Abstract
The content and composition of the plant cell wall polymer lignin affect plant fitness, carbon sequestration potential, and agro-industrial processing. These characteristics, are heavily influenced by the supply of hydroxycinnamyl alcohol precursors synthesized by the enzyme cinnamyl alcohol dehydrogenase (CAD). In angiosperms, CAD is encoded by a multigene family consisting of members thought to have distinct roles in different stages of plant development. Due to the high sequence similarity among CAD genes, it has been challenging to identify and study the role of the individual genes without a genome sequence. Analysis of the recently released sorghum genome revealed the existence of 14 CAD-like genes at seven genomic locations. Comparisons with maize and rice revealed subtle differences in gene number, arrangement, and expression patterns. Sorghum CAD2 is the predominant CAD involved in lignification based on the phylogenetic relationship with CADs from other species and genetic evidence showing that a set of three allelic brown midrib (bmr) lignin mutants contained mutations in this gene. The impact of the mutations on the structure of the protein was assessed using molecular modeling based on X-ray crystallography data of the closely related Arabidopsis CAD5. The modeling revealed unique changes in structure consistent with the observed phenotypes of the mutants.
THE biogenesis of the lignified plant cell wall is a complex metabolic process in which the formation and modification of several cell wall polymers need to be coordinated. Lignin is synthesized via the oxidative coupling of several phenolic monomers. The traditional view holds that lignin is derived from the monolignols p-coumaryl, coniferyl, and sinapyl alcohol, which give rise to p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) residues, respectively. Small amounts of other phenolic molecules can be incorporated as well, as a function of the species, genetic make-up, and tissue (reviewed by Ralph et al. 2004). The last step in monolignol biosynthesis is the reduction of cinnamyl aldehyde precursors (reviewed by Humphreys and Chapple 2002), catalyzed by the enzyme cinnamyl alcohol dehydrogenase (CAD; EC 1.1.1.95; Mansell et al. 1974). Two enzymes with cinnamyl alcohol dehydrogenase activity were identified in Eucalyptus gunnii, and named EgCAD1 and EgCAD2 (Goffner et al. 1992). The enzymes are structurally unrelated and share no sequence similarity. EgCAD1 is more similar to cinnamoyl-CoA reductase (EC 1.2.1.44), its active form is a monomer, and it is active toward a range of substrates, including coniferaldehyde and benzaldehyde derivatives, but not sinapaldehyde (Goffner et al. 1992, 1998; Hawkins and Boudet 1994). EgCAD2 is active as a homo- or heterodimer (Hawkins and Boudet 1994; Boudet et al. 2004) and can synthesize all three monolignols. EgCAD2 was considered the predominant CAD involved in lignification (Goffner et al. 1992), and was the basis for studies on the role of CAD in lignification in other species. Recently, however, downregulation of the tobacco ortholog of EgCAD1 was shown to have a subtle effect on the synthesis of coniferyl alcohol (Damiani et al. 2005). Unless stated differently, further mention of CAD refers to EgCAD2-type enzymes.
In gymnosperms, which contain lignin composed primarily of G residues, the CAD activity is strongly coniferaldehyde specific. Several CAD isoforms have been identified in gymnosperm species, although it is still unclear whether those isoforms represent alleles of a single locus or paralogs (Galliano et al. 1993; MacKay et al. 1997; Schubert et al. 1998). In angiosperm species CAD is encoded by a multigene family (Dixon et al. 2001; Li et al. 2001; Lynch et al. 2002; Sibout et al. 2003; Tobias and Chow 2005). The Arabidopsis and rice CAD gene families consist of 9 and 12 members, respectively (Tavares et al. 2000; Sibout et al. 2003; Tobias and Chow 2005). Angiosperm CAD proteins are multifunctional enzymes capable of catalyzing the reduction of p-coumaryl, coniferyl, and sinapyl aldehydes to their corresponding alcohols (Hawkins and Boudet 1994; Kim et al. 2004). The greater variation in lignin composition in angiosperms related to maturity and tissue type, together with the presence of multiple CAD genes, has led to the hypothesis that specialized CAD proteins with differential affinities for their aldehyde substrates play a role in regulating lignin composition during development (Li et al. 2001). The identification of a sinapyl alcohol dehydrogenase (SAD) in aspen (Populus tremuloides), which has greater affinity for sinapaldehyde than coniferaldehyde (Li et al. 2001), provides support for this hypothesis. However, no specific requirement for SAD activity for the synthesis of S units has been identified in Arabidopsis (Kim et al. 2004; Sibout et al. 2005).
Phylogenetic analyses of the Arabidopsis and rice CAD gene families reveal several subgroups (Tavares et al. 2000; Raes et al. 2003; Sibout et al. 2003; Tobias and Chow 2005), one of which displays strong similarity to the gymnosperm CAD genes. This group contains the genes that have been implicated in lignification of the vascular tissue by biochemical, expression, and mutant studies, including AtCAD4 and AtCAD5 (Sibout et al. 2003; Kim et al. 2004; Sibout et al. 2005), ZmCAD2 (Halpin et al. 1998; Guillaumie et al. 2007a), and OsCAD2 (Tobias and Chow 2005; Zhang et al. 2006). The enzymes encoded by the genes belonging to other groups have lower sequence similarity to these bona fide CADs and present wider substrate preferences and varying degrees of enzymatic activity and expression (for review see Raes et al. 2003; Kim et al. 2004; Guillaumie et al. 2007a). It is likely that some of the genes are involved in defense response (Kiedrowski et al. 1992; Brill et al. 1999) or metabolic processes not related to the lignification of the vascular tissue.
Although modifications in lignin content and composition have been observed in the vascular tissue of mutants and transgenics with reduced CAD activity (Halpin et al. 1994, 1998; Baucher et al. 1996, 1999; Chabannes et al. 2001; Sibout et al. 2005), it has been difficult to assign unique functions to individual CAD genes, underscoring the complementation capacity of the CAD multienzyme network. Without clear differences in substrate specificity, the spatiotemporal control of lignin biosynthesis in Arabidopsis and maize may therefore be the result of the regulated expression of tissue-specific CAD genes (Raes et al. 2003; Guillaumie et al. 2007a).
The maize brown midrib1 (bm1) mutant, which has reduced CAD activity (Halpin et al. 1998), reportedly has lower expression of several CAD genes (Guillaumie et al. 2007b) and several other monolignol biosynthetic genes. This led to the hypothesis that the Bm1 gene may encode a transcription factor. Regulatory genes implicated in the regulation of lignification were shown to be reduced in expression (Guillaumie et al. 2007b). In contrast, a phenotypically similar mutant in rice, the golden hull and internode2 (gh2) mutant, is caused by a mutation in the OsCAD2 gene, the ortholog of the maize ZmCAD2 gene identified by Halpin et al. (1998). Genetic complementation experiments demonstrated that expression of the wild-type OsCAD2 gene was sufficient to restore normal cell wall composition of gh2 plants (Zhang et al. 2006). The brown midrib (bmr) mutants of sorghum (Porter et al. 1978) are similar to the bm mutants of maize. The abbreviation bmr was adopted to distinguish it from bm, already in use for the sorghum bloomless mutants (Ayyangar and Ponnaiya 1941). There are at least four independent bmr loci (Bittinger et al. 1981; Saballos et al. 2008). One of the mutants, bmr6, displays decreased CAD activity (Bucholtz et al. 1980; Palmer et al. 2008) and contains cell walls with higher levels of cinnamaldehydes (Pillonel et al. 1991; Saballos et al. 2008). Aided by the release of the sorghum genome sequence (www.phytozome.net/sorghum), we report here the systematic analysis of the sorghum CAD gene family, its relationship with the gene families from other species, and the identification of the Bmr6 gene.
MATERIALS AND METHODS
Genome analysis:
CAD-like genes were identified by performing a BLAST homology search (Altschul et al. 1990) of the sorghum genome sequence database (http://www.phytozome.net/sorghum) using as queries the DNA or deduced amino acid sequences of the rice OsCAD2 (AK105011; Tobias and Chow 2005), Arabidopsis AtCAD4 and AtCAD5 (At3g19450 and At4g34230, respectively; Sibout et al. 2003), and maize ZmCAD2 (AJ005702.1; Halpin et al. 1998; Guillaumie et al. 2007b). The deduced amino acid sequences were used to query the translation of the sorghum genome in all six frames. The deduced amino acid sequence of the best DNA sequence match was used as a new query in both the sorghum and Arabidopsis (www.arabidopsis.org) genomes. This process was repeated until no additional CAD-like sequences were identified. Gene structure was predicted using Fgenesh+ (Salamov and Solovyev 2000), and predicted intron/exon structures were compared with available EST sequences of rice and other grass species. Phylogenetic analysis included all rice and Arabidopsis CAD protein family members (Kim et al. 2004; Tobias and Chow 2005). Two sorghum CAD genes (genomic accession nos. Sb02g024200.1 and Sb10g006310.1) were excluded from the analysis, because their sequences did not appear to encode functional proteins. Multiple sequence alignments were generated using CLUSTAL W version 3.2 (Thompson et al. 1994) with default parameters (http://workbench.sdsc.edu). A Bayesian phylogenetic analysis was performed using MrBayes software (http://mrbayes.csit.fsu.edu/index.php; Huelsenbeck and Ronquist 2001) with the amino acid model set to pr = mixed, parameters lset rates = gamma, number of generations = 256,000 with sampling every 100 generations (Ronquist et al. 2005). The output file was visualized for analysis using TreeView (Page 1996).
Plant materials and nucleic acid extraction:
The brown midrib mutants bmr6-ref, bmr6-3, and bmr6-27, belonging to the bmr6 group (Saballos et al. 2008) and their normal isolines, were planted in 2004 and 2008 at the Purdue Agronomy Center for Research and Education (ACRE, West Lafayette, IN). DNA was extracted from leaf tissue by a modified CTAB protocol (Bernatzky and Tanksley 1986). Three bmr6-ref near-isogenic lines (NILs) in the inbred lines BTx623, RTx430, and Kansas Collier (Pedersen et al. 2006a,b) were planted in the greenhouse at the University of Florida (Gainesville, FL). Young leaf tissue was harvested and extracted using MoBio Power Plant DNA isolation kit (Carlsbad, CA).
A population of recombinant inbred lines (RILs) were created by crossing Brown County, a non-bmr sweet sorghum cultivar, with the bmr6-ref line. The F1 generation was selfed to produce an F2 population of 218 plants. Individual selfed F2 plants were advanced to the F6 generation using the single seed descent method. The phenotype (bmr vs. wild type) was recorded, and DNA of the individual F6 RILs was extracted from leaf tissue using a modified CTAB method (Bernatzky and Tanksley 1986).
For expression assays, BTx623 and BTx623-bmr6-ref lines were planted in the greenhouse at the University of Florida. Tissue was collected at developmental stage 2 (Vanderlip 1993). At this stage, the stalk is small and covered by the leaf sheaths; therefore, leaves and stacked internodes were harvested together to represent the shoot. In addition, the complete root ball was harvested. Tissue was also collected at stage 4 (growing point differentiation) from the apex of developing roots, the first internode (counting from the top of the stem), and the whorl. For both stages, each line was harvested in triplicate and assayed separately. The tissue was frozen in liquid nitrogen and stored at −80° until RNA extraction with the RNeasy Plant RNA extraction kit with in-column DNase digestion (QIAGEN, Carlsbad, CA). First-strand cDNA was synthesized with the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Tissue from the bmr6-3 mutant and the normal isoline N3, planted in the greenhouse (West Lafayette, IN), was harvested at stage 2 and processed as described above.
PCR and cloning:
A homolog to the maize ZmCAD2 gene was identified in the sorghum genome (accession no. Sb04g005950.1) and used to design primers for cloning and gene expression experiments. The identified genomic sequence corresponded to the cDNA sequence identified by Tsuruta et al. (2007) (GenBank accession no. AB288109). The SbCAD2 gene was cloned from bmr6 lines and their respective wild-type isolines. Primer pair U168 (5′ CGG CTC CGT CGT CGT GTT C 3′) and L3923 (Table 1) was used to amplify a 710-bp fragment from the 5′ end of the gene using cDNA as template, with an annealing temperature of 62°. cDNA was used as template due to the technical difficulties of amplifying and sequencing through the first intron of the gene. A 2516-bp 3′ fragment (with partial overlap to the 5′ fragment) was amplified from genomic DNA using primers U2237 (5′ TTT CAG CAC CTT TGG GAT CGA GT 3′) and L4753 (5′ GCA ACA TTT GGG CAT ACG TCT 3′) as above, except that the annealing temperature used was 61°.
TABLE 1.
Primer sequences used for SbCAD gene expression experiments
| Genomic position
|
Sequence
|
|||
|---|---|---|---|---|
| Gene | Upper | Lower | Upper | Lower |
| SbCAD2 | 2931 | 3923 | AAGATCTGGTCCTACAACGA | GGACGACGAGCTGATCAC |
| SbCAD4-2 | 1257 | 2104 | CACCTCTTCTGTGTGCTGGGATAAC | GAAATGGATCAAACGGATGGTC |
| SbCAD4-3 | 984 | 3039 | GTTGTCTGGACACGGAATATGCACC | GCCGCACAGAAGTTAACCAT |
| SbCAD5 | 51 | 2574 | CCCAGCGCTCCTCCGATCCTT | GTCACCACTCCGGCGATCTC |
| SbCAD6 | 11 | 517 | CACCAAACCACACGCAGAC | TCACCTTGTCGCAGTAGTTCT |
| SbCAD7 | 148 | 2693 | CGGCGCAACGATTATGG | GTGGCATATCCCGCAGTACAG |
| SbCAD8-1 | 255 | 1019 | CGGCGTTCGGSTGGG | ACGCCGGTCACCACG |
| Ubiquitin | 221 | 351 | GGTTCGGGAGGTGGCATAGGT | AGCATGTACATTCCCAGCGGTAG |
All primers are listed in 5′–3′ direction.
PCR was performed with Jump Start RedAccuTaq (Sigma, St. Louis) according to the manufacturer's instructions, using a three-step program. The products were analyzed by gel electrophoresis, purified from the gel using the MinElute gel purification kit (QIAGEN), cloned in vector pT7-Blue3 (EMDBiosciences, Gibbstown, NJ), and introduced in Escherichia coli NovaBlue cells (EMDBiosciences). Plasmids with inserts were sequenced in both directions at the University of Florida's Interdisciplinary Center for Biotechnology Research on an Applied Biosystems 3700 capillary electrophoresis sequencer. The sequence from a minimum of three plasmids for each insert was compared to eliminate discrepancies in the sequences due to PCR and sequencing errors. Sequence analysis was performed using Biology Workbench 3.2 software.
CAPS analysis:
CAPS markers (Konieczny and Ausubel 1993) were based on amplification of the SbCAD2 gene with primers U2931 and L3923 (Table 1), and JumpStart AccuTaq ReadyMix PCR reaction mixes (Sigma), using a three-step PCR program with annealing temperature of 59°. The PCR products were purified using AccuPrep PCR purification kit (Bioneer, Seoul, South Korea) and digested with restriction endonucleases according to manufacturer's directions. The enzymes TaiI and TauI (Fermentas, Glen Burnie, MD) were used for the identification of the bmr6-ref and bmr6-3 alleles, respectively. The digested DNA was separated on a 2% agarose gel.
Structural modeling of SbCAD2 and its mutant versions:
The crystal structure of the AtCAD5 (PDB ID: 2CF6, Youn et al. 2006) was used to build a model for SbCAD2 and the bmr6-3 and bmr6-27 mutant versions. The substitutions, insertions, and deletions were superimposed on the coordinates of AtCAD5, followed by a quick energy minimization by CNS v1.1 (Brünger et al. 1998) using potential function parameters of CHARMM19 as described previously (Kim et al. 2004). Substrate position was generated through the solid docking module on QUANTA (BioSYM/Micron Separations), which is based on conformational space, followed by a quick energy minimization by CNS v1.1.
Expression studies of the CAD-like family of genes in normal, bmr6-ref and bmr6-3 mutants of sorghum:
Gene-specific primer pairs flanking introns were designed to differentiate amplification resulting from cDNA and residual genomic DNA (Table 1). cDNA from shoots and roots of three plants each of BTx623/BTx623 bmr6-ref NILs at stages 2 and 4, and from roots and shoots of N3/bmr6-3 NILs at stage 2 was tested for amplification of seven different sorghum CAD genes. Amplification of the Ubiquitin cDNA was used to control for cDNA loading. The Ubiquitin primers were based on the maize cDNA sequence (GenBank accession no. BT016881). The reactions were carried out using RedTaq Readymix PCR reaction mix (Sigma) in a three-step PCR program consisting of 28 cycles. This was empirically determined to be within the exponential phase of amplification. PCR products were visualized in a 2% agarose gel.
RESULTS
The CAD family in sorghum:
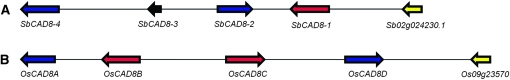
The availability of the sorghum genome sequence has made it relatively straightforward to identify the different members of the CAD gene family. An iterative search of the published sorghum genome sequence using as query the maize, rice, and Arabidopsis bona fide CAD sequences resulted in the identification of 14 CAD-like genes at seven different loci in the sorghum genome. Two of the loci (SbCAD4 and SbCAD8) contain clusters of genes (SbCAD4-1 to -5, and SbCAD8-1 to -4). The DNA sequence of two members of these groups, SbCAD4-1 and SbCAD8-3, are interspersed with retroelements, appear to not encode full-length proteins, and are likely pseudogenes. Sorghum genes in cluster SbCAD8 are very similar to the genes in the rice cluster OsCAD8, but they differ in the gene arrangement. The comparison of the sorghum and rice CAD8 gene clusters is presented in Figure 1. The chromosomal location and EST information of the entire CAD family is presented in Table 2.
Figure 1.—
Organization of the sorghum (A) and rice (B) CAD8 gene clusters. Block arrows represent the genes and their orientation. Arrows with the same color represent genes more similar to each other than to the other members of the cluster. The yellow arrows represent a gene downstream of the CAD8 cluster present in both the rice and sorghum genomes. The scale is approximate and based on the sorghum and rice physical map. The represented sorghum and rice fragments span ∼21.5 and 26 kb, respectively.
TABLE 2.
Genomic location of the sorghum CAD genes and expression data based on ESTs reported in the sorghum genome database
| Gene | Accession no. in genome | No. of reported ESTs | Position in genome |
|---|---|---|---|
| SbCAD2 | Sb04g005950.1 | 21 | 4:5781208..5776540 |
| SbCAD4-1 | Sb10g006310.1 | 0 | 10:5750788..5749847 |
| SbCAD4-2 | Sb10g006300.1 | 0 | 10:5745022..5747508 |
| SbCAD4-3 | Sb10g006290.1 | 5 | 10:5737327..5740568 |
| SbCAD4-4 | Sb10g006280.1 | 3 | 10:5730469..5733611 |
| SbCAD4-5 | Sb10g006270.1 | 1 | 10:5711950..5714251 |
| SbCAD5 | Sb07g006090.1 | 12 | 7:8643832..8640087 |
| SbCAD6 | Sb06g001430.1 | 2 | 6:2167025..2168295 |
| SbCAD7 | Sb06g028240.1 | 5 | 6:57074661..57079263 |
| SbCAD8-1 | Sb02g024220.1 | 19 | 2:58410576..58408367 |
| SbCAD8-2 | Sb02g024210.1 | 13 | 2:58404166..58406013 |
| SbCAD8-3 | Sb02g024200.1 | 0 | 2:58401666..58401105 |
| SbCAD8-4 | Sb02g024190.1 | 9 | 2:58396113..58394105 |
| SbCAD10 | Sb08g016410.1 | 0 | 8:43662072..43660817 |
Phylogenetic analysis:
To understand the structure of the CAD gene family and the possible functional divergence of the individual members, a phylogenetic analysis was performed using deduced amino acid sequences. The nomenclature of sorghum CAD genes was based on similarity with the corresponding rice orthologs (Tobias and Chow 2005). In general, good correspondence between the rice and sorghum family structure was observed, with the exception of OsCAD1, OsCAD3, and OsCAD9, for which no sorghum orthologs were found, and SbCAD10, for which no unambiguous rice ortholog was identified. The predicted length of the proteins varied between 342 and 383 residues and had a maximum and a minimum identity to maize CAD2 of 95% (SbCAD2) and 40% (SbCAD4-2), respectively.
All of the sorghum CADs contain the conserved GHE(X)2G(X)5G(X)2V motif involved in the binding of the catalytic Zn2+, the GD(X)10C(X)2C(X)2(X)7C structural Zn2+ binding domain, and the conserved glycine-rich repeat G(X)GGV(L)G involved in NADP(H) cofactor binding. On the basis of these motifs, these proteins appear to be zinc-dependent alcohol dehydrogenases and members of the medium-chain dehydrogenase/reductase (MDR) superfamily (Persson et al. 1994; Riveros-Rosas et al. 2003). Youn et al. (2006) identified 12 amino acids that constitute the proposed substrate binding site of the bona fide CADs in a number of angiosperms and gymnosperms. Of the 14 identified CAD-like proteins, SbCAD2 contains 10 of these 12 conserved amino acids (Figure 2). All other family members contained a varying number of amino acid substitutions in these residues, as is the case for other CAD family members in Arabidopsis, poplar, and rice (Li et al. 2001; Raes et al. 2003; Bomati and Noel 2005; Tobias and Chow 2005).
Figure 2.—
Sequence comparison of the translated sequences of Sorghum bicolor CAD family members, maize ZmCAD2 (Zea mays sequence AJ005702.1), and Arabidopsis AtCAD5 (Arabidopsis thaliana sequence At4g34230). The conserved amino acids identified by Youn et al. (2006) as part of the substrate binding pocket are indicated by asterisks above the sequences. The conserved glycine-rich motif G(X)GGV(L)G is indicated by a dotted line below the sequences. Italicized letters indicate completely conserved residues across all 14 sequences.
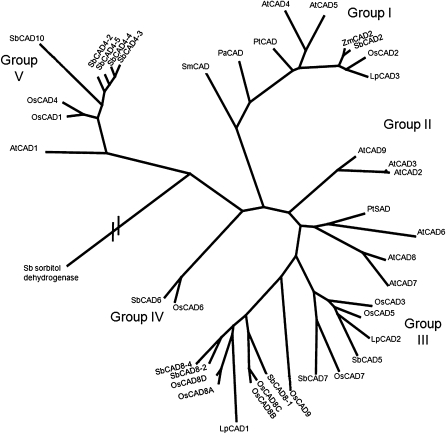
An unrooted phylogenetic tree (Figure 3) constructed from the deduced amino acid sequences shows the relationship between the sorghum CAD family members and previously reported CADs from other species. Sorghum sequence Sb07g025220.1, the closest sequence outside the CAD family, was included in the analysis for comparison. The CAD proteins included in this analysis cluster in five groups.
Figure 3.—
Phylogenic relationship between the sorghum CAD family and CAD proteins from other species. The branch lengths indicate the relative distance between sequences, with the exception of the putative sorghum sorbitol dehydrogenase, which is distantly related to the CAD family (accession no. Sb07g025220.1). The sequences used to build the tree were: Selaginella moellendorffii SmCAD1-1 (protein ID 450958 of the JGI Selanginella genome database); Picea abies PtCAD (O82035); Zea mays ZmCAD2 (CAA06687); Arabidopsis thaliana AtCAD1 (At1g72680), AtCAD2 (At2g21730), AtCAD3 (At2g21890), AtCAD4 (At3g19450), AtCAD5 (At4g34230), AtCAD6 (At4g37970), AtCAD7 (At4g37980), AtCAD8 (At4g37990), and AtCAD9 (At4g39330); Populus tremuloides PtSAD (AAK58693), PtCAD (AAF43140); Oryza sativa OsCAD1 (NP_001064283.1), OsCAD2 (NP_001046132.1), OsCAD3 (AAP53892), and OsCAD4 (NP_001068303.1). OsCAD5 (BK003971), OsCAD6 (CAD39907), OsCAD8 A-C (BK003972), and OsCAD9 (AAN05338). Sorghum sequences are those described in Table 2. The rice CAD gene nomenclature follows Tobias and Chow (2005).
Group I corresponds to class I of Raes et al. (2003), and contains the sequences identified as the bona fide CAD enzymes involved in the reduction of coniferyl and sinapyl aldehydes to their corresponding alcohols for the formation of lignin in lycophytes, gymnosperms, and monocot and dicot angiosperms. SbCAD2 belongs to this group and it is closely related to the maize ZmCAD2 and rice OsCAD2 proteins. Group II contains AtCAD2, AtCAD3, AtCAD9 [following the nomenclature of Kim et al. (2004)], designated by Raes et al. (2003) as class III. Group III encompasses enzymes of diverse substrate specificity, including the proposed poplar SAD (Li et al. 2001) and Arabidopsis enzymes belonging to class II of Raes et al. (2003), involved in pathogen defense (Somssich et al. 1996). Group IV contains the SbCAD6 and OsCAD6 sequences (Tobias and Chow 2005). Both the rice and the sorghum sequences contain the SKL peroxisome targeting sequence at their C terminus. Group V contains monocot and dicot CADs. Even though there appear to be groups specific to monocots (group IV) and dicots (group II), sequence resources are probably still too limited for definitive conclusions.
The bmr6 mutation affects the SbCAD2 gene:
The increased levels of cinnamaldehyde endgroups in the lignin of the bmr6-ref mutant are consistent with reduced CAD activity (Bucholtz et al. 1980; Pillonel et al. 1991; Palmer et al. 2008; Saballos et al. 2008). On the basis of the clustering of SbCAD2 in group I with the bona fide CADs, we hypothesized that the SbCAD2 gene is the main CAD gene in sorghum and that a mutation in this gene would affect lignification.
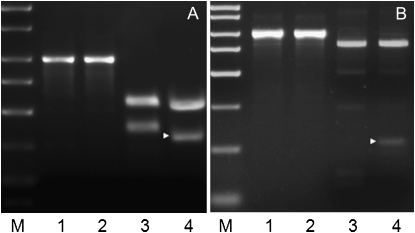
The SbCAD2 alleles of three different inbred lines in which the bmr6-ref allele had been introgressed revealed a C-to-T transition at position 2800 of the SbCAD2 genomic sequence. This particular nucleotide change creates a premature stop codon. The resulting predicted protein contains only 131 amino acids and lacks the nucleotide binding site and most of the residues that form the substrate binding site. Such a truncated protein is likely to be nonfunctional even if produced. The mutation creates a novel restriction site for endonuclease TaiI (ACGC → ACGT). Restriction digest of a fragment of the gene (nucleotides 2931–3923 of the genomic DNA) resulted in three fragments (585, 407, and 24 bp) from the wild-type alleles, whereas the bmr6-ref allele produced four fragments (508, 363, 44, and 24 bp). This CAPS marker is therefore diagnostic for the presence of the bmr6-ref allele. The mutation was confirmed with the CAPS marker using independently isolated DNA from N6 and bmr6-ref (Figure 4). Furthermore, this CAPS marker was used in a cosegregation analysis of an F6 RIL population derived from a cross between Brown County × bmr6, and indicated complete linkage between the mutated SbCAD2 gene (bmr6-ref allele) and the bmr phenotype.
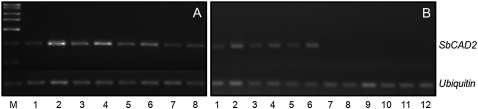
Figure 4.—
Molecular markers can distinguish between the wild-type and mutant SbCAD2 alleles. (A) CAPS marker specific for the bmr6-ref allele. Lanes 1 and 2 show the undigested 992-bp PCR product from N6 and bmr6-ref DNA, respectively. Lanes 3 and 4 show the respective PCR products after digestion with the restriction enzyme TaiI. The 363-bp diagnostic fragment is indicated with a solid triangle. (B) CAPS marker specific for the bmr6-3 allele. Lanes 1 and 2 show the undigested PCR product from N3 and bmr6-3 DNA, respectively. Lanes 3 and 4 show the respective PCR products after digestion with the restriction enzyme TauI. The 138-bp diagnostic fragment is indicated with a solid triangle.The size marker is Sigma PCR Marker [catalog (cat.) no. P9577].
In the absence of a facile transformation protocol for sorghum, proving beyond reasonable doubt that the SbCAD2 gene is the Bmr6 gene requires the identification of additional independent mutations in this gene in mutants that are allelic to bmr6-ref. Indeed, sequencing of the bmr6-3 allele revealed a G-to-A transition at position 3875 of the genomic sequence. This mutation results in an amino acid substitution from Gly (G) to Ser (S) at position 191 of the protein sequence. The glycine-rich motif 188G(X)GGV(L)G193 is highly conserved among studied MDR proteins. The motif is located at the first β-α-β-unit of the cofactor binding site and is presumed to participate in the binding of the pyrophosphate group of the NADP+-cofactor (Youn et al. 2006). Although the mutated allele would encode a full-length protein, its catalytic activity may be compromised. A rice mutant with a single mutation in this same region, 185 G → D, results in complete loss of enzymatic activity, and is the causal mutation of the gh2 phenotype (Zhang et al. 2006).
The base substitution in the bmr6-3 allele eliminates a restriction site for the enzyme TauI. A restriction digest of the amplified gene fragment 2931–3923 with TauI resulted in four fragments in the bmr6-3 allele (796, 138, 54, and 5 bp) with a diagnostic fragment of 138 bp. This product was cleaved into two fragments (92 and 46 bp) in the wild-type allele. The mutation was confirmed with this marker using independently isolated DNA from N3 and bmr6-3 (Figure 4).
The bmr6-27 mutation is a nucleotide insertion after position 4288 (+G), which causes a frameshift that results in the formation of a protein with nonconservative substitutions in the 31 C-terminal residues. The mutation also created a premature stop codon that renders the protein four residues shorter than the wild-type protein. In addition to the mutation responsible for the phenotype, the bmr6-27 allele also contains a deletion of base 3077 (−T), but this is a silent mutation because of its location in the third intron of the gene. This mutation does not cause splicing abnormalities, as sequencing of the CAD2 cDNA from wild type and bmr6-27 showed no sequence variation in this region.
Structural comparison of SbCAD2 and its mutant versions with AtCAD5:
The amino acid sequence comparison between SbCAD2 and AtCAD5 (Figure 2) revealed 93 residue changes in total. Among those, 48 are conservative changes and 45 are nonconservative changes. A close inspection of the 3D structure of AtCAD5 (PDB ID: 2CF6) revealed that 52 of these changes are surface residues, whereas 39 occur in the secondary structure. In general, the 3D model of SbCAD2 remains very similar to that of AtCAD5, with many of the changes not having a significant impact on the overall structure (Figure 5). All the secondary structural elements observed in AtCAD5 are maintained in SbCAD2. The functional unit of AtCAD5 is a dimer tightly associated through two twofold-related β-strands. SbCAD2 very likely maintains the dimer character on the basis of the intact nature of βD, βE, and βF. There is a minor perturbation caused by L271H in βE, which is due to a delocalization of the imidazole ring of residue 271 by the neighboring D250 and H256 residues, but the magnitude of the impact is not enough to disrupt the dimer formation at the βF contact points.
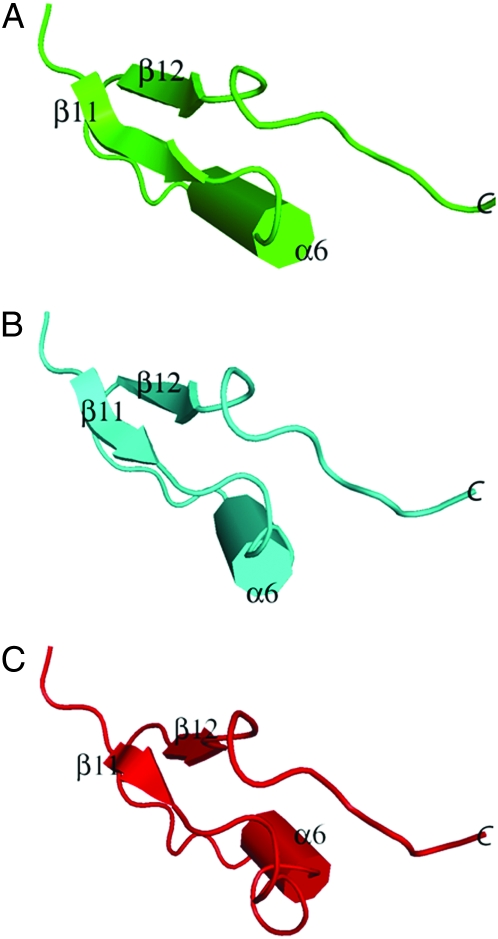
Figure 5.—
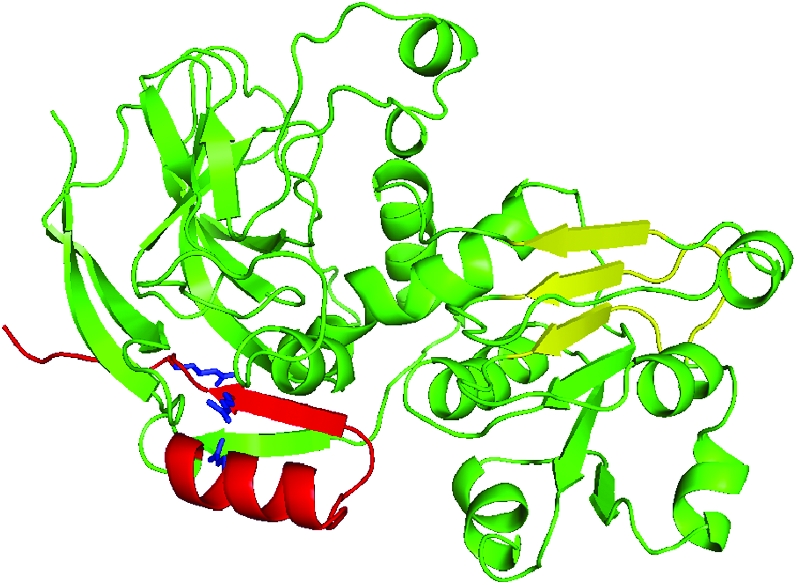
Ribbon diagram representation for the structure of SbCAD2. The significant arginine mutations found in SbCAD2_27b are indicated in blue. The dimer contact positions for βD–F are highlighted in yellow. The three secondary structural elements, β11, β12, and α6 are not maintained in the CAD model of bmr6-27 mutant due to the degenerative hydrophobic interactions caused by the arginine residues highlighted in blue.
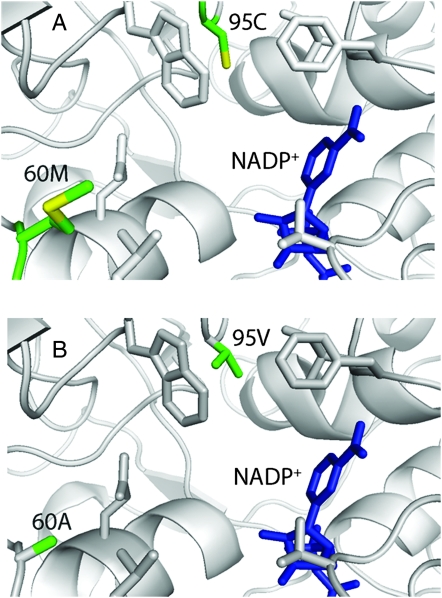
Two of the amino acid changes between AtCAD5 and SbCAD2 are in positions that may alter the enzymatic activity (Figure 6). The first of those is residue 60 M → A. This residue resides near the surface of SbCAD2 and may possibly affect the entry of substrate. The methionine residue can act as a hydrophobic gating mechanism in concert with surrounding side chains. Thus, by replacing it with alanine, the specificity of this potential gate becomes less stringent. The second change, residue 95 C → V, occurs in close proximity of the substrate-binding pocket. The amino acid change generates a more hydrophobic character to this pocket, which is filled with water molecules in the apo-form AtCAD5. Consequently, there might be fewer water molecules in the substrate-binding pocket of SbCAD2, resulting in a significant reduction of the entropic contribution to the free energy associated with substrate binding. Therefore, it is very likely that SbCAD2 has not only reduced efficiency of substrate binding, but also altered substrate specificity compared to AtCAD5.
Figure 6.—
Graphical comparison of active site residues interacting with the NADP+ cofactor of (A) AtCAD5 and (B) SbCAD2. Residue M60 and C95 change to A and V, respectively, whereas the other residues are conserved. This will potentially affect substrate binding and specificity.
SbCAD2 in the bmr6-3 and bmr6-27 mutants:
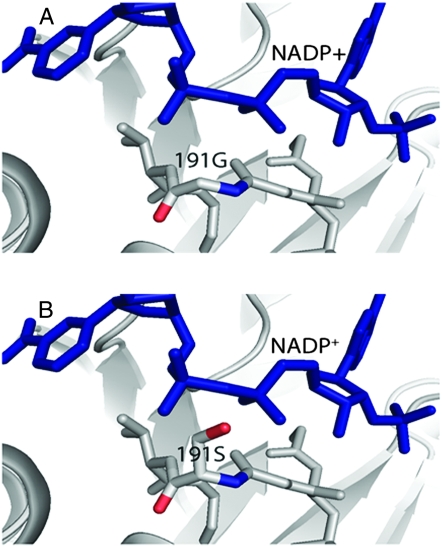
The only amino acid change observed in the bmr6-3 mutant is 191 G → S, which is part of the highly conserved 188GXGGV(L)G193 motif. A serine residue at this site is a very rare alteration. Among the Zn-dependent medium-chain dehydrogenases in the database, this site is highly conserved with only hydrophobic residues. The unique serine substitution at position 191 in the bmr6-3 mutant will change the characteristics of this motif and consequently affect the binding affinity for the NADP+ cofactor (Figure 7).
Figure 7.—
Effect of the bmr6-3 mutation on the structure of SbCAD2. (A) G(X)GGV(L)G motif and its interaction with the phosphate backbone of NADP+. (B) SbCAD2 bmr6-3 variant at residue 191. The G191S mutation will affect the interaction between this flexible motif and phosphate groups.
In the bmr6-27 mutant, there is a total of 27 amino acid changes, all within the C terminus, spanning residues 331–361. Since the mutation affects a relatively small portion of the total sequence, and does not directly affect the active site of the enzyme or its dimerization domain, it is important to determine whether the changes produced structural alterations that would affect its activity. Among those 27 mutations, only 24 can be modeled in because the remaining C-terminal three residues are disordered in the AtCAD5 structure. Those remaining mutations are distributed through the secondary structures β12 (346–349) and α6 (330–338). In AtCAD5, those secondary structural elements, together with β11 (323–326) in the C terminus, maintain their interface through hydrophobic interactions. Those interfaces among β11 and β12 and α6 appear to be significantly affected by several arginine substitutions, which include 333 A → R, 348 V → R, and 350 V → R. Specifically, 333 A → R disrupts the hydrophobic area that maintains β11, 348 V → R disrupts the hydrophobic integrity of α6, while 350 V → R perturbs a hydrophobic pocket generated by V40, P67, G134, and F135. In addition, the 346 F → P substitution disrupts the pocket, which in turn affects the integrity of β12.
The secondary structural prediction algorithms show that both α6 (330–338) and β12 (346–349) are no longer maintained. However, the amino acid changes are unlikely to affect the ability to dimerize due to their distal locations from βD, βE, and βF (Figure 5). Another significant substitution is 342 V → R, which approaches up to 5 Å toward the Sγ-atom of 47C, and could thus alter the coordination of the catalytic Zn2+. The various amino acid substitutions present in bmr6-27 could result in either serious disruption of the catalytic domain or expansion of the intersecondary distances among β11, β12, and α6 (Figure 8). Even though we performed modeling for the bmr6-27 variant of SbCAD2, the number of amino acid substitutions and the consequent potential disruption due to the change in chemical properties is beyond the capabilities of the energy minimization-modeling algorithm. Additional experiments, such as circular dichroism spectroscopy or X-ray crystallography, are required to validate the correct model for this mutant.
Figure 8.—
Representation of the configurations of the secondary structural elements β11, α6, and β12 in (A) wild-type SbCAD2 and (B and C) the bmr6-27 variant of SbCAD2. B and C represent two possible configurations of the corresponding secondary structure. In B, significant repositioning of the α6-helix occurred. In C, a significant reduction of the secondary structure in all three structural elements occurred.
Semiquantitative RT–PCR analysis of SbCAD family members in normal and bmr plants:
Comparison of the transcript levels of SbCAD2 in the near-isogenic pair BTx623/BTx623-bmr6-ref showed that in the latter the transcript levels of SbCAD2 were downregulated (Figure 9B). Such downregulation of transcripts with premature stop codons is known to occur as a result of nonsense-mediated RNA decay (Bout and Vermerris 2003; Conti and Izaurralte 2005). It is likely that the bmr6-ref allele is a null mutation. In contrast, missense mutations are unlikely to trigger mRNA degradation. Indeed, expression analysis showed that the transcript levels of SbCAD2 were not severely reduced in the bmr6-3 mutant compared to its wild-type isoline N3 (Figure 9A).
Figure 9.—
The bmr6-ref, but not the bmr6-3 mutation, causes downregulation of SbCAD2 expression in 2-week-old plants. (Top lane) Products from RT–PCR with primers specific for a fragment of the cDNA encoding SbCAD2. (A) 1–4, N3 cDNA; 5–8, bmr6-3 cDNA. (B) 1–6, BTx623 cDNA; 7–12, BTx623 bmr6-ref cDNA. Odd numbers correspond to cDNA obtained from shoots; even numbers cDNA obtained from roots. (Bottom lane) Products from RT–PCR with primers for Ubiquitin gene as a cDNA loading control. The size marker is Bioline Hyperladder II (cat. no. 33054).
The expression of the CAD family genes varied according to tissue type and stage of development of the plant. Table 3 summarizes the expression differences among the growth stages and tissues tested. The presence of the bmr6-ref or bmr6-3 allele of the SbCAD2 gene in homozygous form did not alter the pattern of expression of the other CAD family members, with the exception of SbCAD5. This gene was upregulated in the shoot of all the bmr6-ref plants, and two of the bmr6-3 plants at stage 2. However, one of the wild-type plants also showed upregulation of this gene. Additional expression experiments with field-grown N6 and bmr6-ref plants confirmed strong plant-to-plant variation in the level of expression of the SbCAD5 gene (data not shown), but in general, this gene was more frequently upregulated in bmr6-ref plants than in wild-type plants.
TABLE 3.
Gene expression of the sorghum CAD-like family based on semi-quantitative RT–PCR experiments
| Stage 2
|
Stage 4
|
||||
|---|---|---|---|---|---|
| Gene | Shoot | Root | Leaves | Stem | Root |
| SbCAD2 | Medium | Strong | Very strong | Very strong | Strong |
| SbCAD4-2 | Very weak | Weak | Very weak | Very weak | Very weak |
| SbCAD4-3 | Weak | Medium | Medium | Medium | Medium |
| SbCAD5 | Variable | Very weak | Weak | Weak | ND |
| SbCAD6 | Weak | Weak | ND | ND | Medium |
| SbCAD7 | Medium | ND | Weak | Weak | ND |
| SbCAD8-1 | Strong | Weak | Medium | Medium | ND |
ND, not detectable.
DISCUSSION
As previously shown in other plants species, CAD is encoded by a small gene family in sorghum. Several genes with high similarity to the bona fide CAD genes are present in the sorghum genome. Of the 14 CAD genes identified with homology searches, there is evidence that at least 10 genes are expressed, on the basis of the presence of corresponding ESTs and expression analysis. Two loci in the genome contain closely related groups of CAD genes. The SbCAD8 1-4 cluster is orthologous with the rice OsCAD8 cluster described by Tobias and Chow (2005). Although the rice and sorghum genes within this cluster are highly similar, the gene arrangement differs between the species, raising the question whether the duplication event occurred prior to the divergence of the species or whether the clusters are the result of independent duplication events. The cluster is divided in two main branches, each of which contains sorghum and rice sequences. This suggests two duplication events, one before and one after speciation. This may explain the different orientation of the rice and sorghum genes. A homology search of the recently published maize draft genome sequence (http://maizesequence.org) with the sorghum SbCAD8 genomic sequences revealed a single ortholog (AC197840.3_FG008). This maize gene is located on chromosome 7, which is syntenic with sorghum chromosome 2 (Wilson et al. 1999) on which the SbCAD8 cluster resides. The presence of a single gene in maize, a species more closely related to sorghum than rice, is not consistent with the hypothesis that a duplication event occurred prior to the divergence of these species unless additional paralogs of AC197840.3_FG008 or remnants thereof are identified as the maize genome sequence reaches completion. The second gene cluster in sorghum (SbCAD4 1-5) has no reported orthologs in other species for which the genome sequence is available at this time. The closest rice sequence to the members of this group is a single gene, OsCAD4.
On the basis of the phylogenetic analysis, the sorghum CAD genes cluster into four groups. SbCAD2 groups together with the bona fide CADs from other species. The high sequence similarity and conservation of important substrate binding pocket residues within this group point to a conservation of the gene function. Indeed, mutations or downregulation of the expression of genes belonging to this group have significant impact in lignin content and composition in tobacco, loblolly pine, poplar, and Arabidopsis (Halpin et al. 1994; MacKay et al. 1997; Ralph et al. 1997; Baucher et al. 1999; Lapierre et al. 1999; Pilate et al. 2002; Sibout et al. 2003). Modeling SbCAD2 over the crystal structure of AtCAD5 revealed that even though SbCAD2 contains a large number of residue changes, it remains similar in overall structure, with secondary structure preserved throughout. The close relationship of SbCAD2 with the other members of this group supports the idea that it is the main sorghum CAD involved in lignification. Consistent with this hypothesis, we observed that SbCAD2 is the predominantly expressed CAD gene in sorghum. In addition, a mutation in this gene is linked to the bmr phenotype in three independently introgressed isolines and it cosegregates with the bmr phenotype in a RIL population. Furthermore, sequencing of three independent bmr6 alleles all revealed nonconservative mutations in the SbCAD2 gene. Therefore, the evidence strongly suggests that mutations in this gene are responsible for the phenotype of the bmr6 mutants.
Members of the other groups present much more diverse substitutions in the substrate binding pocket. These substitutions would allow for a wider variety of substrate specificity by changing the size, conformation, and hydrophobicity of the binding pocket. The role of these enzymes in lignification has not yet been elucidated. Only the Populus SAD has been implicated directly in lignification, catalyzing the reduction of sinapyl aldehyde to sinapyl alcohol (Li et al. 2001; Bomati and Noel 2005). Wound-inducible genes in parsley (Petroselinum crispum) and Arabidopsis cluster together in group III (Schmelzer et al. 1989; Trezzini et al. 1992; Somssich et al. 1996), suggesting that the genes in this group may be involved in the production of defense-related compounds (Hawkins and Boudet 2003). Kinetic analysis of the Arabidopsis genes in groups II, III, and V have shown that the activity against phenylpropanoid aldehydes is much lower than the Arabidopsis CADs belonging to group I. AtCAD7 has been shown to behave as an aromatic alcohol oxidoreductase of wide substrate affinity (Somssich et al. 1996). Consequently, these CAD-like genes are currently annotated as putative mannitol or benzyl alcohol dehydrogenases. However, it is evident that there is some redundancy in the CAD metabolic network. Compositional studies of Arabidopsis, rice, and sorghum mutants in which the bona fide CAD genes are downregulated have shown that their lignin still contains a certain amount of the normal G and S residues (Sibout et al. 2003, 2005; Zhang et al. 2006; Saballos et al. 2008). Reporter gene studies of the Arabidopsis CAD family showed that AtCAD7 and AtCAD8 are expressed in lignifying tissues (Kim et al. 2007). In the Arabidopsis double mutant Atcad5/Atcad4, which lacks activity of the two bona fide CAD proteins, the AtCAD1 gene is highly upregulated (Sibout et al. 2005), suggesting it may act to compensate for the lack of the CAD activity from AtCAD5 and AtCAD4. Transgenic expression of the AtCAD9 gene under control of the AtCAD5 promoter was able to partially rescue the phenotype of the double mutant (Eudes et al. 2006). In our study, we did not find consistent upregulation of any of the SbCAD genes of groups III, IV, or V in the bmr6 lines, suggesting that the steady-state level of the genes in the CAD family are sufficient to produce the observed levels of G and S units in the mutant. The SbCAD8-1 gene is highly expressed in the aerial part of the plant. In the seedling (stage 2) it is the most highly expressed CAD gene. Guillaumie et al. (2007a) analyzed the expression of the CAD genes in maize and its relation with tissue-specific lignification patterns. The EST sequence 3071507.2.1II is the likely ortholog of SbCAD8. The spatiotemporal expression of this gene agrees with our results, but it differs in that it is not the main CAD expressed in the aerial portion of the plant at the seedling stage. Of the identified CAD-like genes, the genes in the SbCAD8 cluster are the most similar to SbCAD2 (52–48% identity, except SbCAD8-3). To our knowledge, the role of the SbCAD8 genes in sorghum has not been investigated. On the basis of the high level of expression of these genes, we hypothesize that the SbCAD8 enzymes may be responsible for the production of S and G residues observed in the bmr6 mutants. Studies of a Sbcad2/Sbcad8 double mutant could help determine the role of SbCAD8 in lignification.
At this moment, it is difficult to attribute specific functions to the other sorghum CAD-like genes from groups III and V on the basis of phylogenetic relationships. We observed that the expression of the CAD-like genes in sorghum varies with the stage of development and tissue type, in agreement with what was observed in maize (Guillaumie et al. 2007a). Given the evolutionary relationship and the general agreement of gene expression patterns between maize and sorghum, it is likely that their functions are conserved. One exception may be SbCAD5. The maize ortholog, EST 4424417.2.1III, is the predominantly expressed CAD in leaves and stems of maize seedlings. In contrast, SbCAD5 showed variable expression levels in the leaves and stems of individual plants, both in seedlings and preflowering (stage 4) plants. More bmr6-ref and bmr6-3 plants showed upregulation of SbCAD5 than their wild-type isolines. It would be tempting to speculate that the gene is upregulated to compensate for the loss of SbCAD2 activity, but the lack of consistent upregulation in all bmr plants does not support this conclusion. Another possible explanation is that SbCAD5 gene is upregulated as part of a pathogen- or other stress-response mechanism, as it clusters in the same group as the Arabidopsis pathogen-inducible defense genes AtCAD7 and AtCAD8. The bmr phenotype is associated with decreased lignin content and results in a fitness penalty in the plant (Oliver et al. 2005a,b; Pedersen et al. 2005), and it is possible that the bmr plants are under increased stress compared to wild-type plants. It is interesting that while in the aerial parts of both maize and sorghum seedlings the bona fide CAD is not the most highly expressed CAD gene, the predominant CAD genes in those tissues represent nonorthologous paralogs in the two species.
To our knowledge, no orthologs of SbCAD6 and OsCAD6 (group IV) have been described in the literature. SbCAD6 is mainly expressed in the roots of stage 4 plants, although at low levels. Both SbCAD6 and OsCAD6 (Tobias and Chow 2005) contain a predicted peroxisomal targeting signal at their C terminus. Dehydrogenase activities in the peroxisome are involved in the metabolism of alcohol, branched amino acids, fatty acids, and plant hormones (Reumann et al. 2004). Detailed biochemical and expression studies are needed to assign a physiological function to this gene.
We have described three different mutant alleles of SbCAD2: bmr6-ref, a likely null allele, bmr6-3, which produces a full-length protein with a single disruption in the cofactor binding site, and bmr6-27, encoding an enzyme with a disrupted secondary structure outside the active site. Although all three mutants produce the visual brown midrib phenotype, it is likely that the mutations have different effects on enzyme activity. There is evidence that the bmr6-3 allele has a stronger effect on enzymatic saccharification of stover than the bmr6-ref allele, wheras the latter allele results in higher coniferaldehyde levels in the midrib tissue than either the bmr6-3 or bmr6-27 allele (Saballos et al. 2008). Now that the molecular basis of the mutations is known, variation in penetrance can be investigated. Tailoring of the effect of the activity of the enzyme may be achieved on the basis of the choice of allele introgressed in a particular sorghum line. The development of molecular markers specific for each of those variants will greatly facilitate the use of the alleles in genetic studies and breeding programs. There are at least six additional known bmr6 alleles available (Saballos et al. 2008), which opens the possibility of finding more useful variants.
Acknowledgments
The authors thank Jeff Pedersen (U.S. Department of Agriculture (USDA)-Agricultural Research Service, Lincoln NE) for providing seeds of the bmr6-ref mutants in different inbred lines. We thank Terry Lemming for excellent management of the field plots. This research was supported by the Office of Science (BER), U.S. Department of Energy, grant DE-FG02-07ER64458 (W.V. and G.E.), and National Research Initiative of the USDA grant 2006-35318-17454 (C.H.K.), with additional funding from Purdue University Agricultural Research Programs, Purdue Research Foundation, and the University of Florida.
References
- Altschul, S., W. Gish, W. Miller, E. Myers and D. Lipman, 1990. Basic local aligment search tool. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Ayyangar, G. N. R. and B. W. X. Ponnaiya, 1941. The occurrence and inheritance of a bloomless sorghum. Curr. Sci. 10 408–409. [Google Scholar]
- Baucher, M., M. A. Bernard-Vailhé, B. Chabbert, J.-M. Besle, C. Opsomer et al., 1999. Down-regulation of cinnamyl alcohol dehydrogenase in transgenic alfalfa (Medicago sativa L.) and the effect on lignin composition and digestibility. Plant Mol. Biol. 39 437–447. [DOI] [PubMed] [Google Scholar]
- Baucher, M., B. Chabbert, G. Pilate, J. Van Doorsselaere, M.-T. Tollier et al., 1996. Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar. Plant Physiol. 112 1479–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatzky, R., and S. D. Tanksley, 1986. Towards a saturated linkage map in tomato based on isozymes and random cDNA sequences. Genetics 112 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittinger, T. S., R. P. Cantrell and J. D. Axtell, 1981. Allelism tests of the brown-midrib mutants of sorghum. J. Hered. 72 147–148. [Google Scholar]
- Bomati, E. K., and J. P. Noel, 2005. Structural and kinetic basis for substrate selectivity in Populus tremuloides sinapyl alcohol dehydrogenase. Plant Cell 17 1598–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet, A. M., S. Hawkins and S. Rochange, 2004. The polymorphism of the genes/enzymes involved in the last two reductive steps of monolignol synthesis: What is the functional significance? C. R. Biol. 327 837–845. [DOI] [PubMed] [Google Scholar]
- Bout, S. and W. Vermerris, 2003. A candidate-gene approach to clone the sorghum Brown midrib gene encoding caffeic acid O-methyltransferase. Mol. Genet. Genomics 269 205–214. [DOI] [PubMed] [Google Scholar]
- Brill, E. M., S. Abrahams, C. M. Hayes, C. L. D. Jenkins and J. M. Watson, 1999. Molecular characterisation and expression of a wound-inducible cDNA encoding a novel cinnamyl-alcohol dehydrogenase enzyme in lucerne (Medicago sativa L.). Plant Mol. Biol. 41 279–291. [DOI] [PubMed] [Google Scholar]
- Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros et al., 1998. CNS: a system for crystallography and NMR: A new software suite for macromolecular structure determination. Acta Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Bucholtz, D. L., R. P. Cantrell, J. D. Axtell and V. L. Lechtenberg, 1980. Lignin biochemistry of normal and brown midrib mutant sorghum. J. Agric. Food Chem. 28 1239–1241. [Google Scholar]
- Chabannes, M., A. Barakate, C. Lapierre, J. M. Marita, J. Ralph et al., 2001. Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 28 257–270. [DOI] [PubMed] [Google Scholar]
- Conti, E., and E. Izaurralde, 2005. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 17 316–325. [DOI] [PubMed] [Google Scholar]
- Damiani, I., K. Morreel, S. Danoun, G. Goeminne, N. Yahiaoui et al., 2005. Metabolite profiling reveals a role for atypical cinnamyl alcohol dehydrogenase CAD1 in the synthesis of coniferyl alcohol in tobacco xylem. Plant Mol. Biol. 59 753–769. [DOI] [PubMed] [Google Scholar]
- Dixon, R. A., F. Chen, D. Guo and K. Parvathi, 2001. The biosynthesis of monolignols: A “metabolic grid,” or independent pathways to guaiacyl and syringyl units? Phytochemistry 57 1069–1084. [DOI] [PubMed] [Google Scholar]
- Eudes, A., B. Pollet, R. Sibout, C.-T. Do, A. Séguin et al., 2006. Evidence for a role of AtCAD 1 in lignification of elongating stems of Arabidopsis thaliana. Planta 225 23–39. [DOI] [PubMed] [Google Scholar]
- Galliano, H., M. Cabané, C. Eckerskorn, F. Lottspeich, H. Sandermann, Jr. et al., 1993. Molecular cloning, sequence analysis and elicitor-/ozone-induced accumulation of cinnamyl alcohol dehydrogenase from Norway spruce (Picea abies L.). Plant Mol. Biol. 23 145–156. [DOI] [PubMed] [Google Scholar]
- Goffner, D., I. Joffroy, J. Grima-Pettenati, M. E. Knight, W. Schuch et al., 1992. Purification and characterization of isoforms of cinnamyl alcohol dehydrogenase from Eucalyptus xylem. Planta 188 48–53. [DOI] [PubMed] [Google Scholar]
- Goffner, D., J. Van Doorsseaere, N. Yahiaoui, J. Samaj, J. Grima-Pettenati et al., 1998. A novel aromatic alcohol dehydrogenase in higher plants: molecular cloning and expression. Plant Mol. Biol. 36 755–765. [DOI] [PubMed] [Google Scholar]
- Guillaumie, S., H. San-Clemente, C. Deswarte, Y. Martinez, C. Lapierre et al., 2007. a MAIZEWALL. Database and developmental gene expression profiling of cell wall biosynthesis and assembly in maize. Plant Physiol. 143 339–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumie, S., M. Pichon, J.-P. Martinant, M. Bosio, D. Goffner et al., 2007. b Differential expression of phenylpropanoid and related genes in brown-midrib bm1, bm2, bm3, and bm4 young near-isogenic maize plants. Planta 226 235–250. [DOI] [PubMed] [Google Scholar]
- Halpin, C., K. Holt, J. Chojecki, D. Oliver, B. Chabbert et al., 1998. Brown-midrib maize (bm1): a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J. 14 545–553. [DOI] [PubMed] [Google Scholar]
- Halpin, C., M. E. Knight, G. A. Foxon, M. M. Campbell, A. M. Boudet et al., 1994. Manipulation of lignin quality by downregulation of cinnamyl alcohol dehydrogenase. Plant J. 6 339–350. [Google Scholar]
- Hawkins, S., and A. M. Boudet, 1994. Purification and characterization of cinnamyl alcohol dehydrogenase isoforms from the periderm of Eucalyptus gunnii Hook. Plant Physiol. 104 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins, S., and A. M. Boudet, 2003. ‘Defence lignin’ and hydroxycinnamyl alcohol dehydrogenase activities in wounded Eucalyptus gunnii. Forest Pathol. 33 91–104. [Google Scholar]
- Huelsenbeck, J. P., and F. Ronquist, 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17 754–755. [DOI] [PubMed] [Google Scholar]
- Humphreys, J. M., and C. Chapple, 2002. Rewriting the lignin road map. Curr. Opin. Plant Biol. 5 224–229. [DOI] [PubMed] [Google Scholar]
- Kiedrowski, S., P. Kawalleck, K. Hahlbrock, I. E. Somssich and J. L. Dangl, 1992. Rapid activation of a novel plant defense gene is strictly dependent on the Arabidopsis RPM1 disease resistance locus. EMBO J. 11 4677–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. J., K. W. Kim, M. H. Cho, V. R. Franceschi, L. B. Davin et al., 2007. Expression of cinnamyl alcohol dehydrogenases and their putative homologues during Arabidopsis thaliana growth and development: Lessons for database annotations? Phytochemistry 68 1957–1974. [DOI] [PubMed] [Google Scholar]
- Kim, S. J., M. R. Kim, D. L. Bedgar, S. G. Moinuddin, C. L. Cardenas et al., 2004. Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc. Natl. Acad. Sci. USA 101 1455–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and F. Ausubel, 1993. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Lapierre, C., B. Pollet, M. Petit-Conil, G. Toval, J. Romero et al., 1999. Structural alterations in lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have an opposite impact on the efficiency of industrial Kraft plulping. Plant Physiol. 119 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., X. F. Cheng, J. Leshkevich, T. Umezawa, S. A. Harding et al., 2001. The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. Plant Cell 13 1567–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, D., A. Lidgett, R. McInnes, H. Huxley, E. Jones et al., 2002. Isolation and characterisation of three cinnamyl alcohol dehydrogenase homologue cDNAs from perennial ryegrass (Lolium perenne L.). J. Plant Physiol. 159 653–660. [Google Scholar]
- MacKay, J. J., D. M. O'Malley, T. Presnell, F. L. Booker, M. M. Campbell et al., 1997. Inheritance, gene expression, and lignin characterization in a mutant pine deficient in cinnamyl alcohol dehydrogenase. Proc. Natl. Acad. Sci. USA 94 8255–8260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansell, R. L., G. G. Gross, J. Stoeckint, H. Franke, and M. H. Zenk, 1974. Purification and properties of cinnamyl alcohol dehydrogenase from higher plants involved in lignin biosynthesis. Phytochemistry 13 2427–2435. [Google Scholar]
- Oliver, A. L., J. F. Pedersen, R. J. Grant and T. J. Klopfenstein, 2005. a Comparative effects of the sorghum bmr-6 and bmr-12 genes: I. Forage sorghum yield and quality. Crop Sci. 45 2234–2239. [Google Scholar]
- Oliver, A. L., J. F. Pedersen, R. J. Grant, T. J. Klopfenstein and H. D. Jose, 2005. b Comparative effects of the sorghum bmr-6 and bmr-12 genes: II. Grain yield, stover yield, and stover quality in grain sorghum. Crop Sci. 45 2240–2245. [Google Scholar]
- Page, R., 1996. Tree View: an application to display phylogenetic trees on personal computers. Bioinformatics 12 257–258. [DOI] [PubMed] [Google Scholar]
- Palmer, N., S. E. Sattler, A. J. Saathoff, D. Funnell, J. F. Pedersen et al., 2008. Genetic background impacts soluble and cell wall-bound aromatics in brown midrib mutants of sorghum. Planta 229 115–127. [DOI] [PubMed] [Google Scholar]
- Pedersen, J. F., D. L. Funnell, J. J. Toy, A. L. Oliver and R. J. Grant, 2006. a Registration of seven forage sorghum genetic stocks near-isogenic for the brown midrib genes bmr-6 and bmr-12. Crop Sci. 46 490–491. [Google Scholar]
- Pedersen, J. F., D. L. Funnell, J. J. Toy, A. L. Oliver and R. J. Grant, 2006. b Registration of twelve grain sorghum genetic stocks near-isogenic for the brown midrib genes bmr-6 and bmr-12. Crop Sci. 46 491–492. [Google Scholar]
- Pedersen, J. F., K. P. Vogel and D. L. Funnell, 2005. Impact of reduced lignin on plant fitness. Crop Sci. 45 812–819. [Google Scholar]
- Persson, B., J. S. Zigler Jr. and H. Jörnvall, 1994. A super-family of medium-chain dehydrogenases/reductases (MDR): sub-lines including ζ-crystallin, alcohol and polyol dehydrogenases, quinone oxidoreductases, enoyl reductases, VAT-1 and other proteins. Eur. J. Biochem. 226 15–22. [DOI] [PubMed] [Google Scholar]
- Pilate, G., E. Guiney, K. Holt, M. Petit-Conil, C. Lapierre et al., 2002. Field and pulping performances of transgenic trees with altered lignification. Nat. Biotechnol. 20 607–612. [DOI] [PubMed] [Google Scholar]
- Pillonel, C., M. M. Mulder, J. J. Boon, B. Forster and A. Binder, 1991. Involvement of cinnamyl-alcohol dehydrogenase in the control of lignin formation in Sorghum bicolor (L.) Moench. Planta 185 538–544. [DOI] [PubMed] [Google Scholar]
- Porter, K. S., J. D. Axtell, V. L. Lechtenberg and V. F. Colenbrander, 1978. Phenotype, fiber composition, and in vitro dry matter disappearance of chemically induced brown midrib (bmr) mutants of sorghum. Crop Sci. 18 205–208. [Google Scholar]
- Raes, J., A. Rohde, J. H. Christensen, Y. Van de Peer and W. Boerjan, 2003. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133 1051–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph, J., K. Lundquist, G. Brunow, F. Lu, H. Kim et al., 2004. Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl propanoids. Phytochem. Rev. 3 29–60. [Google Scholar]
- Ralph, J., J. J. MacKay, R. D. Hatfield, D. M. O'Malley, R. W. Whetten et al., 1997. Abnormal lignin in a loblolly pine mutant. Science 277 235–238. [DOI] [PubMed] [Google Scholar]
- Reumann, S., C. Ma, S. Lemke and L. Babujee, 2004. AraPerox: a database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 13 2587–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveros-Rosas, H., A. Julian-Sanchez, R. Villalobos-Molina, J. P. Pardo and E. Pina, 2003. Diversity, taxonomy and evolution of medium-chain dehydrogenase/reductase superfamily. Eur. J. Biochem. 270 3309–3334. [DOI] [PubMed] [Google Scholar]
- Ronquist, F., J. P. Huelsenbeck and P. Van Der Mark, 2005. MrBayes 3.1 Manual. http://mrbayes.csit.fsu.edu/manual.php.
- Saballos, A., W. Vermerris, L. Rivera and G. Ejeta, 2008. Allelic association, chemical characterization and saccharification properties of brown midrib mutants of sorghum (Sorghum bicolor (L.) Moench). Bioenerg. Res. 1 193–204. [Google Scholar]
- Salamov, A. A., and V. V. Solovyev, 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer, E., S. Kruger-Lebus and K. Hahlbrock, 1989. Temporal and spatial patterns of gene expression around sites of attempted fungal infection in parsley leaves. Plant Cell 10 993–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, R., C. Serisen, G. Müller-Starck, S. La Scala, D. Ernst et al., 1998. The cinnamyl alcohol dehydrogenase gene structure in Picea abies (L.) Karst.: genomic sequences, Southern hybridization, genetic analysis and phylogenetic relationships. Trees Structure and Function 12 453–463. [Google Scholar]
- Sibout, R., A. Eudes, G. Mouille, B. Pollet, C. Lapierre et al., 2005. CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17 2059–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout, R., A. Eudes, B. Pollet, T. Goujon, I. Mila et al., 2003. Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis. Isolation and characterization of the corresponding mutants. Plant Physiol. 132 848–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich, I. E., P. Wernert, S. Kiedrowski and K. Hahlbrock, 1996. Arabidopsis thaliana defense-related protein ELI3 is an aromatic alcohol: NADP(+) oxidoreductase. Proc. Natl. Acad. Sci. USA 93 14199–14203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares, R., S. Aubourg, A. Lecharny and M. Kreis, 2000. Organization and structural evolution of four multigene families in Arabidopsis thaliana: AtLCAD, AtLGT, AtMYST and AtHD-GL2. Plant Mol. Biol. 42 703–717. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias, C. M., and E. K. Chow, 2005. Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification. Planta 220 678–688. [DOI] [PubMed] [Google Scholar]
- Trezzini, G. F., A. Horrichs and E. Somssich, 1992. Isolation of putative defense-related genes from Arabidopsis thaliana and expression in fungal elicitor-treated cells. Plant Mol. Biol. 21 385–389. [DOI] [PubMed] [Google Scholar]
- Tsuruta, S., M. Ebina, H. Nakagawa, O. Kawamura and R. Akashi, 2007. Isolation and characterization of cDNA encoding cinnamyl alcohol dehydrogenase (CAD) in sorghum (Sorghum bicolor (L.) Moench). Grassland Sci. 53 103–109. [Google Scholar]
- Vanderlip, R. L., 1993. How a Grain Sorghum Plant Develops. Kansas State University, Manhattan, KS.
- Wilson, W., S. Harrington, W. Woodman, M. Lee, M. Sorrels and S. McCouch, 1999. Inferences on the genome structure of progenitor maize through comparative analysis of rice, maize and the domesticated panicoids. Genetics 153 453–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn, B., R. Camacho, S. G. A. Moinuddin, C. Lee, L. B. Davin et al., 2006. Crystal structures and catalytic mechanism of the Arabidopsis cinnamyl alcohol dehydrogenases AtCAD5 and AtCAD4. Org. Biomol. Chem. 4 1687–1697. [DOI] [PubMed] [Google Scholar]
- Zhang, K., Q. Qian, Z. Huang, Y. Wang, M. Li et al., 2006. GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol. 140 972–983. [DOI] [PMC free article] [PubMed] [Google Scholar]