Abstract
Ubiquitin-dependent protein degradation has emerged as a major pathway regulating eukaryotic biology. By employing a variety of ubiquitin ligases to target specific cellular proteins, the ubiquitin-proteasome system controls physiological processes in a highly regulated fashion. Recent studies on a plant hormone auxin have unveiled a novel paradigm of signal transduction in which ubiquitin ligases function as hormone receptors. Perceived by the F-box protein subunit of the SCFTIR1 ubiquitin ligase, auxin directly promotes the recruitment of a family of transcriptional repressors for ubiquitination, thereby activating extensive transcriptional programs. Structural studies have revealed that auxin functions through a “molecular glue” mechanism to enhance protein-protein interactions with the assistance of another small molecule cofactor, inositol hexakisphosphate. Given the extensive repertoire of similar ubiquitin ligases in eukaryotic cells, this novel and widely adopted hormone-signaling mechanism in plants may also exist in other organisms.
Keywords: auxin, phytohormone, hormone receptor, F-box protein, ubiquitin ligase, protein degradation
in multicellular organisms, hormones are produced and released by one tissue and transported to another to regulate cell growth, metabolism, and a myriad of cellular responses. Past studies have identified several families of receptor proteins in the target cells responsible for sensing various hormones in different chemical forms. These classic receptors include cell surface receptors such as G protein-coupled receptors and receptor tyrosine kinases, which trigger second-messenger and protein phosphorylation-mediated signal transduction, and intracellular receptors such as steroid and thyroid hormone receptors, which function as transcription factors directly regulating gene expression via a DNA-binding domain. Recent advances in plant biology have revealed yet another paradigm of hormone perception in which the hormone signal is sensed by components of the ubiquitin-proteasome system (UPS) that is commonly found in all eukaryotic cells. Structural studies of the prototypical plant hormone receptor unravel a novel mechanism of hormone action and an unexpected small molecule cofactor.
Ubiquitin Ligases and UPS
Cellular protein homeostasis is controlled by the balance between protein synthesis and degradation. Although protein synthesis based on gene transcription and translation has long been recognized as a key process regulating every physiological function, the protein degradation process has gained attention only in the last 20 yr. We now appreciate that the UPS is a major route of protein degradation in eukaryotes and actively regulating a wide spectrum of cellular processes such as gene transcription, signal transduction, metabolism, DNA repair, and replication (9). The UPS consists of a three-enzyme cascade (E1, E2, and E3) and a multicomponent proteolytic apparatus, the 26s proteasome (17). E1, or the ubiquitin-activating enzyme, activates ubiquitin by adenylating its COOH terminus and conjugates the activated ubiquitin to the active site cysteine. E1 then transfers ubiquitin to the E2, or ubiquitin-conjugating enzyme, which also uses a cysteine residue to form a thiolester bond with ubiquitin. At the last step, the E2-conjugated ubiquitin is transferred to a lysine residue of the substrate protein, which is brought to the vicinity of E2 by a specific E3 or ubiquitin ligase. This process repeats by adding ubiquitin to a lysine residue of the previously conjugated ubiquitin, thereby generating a chain of polyubiquitin linked to the substrate protein. Proteasome recognizes the ubiquitin chain pattern and recruits the ubiquitinated protein for proteolysis. In this complex system, the specificity and temporal control are afforded mainly by the large repertoire of E3s, hundreds of which exist in humans. For example, certain E3s recognize specifically phosphorylated peptide sequences of the protein substrate (9). Indeed, substrate phosphorylation has emerged as a major regulatory mechanism of protein ubiquitination, integrating protein degradation with diverse signaling pathways mediated by protein kinases.
Since the protein synthesis process, especially transcription, is under the control of various hormones, one is prompted to ask whether the opposite process, protein degradation, is also associated with hormone signaling. An obvious place to look for this connection is the hormone receptor protein. Not surprisingly, a number of receptors, such as thyroid hormone receptor and growth hormone receptor, are directly regulated by ubiquitination and proteasomal degradation (1, 25). In other cases, such as epidermal growth factor receptor and growth hormone receptor, the availability of the membrane-associated receptors is fine-tuned by ubiquitin-mediated endocytosis and lysosomal breakdown (11). In these two scenarios, the receptors are the substrates of the ubiquitination reaction. The net result of ubiquitination is downregulation of the receptors and thus the hormone responses. In these cases, hormones are not directly involved in the ubiquitination events.
More recently, a surprising new paradigm was revealed in the signaling pathway of an important plant hormone, auxin, in which the hormone takes direct control of an E3 ligase and signals by promoting the ubiquitination and degradation of a set of transcriptional repressors (2, 13). In other words, the hormone receptor is a ubiquitin ligase, whose activity is dependent on the presence of the hormone molecule. This review focuses on the mechanism of this new signaling paradigm and its implications.
Transport Inhibitor Response Protein 1 Ubiquitin Ligase as An Auxin Receptor
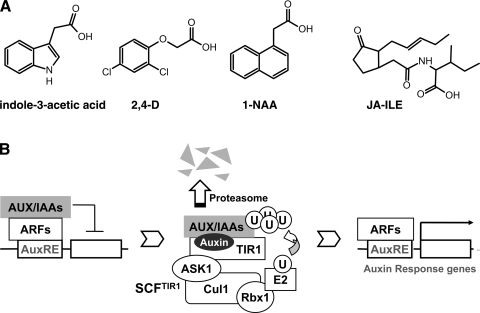
The natural auxin indole-3-acetic acid, the best-studied plant hormone, regulates virtually every aspect of plant growth and development from embryo patterning and growth responses to fruit development (Fig. 1A) (26). Similarly to many animal hormones, auxin achieves its multitude of physiological effects through transcriptional regulation. In response to auxin, hundreds of genes are activated by a family of transcriptional factors called auxin response factors (ARFs), which recognize specific promoter elements characteristic of auxin response genes (7, 24). In the absence of auxin, the protein products of a family of genes named AUX/IAAs (auxin/indole-3-acetic acids) directly interact with ARFs and inhibit their transcriptional activity (Fig. 1B) (19, 22). In the presence of auxin, AUX/IAAs are rapidly removed, abrogating their inhibition of ARFs and the expressions of auxin response genes (28). Interestingly, the transcriptions of some AUX/IAAs are also activated by auxin, providing a negative feedback mechanism to modulate auxin activity (14).
Fig. 1.
The Skp1-Cullin1-F-box transport inhibitor response protein 1 (SCFTIR1) ubiquitin ligase functions as the receptor for the plant hormone auxin. A: chemical structures of the natural auxin, 2 auxin analogs [2,4-dichlorophenoxyacetic acid (2,4-D) and 1-naphthalene acetic acid (1-NAA)], and jasmonate-isoleucine (JA-Ile). B: the role of SCFTIR1 ubiquitin ligase in the auxin-signaling pathway in plants. In the absence of auxin, auxin/indole-3-acetic acids (AUX/IAAs) repress the expression of auxin response genes (AuxRE) by binding to the transcription activator auxin response factors (ARFs) (left). In the presence of auxin, the multisubunit SCFTIR1 ubiquitin ligase recognizes the hormone and promotes the polyubiquitination and proteasomal degradation of AUX/IAAs in a hormone-dependent manner (middle). Auxin-induced degradation of AUX/IAAs frees ARFs, which activate the transcription of auxin response genes (right). U, ubiquitin protein.
The plant UPS plays a central role in auxin-induced degradation of AUX/IAA. An Arabidopsis gene called transport inhibitor response protein 1 (TIR1), which encodes a subunit of a ubiquitin ligase complex, was shown to be a positive regulator of auxin response (20). TIR1 belongs to the F-box family of proteins that function as interchangeable substrate receptor subunits of the Skp1-Cullin1-F-box protein (SCF) ubiquitin ligase complexes, which belong to the largest superfamily of multisubunit Cullin-RING E3s in eukaryotes (16). Mutations in TIR1 cause decreased auxin sensitivity in plants, suggesting that its polyubiquitinated substrates are negative regulators of auxin signaling. AUX/IAAs, which can directly interact with TIR1 through a short stretch of conserved region (domain II), are the obvious substrate candidates (18). In fact, auxin stimulates AUX/IAA-TIR1 binding and accelerates SCFTIR1-catalyzed ubiquitination and degradation of AUX/IAAs (6).
How is the auxin hormone signal perceived and transduced to the TIR1 ubiquitin ligase? Since phosphorylation or other types of covalent posttranslational modifications of the substrate by an upstream signaling molecule are often required for substrate-ubiquitin ligase interactions, it was thought that identification of the AUX/IAA modification prerequisite for TIR1 interaction could lead to the upstream modifying enzyme, the auxin-signaling pathway, and eventually the auxin receptor, whose identity had been elusive for decades. However, phosphorylation of AUX/IAAs was soon ruled out as the determinant for auxin-induced interactions between TIR1 and AUX/IAAs in a cell-free system (3, 12). In 2005, two groups simultaneously reported their surprising finding that there is nothing else in between the plant hormone and the TIR1 ubiquitin ligase; TIR1 is the long-sought auxin receptor (2, 13). Auxin itself provides the determining “modification” for the AUX/IAA substrates to interact with the E3 ligase. Upon interacting with TIR1, auxin promotes the binding of AUX/IAAs to the SCFTIR1 ubiquitin ligase complex and accelerates the polyubiquitination of AUX/IAAs (Fig. 1B). The subsequent degradation of AUX/IAAs alleviates their inhibition on ARFs, which then activate the auxin-induced transcription programs.
The surprising mechanism by which the plant hormone auxin signals provides the first example of hormone perception by a ubiquitin ligase and establishes the first case of a naturally occurring small molecule controlling the E3 activity of an SCF ubiquitin ligase complex. How does the auxin molecule modulate the interactions between the SCFTIR1 ubiquitin ligase and its substrate AUX/IAAs? This question becomes particularly puzzling given the fact that there are more than 200 auxin analogs with different chemical structures known to have the common auxin activity. The only common features shared by these compounds are an unsaturated planar ring structure and a carboxyl side chain (Fig. 1A). How could they all be recognized by the auxin receptor TIR1 and in turn regulate the ubiquitin ligase activity of the E3 enzyme?
Mechanism of Auxin Action
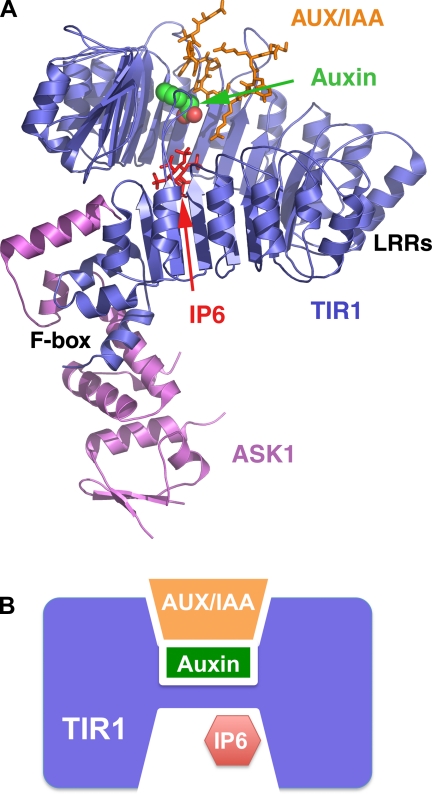
The recently reported X-ray crystal structures of TIR1 in different functional states reveal the mechanisms of auxin action and auxin perception by TIR1 (21). In this study, TIR1 was cocrystallized with its binding partner Arabidopsis SKP1 (ASK1) protein, which is the adaptor subunit of SCF, linking TIR1 to the E3 complex. TIR1 and ASK1 overall adopt a mushroom shape, with the F-box domain of TIR1 together with ASK1 forming the stem of the mushroom and the COOH-terminal 18 leucine-rich repeats (LRRs) of TIR1 forming the mushroom cap in a horseshoe-like solenoid fold (Fig. 2A). Near the center space of the TIR1 horseshoe-shaped solenoid, three long loops of the TIR1 LRRs, together with the inner wall of the solenoid, give rise to a large single surface pocket for both hormone and substrate binding. Occupying the bottom portion of the TIR1 surface pocket, auxin uses its conserved carboxyl group to anchor itself to the pocket floor via a salt bridge, and hydrogen bonds with an arginine residue of TIR1. Meanwhile, the aromatic ring structure of auxin is accommodated by the hydrophobic “wall” of the pocket through hydrophobic packing and van der Waals interactions. The substrate peptide, corresponding to the conserved domain II sequence of AUX/IAAs, sits on top of auxin and covers up the surface pocket. It directly interacts with both the plant hormone and the hormone receptor. In the same study, structures of TIR1 with different auxin analogs were also determined. Two synthetic auxin analogs, 2,4-dichlorophenoxyacetic acid and 1-naphthalene acetic acid (Fig. 1A), bind to TIR1 in a similar mode as auxin, albeit with different potencies due to their different aromatic ring structures. This partial promiscuity of the auxin-binding site offers the structural basis for the recognition of diverse auxin analogs by TIR1.
Fig. 2.
The structure and mechanism of auxin action. A: crystal structure of Arabidopsis SKP1 (ASK1)-TIR1-inositol hexakisphosphate (IP6) in complex with auxin and an AUX/IAA domain II peptide. Both ASK1 (magenta) and TIR1 (blue) are shown in ribbon. Auxin is shown in space-filled model (green and red spheres). The AUX/IAA peptide (orange) and IP6 (red) are shown in sticks. B: a schematic diagram showing how auxin is sensed by TIR1 with the assistance of IP6 and how the hormone promotes TIR1-AUX/IAA interactions. LRRs, leucine-rich repeats.
The action mode of auxin is in drastic contrast to the common mechanism of hormone action. In most, if not all, previously known cases, a hormone binds to an allosteric site in the receptor and causes conformational change of the receptor to elicit downstream effects. Comparison of the structures of free and auxin-bound TIR1 shows that auxin binding does not change the conformation of TIR1. Rather, auxin alters the protein-protein interaction by filling a hydrophobic gap between the two proteins (Fig. 2B). In other words, it sticks two proteins together like a “molecular glue” instead of a conformational switch. This mechanism is consistent with the partial promiscuity of the auxin-binding pocket, because any hydrophobic moiety that can fit in the pocket without causing steric hindrance to substrate binding would be able to increase the affinity between TIR1 and the substrate. In addition, this mechanism bears a certain similarity to the widespread phosphorylation-dependent recruitment of ubiquitination substrates to their specific ubiquitin ligases. In those cases, phosphate groups also do not change the conformation of the modified proteins (8, 27). Rather, they enhance protein-protein association by providing additional interactions and extending the interface, just like auxin does. The only difference is that phosphorylation involves covalently linked phosphate groups catalyzed by kinases, whereas auxin functions through noncovalent hydrophobic interactions, which might have an advantage of rapid response to environmental stimuli.
Inositol Hexakisphosphate As a Cofactor of the Auxin Receptor TIR1
In addition to the novel mechanism of auxin action, another surprise from the structure is a previously unknown small molecule, inositol hexakisphosphate (IP6), copurified with TIR1 (21). The crystal structures reveal that IP6 binds in the core of the TIR1 LRR domain, right beneath the auxin-binding site (Fig. 2, A and B). The strict conservation of eight of the 10 IP6-interacting TIR1 residues in plants implies that the IP6 molecule is not a nonspecifically bound contaminating factor. Close examination of the IP6-binding site further shows that two of the key residues that form the floor of auxin-binding site are buttressed by the IP6 molecule, suggesting an important structural role of IP6 in supporting TIR1's function as an auxin receptor. Although it remains unclear whether IP6 has any signaling function, its close connection to other inositol phosphates indicates that TIR1-mediated auxin signaling might be controlled by additional pathways. Overall, the ability of TIR1 to employ two small molecule ligands, auxin and IP6, showcases the remarkable potential of a ubiquitin ligase to sense and integrate signals in signal transduction.
Ubiquitin Ligases as Hormone Receptors
How pervasive is this new paradigm of ubiquitin ligase-mediated hormone perception and signal transduction? At least in the plant kingdom, this is not a singular case. TIR1 is found in almost all plant species from moss to higher plants. In Arabidopsis thaliana, five paralogs of TIR1 protein (AFB1–5) have been shown to collectively function as auxin receptors (4). Among the six receptors, there might be some specificity in terms of recognition of different auxin analogs and possibly different substrates, but their general mechanisms of action are conserved. Interestingly, coronatine insensitive 1 (COI1), a distant homolog of TIR1, has recently been shown to perceive a very different plant hormone, jasmonate and its amino acid derivatives (Fig. 1A), and induce the degradation of another family of transcriptional repressors (10). Although the molecular mechanism of jasmonate action has not been determined, its action in promoting the binding between COI1 and its ubiquitination substrate polypeptides and the subsequent transcriptional activation is highly similar to auxin. In addition to auxin and jasmonate, gibberellin is yet another plant hormone whose perception involves an SCF ubiquitin ligase complex. Different from auxin and jasmonate, gibberellin is not directly sensed by an F-box protein but instead by an extension molecule of the SCF substrate receptor subunit (23). Upon perceiving gibberellin, the plant protein gibberellin insensitive dwarf (GID)1 recruits a family of transcription regulators named DELLA proteins to the GID2/SLY1 F-box protein for ubiquitination. Given the 700 F-box proteins and the great complexity of chemicals in plants (5), new examples of small molecule sensors like TIR1 will almost certainly emerge in the future.
Are there ubiquitin ligases functioning as hormone receptors in organisms other than plants? At molecular level, there is no obvious reason why this is not possible. With common precursors and synthetic pathways, many hormones in plants and animals share strikingly similar overall structures. Derived from tryptophan, the plant hormone auxin is reminiscent of animal hormones serotonin and melatonin. With fatty acids as precursors, jasmonate in plants and prostaglandin in animals are both cyclopentanone derivatives. On the ubiquitin ligase side, TIR1-like F-box proteins and the SCF E3 complexes are universally found in all eukaryotic cells (16). Expansion of the SCF-like cullin-RING ubiquitin ligases occurred in both the plant and animal kingdoms. In fact, a mammalian SCF-like Cullin-RING E3 complex has recently been shown to be a receptor for an exogenous chemical, dioxin (15). With the nearly 1,000 predicted human ubiquitin ligases, it would not be surprising if some of them employ a similar strategy to sense and transduce endogenous hormone signals. The fact that we have yet to discover a TIR1-like hormone receptor in humans might be to due to the same reason that it took so long to find the auxin receptor in plants. Unlike conventional hormones, auxin does not bind its receptor TIR1 with very high affinity. Upon docking to auxin-bound TIR1, AUX/IAAs cover up auxin and contribute to the formation of the complete auxin-binding site. Although the presence of AUX/IAAs secures auxin binding, AUX/IAAs get rapidly ubiquitinated and degraded, making it difficult to capture this coreceptor complex. In the same vein of reasoning, traditional experiments in the past could have missed the opportunity to isolate a hormone receptor complex in other systems if the high-affinity hormone receptor is a ubiquitin ligase-substrate pair. Now, with the first example of this new paradigm of hormone perception revealed in plants, we will have “the prepared mind” for potentially finding a similar case in other organisms.
Despite the long stride made in past years, the picture of auxin perception in plants is far from complete. For example, it remains unclear whether TIR1 and its five paralogs AFB1–5 show any specificity toward different natural auxin compounds and some of the AUX/IAA substrate proteins. Furthermore, when, where, and in what form are AUX/IAAs ubiquitinated by SCFTIR1 upon auxin sensing? Is IP6 simply a constitutively bound structural cofactor of TIR1, or is it a signaling molecule connecting auxin signaling with other cellular pathways through TIR1? Finally, are there non-TIR1-like auxin receptors transducing the auxin signal for other outputs? Further studies on these questions will allow us to fully recognize the multilayers of specificity behind this new paradigm of hormone signaling and at least partially explain the multitude of diverse effects of auxin.
GRANTS
This work was supported by grants from the National Institutes of Health and the Pew Scholar Program. N. Zheng is a Howard Hughes Medical Institute Investigator.
Acknowledgments
We thank Nabiha H. Saifee for critical reading of the manuscript.
REFERENCES
- 1.Dace A, Zhao L, Park KS, Furuno T, Takamura N, Nakanishi M, West BL, Hanover JA, Cheng S. Hormone binding induces rapid proteasome-mediated degradation of thyroid hormone receptors. Proc Natl Acad Sci USA 97: 8985–8990, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445, 2005. [DOI] [PubMed] [Google Scholar]
- 3.Dharmasiri N, Dharmasiri S, Jones AM, Estelle M. Auxin action in a cell-free system. Curr Biol 13: 1418–1422, 2003. [DOI] [PubMed] [Google Scholar]
- 4.Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119, 2005. [DOI] [PubMed] [Google Scholar]
- 5.Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276, 2001. [DOI] [PubMed] [Google Scholar]
- 7.Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385, 2002. [PubMed] [Google Scholar]
- 8.Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, Pagano M, Pavletich NP. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol Cell 20: 9–19, 2005. [DOI] [PubMed] [Google Scholar]
- 9.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 67: 425–479, 1998. [DOI] [PubMed] [Google Scholar]
- 10.Katsir L, Chung HS, Koo AJ, Howe GA. Jasmonate signaling: a conserved mechanism of hormone sensing. Curr Opin Plant Biol 11: 428–435, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol 3: 893–905, 2002. [DOI] [PubMed] [Google Scholar]
- 12.Kepinski S, Leyser O. Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc Natl Acad Sci USA 101: 12381–12386, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451, 2005. [DOI] [PubMed] [Google Scholar]
- 14.Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387–400, 2002. [PubMed] [Google Scholar]
- 15.Ohtake F, Baba A, Takada I, Okada M, Iwasaki K, Miki H, Takahashi S, Kouzmenko A, Nohara K, Chiba T, Fujii-Kuriyama Y, Kato S. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature 446: 562–566, 2007. [DOI] [PubMed] [Google Scholar]
- 16.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6: 9–20, 2005. [DOI] [PubMed] [Google Scholar]
- 17.Pickart CM Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533, 2001. [DOI] [PubMed] [Google Scholar]
- 18.Ramos JA, Zenser N, Leyser O, Callis J. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13: 2349–2360, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reed JW Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6: 420–425, 2001. [DOI] [PubMed] [Google Scholar]
- 20.Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev 12: 198–207, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645, 2007. [DOI] [PubMed] [Google Scholar]
- 22.Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M. Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol 58: 183–198, 2007. [DOI] [PubMed] [Google Scholar]
- 24.Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA 96: 5844–5849, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Kerkhof P, Putters J, Strous GJ. The ubiquitin ligase SCF(betaTrCP) regulates the degradation of the growth hormone receptor. J Biol Chem 282: 20475–20483, 2007. [DOI] [PubMed] [Google Scholar]
- 26.Woodward AW, Bartel B. Auxin: regulation, action, interaction. Ann Bot (Lond) 95: 707–735, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell 11: 1445–1456, 2003. [DOI] [PubMed] [Google Scholar]
- 28.Zenser N, Ellsmore A, Leasure C, Callis J. Auxin modulates the degradation rate of Aux/IAA proteins. Proc Natl Acad Sci USA 98: 11795–11800, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]