Abstract
Reactive oxygen species (ROS) generated in hypoxic pulmonary artery endothelial cells cause transient oxidative base modifications in the hypoxic response element (HRE) of the VEGF gene that bear a conspicuous relationship to induction of VEGF mRNA expression (FASEB J 19: 387-394, 2005). If such base modifications are indeed linked to transcriptional regulation, then they should be detected in HRE sequences associated with transcriptionally active nucleosomes. Southern blot analysis of the VEGF HRE associated with nucleosome fractions prepared by micrococcal nuclease digestion indicated that hypoxia redistributed some HRE sequences from multi-nucleosomes to transcriptionally-active mono- and di-nucleosome fractions. A simple PCR method revealed that VEGF HRE sequences harboring oxidative base modifications were found exclusively in mono-nucleosomes. Inhibition of hypoxia-induced ROS generation with myxathiozol prevented formation of oxidative base modifications but not the redistribution of HRE sequences into mono- and di-nucleosome fractions. The histone deacetylase inhibitor, trichostatin A, caused retention of HRE sequences in compacted nucleosome fractions and prevented formation of oxidative base modifications. These findings suggest that the hypoxia-induced oxidant stress directed at the VEGF HRE requires the sequence to be repositioned into mono-nucleosomes, and support the prospect that oxidative modifications in this sequence are an important step in transcriptional activation.
Keywords: Hypoxia, reactive oxygen species, oxidative DNA modifications, chromatin remodeling, nucleosomes, gene expression
INTRODUCTION
Hypoxia is a fundamental stimulus in mammalian biology, and similar to many physiological signals appears to employ ROS as second messengers [1–5]. Multiple sources have been incriminated in hypoxia-induced ROS generation, including the membrane NADPH oxidase and mitochondria, with some recent evidence supporting a key role for ROS generated at the Qo site of mitochondrial complex III in stabilizing HIF-1, a key transcription factor driving hypoxia-induced gene expression [6–8]. Another action of hypoxia-generated ROS in pulmonary artery endothelial cells (PAECs) and other lung cell populations is to oxidatively modify specific nucleotides within the hypoxic response elements (HREs) of multiple hypoxia-inducible genes, including the VEGF gene [9–11]. These oxidative base modifications are conspicuous for several reasons. First, they are temporally related to initiation of gene expression [11]. Perhaps more interestingly, an analysis of modifications in the VEGF HRE at single nucleotide resolution revealed that while a number of nucleotides were targeted, the most common was a guanine residing at the 3′ end of the HIF-1 DNA recognition sequence [9, 10]. Mimicking this effect of hypoxia by introducing a base excision repair intermediate – an abasic site – at the hypoxia-modified guanine within an oligonucleotide corresponding to the VEGF HRE increased binding of HIF-1, engendered more robust reporter gene expression [10] and markedly enhanced local sequence flexibility, probably by serving as a substrate for a Ref-1/Ape1-mediated DNA strand break formation [12].
The foregoing observations raise the intriguing possibility that controlled oxidative base damage and repair initiated in the VEGF HRE by ROS generated as part of hypoxic signaling contribute to transcriptional regulation. One event that might be impacted by ROS-mediated base damage occurs at the level of the nucleosome, the repeating structural unit of chromatin consisting of approximately 150 bp wrapped around the nucleosome core particle. While it has been known for many years that promoter DNA was converted to a nuclease sensitive state upon transcriptional activation, it is now appreciated that the acquisition of nuclease sensitivity corresponds to fundamental changes in the apposition between promoter DNA and nucleosome core particles that probably impact on DNA interactions with transcriptional regulators. Nucleosome sliding and nucleosome eviction are both likely mechanisms underlying this required change in DNA-nucleosome contact [13, 14], and very recently it has been shown for estrogen receptor-α (ERα)-mediated transcriptional activation that controlled DNA strand breakage and repair may be a previously unappreciated pathway by which DNA “unwraps” or dissociates from the nucleosome to permit DNA-protein interactions necessary for transcription [15, 16]. Accordingly, to provide further evidence that ROS-mediated oxidative base modifications in hypoxia contribute to transcriptional regulation, the present study used cultured rat PAECs to test the predictions that the base modifications should be prominent in transcriptionally active nucleosomes, and that the propensity for hypoxia-induced oxidative base modifications should bear some relation to the state of chromatin organization.
METHODS
Pulmonary artery endothelial cell culture
Rat pulmonary artery endothelial cells (PAEC) were harvested and cultured as described previously [10]. Control, “normoxic” cells were cultured in incubator purged with air + 5% CO2, while “hypoxic” cells were cultured for the indicated periods in an environment consisting of 2% O2, 5% CO2, and 93% N2.
Nuclei isolation and nuclease treatment
Nuclei from cultured PAECs were isolated and treated with micrococcal nuclease using methods previously described [17] with minor modifications. In brief, cells were washed 3 times with PBS and 2 times with TKM buffer (10 mM Tris-HCl pH 7.8, 25 mM KCl, 5 mM MgCl2), scraped on ice in TSKM buffer (10 mM Tris-HCl pH 7.8, 25 mM KCl, 5 mM MgCl2, 320 mM glucose, 1% Triton X-100) supplied with 1% protease inhibitor cocktail (Sigma-Aldrich, Saint Louis, MO) and 1mM PMSF, and then homogenized with 6–8 strokes in a Dounce homogeniser. Cell homogenates were centrifuged for 8 min at 1000 × g (4° C) and washed again with the same buffer. After subsequent centrifugation pellets were washed in TSKM buffer without Triton X-100 and centrifuged for 10 min under the same conditions. Pelleted nuclei were suspended in digestion buffer (50 mM Tris-HCl pH 7.0, 25 mM KCl, 4 mM MgCl2, 1 mM CaCl2, 300 mM sucrose) to yield 2 mg/ml of DNA, and digested with micrococcal nuclease (Sigma-Aldrich, Saint Louis, MO) at a concentration 500 mU/ml for 5, 15 or 30 min at 37 °C. Reaction was stopped by adding EDTA to final concentration 25 mM and placing samples on ice. Nuclei were pelleted by centrifugation at 1000 × g for 20 min (4 °C). DNA released from nuclei during micrococcal nuclease digestion was isolated from supernatants with DNeasy Blood and Tissue Kit (Qiagen GmbH, Hamburg, Germany) and used for further experiments.
Southern blot detection of the VEGF HRE in nucleosome fractions
Isolated DNA fragments were separated by neutral electophoresis in 1.8% agarose gel into mono- to multi-nucleosomal repeats. Approximately 5 μg of DNA per well was loaded. After ethidium bromide staining, DNA was transferred to Hybond N+ membrane (Amersham plc, Little Chalfont, UK) and hybridized with 32P-labeled probe corresponding to a rat VEGF promoter sequence containing the hypoxic response element. The probe was generated by PCR using rat DNA sequence as a template and the following pair of primers: 5′-TCTGTCTGCCAGCTGTCTCT-3′ and 5′-GAGCTCTTGTCTGATCT TCATAC-3′, to obtain a 230 bp product. After hybridization, the membrane was washed and exposed to BioMax X-ray film (Eastman Kodak Company, Rochester, New York) in combination with intensifying screens.
PCR detection of oxidative base modifications in the VEGF HRE
We used a simple PCR-based assay, described previously [11, 18], to detect base modifications in the VEGF HRE. The basis of the assay is that treatment of DNA with Fpg (New England Biolabs, Beverly, MA) results in strand cleavage at sites of oxidized purines, thereby creating single strand breaks that block PCR amplification. Differences in PCR amplification between Fpg-treated and untreated DNA are thus a specific indicator of the presence of oxidative base damage. After electrophoresis of DNA fragments as described above, bands of nucleosome ladder were cut from the gel and DNA was extracted using QIAquick columns (Qiagen GmbH, Hamburg, Germany). Purified DNA from each band was treated with or without 8 units of Fpg in 1X NEBuffer 1 (10mM Bis Tris Propane-HCl pH 7.0, 10 mM MgCl2, 1mM DTT) and 100 μg/ml BSA in a volume of 20 μl. Incubations were carried out at 37°C for 16 hr. Fpg was then inactivated by heating at 65 °C for 5 min. A 2 μl aliquot was then used for the PCR assay to detect Fpg-sensitive cleavage sites. Primers used to amplify a sequence of the VEGF HRE were 5′-TCTGTCTGCCAGCTGTCTCT-3′ and 5′-GGAAGCCGAGCAGTTAGTCA-3′. PCR-generated 107 bp product was revealed by electrophoresis and ethidium bromide staining. Data are presented as the ratio of PCR amplification in the presence and absence of Fpg treatment.
Detection of ROS production using DCF fluorescence
Reactive oxygen species production in PAECs cultured in normoxia and hypoxia was detected using dichlorofluoroscein (DCF; Invitrogen, Carlsbad, CA) fluorescence as described previously [4].
Drugs and treatment regimens
Myxothiazol and trichostatin A, both acquired from Sigma-Aldrich Chemical Co (St. Louis, MO), were added to culture media to obtain final concentrations of 2 μM and 100 nM, respectively. Hypoxic exposure was initiated 30 min after addition of the selected drug.
RESULTS
Effect of hypoxia on distribution of the VEGF HRE within nucleosome fractions
We examined micrococcal nuclease-derived nucleosome fractions and the distribution of the VEGF HRE within these fractions after 3h of culture under hypoxic conditions. This duration was selected based on our previous finding in PAECs and other lung cell types that Fpg-detectable base oxidation products within the VEGF HRE peaked at 3h of initiation of hypoxia. The representative ethidium bromide-stained gel shown in Figure 1 indicates that treatment of nuclei from normoxic and 3h hypoxic PAECs with micrococcal nuclease releases DNA in aggregates ranging from mono-nucleosomes to highly compacted, large-sized chromatin structures. Hypoxia has no apparent effect on the distribution of total DNA within the micrococcal nuclease-derived nucleosomal fractions.
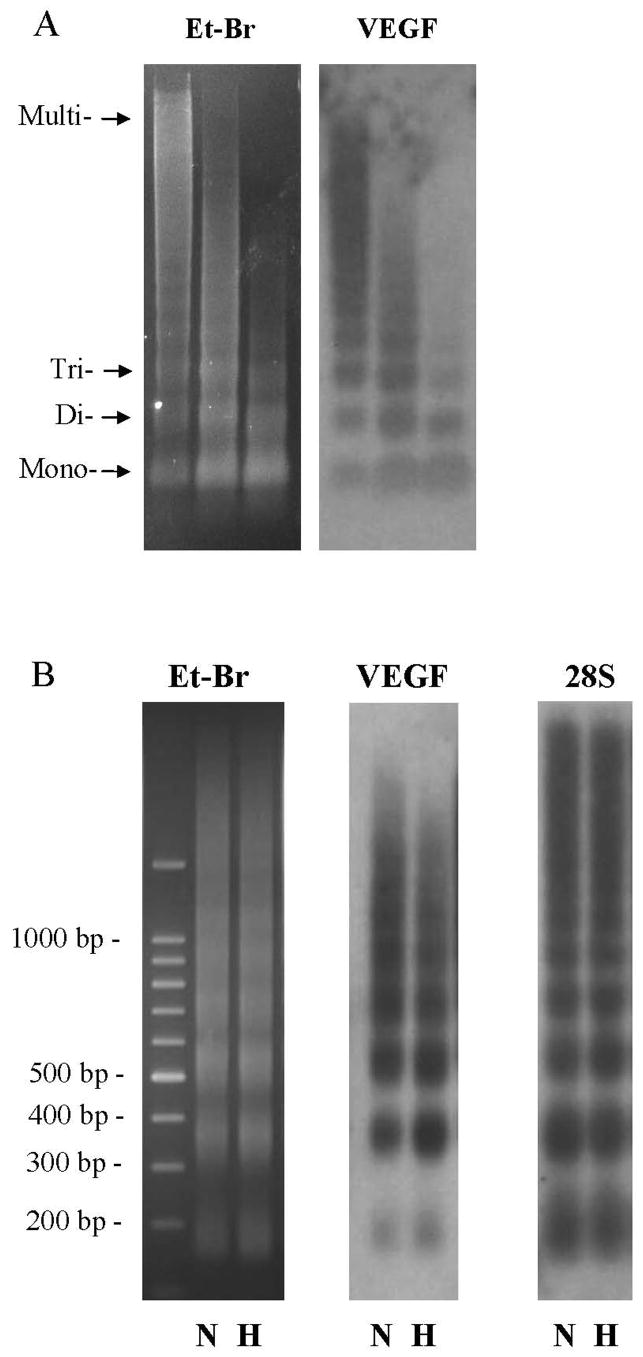
FIGURE 1.
(A) Ethidium bromide-stained gel (Et-Br) and Southern blot analysis of the VEGF hypoxic response element (VEGF) in DNA fragments released from PAEC nuclei after MN digestion for 5, 15 and 30 min. (B) Ethidium bromide-stained gel (Et-Br) and Southern blot analysis of the VEGF hypoxic response element (VEGF) and 28S rRNA (28S) in DNA fragments released from MN-treated (15 min) nuclei of normoxic (N) PAECs and cells after 3h of hypoxia (H). Note the shift of hybridization intensity of VEGF HRE to mono- and di-nucleosome fractions after hypoxia.
The gels were then transferred to a nylon membrane and hybridized with a 230 bp probe for a sequence of the VEGF HRE encompassing the HIF-1 DNA binding site. The resulting Southern blot, also shown in Figure 1, reveals that HRE sequences were detected in all nucleosome fractions in both normoxic and hypoxic cells. Importantly, 3h hypoxic culture regularly caused a subtle enrichment in the abundance of HRE sequences detected in mono- and di-nucleosome fractions accompanied by a similar diminution in the abundance of HRE sequences associated with higher-ordered nucleosome structures. Thus, hypoxia appears to redistribute a portion of nucleosomes and associated HRE sequences such that they are more sensitive to release by micrococcal nuclease digestion. As a negative control, we also probed for a sequence of the constitutively-expressed 28S rRNA gene. Little of this sequence could be localized to the mono-nucleosome fraction, and no redistribution of the sequence was noted after 3h hypoxic exposure.
Detection of hypoxia-induced oxidative base products within micrococcal nuclease-derived nucleosome fractions
Nucleosome fractions released by micrococcal nuclease digestion were excised from the gel. DNA recovered from mono-, di-, tri-, and multi-nucleosome fractions was treated for 16h with Fpg, a bacterial DNA glycosylase that cleaves at sites of 8-oxoG and related base oxidation products after which sequences of the VEGF HRE or the 28S rRNA gene were amplified by PCR. If oxidative base products are present in these sequences, then treatment of the DNA with Fpg will diminish PCR amplification of the indicated sequence relative to amplification of the non-Fpg-treated DNA sequence.
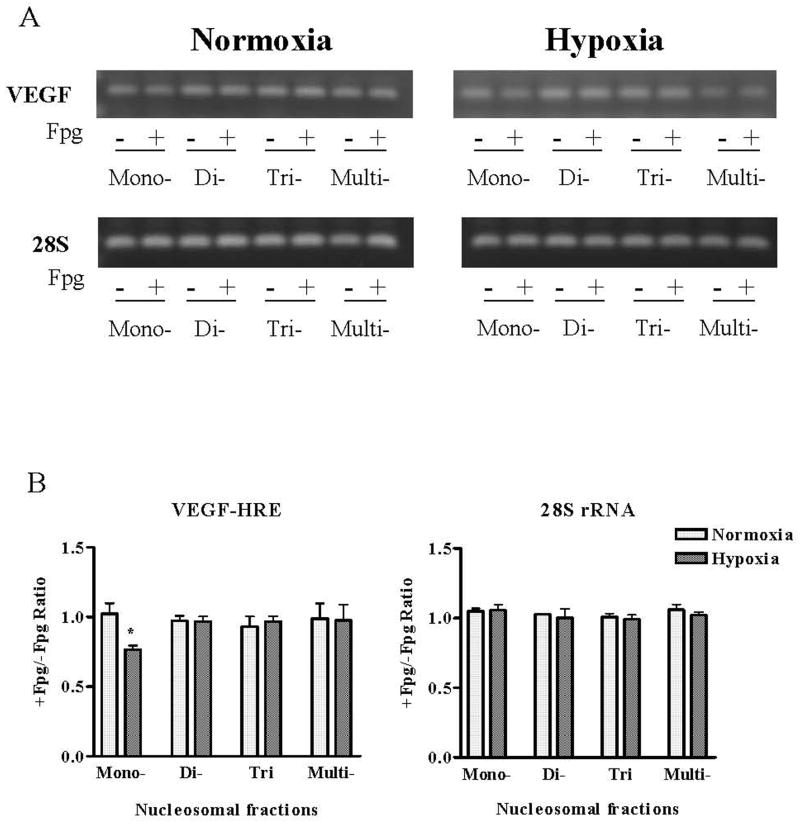
Representative gels of PCR amplifications +/− Fpg along with the pooled data shown in Figure 2 indicate that Fpg reduces PCR amplification of the VEGF HRE only in mono-nucleosome fractions. Despite a slight increase in the abundance of di-nucleosome-associated VEGF HRE in hypoxia, these sequences were insensitive to Fpg treatment. Thus, hypoxia seems to cause formation of oxidized base products in VEGF HRE localized to mono-nucleosomes but not elsewhere in micrococcal nuclease-releasable fractions. Fpg-sensitive base oxidation products were not detected in any nucleosomal sequence of the 28S rRNA gene.
FIGURE 2.
(A) PCR of treated and untreated with Fpg DNA fragments isolated from monomer, dimer, and trimer nucleosomal repeates and multi-nucleosomal zone with primers specific for the HRE of the VEGF promoter and 28S rRNA. (B) Pooled data for +/−Fpg PCR band intensity ratio. N = 6–8, *Different from normoxic controls at p<0.05.
Relation of hypoxia-induced base modifications in the VEGF HRE to mitochondria-derived ROS and to the general state of chromatin remodeling
Several groups have shown that hypoxia increases ROS generation from mitochondria [5, 7]. One line of evidence supporting this contention is that the complex III inhibitor, myxothiazol, suppresses hypoxia-induced ROS production. We used a similar approach to determine if mitochondrial-derived ROS contribute to the formation of hypoxia-induced oxidative base modifications in the VEGF HRE. PAECs were treated with myxothiazol after which ROS production was detected in terms of DCF fluorescence, VEGF mRNA was quantified by RT-PCR, the disposition of VEGF HRE in nucleosome fractions was determined after micrococcal nuclease digestion, and finally, the presence of the oxidative base modification in mono-nucleosome localized VEGF HRE was evaluated by PCR +/− Fpg.
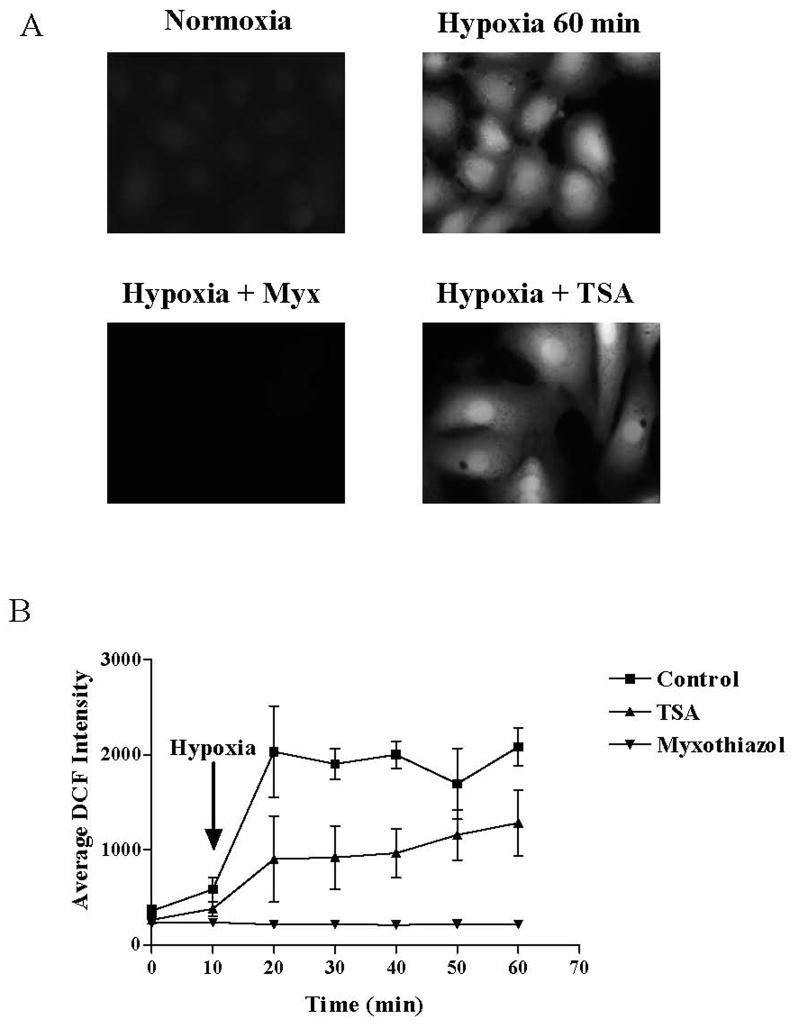
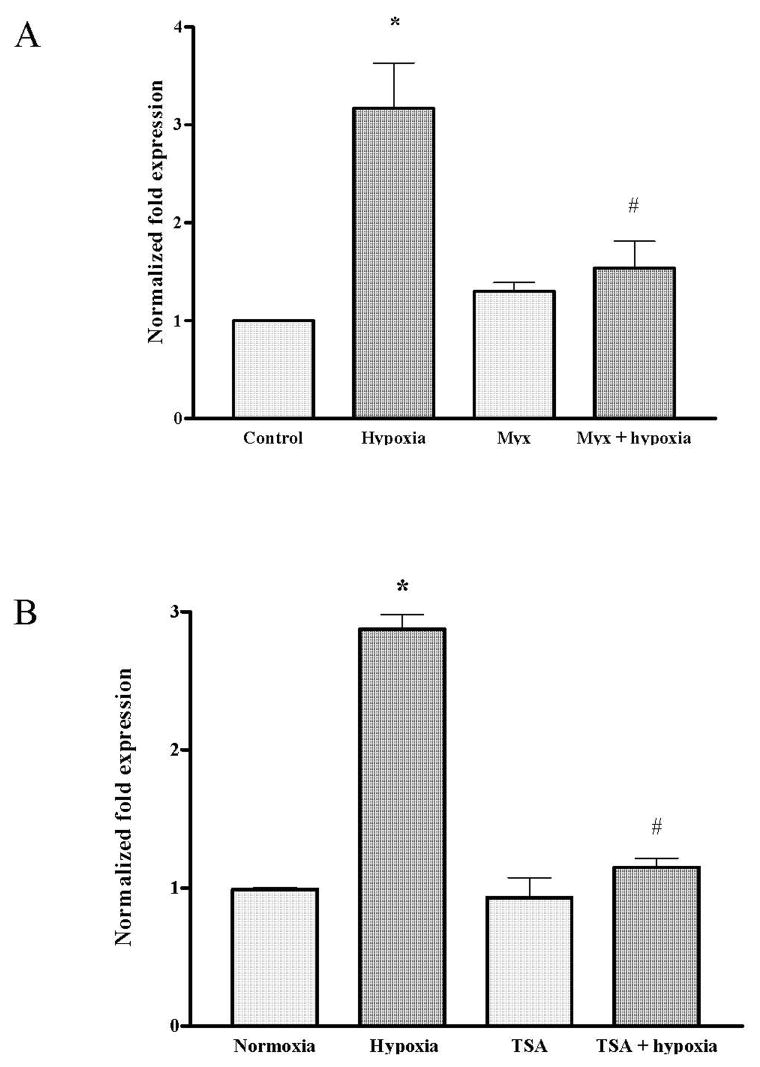
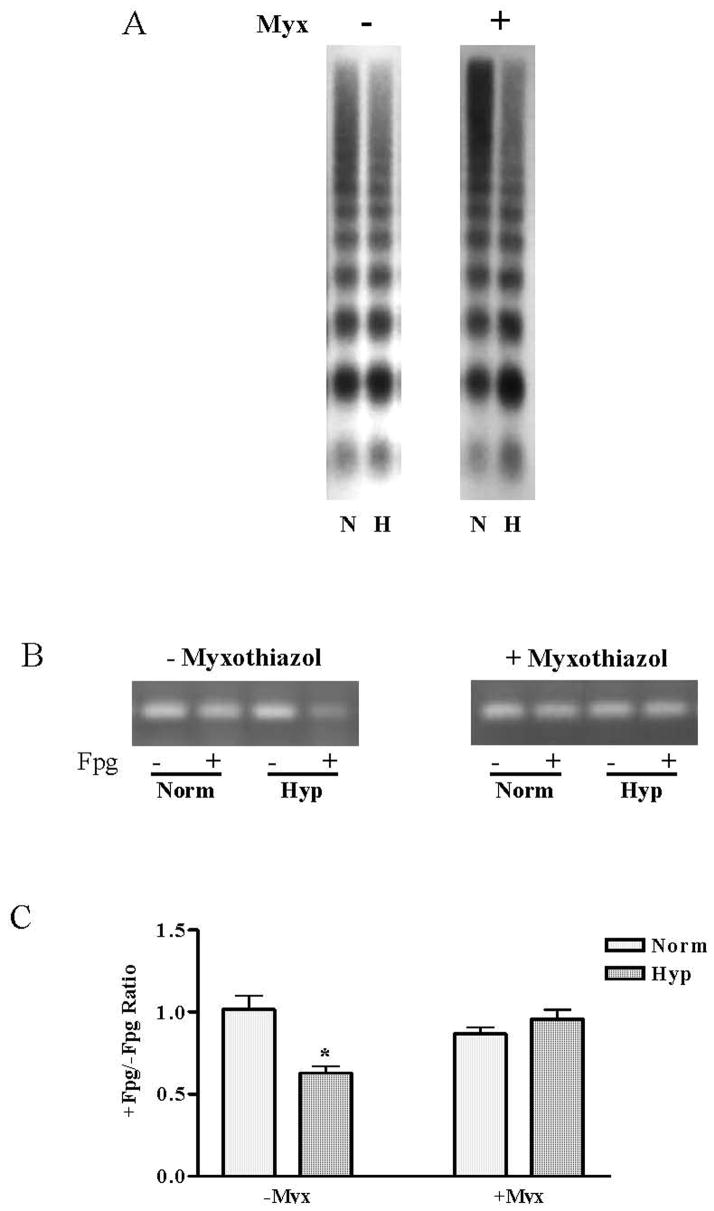
Data shown in Figure 3 indicate that myxothiazol suppressed both the acute increase in DCF fluorescence in PAECs cultured in hypoxia for up to an hour and, as shown in Figure 4A, prevented the increase in VEGF mRNA present after 3h of hypoxic culture. More interesting in the context of the present study, the representative Southern blot for the VEGF HRE displayed in Figure 5A show that despite suppression of mitochondrial ROS production with myxothiazol, the hypoxia-induced repositioning or remodeling of the VEGF HRE into the mono- and di-nucleosome fractions persisted. In contrast, and as shown by the representative ethidium-stained gels and pooled data depicted in Figure 5B and C, respectively, myxothiazol prevented formation of Fpg-detectable base oxidation products in mono-nucleosome-associated VEGF HRE sequences.
FIGURE 3.
(A) Representative microphotographs of DCF fluorescence of normoxic PAECs and cells exposed to hypoxia for 50 min. In experiments with myxothiazol or TSA, cells were pretreated with respective agent for 30 min before exposure to hypoxia. (B) Time course of DCF fluorescence in untreated PAECs and cells pretreated with myxothiazol or TSA and exposed to hypoxia for 50 min. N = 4. *Different from normoxic controls at p<0.05.
FIGURE 4.
Relative VEGF mRNA accumulation in PAECs exposed to hypoxia for 3 hours in absence or presence of myxothiazol (Panel A, N = 6) and TSA (Panel B, N = 5–13). *Different from normoxic controls at p<0.05.
FIGURE 5.
(A) Southern blot analysis of the VEGF hypoxic response element of DNA fragments released from MN-digested (15 min) nuclei of normoxic (N) and hypoxic (H) PAECs with or without myxothiazol treatment. (B) PCR analysis of treated and untreated with Fpg DNA fragments recovered from mono-nucleosomal fraction with primers specific for the HRE of the VEGF promoter. (C) Pooled data for +/−Fpg PCR band intensity ratio. N = 6. *Different from normoxic controls at p<0.05.
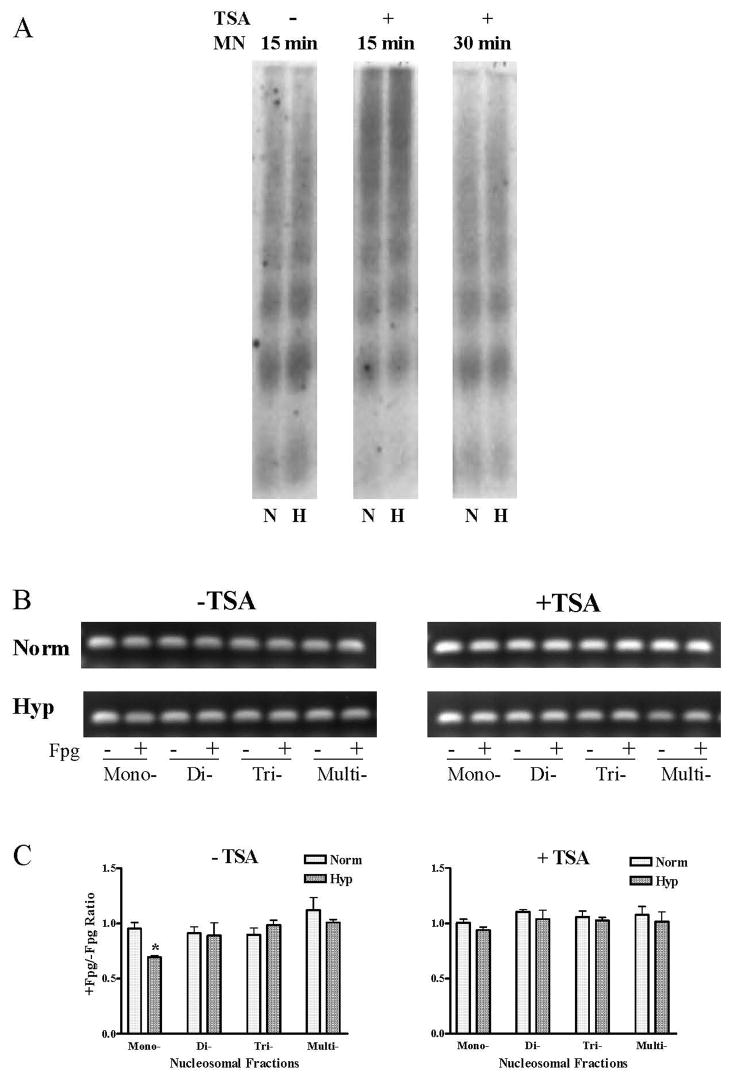
We used a similar approach to examine the effects of the broad-spectrum histone deacetylase inhibitor, TSA, on hypoxia-induced ROS production and on the disposition of the HRE and oxidative base lesions within micrococcal nuclease-resolved nucleosome fractions. Data shown in Figure 3 indicate that TSA attenuated but did not abolish hypoxia-induced ROS production in PAECs. TSA did, however, prevent the hypoxia-induced increase in VEGF mRNA (Figure 4B). As illustrated by the representative Southern blot shown in Figure 6A, HDAC inhibition with TSA resulted in the formation of highly compacted chromatin from which protracted incubation with micrococcal nuclease released only small amounts of mono- and di-nucleosome fractions and associated VEGF HRE sequences. As shown in Figures 6B and C, when nucleosome-associated VEGF HRE sequences obtained after more extensive micrococcal nuclease digestion was subjected to PCR +/− Fpg, no Fpg-mediated decrements in PCR amplification were detected in either hypoxic or normoxic DNA associated with any of the nucleosomal fractions. These observations indicate that TSA-induced compaction of chromatin was associated with prevention of hypoxia-induced formation of base oxidation products in the VEGF HRE.
FIGURE 6.
(A) Southern blot analysis of the VEGF hypoxic response element of DNA fragments released from MN-digested (15 or 30 min) nuclei of normoxic (N) and hypoxic (H) PAECs with or without TSA treatment. (B) PCR analysis of treated and untreated with Fpg DNA fragments recovered from gel with primers specific for the HRE of the VEGF promoter. (C) Pooled data for +/−Fpg PCR band intensity ratio. N = 3–8. *Different from normoxic controls at p<0.05.
DISCUSSION
Multiple lines of indirect evidence suggest that ROS-mediated changes in DNA integrity might contribute to hypoxia-regulated transcription. First, hypoxia seems to increase ROS generation as part of signaling [1–5]. Second, ROS generated in hypoxia cause nucleotide-specific oxidative modifications in promoters of hypoxia-inducible genes [9–11]. Third, mimicking the hypoxia-induced, ROS-mediated base oxidation by incorporating an abasic site at the hypoxia-modified guanine located at the 3′ end of the HIF-1 DNA recognition sequence in an oligonucleotide corresponding to the VEGF HRE increased its association with HIF-1 and engendered more robust reporter gene expression [10]. Fourth, the presence of an abasic site at the hypoxia-modified guanine in the model VEGF HRE also engendered profound increases in local sequence flexibility – another postulated event in transcriptional activation - probably caused by the Ref-1/Ape1-mediated formation of a single strand break [12]. Finally, and as discussed below, the current finding that hypoxia-induced oxidative base modifications are restricted to micrococcal nuclease-releasable, transcriptionally-active sequences of the VEGF HRE also supports the contention that controlled DNA “damage” and repair contributes to transcriptional activation.
The proposal that oxidative base modifications in hypoxia-inducible promoters modulate transcriptional activation is similar in several respects to recently described models of ERα-mediated gene expression. Like hypoxia, activation of the ERα-also stimulates ROS production. However, in the case of ligated ERα binding to estrogen responsive elements (ERE), ROS are generated by histone-bound lysine-specific DNA methylase, LSD1 [19], whereas hypoxia likely causes ROS generation from mitochondria [7, 8]. Another feature common to both ERα– and hypoxia-induced transcription is that the oxidant stress targets functionally relevant promoter sequences. In the case of the ERα, nucleotides in the vicinity of the ERE are targeted [19] while, as noted above, specific nucleotides in the HREs of multiple genes are oxidatively modified in hypoxic cells. Finally, alterations in DNA flexibility also seem to occur as a consequence of ROS-mediated DNA modifications in ERα-mediated transcriptional activation. Upstream promoter and ERE enhancer regions form a looped structure in ERα-activated genes that are dependent on oxidative modifications and strand breaks in specific sequences [19]. Thus, the involvement of ROS in signaling, oxidative modifications to specific DNA sequences, and apparent changes in local sequence flexibility associated with the oxidant stress are all properties shared by ERα-mediated activation of estrogen responsive genes and the hypoxia-induced activation of VEGF expression.
A key line of evidence supporting the concept that controlled DNA “damage” and repair is important for transcription initiated by the ERα - namely, that the DNA strand break initiated by oxidative damage occurs in the context of transcriptionally-active nucleosomes [15] - is lacking from the model proposed for hypoxic VEGF expression. Accordingly, the present study determined the presence of base oxidation lesions in VEGF HREs resolved into nucleosome fractions by micrococcal nuclease digestion. As ancillary but related issues, we also performed an initial exploration of the relationship between promoter oxidative base modifications, the positioning of the VEGF HRE into micrococcal nuclease-releasable mono-nucleosomes, and the general state of chromatin structure in hypoxic PAECs.
In both normoxic and hypoxic PAECs, we found that VEGF HRE sequences were distributed throughout all of the micrococcal nuclease-releasable DNA fractions, from mono-nucleosomes to highly compacted chromatin structures. Since highly compacted chromatin is generally believed to be transcriptionally repressed, the wide distribution of VEGF HRE amongst nucleosome fractions likely means that only a portion of the cells are actively transcribing VEGF mRNA at any one time. In hypoxia, however, we found small but predictable redistribution of the VEGF HRE from nuclease-resistant DNA aggregates to sequences associated with mono- and di-nucleosomes in hypoxic PAECs relative to normoxic control cells. This effect of hypoxia was not reflected in the nucleosomal distribution of total cellular DNA, which was not demonstrably different between normoxic and hypoxic cells, nor by the nucleosome distribution of a sequence of the constitutively transcribed 28S rRNA gene, which was also unaffected by hypoxia. That the distribution of total cell DNA or of a constitutively transcribed gene would be unaffected by hypoxia is not surprising. After all, a small fraction of the genome - about 600 genes - are differentially expressed in hypoxic cells, with roughly half increased and half decreased in comparison to controls [20]; changes in the distribution of total DNA released by nuclease treatment would not be expected.
Only those VEGF HRE sequences that were highly sensitive to release by micrococcal nuclease – sequences associated with mono-nucleosomes – were oxidatively modified in hypoxia. VEGF HRE sequences associated with di-nucleosomes, whose abundance also was increased in hypoxia, and VEGF HRE sequences detected in more compacted nucleosome structures all failed to exhibit oxidative base lesions. The reason why only mono-nucleosome-associated HREs are targeted by hypoxia-generated ROS is unclear. Traditional concepts hold that transcriptionally active, nuclease-releasable DNA is more sensitive to oxidative damage than non-transcribed DNA because it is not “protected” by histones. However, accumulating data suggest that the mechanism underlying the oxidant sensitivity is probably more complex than mere “packaging.” In this context, Barton and colleagues have demonstrated that long-range DNA charge transport occurs in the nucleus of living cells [21], including DNA sequences associated with nucleosome core particles [22], and that such charge transport may be responsible for driving non-uniform distribution of oxidative DNA lesions to sequences harboring doublets or triplets of guanine [23]. This would explain why the HRE, and the HIF-1 DNA recognition sequence in particular, is prone to oxidative modifications in hypoxia, but the reason that only VEGF HRE sequences sensitive to micrococcal nuclease release are prone to attack requires additional explanation. In this regard, it is known that interactions between DNA and proteins that cause structural alterations in the DNA base-pair p stack or that change the electronic properties of specific bases can disrupt charge transport, and thereby have the potential to insulate certain sequences despite their being rich in guanine doublets or triplets [23]. It is thus tempting to speculate that alterations in DNA-protein interactions required for chromatin remodeling may increase sensitivity of transcriptionally active VEGF HREs to oxidative modifications evoked by hypoxia-generated ROS while inactive VEGF HRE sequences are in effect insulated from charge transport and oxidant attack.
We examined the effects of the mitochondrial complex III inhibitor, myxothiazol, first, to determine if mitochondria-derived ROS contributed to hypoxia-induced base modifications, and second, to examine the link between the oxidative base modifications and hypoxia-induced repositioning or remodeling of the VEGF HRE from compacted chromatin structure to mono-and di-nucleosomes. With regard to the first issue, inhibition of base modifications by myxothiazol incriminates mitochondrial complex III. We do not wish to suggest, however, that mitochondria-derived ROS are the direct mediators of oxidative base damage in hypoxia. It is difficult to envision how only certain genes are targeted for oxidative modifications [11] by ROS that must diffuse into the nucleus from an anatomically distant source. It may be, however, that mitochondrial-derived ROS serve to trigger secondary ROS production from a source closely associated with hypoxia-inducible genes, much like the role of LSD1 in ERα-mediated transcription [19]. While there is a precedent for membrane NADPH oxidase-mediated activation of mitochondrial ROS production after angiotensin II [24] or hypoxia [25], a similar connection between mitochondria-derived ROS production in hypoxia and a nuclear source of ROS generation has yet to be reported.
We also found that myxothiazol, which prevented formation of base oxidation products in mono-nucleosome-associated VEGF HRE, failed to suppress accumulation of the sequence in nuclease-releasable mono-and di-nucleosome fractions. In some ways, this is surprising. Myxothiazol as well as other drugs or interventions attenuating ROS production prevent accumulation of HIF-1 and attendant gene expression in hypoxic cells [26]. Since transcription factor binding is believed to initiate nucleosome eviction and chromatin remodeling [13], it was expected that myxothiazol would suppress the accumulation of nuclease-releasable VEGF HRE sequences. The failure of myxothiazol to do so implies that neither the key transcription factor in hypoxia, HIF-1, nor the introduction of oxidative base modifications are required for repositioning or remodeling of VEGF HRE into the nuclease-sensitive DNA fractions.
As indicated above, highly compacted DNA is believed to be relatively protected from oxidant attack, and results of our study tend to support this contention. When histone deacetylases were suppressed by the broad-spectrum inhibitor TSA, nucleosome fractions released by prolonged micrococcal nuclease digestion were not repositioned or remodeled during hypoxia, VEGF mRNA expression was suppressed, and importantly, oxidative base modifications in the VEGF HRE were not detected. Surprisingly, however, we found that TSA attenuated but did not abolish hypoxia-induced ROS generation, thus raising the possibility that the inhibition of base modifications was linked in part to a diminished hypoxia-induced oxidant stress. However, because TSA-treated PAECs still generated considerable levels of DCF-detectable ROS, a more likely explanation for the suppression of base modifications in the VEGF HRE is that the sequence must be repositioned into nuclease-releasable structures to acquire sensitivity to oxidant attack.
In summary, the present study shows that the oxidant stress associated with hypoxic signaling and targeted to nucleotides within the VEGF HRE is restricted to sequences that are repositioned into the nuclease-releasable, mono-nucleosome fraction believed to be transcriptionally active. These findings, together with previous observations, add to the growing evidence that controlled DNA “damage” and repair play a role in guiding productive transcription. In addition, repositioning or remodeling of the HRE sequence into mono-nucleosome fractions is not dependent on the hypoxia-induced oxidant stress per se or on the introduction of base modifications in the VEGF HRE. On the other hand, for the VEGF HRE to be sensitive to hypoxia-induced base modifications, it cannot be situated in chromatin structures that are highly compacted. The precise mechanism by which hypoxia-induced base modifications contribute to regulation of VEGF mRNA expression remains to be defined, but it is tempting to speculate that they direct some aspect of the interactions between DNA, nucleosomes, and transcriptional activators and co-activators.
LIST OF ABBREVIATIONS
- ROS
Reactive oxygen species
- PAECs
Pulmonary artery endothelial cells
- HRE
Hypoxic response element
- HIF-1
Hypoxia inducible factor-1
- VEGF
Vascular endothelial growth factor
- ERα
Estrogen receptor alpha
- ERE
Estrogen responsive elements
- Fpg
Formamidopyrimidine DNA glycosylase
- TSA
Trichostatin A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Marshall C, Mamary AJ, Verhoeven AJ, Marshall BE. Pulmonary artery nadph-oxidase is activated in hypoxic pulmonary vasoconstriction. Am J Respir Cell Mol Biol. 1996;15:633–644. doi: 10.1165/ajrcmb.15.5.8918370. [DOI] [PubMed] [Google Scholar]
- 2.Liu JQ, Zelko IN, Erbynn EM, Sham JS, Folz RJ. Hypoxic pulmonary hypertension: Role of superoxide and nadph oxidase (gp91phox) Am J Physiol Lung Cell Mol Physiol. 2006;290:L2–10. doi: 10.1152/ajplung.00135.2005. [DOI] [PubMed] [Google Scholar]
- 3.Ward JP. Point: Hypoxic pulmonary vasoconstriction is mediated by increased production of reactive oxygen species. J Appl Physiol. 2006;101:993–995. doi: 10.1152/japplphysiol.00480.2006. discussion 999. [DOI] [PubMed] [Google Scholar]
- 4.Killilea DW, Hester R, Balczon R, Babal P, Gillespie MN. Free radical production in hypoxic pulmonary artery smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2000;279:L408–412. doi: 10.1152/ajplung.2000.279.2.L408. [DOI] [PubMed] [Google Scholar]
- 5.Schumacker PT. Current paradigms in cellular oxygen sensing. Adv Exp Med Biol. 2003;543:57–71. doi: 10.1007/978-1-4419-8997-0_5. [DOI] [PubMed] [Google Scholar]
- 6.Bell EL, Klimova TA, Eisenbart J, Moraes CT, Murphy MP, Budinger GR, Chandel NS. The qo site of the mitochondrial complex iii is required for the transduction of hypoxic signaling via reactive oxygen species production. J Cell Biol. 2007;177:1029–1036. doi: 10.1083/jcb.200609074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klimova T, Chandel NS. Mitochondrial complex iii regulates hypoxic activation of hif. Cell Death Differ. 2008;15:660–666. doi: 10.1038/sj.cdd.4402307. [DOI] [PubMed] [Google Scholar]
- 8.Guzy RD, Mack MM, Schumacker PT. Mitochondrial complex iii is required for hypoxia-induced ros production and gene transcription in yeast. Antioxid Redox Signal. 2007;9:1317–1328. doi: 10.1089/ars.2007.1708. [DOI] [PubMed] [Google Scholar]
- 9.Grishko V, Solomon M, Breit JF, Killilea DW, Ledoux SP, Wilson GL, Gillespie MN. Hypoxia promotes oxidative base modifications in the pulmonary artery endothelial cell vegf gene. Faseb J. 2001;15:1267–1269. doi: 10.1096/fj.00-0755fje. [DOI] [PubMed] [Google Scholar]
- 10.Ziel KA, Grishko V, Campbell CC, Breit JF, Wilson GL, Gillespie MN. Oxidants in signal transduction: Impact on DNA integrity and gene expression. Faseb J. 2005;19:387–394. doi: 10.1096/fj.04-2805com. [DOI] [PubMed] [Google Scholar]
- 11.Pastukh V, Ruchko M, Gorodnya O, Wilson GL, Gillespie MN. Sequence-specific oxidative base modifications in hypoxia-inducible genes. Free Radic Biol Med. 2007;43:1616–1626. doi: 10.1016/j.freeradbiomed.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 12.Breit JF, Ault-Ziel K, Al-Mehdi AB, Gillespie MN. Nuclear protein-induced bending and flexing of the hypoxic response element of the rat vascular endothelial growth factor promoter. Faseb J. 2008;22:19–29. doi: 10.1096/fj.07-8102com. [DOI] [PubMed] [Google Scholar]
- 13.Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 14.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase iibeta-mediated dsdna break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 16.Haince JF, Rouleau M, Poirier GG. Transcription. Gene expression needs a break to unwind before carrying on. Science. 2006;312:1752–1753. doi: 10.1126/science.1129808. [DOI] [PubMed] [Google Scholar]
- 17.Faulkner RD, Bhatnagar YM. A protease activity is associated with testicular chromatin of the mouse. Biol Reprod. 1987;36:471–480. doi: 10.1095/biolreprod36.2.471. [DOI] [PubMed] [Google Scholar]
- 18.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 19.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by h3k9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 20.Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL. Transcriptional regulation of vascular endothelial cell responses to hypoxia by hif-1. Blood. 2005;105:659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- 21.Nunez ME, Holmquist GP, Barton JK. Evidence for DNA charge transport in the nucleus. Biochemistry. 2001;40:12465–12471. doi: 10.1021/bi011560t. [DOI] [PubMed] [Google Scholar]
- 22.Nunez ME, Noyes KT, Barton JK. Oxidative charge transport through DNA in nucleosome core particles. Chem Biol. 2002;9:403–415. doi: 10.1016/s1074-5521(02)00121-7. [DOI] [PubMed] [Google Scholar]
- 23.Merino EJ, Boal AK, Barton JK. Biological contexts for DNA charge transport chemistry. Curr Opin Chem Biol. 2008;12:229–237. doi: 10.1016/j.cbpa.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ricci C, Pastukh V, Leonard J, Turrens J, Wilson G, Schaffer D, Schaffer SW. Mitochondrial DNA damage triggers mitochondrial-superoxide generation and apoptosis. Am J Physiol Cell Physiol. 2008;294:C413–422. doi: 10.1152/ajpcell.00362.2007. [DOI] [PubMed] [Google Scholar]
- 25.Rathore R, Zheng YM, Niu CF, Liu QH, Korde A, Ho YS, Wang YX. Hypoxia activates nadph oxidase to increase [ros](i) and [ca(2+)](i) through the mitochondrial ros-PKCε signaling axis in pulmonary artery smooth muscle cells. Free Radic Biol Med. 2008 doi: 10.1016/j.freeradbiomed.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emerling BM, Platanias LC, Black E, Nebreda AR, Davis RJ, Chandel NS. Mitochondrial reactive oxygen species activation of p38 mitogen-activated protein kinase is required for hypoxia signaling. Mol Cell Biol. 2005;25:4853–4862. doi: 10.1128/MCB.25.12.4853-4862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]