Abstract
Changes in near UV circular dichroism (CD) and fluorescence spectra of site-specifically placed pairs of 2-aminopurine residues have been used to probe the roles of the RNA hairpin and the RNA-DNA hybrid in controlling intrinsic termination of transcription. Functional transcription complexes were assembled directly by mixing preformed nucleic acid scaffolds of defined sequence with T7 RNA polymerase (RNAP). Scaffolds containing RNA hairpins immediately upstream of a GC-rich hybrid formed complexes of reduced stability, whereas the same hairpins adjacent to a hybrid of rU-dA base pairs triggered complex dissociation and transcript release. 2-Aminopurine probes at the upstream ends of the hairpin stems show that the hairpins open on RNAP binding and that stem re-formation begins after one or two RNA bases on the downstream side of the stem have emerged from the RNAP exit tunnel. Hairpins directly adjacent to the RNA-DNA hybrid weaken RNAP binding, decrease elongation efficiency, and disrupt the upstream end of the hybrid as well as interfere with the movement of the template base at the RNAP active site. Probing the edges of the DNA transcription bubble demonstrates that termination hairpins prevent translocation of the RNAP, suggesting that they transiently “lock” the polymerase to the nucleic acid scaffold and, thus, hold the RNA-DNA hybrid “in frame.” At intrinsic terminators the weak rU-dA hybrid and the adjacent termination hairpin combine to destabilize the elongation complex sufficiently to permit significant transcript release, whereas hairpin-dependent pausing provides time for the process to go to completion.
Transcription termination represents one of several alternative reaction pathways that potentially compete with elongation at every position along the DNA template (1). Intrinsic termination of transcription in bacteria involves a common mechanism characterized by two sequence motifs in the RNA that are required for the release of the transcript. These are (i) formation of a stable stem-loop (terminator hairpin) structure in the nascent RNA (2, 3) followed by (ii) a run of 8–10 nucleotide residues (nt),2 consisting primarily of uridine residues, that is located immediately downstream of the hairpin and at the 3′-end of the released RNA (4). In the absence of protein factors these nucleic acid elements act together to form a termination signal that brings about RNA release and polymerase dissociation from the DNA template.
These two elements of the intrinsic termination signal, functioning separately, are also responsible for the two major (Class I and II) pausing mechanisms in Escherichia coli that lead the transcription complex into reaction pathways that compete with elongation (1). Working together as an intrinsic terminator, these elements undermine (directly or indirectly) two of the interactions that are responsible for the extraordinary stability of elongation complexes at non-terminator positions. These are (i) the stable RNA-DNA hybrid duplex of the elongation complex, which is replaced at intrinsic terminators by a much weaker hybrid containing primarily rU-dA base pairs and (ii) the interaction between the so-called “single-stranded RNA binding site” of the polymerase and the single-stranded transcript sequence formed at the “strand separator” just upstream of the 5′RNA end of the hybrid (5, 6). This interaction is at least partially disrupted at intrinsic terminators by the formation of the termination hairpin. The detailed mechanisms by which this hairpin and the weak rU-dA hybrid collaborate to bring about intrinsic termination remain unresolved, but several models, based primarily on mechanistic studies of transcription complexes containing E. coli RNA polymerase (RNAP), have been proposed.
An early “thermodynamic destabilization model” dealt quantitatively with the nucleic acid components of the complex (7). Here, standard free energy data for the DNA, RNA, and RNA-DNA base pairs of the nucleic acid framework of the transcription complex were used to calculate that the presence of the weak rU-dA hybrid at intrinsic terminator sites, in combination with the invasion or distortion of the upstream end of the hybrid by the termination hairpin, should result in destabilization of the elongation complex sufficient to bring about significant transcript release and elongation complex dissociation. The subsequent determination of the structural details of transcription complexes by x-ray crystallography added protein-based elements, suggesting that specific conformational changes induced within the polymerase by contact with the termination hairpin might be transmitted “allosterically” to the active site of the RNAP to trigger complex destabilization and transcript release (8). A “forward translocation” model suggested specifically that the formation of the stem-loop of the termination hairpin at intrinsic terminators might obstruct or compete with the single-stranded RNA binding domain of the polymerase and “pull” (without synthesis) the RNA out of the transcription complex by forward translocation of the complex along the template DNA (9).
T7 RNA polymerase is a single subunit enzyme that performs all the essential steps of template-directed transcription characteristic of the more complex multi-subunit RNAPs of bacteria and higher organisms. A major class of intrinsic terminators in T7 closely resembles those of E. coli in that they also encode a stable stem-loop hairpin in the RNA followed by a run of rU residues (10). In addition, T7 RNAP can terminate successfully at bacterial intrinsic terminators, suggesting a common mechanism (11, 12). This sequence conservation between the intrinsic terminators of E. coli and of T7 combined with the fact that both transcription complexes manifest essentially the same transcription bubble size, hybrid length, and transcript elongation mechanism (13, 14) suggests that these intrinsic terminators of T7 are likely to function in essentially the same way as those of the more extensively studied E. coli transcription system.
We have used the near UV spectral properties of pairs of 2-aminopurine (2-AP) bases, site-specifically placed into a synthetic RNA-primed transcription bubble scaffold to which T7 RNAP was then added to follow the conformational changes of the nucleic acid framework that accompany the various steps of the elongation reaction (15). Earlier studies in our laboratory and others had shown that fully functional elongation complexes can be formed directly (without going through the normal promoter-dependent transcription initiation process) by mixing nucleic acid scaffold constructs with either E. coli or T7 RNA polymerase (16, 17). In addition it has recently been shown that hairpin-stabilized paused complexes containing E. coli RNAP reconstituted with a non-complementary DNA bubble also exhibit key features of promoter-initiated hairpin-induced paused states, thus confirming that such “synthetic” constructs can be used to study pausing as well as elongation and termination (18).
2-AP is a structural isomer of adenine in which the amino group is moved from position 6 to position 2 of the purine ring. Like adenine, 2-AP forms base pairs with thymine (in DNA) and uridine (in RNA), and the substitution of 2-AP for adenine does not significantly perturb the structure or stability of the resultant helical duplexes. Because the major CD and fluorescence signals for the 2-AP probes fall in the near UV (wavelengths >300 nm), a spectral region in which the normal protein and nucleic acid components of transcription complexes are transparent, we have been able to develop this type of spectroscopy (19) into a quantitative technique for studying local conformational changes at specific positions within the nucleic acid frameworks of a variety of macromolecular machines functioning in solution (15, 20). Here we use these methods to examine T7 transcription complexes with scaffolds containing (either separately or together) the nucleic acid elements that define intrinsic terminators and E. coli Class I and Class II pause sites. Detailed spectral observations at defined positions within the nucleic acid scaffolds of transcription complexes located at intrinsic terminators on the template are used to evaluate and modify structural models for termination that have been put forward in the literature.
EXPERIMENTAL PROCEDURES
Materials
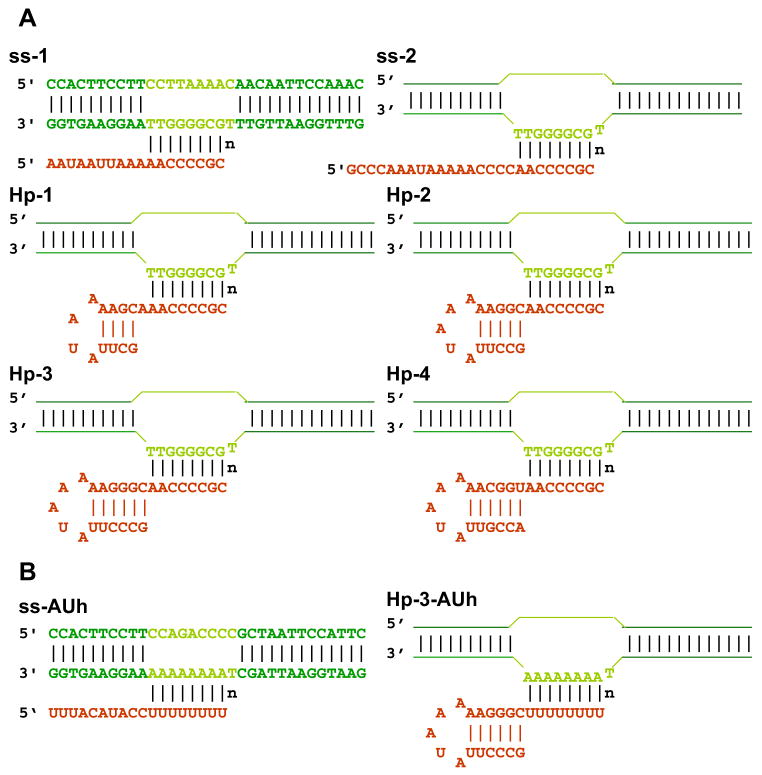
Unlabeled and 2-AP-labeled DNA oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA) and from Operon (Huntsville, AL). 2-AP-labeled and unlabeled RNA were from Dharmacon (Lafayette, CO). Oligonucleotide concentrations were determined using absorbance at 260 nm and extinction coefficients furnished by the manufacturer. RNase H was purchased from Epicenter Biotechnologies (Madison, WI). His-tagged T7 RNA polymerase was purified from clone pBH161 (21), kindly provided by William McAllister. Concentrations of T7 RNAP were estimated using a molar extinction coefficient of 1.4 × 10−5 at 280 nm. The DNA scaffold constructs containing non-complementary bubble sequences were formed and annealed by heating a solution containing equimolar concentrations of template and non-template strands at 90 °C for 4 min and then gradually cooling to room temperature over a period of 2 h. Equimolar concentrations of the relevant RNA construct were then added to the annealed DNA bubble, heated to 35 °C for 5 min, and gradually cooled to room temperature to form the RNA-primed bubble structures used as nucleic acid framework constructs in our study. The various final RNA-primed DNA bubble constructs are shown in Fig. 1.
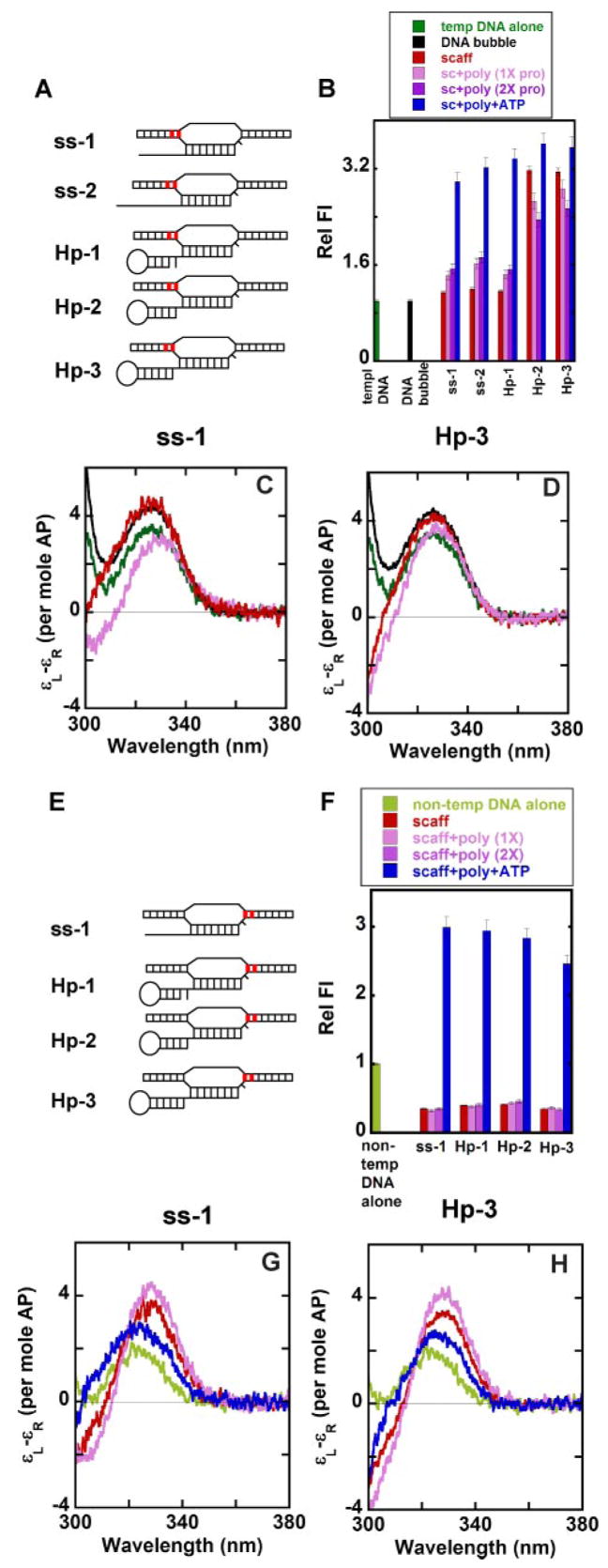
FIGURE 1. Nucleic acid sequences and nomenclature of the scaffold constructs used in this study.
A, scaffolds with GC-rich RNA-DNA hybrid. B, scaffolds with a rU-dA hybrid. Dark green, duplex region of the DNA bubble framework; light green, non-complementary region of the DNA bubble; red, RNA transcript. n represents the active site template base.
All experiments were performed at 25 °C in buffer containing 20 mM Hepes (pH 7.9), 100 mM sodium acetate, 10 mM magnesium acetate, 1 mM dithiothreitol, 0.1 mM EDTA, 0.05% Tween 20 (protein grade, from Calbiochem), and 1% glycerol. Unless otherwise stated, 3 μM (equimolar) concentrations of T7 RNAP and nucleic acid framework constructs were used in all experiments. 20 μM NTP substrate concentrations were used to form the stalled T7 elongation complexes, whereas1 mM NTP concentrations were used for the termination and run-off transcription experiments.
Fluorescence Measurements
Fluorescence spectra were measured using a Jobin-Yvon Fluorolog spectrofluorimeter. Samples were excited at 315 nm, and emission spectra were recorded from 310–420 nm. The fluorescence intensities presented in the figures represent values measured at 370 nm under identical conditions for the different scaffolds. For stoichiometry titrations of 2-AP-labeled scaffold constructs with RNAP, 3 μM concentrations of scaffold were titrated with increasing amounts of protein and monitored by fluorescence (Fig. 2A). Results with twice the polymerase concentration (6 μM) are also shown in some figures to confirm that RNAP-scaffold binding is indeed effectively stoichiometric in these experiments.
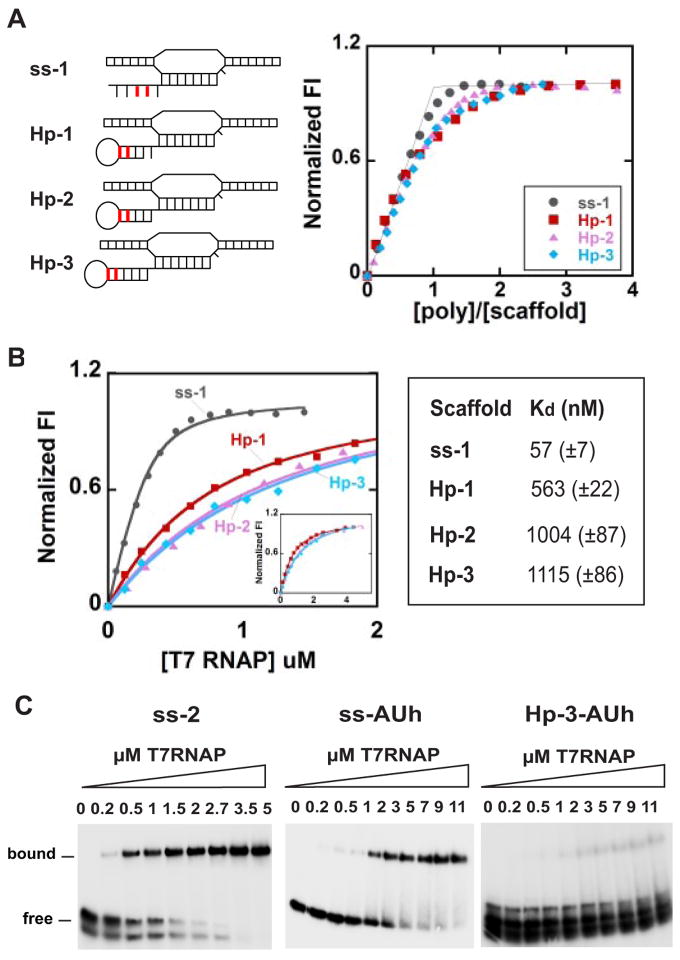
FIGURE 2. Fluorescence and gel-shift titrations with T7 RNAP of nucleic acid scaffolds containing GC-rich and rU-dA hybrids.
A, fluorescence titration curves showing the stoichiometry of binding of T7 RNAP to scaffolds containing GC-rich hybrids (the scaffold concentration was 3 μM). The 2-AP base(s) within the RNA strands of the scaffolds used is shown in red. B, fluorescence titration of the same scaffolds at 300 nM and the apparent binding affinities of T7 RNAP to these scaffolds. The inset shows the full titrations for the hairpin scaffolds. C, gel-shift assays showing binding of the ss-2, ssAUh, and Hp-3-AUh scaffolds with RNAP. Numbers at the top of the gels represent the concentration of T7 RNAP (in μM) used in the experiments shown in each lane. Scaffold concentrations were 3 μM throughout.
CD Measurements
CD spectra were measured at 300–400 nm using a Jasco model J-720 CD spectrophotometer equipped with a temperature-controlled cell holder. The CD spectra shown represent the average of 8–10 measurements. Control spectra measured with identical concentrations of unlabeled oligonucleotides (and RNAP, NTPs, etc.) were subtracted from the probe-containing low energy spectra of the polymerase-scaffold complexes to correct for any non-probe-related optical effects. CD spectra were reported as extinction coefficient differences (εL–εR) per mole of 2-AP (units of M−1 cm−1).
Gel Shift Assays
These assays were performed under the same conditions as the fluorescence titrations. For detection, the RNA components of the nucleic acid scaffolds were labeled at the 5′-terminus with [γ-32P]ATP. Scaffolds at 3 μM concentrations were titrated with RNAP, and the reactions were incubated for 30 min at each concentration. The complexes were then loaded onto 4–20% Tris-boric acid EDTA (TBE) polyacrylamide Redigels (from Bio-Rad) that had been pre-equilibrated with 1× TBE running buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA (pH 8.3) containing 8 mM MgCl2. Gels were run at 4 °C at 100 V and visualized by exposure to a Phosphor-Imager screen with a Storm scanner. To measure binding to RNAP, the bands representing the free scaffold remaining in each lane as a function of increasing RNAP concentration were quantitated using ImageQuaNT software (GE Healthcare).
Transcript Elongation and Termination Assays
Transcription assays were performed in parallel and under the same conditions as the spectral measurements using scaffold constructs in which the RNA had also been 5′-end-labeled with [γ-32P]ATP. Equimolar concentrations of scaffold and RNAP were mixed at total volumes of 10 μl, and transcription was initiated by adding NTP substrate(s). The RNAP was preincubated with the nucleic acid scaffold for 30 min at 25 °C for all transcription reactions followed by 30 min of elongation with the NTPs indicated. RNA products were analyzed on a 12% sequencing gel in 7 M urea and were visualized by exposure to a Phosphor-Imager screen. Unlabeled and 2-AP-containing scaffolds behaved similarly in all transcription assays.
RNase H Digestion of Free Scaffold Constructs and Scaffold-RNAP Complexes
In these experiments the nucleic acid scaffolds (containing 5′-end-labeled RNA as above) and the scaffold-RNAP complexes (equimolar concentrations) were digested with RNase H in the same buffer used for the transcription and spectral measurements. Assays were performed at 3 μM (same as the spectral measurements) and 300 nM scaffold and scaffold-RNAP complex concentrations. RNase H activities of 1 and 0.1 units/μl, respectively, were used with the 3 μM and 300 nM scaffold-RNAP complexes, and digestions were run for 30 min at 37 °C.
RESULTS
Scaffold Sequences, Structures, and Nomenclature
The nucleic acid scaffolds used in this study are shown in Fig. 1. The sequences of the template and non-template DNA strands used were identical for scaffolds containing GC-rich hybrids, whereas the upstream portions of the RNA were varied to include palindromic sequences that can form RNA stem-loop hairpins at the 5′-ends. We note that the downstream ends of the stems of all the constructs containing RNA hairpins (except Hp-1) are located directly adjacent to the first (upstream) bps of the RNA-DNA hybrid, as is characteristic of all “natural” intrinsic terminator hairpins that have been examined (22). The construction of nucleic acid scaffolds with hybrids consisting of rU-dA bps within the non-complementary transcription bubble required alteration of some DNA sequences (Fig. 1B).
Proof of Formation of the RNA-primed DNA Bubble Constructs
To confirm the proper annealing of the RNA transcript to the non-complementary DNA framework, RNA-primed scaffold constructs containing GC-rich hybrids were digested with RNase H, which cleaves only RNA involved in RNA-DNA duplexes (see lanes 2, supplemental Fig. S1, A and B). This showed that the RNA transcripts were annealed correctly in each case, that binding of the RNAP to the constructs protects the RNA of the hybrid from digestion to variable extents for the different scaffolds (lanes 4, supplemental Figs. S1, A and B), and that the hybrids remain partially accessible to RNase H even when bound to RNAP.
The length of the hairpin stems used in the constructs ranged from 4–6 bps, and the loops contained 4–5 nt (Fig. 1). The melting curves of the individual RNA oligomers monitored by UV absorbance at 260 nm show that the designed hairpins do form (at least in isolation) and have the expected stabilities (supplemental Table S1). In addition, control melts of these hairpins performed at two different RNA concentrations (0.3 and 3 μM, data not shown) revealed no concentration dependence of the melting profile, indicating that over this concentration range all RNA hairpins formed intramolecularly. The scaffold constructs of Fig. 1 that do not form hairpins are referred to as single-stranded (ss) and, as previously shown (15), form fully functional elongation complexes when mixed with equimolar concentrations of T7 RNAP. The scaffold constructs labeled with pairs of 2-AP bases behaved identically in all binding and transcription assays to those containing adenine bases in the same positions.
Scaffolds Containing RNA Hairpins and GC-rich Hybrids Form Stable Complexes with T7 RNA Polymerase
Having shown that our synthetic nucleic acid scaffolds have the structures and properties intended, we next examined the changes in these properties that occurred when T7 RNAP was added to form synthetic transcription complexes that contain the nucleic acid elements (termination hairpins and/or rU-dA hybrids) of intrinsic terminators. Fig. 2 shows titration curves obtained by adding increasing concentrations of RNAP to a set of nucleic acid scaffolds containing ss or hairpin-containing RNAs (ss-1, ss-2, Hp-1, Hp-2, and Hp-3) with GC-rich RNA-DNA hybrids to permit us to study first the effects of these hairpins in the absence of an unstable hybrid.
The binding of RNAP to the scaffolds was monitored using fluorescence and gel-shift assays. Hairpin scaffolds with 2-APs located in the upstream ends of the RNA stems showed maximal fluorescence enhancement (38–78%) on binding to RNAP and were used as binding targets in these titrations (Fig. 2A). The binding of RNAP to the ss-1 scaffold was tight and stoichiometric, with a clear breakpoint where the protein and nucleic acid framework concentrations were equimolar. Extrapolation of the high and low concentration data for the hairpin-containing constructs also showed 1:1 binding stoichiometries. Fluorescence titrations of the same scaffolds with RNAP were also performed at a lower scaffold concentration (300 nM) (Fig. 2B), and the apparent binding affinities were determined by fitting these titration curves to single-site binding isotherms (supplemental Equation 1).
The gel-shift assays for scaffolds ss-2 (Fig. 2C) and Hp-1, Hp-2, and Hp-3 (supplemental Fig. S2A) also show that >80% of the scaffolds are bound to RNAP at equimolar concentrations and that the apparent binding affinity of Hp-3 scaffold to RNAP is weaker than that of the ss-2 scaffold construct (ss-1 and ss-2 behave similarly in all our assays). We note that the apparently double gel bands observed for the free scaffolds arise from apparent partial dissociation of RNA from the DNA scaffold construct and are thus likely an electrophoresis artifact. The RNase H digestion experiments show that the nucleic acid scaffolds are essentially completely formed (i.e. no detectable free RNA is present in the reaction), even at scaffold concentrations as low as 300 nM (see supplemental Fig. S1B). We note that the free RNAs migrate as single bands in both denaturing and non-denaturing gels, indicating the absence of alternate conformations of the RNA transcripts.
Scaffolds Containing a rU-dA Hybrid, but No Hairpin, Also Form Relatively Stable Complexes with RNAP
The ssAUh nucleic acid scaffold (Fig. 1B) was used to investigate the role of the rU-dA hybrid component in intrinsic termination. This construct contains a complete rU-dA hybrid sequence but no termination hairpin. This scaffold also bound to RNAP with 1:1 stoichiometry (supplemental Fig. S2C) but with a significantly weaker apparent affinity than that of the ss-2 scaffold containing the GC-rich hybrid (compare the ss-2 gel with the ssAUh gel in Fig. 2C). These results are consistent with other evidence that supports the view that both the hairpin and the rU-dA hybrid elements contribute to the stability decrease of the scaffold-RNAP transcription complex at intrinsic terminators.
Constructs containing Both a Termination Hairpin and a rU-dA Hybrid Form an Unstable Scaffold and an Unstable Complex with RNAP
To examine the binding interactions of a complete intrinsic terminator sequence with RNAP, we combined the rU-dA hybrid of ssAUh and the GC-rich hairpin of Hp-3 to form scaffold construct Hp-3-AUh. Fig. 2C shows that the resulting complex between RNAP and the Hp-3-AUh construct was almost completely unstable at the ~3 μM concentration at which these experiments were performed. In fact, this figure shows that only traces of transcription complex were formed with this “terminator scaffold” at the highest RNAP concentrations tested. We note also that the gel bands for the scaffold alone were significantly broadened and appeared to separate into two components, with the diffuse lower band representing the free RNA and the lighter upper band corresponding to the complete scaffold. This suggests, in accord with earlier free energy calculations (7), that a scaffold containing both intrinsic terminator components is itself quite unstable at these concentrations. A labile dissociation equilibrium of this sort would be expected to broaden the gel bands involved, as seen in Fig. 2C.
Unlike the Hp-1, Hp-2, and Hp-3 nucleic acid scaffolds, neither the free ssAUh nor the free Hp-3-AUh scaffold is susceptible to RNase H digestion. However, as shown, the ssAUh scaffold can form a partially stable complex with RNAP, and this complex can be partially digested by RNase H (supplemental Fig. S2B). This suggests that the rU-dA hybrid in the free scaffold construct is too labile to form an appropriate substrate for a RNase H cleavable complex, whereas weak binding of RNAP to the ssAUh scaffold appears to stabilize the conformation of the rU-dA hybrid component of the construct sufficiently to make it RNase H susceptible. No significant binding of T7 RNAP to RNA alone was observed under our reaction conditions (data not shown). We conclude from these results that the nucleic acid elements of the intrinsic terminator, present either separately or together, do destabilize synthetic transcription complexes as expected and that the decreases in stability induced by the termination hairpin and the “weak” hybrid are approximately additive.
Transcript Elongation on Scaffolds Containing Hairpins and GC-rich RNA-DNA Hybrids
Supplemental Fig. S3 shows that the RNA of RNA-primed scaffolds containing either ssRNA (ss-1) or hairpins (Hp-1, Hp-2, and Hp-3) can be extended in a template fashion by the addition of appropriate NTPs. In the presence of 20 μM ATP a significant fraction of the RNA was elongated by 3 nt on all these scaffolds, as expected from the template sequence, although the observed elongation efficiencies were significantly lower for the hairpin-containing scaffolds. Increasing the concentration of ATP (when present as the sole NTP substrate) beyond 20 μM generated longer RNA chains, probably due to nucleotide misincorporation reactions that could not then be successfully chased (data not shown).
Table 1 shows the percentage of each of the input RNAs that was either not extended or elongated by 3 nt in the presence of 20 μM ATP. We found that the efficiency of transcript elongation on the various hairpin scaffolds decreased in the following order ss-1 > Hp-1 > Hp-2 > Hp-3. The RNA products located at and around position +3 in lanes 3 of supplemental Fig. S3 could be chased (lanes 4), indicating that these transcripts were present largely as stalled complexes rather than as dissociated RNA. These results show that the RNA of all the hairpin-containing scaffolds could be elongated (although with varying efficiency) when paired with a GC-rich hybrid and that the elongation observed was indeed largely template-directed. Lowering the scaffold and RNAP concentrations to 300 nM (conditions where the RNA is still fully annealed to the DNA framework; lanes 2 of supplemental Fig. S1B) resulted in significant decreases in the efficiency of transcription of the hairpin-containing scaffolds (especially Hp-3; Table 1). This result is consistent with reduced RNAP binding affinities to scaffolds that contain hairpins.
TABLE 1. Elongation efficiencies on the single-stranded RNA and hairpin RNA scaffold constructs at 3 μM and 300 nM scaffold-RNAP concentrations.
S.E. of all measurements were ±5%. The ATP concentration was 20 μM.
| Scaffold | Percent (%) extended to position +3 with ATP
|
Percent (%) not extended in the presence of ATP
|
||
|---|---|---|---|---|
| 3 μM | 300 nM | 3 μM | 300 nM | |
| ss-1 | 64 | 69 | 7 | 7 |
| Hp-1 | 58 | 42 | 16 | 32 |
| Hp-2 | 48 | 28 | 26 | 54 |
| Hp-3 | 25 | 1.3 | 44 | 84 |
Transcript Elongation on Scaffolds Containing a rU-dA Hybrid without or with a Termination Hairpin
Fig. 3, A and B, show the transcription products obtained with RNAP bound to a ssAUh and a Hp-3-AUh scaffold, respectively, in the presence of the various substrate NTPs. The addition of 20 μM ATP resulted in the extension of all the RNA primers by 1 to 2 nt, although the elongation efficiency was very low for both RNAP-scaffold complexes. We also found that ~41 and ~50%, respectively, of the transcripts for the ssAUh and Hp-AUh scaffold-containing complexes remained unextended after 30 min of incubation. The addition of ATP and GTP substrates to complexes containing the ssAUh scaffold resulted in the formation of RNA products extended by 4–6 nt (significantly longer than expected from the template sequence) as well as in dramatic increases in elongation efficiency.
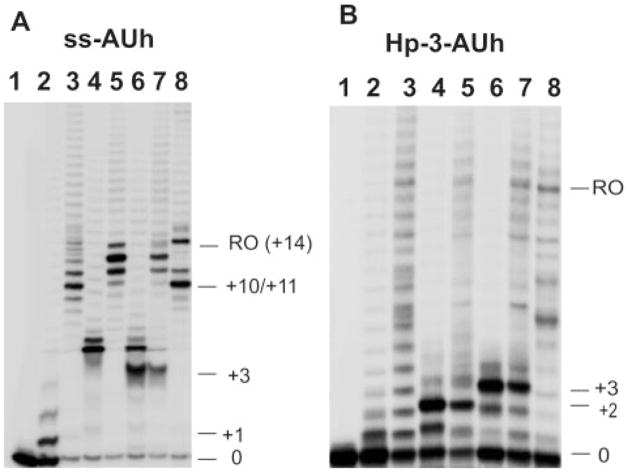
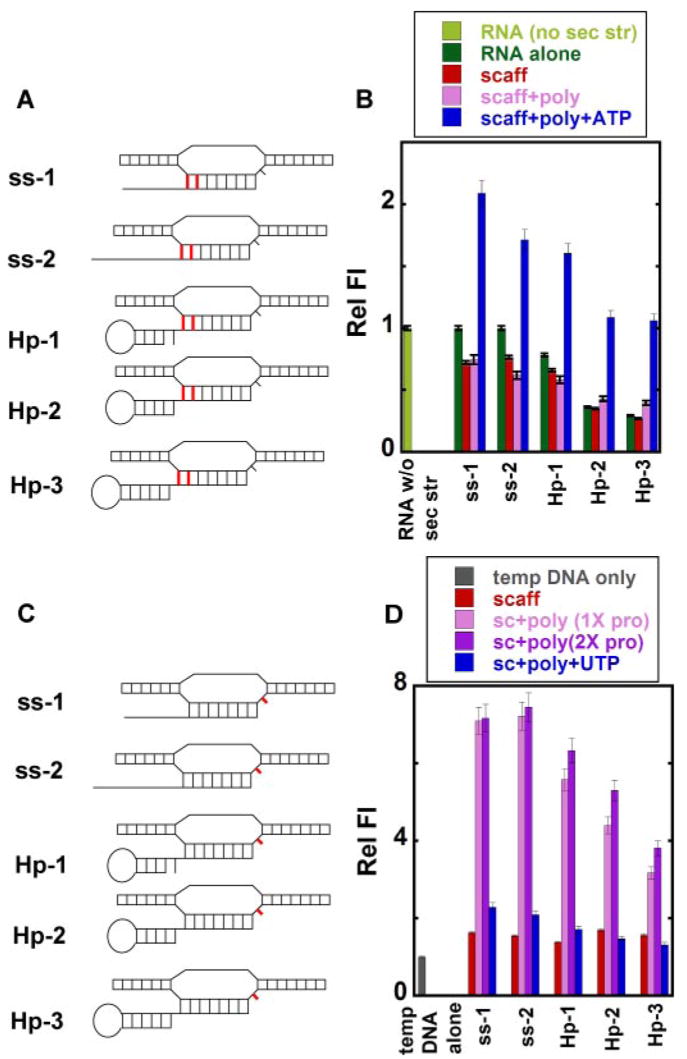
FIGURE 3. Transcription assays with scaffolds containing rU-dA hybrids.
A and B, denaturing gels showing the elongation products formed with different NTPs with ssAUh (A) and Hp-3-AUh (B) scaffolds. Lanes 1, free scaffold; lanes 2, scaffold-RNAP complexes with 20 μM ATP; lanes 3, products in lanes 2 chased with all four NTPs; lanes 4, scaffold-RNAP complexes with 20 μM ATP and GTP; lanes 5, products in lanes 4 chased with all four NTPs; lanes 6, scaffold-RNAP complex with 20 μM ATP, GTP, and CTP; lanes 7, products in lanes 6 chased with all NTPs; lanes 8, scaffold-RNAP complexes with 1 mM concentrations of all four NTPs. The numbers on the right for all gels represent the number of nucleotide residues added to the initial RNA transcript by RNAP. RO indicates the run-off products.
In contrast to the above anomalous transcription behavior seen with the ssAUh-RNAP complex in the presence of ATP and GTP substrates (such transcription has been termed “slippage” synthesis and has been seen before (23)), the Hp-3-AUh-RNAP complex (Fig. 3B) produced only shorter and fully template-directed transcripts. Furthermore, the elongation efficiency for this scaffold-RNAP complex, which contained both nucleic acid elements of the intrinsic terminator, was significantly reduced. Also, as demonstrated by our inability to chase all the elongated products (Fig. 3B, lanes 5 and 7), a large fraction of the elongation products observed in the gel had dissociated from the RNAP. This further demonstrates that the presence of the rU-dA hybrid, in combination with the stem-loop structure, destabilizes the stalled Hp-3-AUh scaffold-RNAP complex significantly relative to the Hp-3-scaffold that contains the Hp-3 hairpin in combination with a GC-rich hybrid. The addition of the next three correct NTPs (ATP, GTP, and CTP) resulted in the extension of the RNA transcripts on both scaffolds by 3 nt, as expected from the template sequence but again with significantly greater release of the RNA products from the Hp-3-AUh scaffold-containing complex, as expected for intrinsic terminators with some read-through.
As indicated above, the fact that transcription with ATP and GTP on complexes containing the ssAUh scaffold resulted in the formation of longer RNA products that did not match the expectations of processive template-directed synthesis suggests that these products result from a slippage synthesis that is only partially template-directed. A detailed examination of transcription products obtained using the ssAUh-RNAP complex with different NTP substrates (added one at a time) shows that these slippage events show a strong preference for the incorporation of GMP residues into the RNA product. By using GTP as the sole elongation substrate we have shown that T7 RNAP can efficiently incorporate 3–6 consecutive GMPs at the 3′-end of the RNA primer of the ssAUh scaffold, although GTP is not the next required nucleotide in terms of template sequence. Digestion with RNase T1 of the longer products obtained on extension with the ATP and GTP substrates also demonstrated that the additional GMPs were incorporated at the 3′-end of the RNA primer (data not shown). We note that this reiterative GMP incorporation into the product RNA is only seen with scaffolds containing the rU-dA hybrid in the absence of a termination hairpin and can be prevented by inserting a termination hairpin into the scaffold construct. This slippage synthesis is also not seen with transcription complexes containing either ss-1 or Hp-3-AUh scaffolds, which show only “normal” patterns of RNA synthesis for transcripts elongated beyond the terminator. The implications of these findings for termination mechanisms are considered further in the “Discussion.”
Interaction of RNAP with Scaffolds Containing Termination Hairpins
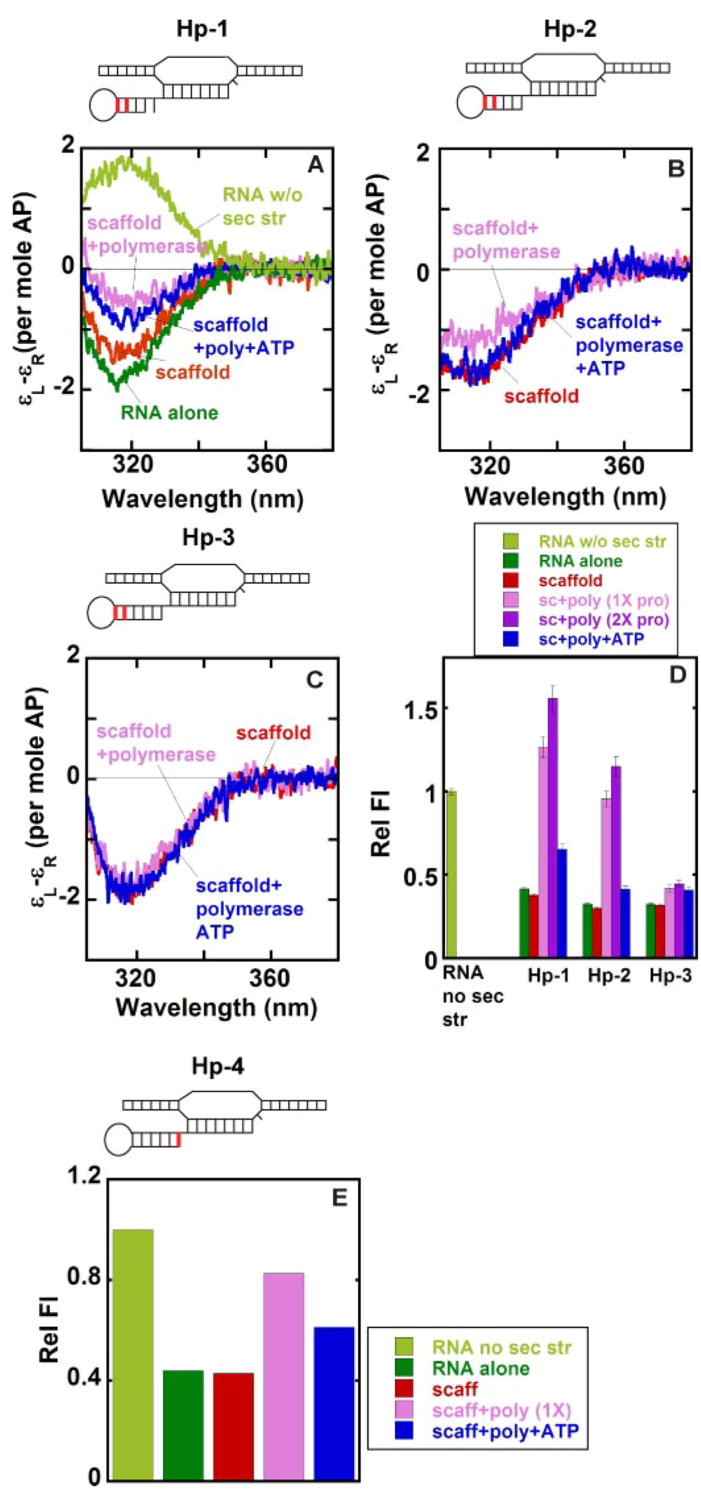
Figs. 4, A–D, show the low-energy CD spectra and the fluorescence intensities at 370 nm measured with scaffold constructs in which the 2-AP dimer probe was placed at the upstream end of the stem (these are the same constructs that were used to measure RNAP binding stoichiometry in Fig. 2A). As expected, the CD spectrum of the Hp-1 RNA hairpin alone showed a trough at 320 nm, demonstrating that the RNA surrounding the 2-AP probes is in the double-stranded A-form conformation. This contrasts with spectra obtained with ssRNAs lacking defined secondary structure in which a peak (rather than a trough) is seen at 320 nm (Fig. 4A; see also Refs. 15 and 20). The other RNA hairpins used in this study also showed troughs centered at 320 nm (data not shown), as did the spectra for the complete hairpin-containing scaffolds (Hp-1, Hp-2, and Hp-3). This confirms that the 2-AP probes located at these stem position were fully base-paired before RNAP addition.
FIGURE 4. Local base conformations within the stems of various free and RNAP-complexed hairpin-containing scaffolds.
The 2-AP base(s) in defined positions within the RNA strand of the scaffolds is shown in red. A–C, low energy CD spectra. D and E, relative fluorescence intensities. Color coding for both the CD spectra and the fluorescence intensities: light green, control RNA without any secondary structure; dark green, hairpin-containing RNA alone; red, RNA-DNA scaffold; pink, scaffold-polymerase complex at equimo-lar concentrations; purple, scaffold-polymerase complex with twice the concentration of RNAP; blue, scaffold-RNAP complex at equimolar concentrations and RNA extended by the addition of ATP.
A significant decrease in the depth of the trough for Hp-1 was observed upon the addition of RNAP to form the scaffold-polymerase complex, as expected if these 2-AP bases were, at least in part, unstacked (and un-base-paired) as a consequence of RNAP binding (15). A small decrease in trough depth was also seen for the Hp-2 scaffold upon the addition of polymerase, whereas for the Hp-3 scaffold the intensity at the trough remains unchanged on RNAP addition (Fig. 4, B and C). Extension of the RNA transcript with ATP results in an increase in the depth of the spectral trough at 320 nm for the Hp-1 and Hp-2 scaffold-RNAP complexes, whereas the trough seen with the Hp-3 scaffold-containing complex showed no change on elongation of the RNA.
These results suggest that the shorter RNA hairpins were partially unstacked as a consequence of RNAP binding, consistent with expectations for these positions at the upstream end of the hairpin stem if the hairpins were indeed opened on polymerase binding to permit the 5′-ends of the RNA primers to be threaded into the RNA exit tunnel. RNA chain elongation would then cause these conformational probes to move beyond the RNA exit tunnel, allowing reformation of the RNA hairpin to begin as the RNAP moves downstream, and RNA residues required to form the downstream side of the termination hairpin emerge from the tunnel. The present results suggest that bases at the upstream end of the putative hairpin begin to restack (and re-base pair) at positions located four or more RNA positions beyond the downstream end of the RNA-DNA hybrid, presumably as a consequence of the nucleation of the RNA stem that occurs on transcript elongation for the shorter stems and without elongation for the longer stem of the Hp-3 scaffold. The hairpin-containing scaffold complexes that had been extended with ATP were not structurally homogeneous (supplemental Fig. S3). However, the CD changes seen on incubation with the ATP substrate do confirm that the complexes we observed spectrally were all transcriptionally active.
As expected, the fluorescence intensities of the 2-AP bases of the RNA component, whether studied alone or within scaffolds carrying stable RNA hairpins at the upstream end of the stem, were significantly quenched, presumably reflecting the base-pairing of these probes within the stem (Fig. 4D). The Hp-1 scaffold exhibited a dramatic increase in fluorescence intensity on RNAP addition, to a level greater than that characteristic of 2-AP probes located within single-stranded RNA sequences (Fig. 4D). Such hyper-fluorescent complexes have been observed previously for fully functional elongation complexes with 2-AP bases placed at similar positions upstream of the RNA-DNA hybrid and have been interpreted as reflecting unstacking (beyond the level seen in free ssRNA) of the 2-AP bases located within the RNA exit tunnel as well as possible non-base-specific ssRNA binding near and within tunnel (15). A slightly smaller increase in fluorescence intensity upon complex formation with RNAP was seen for the Hp-2 scaffold, whereas complex formation with the Hp-3 scaffold (where the 2-AP probes should be entirely out of the tunnel) showed only a very small fluorescence increase. As expected, the addition of ATP substrates and chain extension decreased the fluorescence intensity for Hp-1 and Hp-2 scaffold complexes but resulted in no further change for the Hp-3 scaffold complex. These results suggest that nucleation of a stable stem can begin only after 1 or 2 nt of the second “side” of the stem have emerged from the exit tunnel.
We also examined the downstream end of the RNA stem. For this purpose a single 2-AP residue was added to the 5′-end of the RNA transcript to form the downstream end of a 6-base pair stem (Fig. 4E). The transcription efficiency of this Hp-4 scaffold-polymerase complex was similar to that of the Hp-3 scaffold-RNAP complex (data not shown). As expected, the fluorescence intensity of the RNA alone (which forms a stable hairpin) as well as that of the free scaffold were significantly quenched relative to the fluorescence intensity of a similar RNA that does not form secondary structure. The addition of RNAP resulted in significant unstacking of the single 2-AP residue, suggesting that the downstream end of the stem is unwound in forming the Hp-4-scaffold-RNAP complex. This is expected if the potential rU base-pairing partner of the 2-AP base is located within the RNA tunnel and, thus, not available for complementary base pair formation. As also expected, elongation of the RNA with ATP allows the reformation of the stem, as shown by a decrease in fluorescence.
Probing Local DNA Conformation at the Upstream End of the DNA Duplex
To examine the effect of RNAP binding on the transcription bubble, we incorporated a 2-AP dimer probe into the template DNA at the base-paired upstream end of the bubble in the various scaffold constructs (Fig. 5A). Although the 2-AP bases were paired with the complementary nt of the non-template DNA at these positions, the fluorescence intensity of the DNA bubble did not change relative to that of the single-stranded template alone, whereas the fluorescence intensities of the ss-1, ss-2, and Hp-1 scaffolds were only slightly higher than that of the single-stranded template DNA alone (Fig. 5B). This may reflect some fraying or “breathing” of the dsDNA at this position immediately adjacent to the non-complementary bubble sequence within the scaffold, giving these bases a more “open” character (in terms of fluorescence intensities) than their positioning in the sequence might otherwise lead one to expect. Such breathing has been demonstrated directly in our laboratory3 using 2-AP probes.
FIGURE 5. Local base conformations at the upstream and downstream ends of the noncomplementary DNA bubble of various free and RNAP-complexed nucleic acid scaffolds.
A and E, scaffolds showing the positions of the 2-AP bases (in red) in the template and non-template strands. B and F, relative fluorescence intensities. Panels C and D and panels G and H, low energy CD spectra for ss-1 and Hp-3 scaffolds, respectively. Color coding for both the CD spectra and the fluorescence intensities: dark green, single-stranded template DNA with the 2-AP bases in the template strand (A); black, DNA bubble alone with the 2-AP bases in the template strand; light green, single-stranded non-template DNA with the 2-AP bases in the non-template strand (E); red, RNA-DNA scaffold; pink, scaffold-polymerase complex at equimolar concentrations; purple, scaffold-polymerase complex with twice the concentration of RNAP; blue, scaffold-RNAP complex at equimolar concentrations and RNA extended by the addition of ATP.
We note (Fig. 5B) that the fluorescence intensities of the 2-APs within the scaffolds for the Hp-2 and Hp-3 constructs were significantly higher than those measured with the ssDNA template strand in isolation. However, the amplitudes of the low energy CD peak at 325 nm for all these scaffold constructs as well as for the DNA bubble in the absence of RNA were similar and higher than for the ssDNA template, indicating that the probe bases assumed a similar and more stacked conformation in all these scaffold constructs than in single-stranded sequences in isolation (data for the ss-1 and Hp-3 scaffolds are in Fig. 5, C and D; for the other scaffolds see supplemental Fig. S4). This suggests that the hyper-fluorescent state observed for 2-AP probes within the Hp-2 and Hp-3 scaffolds alone does not involve significant unstacking.
A small increase in fluorescence and a significant decrease in the exciton peak for the ss-1, ss-2, and Hp-1 scaffolds were seen on RNAP addition. This suggests that the 2 bp at the upstream edge of the transcription bubble of the construct were opened as a consequence of elongation complex formation. In contrast, the binding of RNAP to the Hp-2 and Hp-3 scaffolds resulted in a decrease in fluorescence intensity and a slight decrease in the height of the exciton peak, suggesting that the binding of RNAP to these latter scaffolds did not significantly unwind the upstream end of the bubble. This result is consistent with the notion that the presence of the termination hairpins of scaffolds Hp-2 and Hp-3, which are located immediately adjacent to the upstream end of the RNA-DNA hybrid, prevent any upstream translocation (or expansion) of the RNAP along the template on binding, whereas such translocation can and does occur when the termination hairpin is either absent (scaffold constructs ss-1 or ss-2) or not directly contiguous to the hybrid (Hp-1). This point is discussed further below. Control experiments demonstrated that template bases located further upstream from the non-complementary transcription bubble did maintain stacked and base-paired interactions for all scaffold-RNAP complexes examined (data not shown).
An increase in fluorescence for all the scaffold-polymerase complexes was observed on extension of the RNA primer with ATP. The ss-1, ss-2, and Hp-1 scaffolds showed the largest change. The low energy CD spectra of all the scaffolds remained almost unchanged relative to the corresponding scaffold-RNAP complexes (data not shown). This observed increase in fluorescence probably reflects the inability (due to the non-complementary bubble) of the bases just downstream from the 2-AP probes to rewind as the polymerase translocates forward, thus creating a strain on the 2-AP bases at the upstream end of the bubble.
Interactions of RNAP with DNA Bases Downstream of the RNAP Active Site
We examined the fluorescence change of constructs in which pairs of 2-AP bases were incorporated in the non-template strand at the downstream edge of the unpaired transcription bubble (Fig. 5E). Figs. 5, F–H, show the changes in the fluorescence and CD spectra observed on binding the RNAP to the scaffold constructs. As expected (18, 19), the low energy CD band for the single-stranded non-template DNA exhibits a peak at 320 nm for all the scaffolds examined. The free scaffolds also showed the characteristic exciton peak at 325 nm that results from pairing of the 2-AP bases with the template strand (data shown for the ss-1 and Hp-3 scaffolds in Fig. 5, G and H; for scaffolds Hp-1 and Hp-2, see supplemental Fig. S5). As expected, the fluorescence intensity of the scaffold was significantly quenched as a consequence of stacking with the complementary bases of the template (Fig. 5F).
Upon binding to the RNAP, the CD band at 325 nm increased slightly in intensity for all the scaffolds examined, indicating that this complex formation resulted in further stacking of the downstream DNA bases (Figs. 5, G and H, and S-5). The fluorescence intensities of the polymerase-scaffold complexes did not change relative to those of the free scaffold. This clearly showed that the polymerase in the hairpin-containing scaffolds does not translocate forward in the absence of synthesis. With extension of the transcript with ATP, the fluorescence intensity increased dramatically for all the scaffold complexes as the RNAP moved forward along the template and unstacked the 2-AP probes located in the non-template DNA at the downstream edge of the transcription bubble. The low energy CD spectrum also exhibited a decrease in the 325 nm band accompanied by a blue shift, as expected (15, 19) for the formation of a single-stranded unstacked DNA conformation.
Conformation of Bases Located at the 5′-End of the RNA-DNA Hybrid
Scaffold constructs were assembled with the 2-AP dimer in the RNA at the upstream end of the hybrid (Fig. 6A). The fluorescence intensities of the hairpin RNAs alone were quenched to variable extents relative to the fluorescence of ssRNA oligomers without secondary structure even though all the 2-AP probes were in unpaired regions of the hairpin RNAs. The quenching effect was greatest for the RNAs containing the more stable hairpins (Hp-2 and Hp-3; Fig. 6B). The low energy exciton peak for Hp-2 and Hp-3 RNA hairpins alone also showed a slight red shift when compared with RNA lacking any secondary structure (supplemental Fig. S6). Thus, both the fluorescence and CD measurements suggest that the presence of a stem-loop structure at the upstream end of the RNA affects the flexibility of downstream RNA residues, resulting in a more stacked conformation of the 2-AP bases in the free RNA of the hairpin constructs. A significant decrease in the low energy CD peak accompanied by the appearance of a trough at ~315 nm (supplemental Fig. S6) was observed on annealing these hairpin-containing RNA oligomers to the DNA bubble to form the complete scaffolds. The fluorescence intensities for the ss-1, ss-2, and Hp-1 scaffolds decreased slightly relative to those of the free RNAs upon formation of the hybrid, whereas no significant change in fluorescent intensity was observed on forming the Hp-2 and Hp-3 scaffolds (Fig. 6B).
FIGURE 6. Local base conformations at the upstream end of the RNA-DNA hybrid and at the active site in free and RNAP-complexed nucleic acid scaffolds.
A and C, scaffolds showing the positions of 2-AP base(s) (in red) in the RNA and in the template DNA. B and D, relative fluorescence intensities. Light green, control RNA without secondary structure (sec str); dark green, RNA alone with the 2-AP bases in the RNA strand (A); gray, single-stranded template DNA with the 2-AP base in the template DNA (C); red, RNA-DNA scaffold; pink, scaffold-polymerase complex at equimolar concentrations; purple, scaffold-polymerase complex with twice the concentration of RNAP; blue, scaffold-RNAP complex at equimolar concentrations and RNA extended by the addition of ATP (B) or UTP (D).
The addition of RNAP increased the depth of the low energy trough at 315 nm for the ss-1, ss-2, and Hp-1 scaffolds and resulted also in the disappearance of the 320-nm peak. These changes are characteristic of the further stabilization of the RNA-DNA hybrid within the polymerase-scaffold complex (supplemental Fig. S6). However, the addition of RNAP to the Hp-2 scaffold did not result in any significant change in the CD spectrum, suggesting that the binding of RNAP did not induce any further change in the conformation of the hybrid in this scaffold. The CD spectrum of the Hp-3 scaffold-RNAP complex revealed a negative low energy CD band that also extended into the high energy part of the spectrum; we do not fully understand the origin of this signal, but examination of the low wavelength region of the CD spectra suggests some additional interaction of (possibly aromatic) amino acid residues of the polymerase with the 2-APs of the RNA at these positions (data not shown). Mixed interactions of this type are currently being investigated in other protein-nucleic acid systems.4
In contrast to the ss-1, ss-2, and Hp-1-scaffold-RNAP complexes, which showed no change or a slight decrease in fluorescence intensities on RNAP binding, the Hp-2 and Hp-3 scaffolds exhibited increases in fluorescence intensity when RNAP was added (Fig. 6B), suggesting that some disruption of the base stacking at the upstream end of the hybrids occurred in the Hp-2- and Hp-3-scaffold-RNAP complexes. For all the scaffolds the addition of ATP and the consequent RNA extension resulted in further unstacking (as expected) of the 2-AP probes as the RNAP (and the transcription bubble) translocated downstream, leading (Fig. 6B) to an increase in fluorescence intensity and a decrease in the depth of the low energy CD trough (supplemental Fig. S6).
Interactions at the Polymerase Active Site
In the absence of NTPs, the coding template base of the T7 RNAP-containing elongation complex is “flipped out” of the active site (5, 24). This flipped-out conformation results in a dramatic increase in fluorescence intensity when the coding base is replaced with 2-AP (15). We have used this signal change to monitor the conformation of the template base at the active site in the hairpin-containing transcription complexes (Fig. 6C).
Fig. 6D plots the changes in fluorescence intensity observed for a single 2-AP probe inserted at the coding base (n) position of the different scaffolds used in this study. All the scaffolds in which the 2-AP probe was placed opposite a non-complementary template base exhibited slightly higher fluorescence intensities than seen with this probe in a comparable position within an isolated ssDNA oligomer (25). In the scaffold-RNAP complexes, both the ss-1 and the ss-2 scaffolds (lacking any secondary structure in the upstream RNA transcript) exhibited a dramatic increase in fluorescence that was significantly higher than that seen for the single-stranded constructs. On the other hand this increase (relative to the ss-1 and ss-2 scaffolds) in the fluorescence of this 2-AP probe on RNAP addition was relatively smaller for scaffolds containing RNA hairpins. In addition, we also observed a gradual decrease in relative fluorescence intensities as stem lengths were increased (Fig. 6D).
This strongly suggests that the presence of the stem-loop structures significantly interferes with the positioning of the coding template base at the active site in a conformation appropriate for efficient incorporation of the incoming NTP (see also “Discussion”). The addition of UTP (complementary to the coding base 2-AP) to the RNAP-containing complexes resulted in an overall decrease of fluorescence for all the scaffold constructs, presumably due to the restacking of the coding base as the RNAP incorporated the next required NTP.
DISCUSSION
We have used site-specifically placed 2-AP residues (the fluorescent analogue of adenine) to investigate the local conformational changes of RNA and DNA bases at critical positions within the elongation complex when it encounters characteristic nucleic acid elements of intrinsic terminators and pause sites. These elements include stem-loop hairpins within the nascent RNA and weak (rU-dA) RNA-DNA hybrids. A study of such synthetic termination complexes per se is somewhat difficult because the nucleic acid terminator elements destabilize the elongation complex, leading to complex dissociation and transcript release. We have in part been able to bypass these difficulties because the termination hairpin and the weak hybrid work together to bring about intrinsic termination, and although these elements do destabilize and otherwise perturb the elongation complex when present together, they do so less for complexes containing only one or the other. Therefore, we have been able to build reasonably stable nucleic acid scaffold constructs that contain these elements separately, and this has permitted us to examine the role played by each element in isolation before attempting to deal with the more unstable complexes in which both elements are present together.
Structural Effects of a Termination (or Pausing) Hairpin on the Nucleic Acid Framework
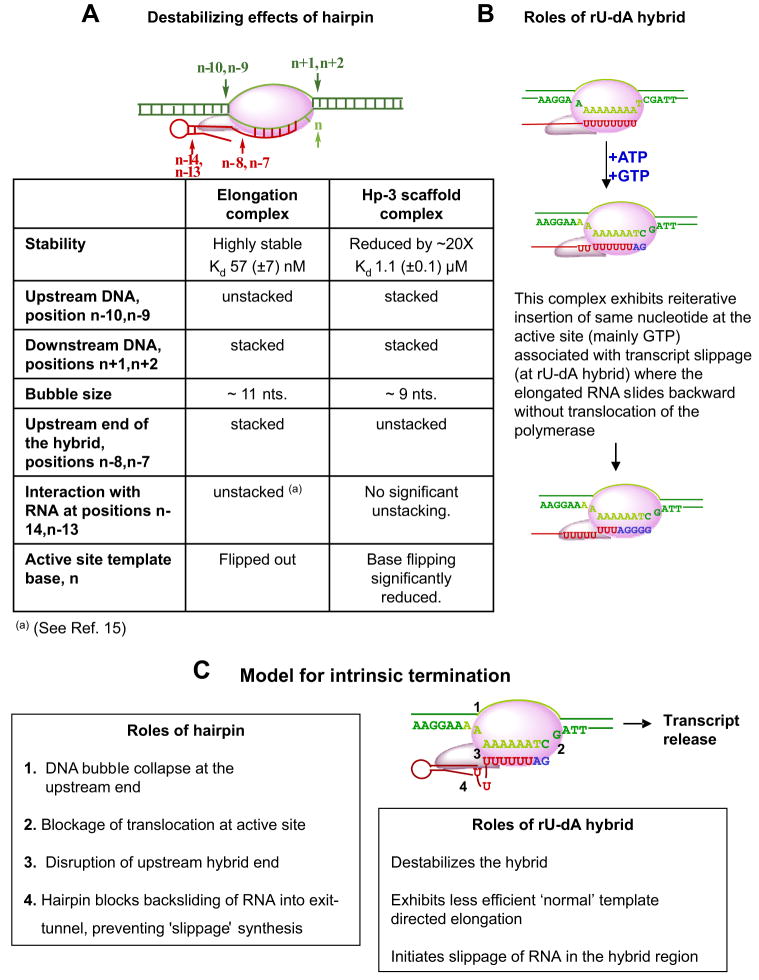
The consequences of the placement of a hairpin containing a 5 (Hp-2)- or 6 (Hp-3)-bp GC-rich stem and a 5-nt loop within the nucleic acid scaffold just downstream of a stable (GC-rich) RNA-DNA hybrid are summarized in Fig. 7A. The following observations have been made. (i) A pair of 2-AP bases placed in the first two DNA base pairs located directly upstream of the non-complementary transcription bubble (Fig. 5, A–D) show that these base pairs open on forming normal elongation complexes and transform the bubble from 9 nt in length on each strand (the size of the original non-complementary bubble) to 11 nt on each strand after RNAP binding. This opening does not occur when the nucleic acid scaffold contains a hairpin located directly upstream of the last bp of the RNA-DNA hybrid, as in scaffolds Hp-2 and Hp-3 (Fig. 1), suggesting that the presence of such hairpins prevents further (upstream) opening of the DNA transcription bubble on RNAP binding. (ii) In contrast, 2-AP probes placed in the first two DNA base pairs located directly downstream of the non-complementary bubble (Fig. 5, E–H) do not open on RNAP addition in either the presence or the absence of the termination hairpin. This result, which appears contrary to expectations from the “forward-translocation” model for intrinsic termination (9), suggests that the presence of the hairpin (in conjunction with a GC-rich hybrid) does not drive the RNAP forward along the nucleic acid scaffold. Rather, bubble collapse at the upstream end of the DNA framework (due to the presence of the hairpin) may partially destabilize the complex by eliminating some of the favorable interactions involved in maintaining the normal elongation bubble size of 11 nt.
FIGURE 7.
A, summary of the changes induced at various positions within the nucleic acid scaffold of the transcription complex by the presence of the termination hairpin. B, effects of the weak rU-dA hybrid. C, mechanistic model for the intrinsic termination process. For explanations, see boxes and “Discussion.”
It has been proposed in a recent study (26) that the dissociation of T7 RNAP elongation complexes halted by NTP limitation might also proceed via forward translocation. This conclusion is not consistent with our results. We note that although the DNA bubble framework used in our assays does not allow closing of the original non-complementary 9-nt bubble, probing of the upstream end of the DNA bubble shows that the bubble size increases here from 9 to 11 nt for the ss-1 scaffold-polymerase complex (Figs. 5, A–B). This provides two upstream bps that can re-close on elongation complex movement downstream for the ss-1-RNAP complex. Nevertheless we do not observe any unwinding of the downstream end of the DNA framework with the ss-1 (elongation) scaffold (Figs. 5, E–G). These results are consistent with the proposal that bubble collapse at the upstream end of the DNA framework does not drive the RNAP forward. (iii) 2-AP probes placed at the upstream end of the RNA-DNA hybrid (Fig. 6, A and B) are base-paired and stacked in the elongation complex but open with partial unstacking in the presence of the termination hairpin. This confirms that the termination hairpin partially destabilizes the upstream end of the hybrid (27). (iv) 2-AP RNA probes placed at the upstream end of the stem of a preformed RNA termination hairpin open and unstack on RNAP binding to the Hp-1 and Hp-2 scaffolds (Figs. 4, A–D). Additional unstacking (above the ssRNA level) was also observed for ss-1 scaffold-RNAP complexes, in which 2-AP probes interact with the RNAP exit tunnel (15). In contrast, 2-AP probes at these positions in the Hp-3 (6-bp stem) scaffold complex remain primarily base-paired on RNAP binding, suggesting that the first four upstream residues of the stem suffice to get the RNA through the tunnel. Previous model-building studies on the T7 RNAP elongation complex suggested that the exit tunnel accommodates ~5 nt of RNA in an extended conformation (24). (v) In elongation complexes formed with the ss-1 and ss-2 scaffolds, the template base located at the RNAP active site is normally flipped out of the axis of the downstream duplex DNA when the next required NTP is absent. This base-flipping is significantly reduced in the presence of the termination (or pausing) hairpin, suggesting that the interaction of this hairpin with the RNAP inhibits normal transcript elongation, possibly by locking the elongation complex in the pre-translocated state. Consistent with these findings, previous studies with E. coli RNAP also suggest that the 3′-end of the RNA is likely to be in the pre-translocated state in hairpin-induced paused complexes (8, 18).
Other aspects of the effects of a pausing or termination hairpin on the transcription complex emerge from these studies. First, it is apparent that to be fully effective in inducing maximal distortion at the upstream end of the RNA-DNA hybrid, the RNA base involved in the downstream end of the hairpin stem must be immediately adjacent to the first (upstream) base pair of the hybrid (Hp-2 and Hp-3). Such hairpins prevent upstream or downstream translocation of the RNAP and, thus, would likely serve to prevent an elongation complex stopped at a “real” terminator from either “backsliding” or forward translocating from a weak (rU-dA) hybrid. As expected (7, 28), the presence of the isolated hairpin component of the intrinsic terminator has been shown to destabilize elongation complexes at these template positions (Fig. 2B). A complex containing a single nt spacer between the end of the stem and the beginning of the hybrid (Hp-1 scaffold) showed significantly greater elongation efficiency than did complexes (Hp-2 and Hp-3) in which the ends of the duplex hairpin stems were directly adjacent to the last bp of the hybrid, thus also demonstrating the importance of this spacing in controlling the stability of the complex and defining the difference between a pausing and a termination hairpin. This is consistent with previous studies that have also shown that this spacing plays a crucial role in triggering transcript release during intrinsic termination (28, 29) and further validates the use of such preformed hairpin scaffolds for intrinsic termination and pausing studies.
The Effects of the Weak rU-dA Hybrid
The presence of the rU-dA hybrid element of an intrinsic terminator also induced the expected effects (Fig. 7B). Thus, both free scaffold and RNAP-scaffold complexes containing rU-dA hybrids (ssAUh) were shown to be less stable than scaffolds and complexes containing GC-rich hybrids. Also normal (template-directed) transcription through the rU-dA hybrid in the presence of the next required NTP was noticeably less efficient than transcription through other hybrids. In addition we found that transcription into the rU-dA hybrid sequence resulted in anomalous, although still template-directed slippage synthesis of the RNA. We suggest that complexes containing the ssAUh scaffold may be somewhat stabilized by random sliding of the slippage-generated RNA products into the exit-tunnel as long as the complex can maintain register with the active site cytosine base that directs incorporation of GMP residues. In contrast the presence of an effective termination hairpin directly adjacent to a weak rU-dA hybrid (scaffold Hp-3-AUh) not only prevented this slippage synthesis, thus significantly reducing apparent transcription levels (Fig. 3B), but also promoted dissociation of the RNA transcripts. This is likely due to a hairpin-RNAP interaction that inhibits passage of the slippage-generated single-stranded RNA into and through the exit tunnel.
Combined Effects of the Termination Hairpin and the rU-dA Hybrid
As shown in Fig. 2C, the presence of both the hairpin and the weak hybrid elements destabilizes the nucleic acid scaffold (Hp-3-AUh) almost completely. Virtually no trace of a fully formed scaffold can be seen when both of these elements are present. However, this “terminator” scaffold is somewhat stabilized by the binding of RNAP (Fig. 2C), and some read-thorough synthesis of the terminator is also observed (Fig. 3B), demonstrating that these synthetic terminators (like their promoter-initiated analogues) are not fully efficient in transcript release.
Our results argue that the main effect controlling transcript and RNAP release at intrinsic terminators is likely to be the thermodynamic instability of the scaffold at these template positions combined with the multiple effects of the terminator hairpin that (i) further destabilize the upstream end of the rU-dA hybrid, (ii) weaken the interaction of the RNAP with the scaffold by competitively inhibiting ssRNA binding within or near to the RNA exit tunnel, (iii) inhibit binding of the next required NTP at the active site of the polymerase, presumably by interfering with the RNAP translocation (relative to the DNA template) that is involved in the single NTP addition cycle, and (iv) “anchor” the RNAP to the scaffold to prevent the “sliding away” of the elongation complex from the terminator position, thus allowing time for the thermodynamically mandated dissociation and release process to go to completion.
Intrinsic Termination Models and Mechanisms
The thermodynamic model (7) proposed that the standard free energy of formation of the elongation complex from its protein and nucleic acid components can be written ΔG°net = ΔG°bubble + ΔG°hybrid + ΔG°polymerase, where ΔG°bubble represents the (unfavorable) standard free energy of formation of the transcription bubble, ΔG°hybrid represents the (favorable) standard free energy of hybrid formation, and ΔG°polymerase represents the (net favorable) standard free energy of interaction of the nucleic acid framework of the elongation complex with the polymerase. Replacing a GC-rich RNA-DNA hybrid with a hybrid consisting of ~8 rU-dA bps has here been directly shown to decrease the stability of the elongation complex significantly, and the presence of an adjacent upstream termination hairpin reduces the stability of the complex still further, presumably in part by unwinding the upstream bps of the already weak hybrid duplex. These elements undermine the stability of the elongation complex at terminator positions on the template and provide a thermodynamic basis for the dissociation of the transcription complex and the release of the nascent RNA.
In addition to these direct thermodynamic changes, our results are consistent with earlier evidence that suggested that an additional role of the termination hairpin might be to prevent the complex from “slipping back” from the terminator position (29, 30). Thus, the hairpin immobilizes the complex at its thermodynamically weakest position on the template either by binding directly to the RNAP or perhaps by preventing the nascent RNA from binding in and slipping back through the exit channel of the polymerase. This effectively locks the nucleic acid framework of the complex to the polymerase and prevents it from moving “out of register” with the terminator sequence, thus providing time for the intrinsic termination (release) process to go to completion.
Finally, we have shown that the binding of the hairpin to the RNAP appears also to prevent the normal flipping of the template (n) base that occurs when the 3′-end of the nascent RNA is positioned in the product binding sub-site of the RNAP (5). This may occur because the locking of the hairpin to the polymerase prevents the translocation step that moves the 3′-end of the nascent transcript back from the substrate to the product binding sub-site of the RNAP and, thus, makes the template base inaccessible to the incoming NTP. This hairpin-induced locking seems also to prevent the slippage synthesis that occurs when the rU-dA hybrid is present in the absence of the termination hairpin, perhaps providing another reason why this directly vicinal hairpin is required for effective termination.
Possible Insights into Pausing Mechanisms
This view of intrinsic termination may also provide some insight into the mechanisms of Class I and II pausing, since the nucleic acid elements of these pausing sites are, respectively, the hairpin and the weak hybrid of the intrinsic terminator. We suggest that a Class I pausing hairpin located upstream of a GC-rich hybrid may transiently inhibit elongation in the same way as a termination hairpin does at an intrinsic terminator, but here pausing results instead of termination because the hairpin alone cannot destabilize the complex sufficiently to induce dissociation. Otherwise pausing and termination hairpins should function identically, and we have demonstrated this experimentally by placing a Class I pausing hairpin next to the rU-dA hybrid of an intrinsic terminator, resulting in effective intrinsic termination as expected.5 Class II-type pausing occurs as an elongation complex transcribes into a weak hybrid in the absence of a hairpin, as in scaffold ssAUh. As suggested previously by Landick and Artsimovitch (31), a transcription complex positioned at such a hybrid may have an increased tendency to “backslide” along the template to position the complex at a thermodynamically more stable hybrid position, resulting in an apparently decreased elongation rate (i.e. a pause) before further random sliding brings the end of the nascent transcript back to the active site and permits transcription to resume.
Acknowledgments
We are pleased to acknowledge helpful discussions with Dr. Neil Johnson of the CNRS in Toulouse, France, as well as with members of our laboratory. We thank Dr. William McAllister of SUNY-Downstate for the His-tagged T7 RNA polymerase clone and Dr. Walter Baase for much helpful advice on measuring CD spectra.
Footnotes
This research was supported in part by National Institutes of Health Grants GM-15792 and GM-29158 (to P. H. vH.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6, Table S1, and Equation 1.
The abbreviations used are: nt, nucleotide(s); RNAP, RNA polymerase; 2-AP, 2-aminopurine; ss, single-stranded.
D. Jose, unpublished experiments.
K. Datta, unpublished results.
C. Conant, unpublished experiments.
References
- 1.Greive SJ, von Hippel PH. Nat Rev Mol Cell Biol. 2005;6:221–232. doi: 10.1038/nrm1588. [DOI] [PubMed] [Google Scholar]
- 2.Arndt KM, Chamberlin MJ. J Mol Biol. 1990;213:79–108. doi: 10.1016/S0022-2836(05)80123-8. [DOI] [PubMed] [Google Scholar]
- 3.Wilson KS, von Hippel PH. Proc Natl Acad Sci U S A. 1995;92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Platt T. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- 5.Tahirov TH, Temiakov D, Anikin M, Patlan V, McAllister WT, Vassylyev DG, Yokoyama S. Nature. 2002;420:43–50. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- 6.Wilson KS, Conant CR, von Hippel PH. J Mol Biol. 1999;289:1179–1194. doi: 10.1006/jmbi.1999.2814. [DOI] [PubMed] [Google Scholar]
- 7.Yager TD, von Hippel PH. Biochemistry. 1991;30:1097–1118. doi: 10.1021/bi00218a032. [DOI] [PubMed] [Google Scholar]
- 8.Toulokhonov I, Artsimovitch I, Landick R. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- 9.Yarnell WS, Roberts JW. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 10.Ma K, Temiakov D, Anikin M, McAllister WT. Proc Natl Acad Sci U S A. 2005;102:17612–17617. doi: 10.1073/pnas.0508865102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartvig L, Christiansen J. EMBO J. 1996;15:4767–4774. [PMC free article] [PubMed] [Google Scholar]
- 12.Jeng ST, Gardner JF, Gumport RI. J Biol Chem. 1992;267:19306–19312. [PubMed] [Google Scholar]
- 13.Ebright RH. J Mol Biol. 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 14.Liu C, Martin CT. J Mol Biol. 2001;308:465–475. doi: 10.1006/jmbi.2001.4601. [DOI] [PubMed] [Google Scholar]
- 15.Datta K, Johnson NP, von Hippel PH. J Mol Biol. 2006;360:800–813. doi: 10.1016/j.jmb.2006.05.053. [DOI] [PubMed] [Google Scholar]
- 16.Daube SS, von Hippel PH. Science. 1992;258:1320–1324. doi: 10.1126/science.1280856. [DOI] [PubMed] [Google Scholar]
- 17.Temiakov D, Anikin M, McAllister WT. J Biol Chem. 2002;277:47035–47043. doi: 10.1074/jbc.M208923200. [DOI] [PubMed] [Google Scholar]
- 18.Kyzer S, Ha KS, Landick R, Palangat M. J Biol Chem. 2007;282:19020–19028. doi: 10.1074/jbc.M701483200. [DOI] [PubMed] [Google Scholar]
- 19.Johnson NP, Baase WA, Von Hippel PH. Proc Natl Acad Sci U S A. 2004;101:3426–3431. doi: 10.1073/pnas.0400591101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson NP, Baase WA, von Hippel PH. J Biol Chem. 2005;280:32177–32183. doi: 10.1074/jbc.M504619200. [DOI] [PubMed] [Google Scholar]
- 21.He B, Rong M, Lyakhov D, Gartenstein H, Diaz G, Castagna R, McAllister WT, Durbin RK. Protein Expression Purif. 1997;9:142–151. doi: 10.1006/prep.1996.0663. [DOI] [PubMed] [Google Scholar]
- 22.d’Aubenton Carafa Y, Brody E, Thermes C. J Mol Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 23.Macdonald LE, Zhou Y, McAllister WT. J Mol Biol. 1993;232:1030–1047. doi: 10.1006/jmbi.1993.1458. [DOI] [PubMed] [Google Scholar]
- 24.Yin YW, Steitz TA. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 25.Gong P, Esposito EA, Martin CT. J Biol Chem. 2004;279:44277–44285. doi: 10.1074/jbc.M409118200. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Y, Navaroli DM, Enuameh MS, Martin CT. Proc Natl Acad Sci U S A. 2007;104:10352–10357. doi: 10.1073/pnas.0606306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komissarova N, Becker J, Solter S, Kireeva M, Kashlev M. Mol Cell. 2002;10:1151–1162. doi: 10.1016/s1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 28.Chan CL, Wang D, Landick R. J Mol Biol. 1997;268:54–68. doi: 10.1006/jmbi.1997.0935. [DOI] [PubMed] [Google Scholar]
- 29.Feng GH, Lee DN, Wang D, Chan CL, Landick R. J Biol Chem. 1994;269:22282–22294. [PubMed] [Google Scholar]
- 30.Komissarova N, Kashlev M. Proc Natl Acad Sci U S A. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Artsimovitch I, Landick R. Proc Natl Acad Sci U S A. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]