, whereas <4% of the D. simulans chose the Morinda toxin medium.
For the microarrays, we performed three replicates of each species for each treatment. Flies were collected and frozen in liquid nitrogen without anesthesia. Only the fly bottle with the majority of flies was collected for each test chamber. Typically there were ≥45 flies per replicate. So that the transcriptional profile of the head could be clearly observed relative to the remaining carcass, the heads of flies were separated for their own analysis.
As a control for aggregation behavior in Drosophila, we also compared the behavior of D. simulans and D. sechellia when presented with “no choice” (i.e., only standard medium). Not surprisingly, the flies spread themselves between the two identical substrates (D. simulans, 54 ± 7%; D. sechellia, 53 ± 8%).
We confirmed that our preference assay mimicked the normal fly response to the fruit of M. citrifolia. We placed ∼1.75 g of ripe Morinda fruit on untainted medium (the resulting medium was 4% Morinda by weight). D. simulans sim6 and D. sechellia Syn A flies were tested as above. Eighty-eight percent of D. sechellia chose the medium with Morinda (N = 52), whereas only 18% of the D. simulans did (N = 73).
RNA preparation for hybridization:
Total RNA from heads or bodies was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) per manufacturer's instructions. RNA was further purified using a Qiagen RNeasy Mini Kit per the manufacturer's protocol (Qiagen Science, Germantown, MD). Contaminating genomic DNA was removed by DNAse treatment (20 units per 100 μg of RNA). mRNA (500 ng from each sample) was amplified using the low RNA input fluorescent linear amplification kit, according to manufacturer's specifications (Agilent Technologies, Santa Clara, CA). Following mRNA purification and quantification, samples were prepared according to the standard operating procedure (The Institute for Genomic Research, www.compbio.dfci.harvard.edu/docs/MicroarrayLabelling.pdf). The cRNA products were pooled, and for each sample to be hybridized 4 μg of cRNA were indirectly labeled with aminoallyl during cDNA synthesis [Promega (Madison, WI) ImProm II enzyme], linked to Cy3 or Cy5 esters, and hybridized overnight following the standard operating procedure (The Institute for Genomic Research). To reduce technical variation, samples were pooled and then split following both the amino-allyl and dye incorporation, prior to combining samples for hybridization. Hybridizations were performed using printed slides treated with 5× SSC, 0.1% SDS, and 1% BSA for 45 min to block nonspecific binding. Hybridization reactions containing labeled cDNA, 5× SSC, polyadenine oligo, 5× Denhardt's solution, 50% formamide, 0.5% SDS, and calf thymus DNA were placed onto the prepared slides and incubated for 20 hr at 42°. A balanced incomplete block design was used, in a full loop configuration with dye swapping (Figure 1). This design avoids confounding any variables with dye effects. For the RNA from both the heads and the bodies, 12 two-channel hybridizations were performed, resulting in 6 replicate hybridizations per treatment (3 per treatment per dye). The slides went through a series of washes (1× SSC/0.2% SDS, 0.1× SSC/0.2% SDS, 0.1× SSC) before being scanned with a ScanArray 4000 (Packard Biosciences).
Figure 1.—
Experimental design for DGRC arrays. Red indicates “Morinda medium”; green indicates “standard medium.” A balanced incomplete block design was used, in a full-loop configuration with dye swapping. This design avoids confounding any variables with dye effects. For the RNA from both the heads and the bodies, 12 two-channel hybridizations were performed, resulting in 6 replicate hybridizations per treatment (3 per treatment per dye). We contrasted gene expression in D. sechellia when these flies were allowed to choose preferred Morinda medium vs. D. sechellia that were forced to use standard medium. We performed a parallel experiment with D. simulans as a control. We expect almost no genes will differ between treatments in D. simulans, although it is possible that the odors from the Morinda medium could cause some minor changes in expression. (Forcing D. simulans on the Morinda medium is problematic because it is toxic to these flies.)
DGRC arrays:
We used version 1 DGRC amplicon transcriptome microarrays (Print run A1.3.12.17), which are spotted with DNA fragments amplified from genomic DNA using gene-specific oligonucleotide primer pairs, covering 88% of the release 4.1 predicted genes. Details of this array are described at http://dgrc.cgb.indiana.edu/. Data were deposited in the GEO database (accession nos. GSE13723, GSE13778, GSE13789, EU584560–EU584564).
Statistical analysis of DGRC array data:
Raw fluorescence intensities (background subtracted) using both the histogram and connected-component spot segmentation algorithms were exported from UCSF SPOT v2.1 (Jain et al. 2002) and log2 transformed. For the analysis, a two-step general linear mixed model was performed using a mixed procedure in SAS version 8.2 (SAS Institute, Cary, NC) (Wolfinger et al. 2001). The first model provides a global linear normalization step for dye (fixed), array (random), and their interaction (random). The residuals from the normalization step represent the relative fluorescence intensity (log2 RFI) for each feature as the fold change in expression intensity relative to the sample mean conditioned on array and dye. These values were then used for the following gene (spot)-specific models,
 |
where Si is the species (sechellia or simulans), Tj is the treatment (choice or no choice), Dl is dye, Am is array, and ɛijk is the residual error. To evaluate the statistical significance controlling for the familywise error rate, we used the false discovery rate (FDR) method (Storey and Tibshirani 2003). q-values were calculated using the q-value library in the R statistical package (Storey and Tibshirani 2003) and were used to limit the expected number of false positive genes to a small number (as discussed in results).
Correction for sequence mismatches:
As Ranz et al. (2003) and Gilad et al. (2005) have noted, sequence divergence between the probes on a microarray and the RNA being hybridized can affect estimates of expression differences. Typically, the greater the divergence is between the sequence of the probe and the sample hybridized, the lower the estimated expression (Gilad et al. 2005). On average the D. melanogaster probes diverge by 6.8% from the D. simulans mRNA sequence that would hybridize to the probe (median divergence: 4.2%), which is in line with prior estimates (Ometto et al. 2005). This leads us to underestimating the changes in gene expression [as the quantitative (q)RT–PCR results show; Ranz et al. 2003; Gilad et al. 2005]. D. sechellia and D. simulans, however, share 80.2% of these divergent bases. With <2% of sites polymorphic, the amount of sequence divergence between D. sechellia and D. simulans is approximately equal to levels of polymorphism seen between geographic disparate populations of D. melanogaster (Ometto et al. 2005). Thus most sequence divergence between the array and our species is irrelevant to any relative comparisons between D. sechellia and D. simulans. Moreover, the long probes of the DGRC arrays are relatively robust to a few mismatches. Nevertheless, as discussed below, we used several approaches to control for any hybridization mismatches (see also Holloway et al. 2007; Kopp et al. 2008; Mezey et al. 2008).
We used BLAST to compare the probe sequences from the arrays to identify homologous regions in D. simulans and D. sechellia. We conduct our analysis on each chromosome arm to minimize spurious matches. Contigs with the best matches to the target sequences were then compared to the probe sequences. Full matches were then extracted, the percentage of match was calculated, and the locations and types of mismatches were noted (available from the authors).
Sequence divergence between D. simulans and D. sechellia is not an issue for this study. The range of sequence divergence between the two species for the features on the array is between 0 and 18%, with a mean of 1.9%. If divergence in the probe sequence affected hybridization, then it would be expected that regressing mean expression difference (between species) onto probe divergence between these two species should explain a significant fraction of the variation for expression difference. However, R2 ≪ 0.01, suggesting that there is little evidence of a linear effect. The Q-Q plot between expression difference and sequence divergence was not consistent with nonlinear effects (Sokal and Rohlf 1995, pp. 118–123). Even the 50 genes that show the greatest degree of interspecific difference in expression show a mean divergence of 2.2%, which was not significantly different from the data set as a whole.
While divergence between D. simulans and D. sechellia is not an issue in our array experiments, the differences between the D. melanogaster-based cDNAs on the array and the mRNA of D. simulans and D. sechellia may affect the intensity of hybridization for any particular gene. Recent work has shown that this effect inflates the variance in hybridization signal (Mezey et al. 2008). This makes detecting a significant difference between “treatments” harder and, therefore, our results will be conservative. Moreover, as our comparison is between D. simulans and D. sechellia, which are equally diverged from D. melanogaster, our results will be robust to any differences between the mRNA from these species and the sequences of the probes. Nevertheless, we confirmed our results with both species-specific qRT–PCR and Affymetrix array data that were masked for any divergent probes.
Quantitative RT–PCR:
For confirmation of the relative expression levels between the two treatments, quantitative RT–PCR was carried out with SYBR Green technology (Quantitect SYBR Green RT–PCR kit, Qiagen catalog no. 204243) on the total RNA used for the microarray analysis. Primers for eight genes were designed to yield 150- to 250-bp products (gene products actin5c, Obp56e, SP50, nompB, jonahfi, arc42, Fad2, and cp36; supplemental Table 5). actin5c was used as the control. The dual reverse transcription and PCR were carried out in duplicate for each sample in an iCycler (Bio-Rad, Hercules, CA) in 25-μl volume as per the Qiagen Quantitect SYBR Green RT–PCR kit protocol. Reverse transcription of RNA was performed at 50° for 30 min and then at 95° for 15 min, followed by 40 cycles of 94° for 15 sec, 55°–60° for 30 sec, and 72° for 30 sec. A melting curve was implemented during the reactions to check for the possibility of mispriming or primer dimer formation. Primer sequences are in supplemental Table 5.
Three replicates for each treatment and/or species were used, with the exception of SP50 for no-choice D. simulans, where only two replicates were used. The direction of the species effect was consistent between our array data (noted in supplemental Tables).
Affymetrix GeneChip expression arrays:
As a validation procedure for the general efficacy of the DGRC expression analysis as well as for concerns regarding sequence divergence, we utilized the Affymetrix GeneChip Drosophila Genome 2.0 Arrays for the hybridization of RNA from the bodies of both D. simulans and D. sechellia. This allowed us to utilize not only a very different technology, but also an independent approach for correcting any interspecific probe mismatches. In particular, all probes that were not perfect three-way matches between D. melanogaster, D. simulans, and D. sechellia were “masked” and not included in the analysis (Holloway et al. 2007; Kopp et al. 2008; Mezey et al. 2008).
We utilized two arrays per species per treatment as described previously for the DGRC arrays. Starting with the same biological materials that were used for the original expression profiling experiment, we followed the manufacturer's protocol for labeling and hybridization. Specifically, 7 μg of total RNA were used to synthesize cDNA. A custom cDNA kit from Life Technologies was used with a T7-(dT)24 primer for this reaction. Biotinylated cRNA was then generated from the cDNA reaction, using the BioArray High Yield RNA Transcript kit. The cDNA was then fragmented in fragmentation buffer (5× fragmentation buffer: 200 mm Tris-acetate, pH 8.1, 500 mm KOAc, 150 mm MgOAc) at 94° for 35 min before the chip hybridization. A total of 15 μg of fragmented cRNA were then added to a hybridization cocktail (0.05 μg/μl fragmented cRNA; 50 pm control oligonucleotide B2, BioB, BioC, BioD, and cre hybridization controls; 0.1 mg/ml herring sperm DNA; 0.5 mg/ml acetylated BSA; 100 mm MES; 1 m [Na+]; 20 mm EDTA; 0.01% Tween 20). A total of 10 μg of cRNA were used for hybridization. Arrays were hybridized for 16 hr at 45° in the GeneChip Hybridization Oven 640. The arrays were washed and stained with R-phycoerythrin streptavidin in the GeneChip Fluidics Station 400. After this, the arrays were scanned with the Hewlett Packard GeneArray Scanner. Affymetrix GeneChip Microarray Suite 5.0 was used for washing, scanning, and initial quality analysis. Sample quality was assessed by examination of 3′–5′ intensity ratios of certain genes.
Analysis of Affymetrix array expression data:
We mitigated the effects of sequence divergence between D. simulans and D. sechellia, for each D. melanogaster probe on the Affymetrix GeneChip, using an approach analogous to that used for the DGRC probes. We identified in D. simulans and D. sechellia the target sequences (available from affymetrix.com) homologous to the D. melanogaster target sequences. Only probes that showed a three-way perfect match between the species were then included in analyses. The mismatch probes on the arrays were not used for background correction or normalization. Instead a global median-based normalization was performed on the log2-transformed data. Following the normalization the gene-specific models
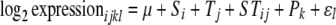 |
were used, where the model terms are as described above, and with P representing the fixed effect of probes.
We examined those genes that were deemed “significant” between species at a nominal level of 0.01 in the DGRC chip and compared the estimated difference in expression between species with the Affymetrix data. For the 343 features represented at this assigned probability level, the correlation between estimated measures of expression difference was quite high (r = 0.82) with a slope of 0.81. After removing features that represent duplications on the DGRC chip or alternate transcripts that could not be properly assigned to both platforms, the 315 features showed approximately the same degree of correlation (r = 0.82) and slope (0.83). Thus the expression differences that are observed are not due to either the platform or issues with sequence divergence between species.
Molecular evolutionary analysis:
We generated multiple alignments of Obps and Arc42 from D. sechellia, D. simulans, and D. melanogaster. We then analyzed these sequences using PAML, which provides a suite of maximum-likelihood-based tools for combining DNA sequence and phylogenetic data to test molecular evolutionary hypotheses (Yang 1997). With PAML, we estimated the ratio of synonymous to nonsynonymous substitutions along both the D. simulans and D. sechellia lineages.
Identification of potential loss-of-function mutations:
For a subset of the differentially expressed genes identified in our analysis, we screened for potential loss-of-function alleles that differed between the two species. Gene models from D. melanogaster genome annotation (v4.3) were mapped on to the genomic sequences of D. sechellia and D. simulans. We used mosaic assembly of D. simulans produced by LaDeana Hillier and colleagues (Washington University Genome Sequencing Center). We coupled these data with the sim6-specific data from the Drosophila Population Genomics Project (http://www.dpgp.org) syntenic assembly (Begun et al. 2007). For D. sechellia, we used the CAF1 assembly. Most gene models were easily adapted across species, although a subset required individual annotation as a result of insertion/deletion polymorphism or gaps in the genomic sequence.
Potential loss-of-function mutations fell into two broad categories of aberration: (1) obvious deletions or insertions that removed a canonical start codon or caused frameshift mutations that led to a premature stop codon and (2) point mutations or single-base-pair insertions or deletions causing premature stop codons. The former we classified as “highly probable”; the latter we classified as “probable.” The mutations were verified by direct resequencing in several cases (such as the Obps; Kopp et al. 2008) and by reanalysis of the assembly from the raw reads and .agp files in all cases. Unless the majority of high-quality reads supported the aberration, we excluded it from analysis. In general, point mutations or single-base-pair indels were often unreliable. Larger indels were always supported.
RNAi knockdown of Obp56e in D. melanogaster:
We crossed w1118; P{UAS-RNAi Obp56e} (hereafter, UAS-RNAi Obp56e) to y1 w1; P{Act5c-GAL4, y+ }/CyO (hereafter, Act5c-GAL4). Act5c-GAL4 ubiquitously expresses GAL4 throughout the fly. We obtained two genotypes from this cross: Act5c-GAL4/UAS-RNAi Obp56e and CyO/UAS-RNAi Obp56e. We assayed both genotypes using our behavioral assay. We verified that Act5c-GAL4 was broadly expressed by crossing to UAS-CD8∷GFP (P{UAS-mCD8∷GFP.L}LL4, y1 w*; PinYt/CyO) and UAS-rpr (w1118 P{UAS-rpr.C}) stocks. Act5c-GAL4/UAS-CD8∷GFP flies express GFP broadly in larval, pupal, and adult tissues (data not shown). As expected, most Act5c-GAL4/UAS-rpr flies died as expected, but not all (Act5c-GAL4/UAS-rpr, N = 69; CyO/UAS-rpr, N = 368).
RESULTS
Interspecific differences in preference behavior:
D. sechellia is attracted to fatty acids commonly found in its host plant, Morinda, whereas D. simulans and D. melanogaster are repelled by these same compounds (R'Kha et al. 1991; Legal et al. 1994, 1999; Farine et al. 1996; Amlou et al. 1998a). The host preference behavior of D. sechellia has been modeled in the lab either with chemotaxis (R'Kha et al. 1991; Higa and Fuyama 1993; Amlou et al. 1998a; Legal et al. 1999) or with oviposition-site preference experiments (R'Kha et al. 1991, 1997; Jones 2004; Matsuo et al. 2007). We developed both types of assay, verified that they produced comparable results, and showed a strong species difference in behavior.
Using the assay of Jones (2004), we compared the oviposition-site preference of D. sechellia to D. simulans and D. melanogaster and confirmed the strong species difference in oviposition-site preference. D. melanogaster and D. simulans avoid ovipositing on medium containing fatty acids found in Morinda fruit (“Morinda medium”), whereas D. sechellia prefers to oviposit on it (D. melanogaster, PI = −0.83, N = 62; D. simulans, PI = −0.86, N = 63; D. sechellia, PI = 0.61, N = 69; Kruskal–Wallis test corrected for ties, H = 108.085, P < 0.0001).
F1 hybrids between D. sechellia and D. simulans show D. simulans-like oviposition preference for Morinda fruit and Morinda medium (Higa and Fuyama 1993; Amlou et al. 1998a). By crossing D. simulans Islamorada females to D. sechellia line 1 males and scoring the oviposition-site preference of the resulting F1 females, we confirmed that F1 hybrid females show D. simulans-like preference (PI = −0.71, N = 60; Mann–Whitney U-test, with the normal approximation, contrasting D. sechellia and F1 hybrids, Z = −8.672, P < 0.0001). However, F1 hybrids are not as extreme in their avoidance as D. simulans (Mann–Whitney U-test contrasting D. simulans and F1 hybrids: Z = −2.303, P = 0.0213). Thus D. sechellia-like behavior is incompletely recessive, relative to D. simulans alleles.
Next, we tested the preference of flies in a choice–no-choice experiment. D. simulans and D. sechellia females were each given a choice between standard medium and medium containing the fatty acids commonly found in Morinda. The choice–no-choice assay differs from the oviposition-site preference assay in that it requires the flies to find and follow an odor plume before encountering the Morinda medium, rather than choosing between two types of media a few centimeters apart. Thus, the choice–no-choice assay tests seeking behavior.
We compared D. simulans sim6 and D. sechellia line 1, using the choice–no-choice assay. If given a “choice,” D. sechellia preferred Morinda medium (in eight of eight replicates the majority of flies were on Morinda medium; fraction of D. sechellia on Morinda medium, 82 ± 8% SE); D. simulans actively avoided the Morinda medium (in zero of eight replicates the majority of D. simulans were on Morinda medium; fraction of D. simulans on Morinda, 4 ± 2% SE). We confirmed this result for several other D. simulans and D. sechellia lines and with actual Morinda fruit (data not shown, see materials and methods).
The results from both the oviposition-site preference assay and the choice–no-choice assay are consistent: D. sechellia shows a species-specific preference for medium containing fatty acids from its host plant.
Transcriptional profiling identified genes differentially expressed between D. sechellia and D. simulans independent of medium:
Our goal is to identify genes important for host preference in D. sechellia. We used whole-genome transcriptional profiling to identify genes induced in D. sechellia by exposure to compounds from its host. In particular, we contrasted gene expression in D. sechellia when these flies were allowed to choose its preferred Morinda medium vs. D. sechellia that were forced to use standard medium. We performed a parallel experiment with D. simulans as a control. Some genes important for host preference may not be induced by Morinda medium (Matsuo et al. 2007; Kopp et al. 2008). We found these loci by identifying genes that differed in expression (independent of choice of medium) between D. sechellia and D. simulans.
Interspecific transcriptional profiling:
We separately contrasted genomewide relative transcript abundance of bodies and heads for D. simulans and D. sechellia in two experiments (described below). These experiments involved hybridization of heterologous cDNA to the D. melanogaster derived probes on the microarrays. We controlled for this complication three ways: bioinformatic masking of divergent probes between D. simulans and D. sechellia, direct comparison of both a short oligonucleotide array (Affymetrix) and a PCR amplicon array (DGRC), and verification of expression differences using qRT–PCR. We found little evidence for an effect of sequence divergence between D. simulans and D. sechellia. Moreover, among significant genes on the Affymetrix and DGRC arrays, the correlation between estimated measures of expression difference was high (r = 0.82) with a slope of 0.81. The expression differences we observed are not due to either the platform used or sequence divergence between species. In the sections that follow, we focus our discussion on the genes on the DGRC array as these are a subset of those on the Affymetrix array and thus can all be confirmed with the Affymetrix data. Additionally, we performed qRT–PCR on eight genes (materials and methods; supplemental Tables 1–4) from the D. sechellia and D. simulans body samples. For all eight genes the direction of the effect was consistent between the array data and the qRT–PCR. The arrays, however, repeatedly underestimate the change in expression in seven of the eight cases (supplemental Tables 1–4).
Genes for egg production are strongly induced in the body of D. sechellia by exposure to host plant compounds:
We compared the effect of the choice–no-choice treatment on both D. sechellia and D. simulans. This comparison identified genes that differed in expression when D. sechellia or D. simulans was on its preferred media. To identify species-specific changes in gene expression, we looked at the interaction of species and treatment. Genes involved with female reproduction were strongly overrepresented and upregulated when D. sechellia was on its preferred host, especially those involved in egg production (Figure 3; Table 1; supplemental Table 1).
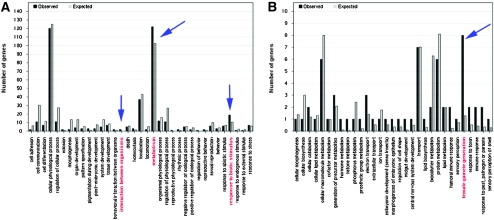
Figure 3.—
Genes involved with biotic interactions, metabolism, and oogenesis are differentially regulated in D. sechellia on media containing compounds from its host plant Morinda. (A) Genes involved in biotic interactions and metabolism are differentially regulated between the choice–no-choice preference assays in the head of D. sechellia. In addition, arrows indicate GO categories that show significant overrepresentation. (B) As suggested by overrepresentation of differentially expressed genes in Gene Ontology (GO) categories, genes involved with oogenesis are upregulated in the bodies of D. sechellia on the media containing Morinda compounds, which is consistent with previously described upregulation of egg production.
TABLE 1.
D. sechellia genes expressed in the body that changed in response to Morinda medium
| Biological rolea | Genesb | Comments |
|---|---|---|
| Egg production | Up: Chorion protein genes (Cp7c, Cp7Fb, Cp18, Cp19, Cp36, Cp38), Femcoat, Vitelline membrane (Vm34Ca, Vm32e) | qRT–PCR suggests that Cp36 expression is induced at least sixfold in D. sechellia on Morinda medium. shd plays a role in a variety of biological processes, including egg chamber growth. |
| Down: shd | ||
| Odor or taste perception | Up: Obp99a | The expression pattern of Obp99a is not known in adult flies.It is expressed in the dorsal organ of larvae (Galindo and Smith 2001). |
| Down: Gr43b, Gr59b, king-tubby | ||
| Digestion and metabolism | Up: Lysozymes (LysE, LysS), CG10163, CG12374, CG17633, CG8560, CG12116 | Lysozymes serve a variety purposes in Drosophila, including a major role as digestive and defensive enzymes (Daffre et al. 1994; Regel et al. 1998). CG10163 is important for lipid metabolism. |
| Down: CG32635 | ||
| Toxin or defense response | Up: Cyp6g1, Lysozymes (LysE, LysS) | Same as above. Cyp6g1 is a cytochrome P450-like gene. CG30437 may be involved in the breakdown of phenols. |
| Down: CG30437 |
Genes with unknown biological process were not analyzed.
As indicated by GO analysis and FlyBase curation.
Full list is in supplemental Table 1.
As expected, D. simulans showed no significant changes in gene expression between treatments, as the medium chosen by D. simulans in the choice experiment is exactly the same medium as is present in the no-choice experiment.
Response to stimulus and metabolism genes are induced in heads of D. sechellia by Morinda medium:
Complex combinations of genes change expression in the heads of D. sechellia and D. simulans when exposed to Morinda medium (Figure 2, B and D; supplemental Table 2). At our FDR q of 0.02, 257 genes are significantly differentially expressed in D. sechellia heads when choosing Morinda medium, of which 14 are upregulated and 233 are downregulated. Three major gene ontology classes were significantly overrepresented in the heads of D. sechellia in the choice experiment. These classes were (1) metabolism, (2) interspecies interactions, and (3) response to biotic stimulus (Figure 3; Table 2). As expected, genes involved in fatty acid metabolism—octanoic acid and hexanoic acid are fatty acids—are also strongly differentially expressed in response to Morinda medium (e.g., CG4500 and CG9914; supplemental Table 2).
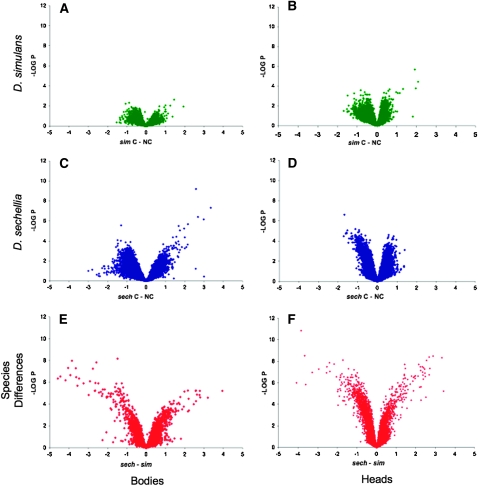
Figure 2.—
Volcano plot showing differences in transcriptional profiles between D. simulans and D. sechellia and in preference assays for each species. (A, C, and E) Data from bodies; (B, D, and F) data from heads. FDR q-value = 0.02 for the between-treatment analysis; FDR q-value = 0.01 for the between-species analysis. As expected, virtually no genes are differentially expressed between the choice (“C”) and no-choice (“NC”) preference assays for D. simulans, which prefers standard media (A and B, green). In contrast, many genes appear to be differentially expressed between the choice and no-choice assays in D. sechellia, which shows a strong preference for the media containing compounds from its host plant Morinda (C and D, blue). (E and F) (red) show differences in gene expression between the two species that are not the results of choice treatments. Heads show more expression changes than bodies. There are substantial expression changes between species. On average, genes are slightly more highly expressed in D. sechellia relative to D. simulans. Despite this, many more genes are strongly downregulated in D. sechellia relative to D. simulans. On the x-axis the difference in log2 expression between factor levels is noted below the axis. The y-axis displays the log10 of the P-value of those differences from the linear mixed model.
TABLE 2.
D. sechellia genes expressed in the head that changed in response to Morinda medium
| Biological rolea | Genesb | Comments |
|---|---|---|
| Odor or taste perception | Up: Obp56e, | Obp56e is normally expressed in antennae (Galindo and Smith 2001). |
| Down: none | ||
| Digestion and metabolism | Up: SP50, Lectin-28c, CG14990, CG7910, CG9914 | CG14990 is a serine proteinase homolog (SPH97) (Ross et al. 2003). CG4500 and CG9914 are genes involved in fatty acid metabolism. |
| Down: Spat, Dmdmc, Prat2, CG11796, CG4500, CG14935, CG6484, CG8234, CG3011, CG3999, CG12030, CG9485, CG5288, CG13795, CG31075, CG33138, CG5288 | ||
| Toxin or defense response | Up: Cyp309a2 | Cyp309a is a cytochrome P450-like gene. Drosomycins are important for fly defense responses. |
| Down: Drosomycins (dro5, Drs), Spat, Tsf1, Def CG18522, CG4302, CG4302 | ||
| Regulation of transcription or translation | Up: Su(z)12, Z4, MTA1-like, RpL8 | Su(z)12 and Z4 both affect chromatin. |
| Down: none |
Results from the 62 most differentially expressed genes are shown. Genes with unknown biological process were not included.
As indicated by GO analysis and FlyBase curation.
Full list is in supplemental Table 1.
Serine proteinase CG32523 (serine proteinase 50, SP50) (Ross et al. 2003) is the most strongly upregulated gene when D. sechellia is on Morinda medium—its expression is increased 3-fold. Strikingly, this gene is normally expressed at 10-fold lower levels in D. sechellia than in D. simulans (below). In D. simulans, SP50 is highly expressed (∼5-fold higher than the median of all D. simulans genes). This large difference in expression is explained by the fact that SP50 is a likely pseudogene in D. sechellia—525 bases, including part of the second exon, are absent. This deletion undoubtedly affects the function and expression of SP50. Although SP50 deletion includes part of the probe sequence of the DGRC array, the Affymetrix probes used in the analysis are unaffected. Both array platforms consistently show lower expression of SP50 in D. sechellia compared to D. simulans and an upregulation of SP50 when D. sechellia is on Morinda medium. Moreover, the choice–no-choice comparison within species is unaffected by differences in the probe.
Obp56e, which is expressed in the antennae and likely has a role in odor perception (Galindo and Smith 2001; Shanbhag et al. 2001; Graham and Davies 2002; Hekmat-Scafe et al. 2002; Kopp et al. 2008), shows a similar pattern to SP50. Obp56e expression is increased 50% in the heads of D. sechellia exposed to Morinda medium relative to expression on the control medium. Like SP50, Obp56e is normally expressed at much lower levels (∼14-fold less) in heads of D. sechellia relative to D. simulans. (The DGRC probe is intact; D. sechellia is 94.8% identical to D. melanogaster; D. simulans is 96.5% identical to D. melanogaster.) Similar to SP50, Obp56e is a likely pseudogene. The beginning of the second exon of Obp56e contains a 7-base deletion that leads to a premature stop codon at the 60th amino acid position, reducing the peptide by 56% (supplemental Figure 1).
None of the other genes upregulated in the head or body appear to be candidate pseudogenes like SP50 and Obp56e. Several of these upregulated genes are involved in DNA binding and signal transduction, suggesting they may play a role in physiological and behavior responses of D. sechellia to Morinda.
As noted above, the control D. simulans choice–no-choice comparison should show little to no differences between treatments. Only 19 genes show a change in expression in the heads of D. simulans (9 downregulated, 10 upregulated; Figure 2, supplemental Table 1). None of these genes overlap with those upregulated in the heads of D. sechellia.
Minimal overlap between gene expression in heads and bodies in response to Morinda medium:
Among the genes that change expression in a species-specific manner in response to Morinda medium, there is little overlap between heads and bodies. Only CaBP1 and CG1648 significantly change expression in both samples (supplemental Tables 1 and 2). CaBP1 is a disulfide oxidoreductase and thus likely a detoxification gene. CG1648 has no known function. Interestingly, in both cases the direction of expression alternates between heads and bodies. Both genes are downregulated in heads and upregulated in bodies.
Gene expression differences between species are asymmetric:
Figure 2 shows that expression differences caused by the choice treatment (Figure 2, A–D) are much less than the “constitutive” gene expression differences between species (Figure 2, E and F). Figure 2, E and F, also shows that a significant subset of genes in D. sechellia is greatly reduced in expression compared to D. simulans (bodies, ratio of the average normalized expression level of statistically significant D. sechellia genes to the average normalized expression level of statistically significant D. simulans genes, 0.66, χ2 = 4.890, P = 0.027; heads, ratio, 0.32, χ2 = 148.929, P < 0.0001). Across all genes—both significant and nonsignificant—D. sechellia genes are somewhat more expressed (bodies, ratio, 1.97, χ2 = 848.031, P < 0.0001; heads, ratio, 1.07, χ2 = 8.723, P = 0.0031), which rules out a bias in our estimates of species-specific expression as a source of the asymmetry.
As before, the patterns of gene expression in the head are more complex than those in the body (Figure 2; supplemental Tables 3 and 4). In the head, genes affecting metabolism, response to stimulus (abiotic, biotic, and external), and response to stress differ between the species. In the body, responses to biotic stimulus—including putative immune, defense, and chemical stimulus response—are the main biological processes different between the species.
Comparison of the genes differentially expressed in the choice–no-choice analysis to the between-species analysis shows that many genes are in both data sets (25 body data, 108 head data). Roughly 80% of these genes “flip-flop” in relative expression between experiments—e.g., genes that are induced in D. sechellia by exposure to its host are normally expressed at lower levels in D. sechellia compared to D. simulans (supplemental Tables 6 and 7).
Genes affecting fatty acid metabolism are highly expressed in D. sechellia relative to D. simulans:
The gene most highly expressed in D. sechellia bodies relative to D. simulans is Fatty acid desaturase (Fad2). Fad2 is expressed at eightfold higher levels in bodies of D. sechellia compared to D. simulans; however, this difference is not observed in the heads. Arc42 (an acyl-CoA dehydrogenase), which mediates the first step in the β-oxidation of fatty acids, is expressed at least fourfold higher in D. sechellia bodies (validated with qRT–PCR; Arc42 has two DGRC probes, both of which suggest increased expression). Likewise in heads, Arc42 is also among the 12 genes showing increased expression in D. sechellia relative to D. simulans. CG9009, which is also involved in fatty acid metabolism, is increased relative to D. simulans in heads (expression is increased in bodies as well, but P = 0.008 does not cross our conservative threshold). The principal toxins in Morinda fruit are fatty acids (Legal et al. 1994, 1999). The upregulation of these genes may be important for detoxification of these fatty acids and/or utilization of these fatty acids as a nutritional resource, and they thus represent excellent candidates for future study for the genetic basis of host plant resistance in D. sechellia.
Several genes differentially expressed between D. simulans and D. sechellia heads and bodies are involved in odor perception:
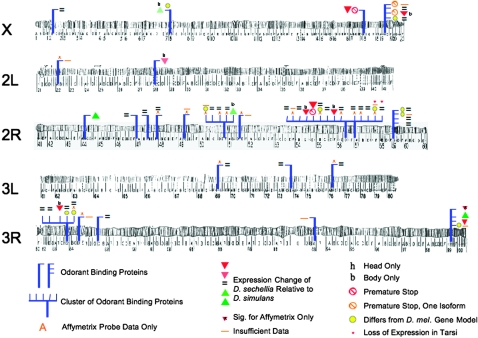
In bodies, two genes involved in perception, Odorant binding protein 99b (Obp99b) and no mechanoreceptor potential B (nompB), are also expressed severalfold higher in D. sechellia. Data from all three DGRC probes of nompB and our qRT–PCR results were consistent with this increase. At Obp99b, data from both DGRC probes suggest increased expression. Obp99b is part of a cluster of Obps on the right arm of chromosome 3 (Figure 4), all of which may play a role in mate recognition and detection of odorants (Mackay et al. 2005; Wang et al. 2007). Two other members of this group are more weakly expressed (Figure 4). Several other genes are also strongly differentially expressed in D. sechellia bodies, but little is known about them (Senescence marker protein-30, CG30419, CG14499, CG15254, CG13183, and CG11669).
Figure 4.—
Location, expression, and functional status of odorant binding proteins (Obps) in D. sechellia. The definitions of the symbols are at the bottom. Additional details can be found in Table 3. All genes were assayed using both the DGRC array and the Affymetrix GeneChip, unless otherwise indicated. With the exception of Obp99a and Obp99c, the gene expression differences were consistent between the DGRC and GeneChip. Gene models with minor differences among species are marked in yellow. Half of these differences from D. melanogaster are shared between D. simulans and D. sechellia, half are specific to D. sechellia, and none are specific to D. simulans. Among Obps, more than twice as many are less expressed in D. sechellia (nine) than in D. simulans (four) and several appear nonfunctional in D. sechellia. Chromosome images are from Lindsey and Zimm (1992).
In heads compared to in bodies, seven times more genes are differentially expressed between species. However, only 32 of these differentially expressed genes are shared between heads and bodies. The most divergently expressed gene is CG9509, a potential choline dehydrogenase. Among the top 12, 4 are predicted genes (CG3699, CG3513, CG9497, and CG18493) that are homologous to genes involved in metabolism, but with little functional information. Also highly divergently expressed are Defensin (Def), Tektin-C, takeout (to), Obp99b, and Attacin-A (AttC). Def and AttC are both normally induced by immune challenges. Tektin-C, although primarily associated with microtubules, is also considered a candidate for smell-impaired 65 (smi65) (Anholt and Mackay 2001). The gene to affects feeding behavior in flies, mosquitoes, and aphids (So et al. 2000; Bohbot and Vogt 2005; Ghanim et al. 2006). Other than a few small in-frame indels, none of these genes are associated with any sequence anomalies.
Loss-of-function alleles are associated with decreased gene expression in D. sechellia:
As noted above, SP50 and Obp56e are strongly induced in heads when D. sechellia is on Morinda, yet are expressed at much lower levels in D. sechellia relative to D. simulans in both bodies and heads and are likely nonfunctional. This pattern is true for several other Obps and related genes (Figure 4; Table 3). Significantly differentially expressed genes are enriched for Obps (vs. genomewide expectation, χ2 = 7.24, P = 0.0071). The majority of Obps show reduced expression in D. sechellia relative to D. simulans. One-third of Obps are only weakly expressed relative to the average gene on our array and thus our ability to detect a difference among these Obps is limited. Of those genes that are differentially expressed between the species, some are associated with potential loss-of-function alleles (Figure 4). Given the role of Obps in olfaction, the olfactory system of D. sechellia has clearly diverged from D. simulans.
TABLE 3.
Expression patterns of genes related to odor perception
| Genea | Expression differenceb | Known expression patternc | Reference(s)d |
|---|---|---|---|
| Gr22c | ND | Tarsi, foreleg | 3 |
| Gr22f | ND | Labial palp, abdomen, leg, wing | 3 |
| Gr28bA | ND | Labellum, cibarial organs | 7 |
| Gr39aA | ND | Labellum, thorax, abdomen, wing | 1 |
| Gr39b | ND | Labellum | 1 |
| Gr43B | Down (body) | Labellum, abdomen, leg, wing | 1 |
| Gr58a | ND | Labellum, thorax | 1 |
| Gr59b | Down (body) | Labial palp | 3 |
| Obp18a | Down (antennae) | Antennae, nonspecific | 5, 9 |
| Obp19a | Down (all) | Antennae | 5, 9 |
| Obp19c | Down (body) | Dorsal organ, LOS, cibarial organs | 5 |
| Obp49a | Up (antennae) | Antennae | 9 |
| Obp44a | Up (all) | Embryonic (stages 13–16), adult | 4, 8 |
| Obp50a | Up (antennae) | Antennae | 9 |
| Obp50e | Up, weakly (body) | Antennae, labellum, whole body | 5 |
| Obp56d | Down (head, antennae) | Dorsal organ, antennae, maxillary palps, wing, tarsi | 5, 9 |
| Obp56e | Down (all) | Antennae, labellum | 5 |
| Obp56h | Down (head) | Broadly expressed | 5 |
| Obp57de | Down (tarsi only) | Tarsi, weak whole body | 5, 6, 8 |
| Obp58c | Up (antennae) | Antennae | 9 |
| Obp59a | Down (antennae) | Antennae | 9 |
| Obp83cd | Down (head) | Labellum | 5 |
| Obp83ef | Up (antennae) | Antennae, nonspecific | 5, 9 |
| Obp99a | Down (Affy only) | Dorsal organ, nonspecific | 5 |
| Obp99b | Up (all) | Antennae, maxillary palps | 5 |
| Obp99c | Down (antennae) | Antennae | 9 |
| Obp99d | Down (antennae) | Antennae | 9 |
| Or9a | Down (antennae) | Antennae, basiconic sensillia (ab8) | 2, 9 |
| Or13a | ND | Antennae, intermediate sensillia (ai1) | 2 |
| Or19a | Down (antennae) | Antennae, tricoid sensillia (at3) | 2, 9 |
| Or22a | Up (antennae) | Antennae, basiconic sensillia (ab3) | 2, 9 |
| Or22b | Down (antennae) | Antennae, basiconic sensillia (ab3) | 2, 9 |
| Or35a | Up (antennae) | Antennae, coeloconic sensillia (ac1) | 2, 9 |
| Or42b | Down (antennae) | Antennae, basiconic sensillia (ab1) | 2, 9 |
| Or65a | Down (antennae) | Antennae, tricoid sensillia (at4) | 2, 9 |
| Or65c | Down (antennae) | Antennae, tricoid sensillia (at4) | 2, 9 |
| Or67a | Down (antennae) | Antennae, basiconic sensillia (ab10) | 2, 9 |
| Or82a | Down (antennae) | Antennae, basiconic sensillia (ab5) | 2, 9 |
| Or85a | Down (antennae) | Antennae, basiconic sensillia (ab2) | 2, 9 |
| Or85b | Up (antennae) | Antennae, not localized | 9 |
| Or98b | ND | Antennae, thin and small sensillia (hypothetical) | 2 |
| Pbprp5 | Down, weakly (head) | Dorsal organ, antennae | 5 |
ND, no data.
Gene symbol of genes with interspecific differences in expression or loss-of-function alleles in D. sechellia (boldface type). Data are from Matsuo et al. (2007), McBride (2007), Kopp et al. (2008), and herein.
Direction and patterns of constitutive expression differences between D. sechellia and D. simulans.
Tissues known to express this gene in D. melanogaster and its relatives. LOS, labral sensory organ.
References for expression patterns: 1, Clyne et al. (2000); 2, Couto et al. (2005); 3, Dunipace et al. (2001); 4, FlyExpress; 5, Galindo and Smith (2001); 6, Matsuo et al. (2007); 7, Scott et al. (2001); 8, herein; and 9, Kopp et al. (2008).
In bodies, our data also suggest that several genes expressed at lower levels in D. sechellia relative to D. simulans are also probable pseudogenes (supplemental Tables 3 and 4). For example, serine peptidase 83 (SP83) is expressed at least 20-fold less in D. sechellia relative to D. simulans. This is likely a result of a 182-base deletion in the 5′ end of SP83 that includes the SP83 start codon (supplemental Figure 2; DGRC probe is not affected). Similarly, δ -Trypsin (δTry) is less expressed in D. sechellia and appears to be nonfunctional as a result of several premature stop codons and a frameshift mutation. A related gene, θ-Trypsin (θTry; SP139) also harbors a confirmed premature stop codon. Jonah99fi (Jon99fi), a member of a large family of trypsin genes, is expressed at least at 20-fold lower levels in D. sechellia relative to D. simulans (supplemental Table 3). The 5′ end of the Jon99fi coding sequence appears deleted in D. sechellia, perhaps removing part of Jon99fii as well. Ten of the 13 Jonah loci that are represented on the array are significantly less expressed in D. sechellia (the other 3 show no significant differences; supplemental Table 3). Similarly, three other trypsin genes from a family of trypsins at 47F4 on chromosome 2 are also significantly downregulated in D. sechellia (none of the other eight genes in this family are significantly different). Another trypsin/chymotrypsin, CG18180, is also among the genes that have decreased expression in D. sechellia. These data suggest that D. sechellia may have dispensed with many proteinases.
The data from heads do not show the same enrichment for non-functional proteinases, although CG8329—the gene with the most decreased expression in D. sechellia heads—is likely a chymotrypsin (SP170). The coding regions of this gene are intact, but 80% of the 3′-UTR is missing in D. sechellia (the DGRC probe is not affected by this deletion). In addition to the Obps mentioned above, only Glutathione-S-transferases (Gsts) and Cytochrome P450s stand out as having rapidly diverged in their expression pattern. Gsts are about fourfold overrepresented (χ2 = 12.554, P = 0.0004), most of which are expressed less in D. sechellia (GstE9, GstE6, GstD9, GstE1, and GstD5). Three of these are also significantly less expressed in D. sechellia bodies (GstE6, GstD9, and GstE1). Of these three, only GstE6 is a putative pseudogene as it is missing 197 bases at the 5′ end of the gene. Gsts are intriguing because of their roles in detoxification and odorant removal (Rogers et al. 1999; Ranson et al. 2001) and recent work suggests that the genomic Gst content is highly labile in Drosophila (Low et al. 2007). Another set of detoxification enzymes, Cytochrome P450s, is significantly differentially expressed between the species (χ2 = 21.16, P < 0.0001). While 11 of 18 are less expressed in D. sechellia, this is likely due to chance (χ2 = 0.889, P = 0.34).
Not all genes that are less expressed in D. sechellia relative to D. simulans appear nonfunctional, for example, several genes involved in egg production—Chorion genes, Femcoat, and yellow-g2. These genes are clearly functional in D. sechellia; the open reading frames of these genes are intact and loss-of-function mutations at Cp36 cause female sterility. The lower expression of these likely reflects the fewer ovarioles and lower rate of egg production in D. sechellia on standard medium (Jones 2004; Orgogozo et al. 2006).
Knockdown of Obp56e activity reduces avoidance response to Morinda fruit medium in D. melanogaster:
Obp56e is unusual in that its expression is substantially lower in the head of D. sechellia compared to D. simulans, yet when D. sechellia is exposed to Morinda medium, the expression of this gene increases relative to control medium. Compared to D. melanogaster and D. simulans, the D. sechellia ortholog harbors a 7-base deletion that results in a premature stop codon (supplemental Figure 1). These observations suggest that the ancestor of D. sechellia harbored a functional copy of Obp56e and that expression of this gene was increased by exposure to the volatiles from Morinda. The ancestor likely avoided Morinda just as D. melanogaster and D. simulans do today. Therefore, loss of a functional Obp56e in D. sechellia may have contributed to its shift to Morinda by removing some of the ancestral avoidance phenotype. This scenario predicts that Obp56e influences avoidance of Morinda in D. melanogaster and D. simulans. We tested this prediction by knocking down expression of Obp56e in D. melanogaster, using RNA interference-based gene silencing (RNAi) (reviewed in Mathey-Prevot and Perrimon 2006) and assaying the knockdown fly behavior relative to that of a genetically similar control. We crossed w1118; P{UAS-RNAi Obp56e} (hereafter, UAS-RNAi Obp56e) to y1 w1; P{Act5c-GAL4, y+}/CyO (hereafter, Act5c-GAL4). Act5c-GAL4 ubiquitously expresses GAL4 throughout the fly. We obtain two genotypes from this cross: Act5c-GAL4/UAS-RNAi Obp56e, which silences Obp56e, and CyO/UAS-RNAi Obp56e, which does not silence Obp56e. We assayed both genotypes using our behavioral assay.
Flies that had reduced Obp56e activity were seven times more likely to choose the Morinda medium than the controls (Fisher's exact test, two-tailed P = 0.0022; power analysis simulation suggests that this difference can be detected 95% of the time). In contrast, 98% of CyO/UAS-RNAi Obp56e flies avoided Morinda medium (N = 2 on Morinda medium; N =86 on regular medium). In contrast, only 85% of Act5c-GAL4/UAS-RNAi Obp56e flies avoid Morinda medium (n = 16 on Morinda medium; n = 90 on regular medium). These data are consistent with the loss of a functional Obp56e in D. sechellia reducing ancestral avoidance of Morinda. We verified that Act5c-GAL4 was expressed broadly. Our data suggest that “ubiquitously” expressed Act5c-GAL4 is only weakly expressed, which suggests that Obp56e influence on avoidance is likely greater than that observed in our experiment.
DISCUSSION
Ecological adaptations often lead to evolutionary diversification. The genetic causes and consequences of these adaptations, however, are not well known. In particular, the genetics of host specialization—a common ecological adaptation among phytophagous insects—is poorly understood. D. sechellia is a host specialist, which has recently evolved to use almost exclusively the fruit of M. citrifolia. We show here that a subset of genes in D. sechellia has dramatically reduced expression relative to D. simulans. This subset includes genes affecting olfaction, gustatory response, and protein metabolism. Surprisingly, among these genes are several that are induced when D. sechellia is exposed to medium containing compounds found in its host plant. These genes and others appear to be loss-of-function alleles. These data imply that reducing the expression of these genes may have been an important step during the evolution of host preference. Indeed our test of one of these genes, Obp56e, showed that it affects fly behavioral response to compounds found in Morinda. If most of these genes harboring loss-of-function alleles have functional consequences like Obp56e, then D. sechellia may have lost its ability to detect and metabolize a broad range of compounds. Unless new genes or functional alleles are formed easily and often, the loss of these genes in D. sechellia may limit its ability to shift to a chemically dissimilar host or return to being a generalist.
Expression changes important to host use:
Prior work shows that antennae and tarsal taste receptors are important for the ability of D. sechellia to find and respond to its host (Dekker et al. 2006; Jones 2007; Matsuo et al. 2007; Kopp et al. 2008). Genes known to affect odor perception, mating behavior, and feeding behavior show D. sechellia-specific changes. For example, takeout (to) expression is sevenfold higher in D. sechellia than in the generalist D. simulans. Hypomorphic mutants of to affect feeding behavior in D. melanogaster (Dauwalder et al. 2002; Meunier et al. 2007). These mutant flies feed indiscriminately and have lost their ability to modulate taste sensitivity (Meunier et al. 2007). Thus it is plausible that the “pickiness” of D. sechellia results in part from the increased expression of to. A similar pattern has been observed in the green peach aphid, Myzus persicae. While apterous adults feed broadly, the alates are highly host specific—they target members of the genus Prunus, especially peaches. The expression of a to homolog is increased in the picky alate stage of the aphid compared to the apterous adult (Ghanim et al. 2006).
Exposure to Morinda increases egg production and oviposition by ∼50% in D. sechellia (R'Kha et al. 1997; Amlou et al. 1998a; Jones 2004). Several key genes involved in egg production are dramatically upregulated when D. sechellia is on Morinda. Increased expression of chorion genes in the ovary follicle cells is achieved by gene amplification during oogenesis. While a number of trans-acting genes affecting this amplification are known, only two of these genes, Geminin and E2F (in heads only), show a marginally significant treatment effect in D. sechellia. Expression of Geminin normally inhibits amplification. Consistent with this role as a negative regulator of amplification, Geminin is downregulated when D. sechellia is on its host. The genetic data on this interspecific difference in egg production suggest a major role for the X chromosome and regions flanking the centromere of chromosome 2 (Jones 2004). Vm34Ca is likely within this same region of chromosome 2. Cp36, Cp38, and Cp7fC reside in cytological band 7F of the X chromosome, but as Jones (2004) did not genetically dissect the X chromosome, it is unclear how close these genes are to the regions causing interspecific differences in egg production.
D. sechellia is also resistant to the toxic levels of fatty acids found in Morinda (R'Kha et al. 1991; Amlou et al. 1998b; Jones 2005). Several genes involved in fatty acid metabolism differ between D. sechellia and D. simulans in expression, but only Arc42 is concordant with the earlier genetic data (Jones 1998, 2001). Arc42 is constitutively expressed at much higher levels in D. sechellia compared to D. simulans. This gene, however, does not show an elevated rate of amino acid substitution (dN/dS = 0.053). Given that Arc42 performs the first step of β-oxidation of fatty acids, the strong expression of Arc42 may contribute to resistance in D. sechellia. Regardless, Arc42—as well as Fad2, which from prior genetic studies is clearly not involved in resistance (Jones 1998, 2001)—is potentially involved in the ability of D. sechellia to take advantage of the nutritional value of the fatty acids in Morinda. The overall changes in transcription of genes involved with fatty acid metabolism and their physiological impacts on metabolism per se are complicated and will require further investigation.
“Genome decay” and the evolution of host preference:
Our data show that several genes with dramatically reduced expression in D. sechellia relative to D. simulans appear to be pseudogenes in D. sechellia. Obps and serine proteinases, in particular, appear to have suffered this fate. A similar pattern has been noted for gustatory receptors (McBride 2007; Table 3). The evolutionary significance of these genetic changes is not clear as there are several possible explanations for this pattern. First, some of these changes may have contributed to the adaptation of D. sechellia to its host. Second, many of these changes in expression or function may have occurred after D. sechellia became a specialist. Third, some of these genetic changes may have occurred as a result of the population bottleneck that accompanied the colonization of the Seychelles by D. sechellia (reviewed in Jones 2005; Gardiner et al. 2008).
Distinguishing among these scenarios is difficult. Obps, for example, move hydrophobic molecules, such as octanoic and hexanoic acid, to olfactory and gustatory receptors (Hallem et al. 2006) and recent work suggests that reduced expression in D. sechellia of Obp57d/e, which is associated with taste perception in tarsi, contributed to the evolution of oviposition-site preference for Morinda (Matsuo et al. 2007). Transgenic experiments in D. melanogaster show that decreased expression of Obp57d/e reduces the repulsion caused by hexanoic and octanoic acid, although reduced expression of Obp57d/e is not sufficient to recapitulate D. sechellia-like behavior. Interestingly, our data show that Obp57d/e is expressed in the whole body of D. sechellia at levels comparable to D. simulans, indicating that the reduced expression of Obp57d/e is limited to the tarsi. As shown in Figure 4, Obp57d/e is not the only Obp that is downregulated in D. sechellia; several neighboring Obps are also downregulated or nonfunctional. Obp56e, for example, has dramatically reduced expression and is clearly a pseudogene. Obp56e is normally induced by the presence of Morinda medium and is expressed in the antennae (Galindo and Smith 2001)—which are critical to host preference behavior. Loss of Obp56e function may have been an important step during the evolution of host preference in D. sechellia if Obp56e was important for avoidance of Morinda in the ancestor of D. sechellia. Consistent with this scenario, we showed that removal of Obp56e expression in D. melanogaster reduces its avoidance of Morinda medium.
Clearly, some Obps affect host preference. Whether the mutations in these genes were the critical changes is not yet known as we cannot prove that these changes are both necessary and sufficient to cause D. sechellia-like preference. For instance, Obp57d/e may have evolved reduced expression in the tarsi after the loss of Gr22c, which is a nonfunctional taste receptor in D. sechellia that is expressed in the same regions as Obp57d/e (Table 3; Dunipace et al. 2001; McBride 2007). (Because the transgenic tests of Obp57d/e were performed only in D. melanogaster, which has a functional Gr22c, we do not know if the presence or absence of Obp57d/e would have any effect in a fly lacking Gr22c.) Similarly, several Ors and Grs in D. sechellia appear to be nonfunctional as a result of premature stop codons or deletions (McBride 2007). These Grs and Ors (and Obps) may have been lost because these genes are no longer needed now that D. sechellia has specialized on Morinda (“relaxed selective constraint”), not because they are important to preference for Morinda in D. sechellia (Table 3).
Genetic drift may also have contributed to the abundance of apparent loss-of-function alleles in D. sechellia. Genetic evidence shows that D. sechellia underwent a strong population bottleneck when the species first colonized the Seychelles, when it shifted to Morinda, or after both events (Jones 2005). Gardiner et al. (2008) have recently suggested that loss-of-function alleles could have drifted to fixation among island endemics, although it is unlikely that all of these alleles would have been segregating in the small founder populations. D. melanogaster and D. simulans populations are often segregating considerable genetic and phenotypic variation in Obps, including loss-of-function alleles (Hekmat-Scafe et al. 2000, 2002; Sanchez-Gracia et al. 2003; Takahashi and Takano-Shimizu 2005; Sanchez-Gracia and Rozas 2007; Wang et al. 2007; Lavagnino et al. 2008; Matsuo 2008). For example, an Obp57e allele with a 10-bp deletion in its coding region is at a high frequency in Japan and found in several other populations (Takahashi and Takano-Shimizu 2005). Although Gardiner et al.'s analysis suggests that island endemicism caused the accumulation of loss-of-function alleles in D. sechellia, the functional effect of Obp56e on preference suggests that some of these loss-of-function alleles were important for the evolution of D. sechellia's preference. Clearly, we cannot yet distinguish between an adaptive explanation for gene loss at Obps (and by extension Grs and Ors) and the loss of these genes due to relaxed selective constraint.
Loss of genetic repertoire may restrict future evolution:
Both our work and that of others suggest that the olfactory repertoire of specialists, such as D. sechellia and D. erecta, contracts following specialization (Clark et al. 2007; Matsuo et al. 2007; McBride 2007; Nozawa and Nei 2007; Vieira et al. 2007; Gardiner et al. 2008; Kopp et al. 2008). Our data show that in D. sechellia this pattern may extend beyond olfactory genes. If this pattern is general to specialists, then specialists may be an evolutionary dead end because they lack the genetic wherewithal to return to being generalists or shift to chemically divergent hosts.
Acknowledgments
We thank Lindy McBride for much discussion and comments. We also thank Robert Anholt and the reviewers for extensive and insightful comments on the manuscript. We thank William Jeck, Erin Gragg, and Sophia Shih for technical assistance. We are both grateful to Greg Gibson and his lab for hosting us during part of this experiment. We also thank Sarah Liljegren for the occasional last-minute loan of reagents. This work benefited from several discussions with Hubi Amrein, Steve Crews, Bob Duronio, Laura Higgins, Artyom Kopp, Lauren McIntyre, and Patti Sheridan. We also thank Joel Kingsolver and Mohamed Noor for comments regarding the manuscript. This work was supported by a National Science and Engineering Research Council post doctoral fellowship (Canada) to I.D., by funds from University of North Carolina–Chapel Hill, and by a grant from the National Science Foundation to C.D.J. (DEB-0212686). We received materials through the Drosophila Genomics Resource Center, and this work was supported in part by a grant from National Institutes of Health (DK56350) to the University of North Carolina Clinical Nutrition Research Unit.
References
- Amlou, M., B. Moreteau and J. R. David, 1998. a Genetic analysis of Drosophila sechellia specialization: oviposition behavior toward the major aliphatic acids of its host plant. Behav. Genet. 28 455–464. [DOI] [PubMed] [Google Scholar]
- Amlou, M., B. Moreteau and J. R. David, 1998. b Larval tolerance in the Drosophila melanogaster species complex toward the two toxic acids of the D. sechellia host plant. Hereditas 129 7–14. [DOI] [PubMed] [Google Scholar]
- Anholt, R. R. H., and T. F. C. Mackay, 2001. The genetic architecture of odor-guided behavior in Drosophila melanogaster. Behav. Genet. 31 17–27. [DOI] [PubMed] [Google Scholar]
- Barker, J. S., W. T. Starmer and J. C. Fogleman, 1994. Genotype-specific habitat selection for oviposition sites in the cactophilic species Drosophila buzzatii. Heredity 72 384–395. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., A. K. Holloway, K. Stevens, L. W. Hillier, Y. P. Poh et al., 2007. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 5 e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernays, E., and M. Graham, 1988. On the evolution of host specificity in phytophagous arthropods. Ecology 69 886–892. [Google Scholar]
- Bernays, E. A., and R. F. Chapman, 1994. Host-Plant Selection by Phytophagous Insects. Chapman & Hall, New York.
- Bohbot, J., and R. G. Vogt, 2005. Antennal expressed genes of the yellow fever mosquito (Aedes aegypti L.); characterization of odorant-binding protein 10 and takeout. Insect Biochem. Mol. Biol. 35 961–979. [DOI] [PubMed] [Google Scholar]
- Bono, J. M., L. M. Matzkin, S. Castrezana and T. A. Markow, 2008. Molecular evolution and population genetics of two Drosophila mettleri cytochrome P450 genes involved in host plant utilization. Mol. Ecol. 17 3211–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carsten, L., T. Watts and T. A. Markow, 2005. Gene expression changes following dietary shifts in Drososphila melanogaster. Mol. Ecol. 14 3203–3208. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., M. B. Eisen, D. R. Smith, C. M. Bergman, B. Oliver et al., 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450 203–218. [DOI] [PubMed] [Google Scholar]
- Cleland, S., G. D. Hocutt, C. M. Breitmeyer, T. A. Markow and E. Pfeiler, 1996. Alcohol dehydrogenase polymorphism in barrel cactus populations of Drosophila mojavensis. Genetica 98 115–117. [DOI] [PubMed] [Google Scholar]
- Clyne, P. J., C. G. Warr and J. R. Carlson, 2000. Candidate taste receptors in Drosophila. Science 287 1830–1834. [DOI] [PubMed] [Google Scholar]
- Couto, A., M. Alenius and B. J. Dickson, 2005. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr. Biol. 15 1535–1547. [DOI] [PubMed] [Google Scholar]
- Daffre, S., P. Kylsten, C. Samakovlis and D. Hultmark, 1994. The lysozyme locus in Drosophila melanogaster: an expanded gene family adapted for expression in the digestive tract. Mol. Gen. Genet. 242 152–162. [DOI] [PubMed] [Google Scholar]
- Dambroski, H. R., C. Linn, Jr., S. H. Berlocher, A. A. Forbes, W. Roelofs et al., 2005. The genetic basis for fruit odor discrimination in Rhagoletis flies and its significance for sympatric host shifts. Evolution 59 1953–1964. [PubMed] [Google Scholar]
- Danielson, P. B., and J. C. Fogleman, 1997. Isolation and sequence analysis of cytochrome P450 12B1: the first mitochondrial insect P450 with homology to 1 alpha,25 dihydroxy-D3 24-hydroxylase. Insect Biochem. Mol. Biol. 27 595–604. [DOI] [PubMed] [Google Scholar]
- Danielson, P. B., R. J. MacIntyre and J. C. Fogleman, 1997. Molecular cloning of a family of xenobiotic-inducible drosophilid cytochrome p450s: evidence for involvement in host-plant allelochemical resistance. Proc. Natl. Acad. Sci. USA 94 10797–10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauwalder, B., S. Tsujimoto, J. Moss and W. Mattox, 2002. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 16 2879–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker, T., I. Ibba, K. P. Siju, M. C. Stensmyr and B. S. Hansson, 2006. Olfactory shifts parallel superspecialism for toxic fruit in Drosophila melanogaster sibling, D. sechellia. Curr. Biol. 16 101–109. [DOI] [PubMed] [Google Scholar]
- Dietzl, G., D. Chen, F. Schnorrer, K. C. Su, Y. Barinova et al., 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 151–156. [DOI] [PubMed] [Google Scholar]
- Dunipace, L., S. Meister, C. McNealy and H. Amrein, 2001. Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Curr. Biol. 11 822–835. [DOI] [PubMed] [Google Scholar]
- Farine, J. P., L. Legal, B. Moreteau and J. L. LeQuere, 1996. Volatile components of ripe fruits of Morinda citrifolia and their effects on Drosophila. Phytochemistry 41 433–438. [Google Scholar]
- Fogleman, J. C., P. B. Danielson and M. R. Frank, 1997. Comparison of Drosophila cytochrome P450 metabolism of natural and model substrates. J. Insect Physiol. 43 953–957. [DOI] [PubMed] [Google Scholar]
- Fogleman, J. C., P. B. Danielson and R. J. MacIntyre, 1998. The molecular basis of adaptation in Drosophila: the role of cytochrome P450s. Evol. Biol. 30 15–77. [Google Scholar]
- Frank, M. R., and J. C. Fogleman, 1992. Involvement of cytochrome P450 in host-plant utilization by Sonoran Desert Drosophila. Proc. Natl. Acad. Sci. USA 89 11998–12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futuyma, D., 1991. Evolution of host specificity in herbivorous insects: genetic, ecological, and phylogenetic aspects, pp. 431–454 in Plant–Animal Interactions: Evolutionary Ecology in Tropical and Temperate Regions, edited by P. Price, T. Lewinsohn, G. Fernandes and W. Benson. John Wiley & Sons, New York.
- Galindo, K., and D. P. Smith, 2001. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics 159 1059–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, A., D. Barker, R. K. Butlin, W. C. Jordan and M. G. Ritchie, 2008. Drosophila chemoreceptor gene evolution: selection, specialization and genome size. Mol. Ecol. 17 1648–1657. [DOI] [PubMed] [Google Scholar]
- Ghanim, M., A. Dombrovsky, B. Raccah and A. Sherman, 2006. A microarray approach identifies ANT, OS-D and takeout-like genes as differentially regulated in alate and apterous morphs of the green peach aphid Myzus persicae (Sulzer). Insect Biochem. Mol. Biol. 36 857–868. [DOI] [PubMed] [Google Scholar]
- Gilad, Y., S. A. Rifkin, P. Bertone, M. Gerstein and K. P. White, 2005. Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res. 15 674–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, L. A., and P. L. Davies, 2002. The odorant-binding proteins of Drosophila melanogaster: annotation and characterization of a divergent gene family. Gene 292 43–55. [DOI] [PubMed] [Google Scholar]
- Hallem, E. A., A. Dahanukar and J. R. Carlson, 2006. Insect odor and taste receptors. Annu. Rev. Entomol. 51 113–135. [DOI] [PubMed] [Google Scholar]
- Hawthorne, D. J., and S. Via, 2001. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature 412 904–907. [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe, D. S., R. L. Dorit and J. R. Carlson, 2000. Molecular evolution of odorant-binding protein genes OS-E and OS-F in Drosophila. Genetics 155 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Scafe, D. S., C. R. Scafe, A. J. McKinney and M. A. Tanouye, 2002. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 12 1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa, I., and Y. Fuyama, 1993. Genetics of food preference in Drosophila sechellia. I. Responses to food attractants. Genetica 88 129–136. [DOI] [PubMed] [Google Scholar]
- Holloway, A. K., M. K. Lawniczak, J. G. Mezey, D. J. Begun and C. D. Jones, 2007. Adaptive gene expression divergence inferred from population genomics. PLoS Genet. 3 2007–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike, J., 1987. Genetics of oviposition-site preference in Drosophila tripunctata. Heredity 59(Pt. 3): 363–369. [DOI] [PubMed] [Google Scholar]
- Jaenike, J., and R. D. Holt, 1991. Genetic-variation for habitat preference: evidence and explanations. Am. Nat. 137 S67–S90. [Google Scholar]
- Jain, A. N., T. A. Tokuyasu, A. M. Snijders, R. Segraves, D. G. Albertson et al., 2002. Fully automatic quantification of microarray image data. Genome Res. 12 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolivet, P., 1992. Insects and Plants: Parallel Evolution and Adaptations. Sandhill Crane Press, Gainesville, FL.
- Jones, C. D., 1998. The genetic basis of Drosophila sechellia's resistance to a host plant toxin. Genetics 149 1899–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, C. D., 2001. The genetic basis of larval resistance to a host plant toxin in Drosophila sechellia. Genet. Res. 78 225–233. [DOI] [PubMed] [Google Scholar]
- Jones, C. D., 2004. Genetics of egg production in Drosophila sechellia. Heredity 92 235–241. [DOI] [PubMed] [Google Scholar]
- Jones, C. D., 2005. The genetics of adaptation in Drosophila sechellia. Genetica 123 137–145. [DOI] [PubMed] [Google Scholar]
- Jones, C. D., 2007. Genetic ablation of antennae or basiconic sensillia reduces Drosophila melanogaster's ability to perceive odors from the fruit of Morinda citrifolia. Dros. Inf. Serv. 90 45–49. [Google Scholar]
- Kliman, R. M., P. Andolfatto, J. A. Coyne, F. Depaulis, M. Kreitman et al., 2000. The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics 156 1913–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp, A., O. Barmina, A. M. Hamilton, L. Higgins, L. M. McIntyre et al., 2008. Evolution of gene expression in the Drosophila olfactory system. Mol. Biol. Evol. 25 1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise, D., and J. F. Silvain, 2004. How two Afrotropical endemics made two cosmopolitan human commensals: the Drosophila melanogaster-D. simulans palaeogeographic riddle. Genetica 120 17–39. [DOI] [PubMed] [Google Scholar]
- Lavagnino, N. J., R. R. H. Anholt and J. J. Fanara, 2008. Variation in genetic architecture of olfactory behaviour among wild-derived populations of Drosophila melanogaster. J. Evol. Biol. 21 988–996. [DOI] [PubMed] [Google Scholar]
- Legal, L., B. Chappe and J. M. Jallon, 1994. Molecular-basis of Morinda-citrifolia (L): toxicity on Drosophila. J. Chem. Ecol. 20 1931–1943. [DOI] [PubMed] [Google Scholar]
- Legal, L., B. Moulin and J. M. Jallon, 1999. The relation between structures and toxicity of oxygenated aliphatic compounds homologous to the insecticide octanoic acid and the chemotaxis of two species of Drosophila. Pestic. Biochem. Physiol. 65 90–101. [Google Scholar]
- Lindsey, D. L., and G. G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, New York.
- Louis, J., and J. R. David, 1986. Ecological specialization in the Drosophila melanogaster species subgroup: a case study of D. sechellia. Acta Oecol. 7 215–229. [Google Scholar]
- Low, W. Y., H. L. Ng, C. J. Morton, M. W. Parker, P. Batterham et al., 2007. Molecular evolution of glutathione S-transferases in the genus Drosophila. Genetics 177 1363–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, T. F. C., S. L. Heinsohn, R. F. Lyman, A. J. Moehring, T. J. Morgan et al., 2005. Genetics and genomics of Drosophila mating behavior. Proc. Natl. Acad. Sci. USA 102(Suppl. 1): 6622–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathey-Prevot, B., and N. Perrimon, 2006. Drosophila genome-wide RNAi screens: Are they delivering the promise? Cold Spring Harbor Symp. Quant. Biol. 71 141–148. [DOI] [PubMed] [Google Scholar]
- Matsuo, T., 2008. Rapid evolution of two odorant-binding protein genes, Obp57d and Obp57e, in the Drosophila melanogaster species group. Genetics 178 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo, T., S. Sugaya, J. Yasukawa, T. Aigaki and Y. Fuyama, 2007. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. 5 e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin, L. M., 2008. The molecular basis of host adaptation in cactophilic Drosophila: molecular evolution of a glutathione S-transferase gene (GstD1) in Drosophila mojavensis. Genetics 178 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin, L. M., T. D. Watts, B. G. Bitler, C. A. Machado and T. A. Markow, 2006. Functional genomics of cactus host shifts in Drosophila mojavensis. Mol. Ecol. 15 4635–4643. [DOI] [PubMed] [Google Scholar]
- McBride, C. S., 2007. Rapid evolution of smell and taste receptor genes during host specialization in Drosophila sechellia. Proc. Natl. Acad. Sci. USA 104 4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor, A. P., V. Orgogozo, I. Delon, J. Zanet, D. G. Srinivasan et al., 2007. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 448 587–590. [DOI] [PubMed] [Google Scholar]
- Meunier, N., Y. H. Belgacem and J. R. Martin, 2007. Regulation of feeding behaviour and locomotor activity by takeout in Drosophila. J. Exp. Biol. 210 1424–1434. [DOI] [PubMed] [Google Scholar]
- Mezey, J. G., S. V. Nuzhdin, F. Ye and C. D. Jones, 2008. Coordinated evolution of co-expressed gene clusters in the Drosophila transcriptome. BMC Evol. Biol. 8 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa, M., and M. Nei, 2007. Evolutionary dynamics of olfactory receptor genes in Drosophila species. Proc. Natl. Acad. Sci. USA 104 7122–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylin, S., G. H. Nygren, J. J. Windig, N. Janz and A. Bergström, 2005. Genetics of host plant preference in the comma butterfly and evolutionary implications. Biol. J. Linn. Soc. 84 755–765. [Google Scholar]
- Ometto, L., S. Glinka, D. De Lorenzo and W. Stephan, 2005. Inferring the effects of demography and selection on Drosophila melanogaster populations from a chromosome-wide scan of DNA variation. Mol. Biol. Evol. 22 2119–2130. [DOI] [PubMed] [Google Scholar]
- Orgogozo, V., K. W. Broman and D. L. Stern, 2006. High-resolution quantitative trait locus mapping reveals sign epistasis controlling ovariole number between two Drosophila species. Genetics 173 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R'Kha, S., P. Capy and J. R. David, 1991. Host-plant specialization in the Drosophila melanogaster species complex: a physiological, behavioral, and genetical analysis. Proc. Natl. Acad. Sci. USA 88 1835–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R'Kha, S., B. Moreteau, J. A. Coyne and J. R. David, 1997. Evolution of a lesser fitness trait: egg production in the specialist Drosophila sechellia. Genet. Res. 69 17–23. [DOI] [PubMed] [Google Scholar]
- Ranson, H., L. Rossiter, F. Ortelli, B. Jensen, X. Wang et al., 2001. Identification of a novel class of insect glutathione S-transferases involved in resistance to DDT in the malaria vector Anopheles gambiae. Biochem. J. 359 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz, J. M., C. I. Castillo-Davis, C. D. Meiklejohn and D. L. Hartl, 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300 1742–1745. [DOI] [PubMed] [Google Scholar]
- Regel, R., S. R. Matioli and W. R. Terra, 1998. Molecular adaptation of Drosophila melanogaster lysozymes to a digestive function. Insect Biochem. Mol. Biol. 28 309–319. [DOI] [PubMed] [Google Scholar]
- Rogers, M. E., M. K. Jani and R. G. Vogt, 1999. An olfactory-specific glutathione-S-transferase in the sphinx moth Manduca sexta. J. Exp. Biol. 202 1625–1637. [DOI] [PubMed] [Google Scholar]
- Ross, J., H. Jiang, M. R. Kanost and Y. Wang, 2003. Serine proteases and their homologs in the Drosophila melanogaster genome: an initial analysis of sequence conservation and phylogenetic relationships. Gene 304 117–131. [DOI] [PubMed] [Google Scholar]
- Sanchez-Gracia, A., and J. Rozas, 2007. Unusual pattern of nucleotide sequence variation at the OS-E and OS-F genomic regions of Drosophila simulans. Genetics 175 1923–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gracia, A., M. Aguade and J. Rozas, 2003. Patterns of nucleotide polymorphism and divergence in the odorant-binding protein genes OS-E and OS-F: analysis in the melanogaster species subgroup of Drosophila. Genetics 165 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, K., R. Brady, Jr., A. Cravchik, P. Morozov, A. Rzhetsky et al., 2001. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell 104 661–673. [DOI] [PubMed] [Google Scholar]
- Shanbhag, S. R., D. Hekmat-Scafe, M. S. Kim, S. K. Park, J. R. Carlson et al., 2001. Expression mosaic of odorant-binding proteins in Drosophila olfactory organs. Microsc. Res. Tech. 55 297–306. [DOI] [PubMed] [Google Scholar]
- Sheck, a. L., and F. Gould, 1993. The genetic-basis of host-range in Heliothis-wirescens: larval survival and growth. Entomol. Exp. Appl. 69 157–172. [Google Scholar]
- Sheck, a. L., and F. Gould, 1995. Genetic-analysis of differences in oviposition preferences of Heliothis-virescens and Heliothis-subflexa (Lepidoptera, Noctuidae). Environ. Entomol. 24 341–347. [Google Scholar]
- So, W. V., L. Sarov-Blat, C. K. Kotarski, M. J. McDonald, R. Allada et al., 2000. takeout, a novel Drosophila gene under circadian clock transcriptional regulation. Mol. Cell. Biol. 20 6935–6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal, R. R., and F. J. Rohlf, 1995. Biometry. W. H. Freeman, New York.
- Storey, J. D., and R. Tibshirani, 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucena, E., and D. L. Stern, 2000. Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proc. Natl. Acad. Sci. USA 97 4530–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, A., and T. Takano-Shimizu, 2005. A high-frequency null mutant of an odorant-binding protein gene, Obp57e, in Drosophila melanogaster. Genetics 170 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsacas, L., and G. Bächli, 1981. Drosophila sechellia, n.sp., huitieme espece du sous-goupe melanogaster des Iles Sechelles [Diptera, Drosophilidae]. Rev. Fr. Entomol. (Nouv. Ser.) 3 146–150. [Google Scholar]
- Via, S., 1990. Ecological genetics and host adaptation in herbivorous insects: the experimental study of evolution in natural and agricultural systems. Annu. Rev. Entomol. 35 421–446. [DOI] [PubMed] [Google Scholar]
- Vieira, F. G., A. Sanchez-Gracia and J. Rozas, 2007. Comparative genomic analysis of the odorant-binding protein family in 12 Drosophila genomes: purifying selection and birth-and-death evolution. Genome Biol. 8 R235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., R. F. Lyman, S. A. Shabalina, T. F. C. Mackay and R. R. H. Anholt, 2007. Association of polymorphisms in odorant-binding protein genes with variation in olfactory response to benzaldehyde in Drosophila. Genetics 177 1655–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfinger, R. D., G. Gibson, E. D. Wolfinger, L. Bennett, H. Hamadeh et al., 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8 625–637. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13 555–556. [DOI] [PubMed] [Google Scholar]