Abstract
Purpose
Identifying individuals with Lynch syndrome (LS) is highly beneficial. However, it is unclear whether microsatellite instability (MSI) or immunohistochemistry (IHC) should be used as the screening test and whether screening should target all patients with colorectal cancer (CRC) or those in high-risk subgroups.
Patients and Methods
MSI testing and IHC for the four mismatch repair proteins was performed on 500 tumors from unselected patients with CRC. If either MSI or IHC was abnormal, complete mutation analysis for the mismatch repair genes was performed.
Results
Among the 500 patients, 18 patients (3.6%) had LS. All 18 patients detected with LS (100%) had MSI-high tumors; 17 (94%) of 18 patients with LS were correctly predicted by IHC. Of the 18 probands, only eight patients (44%) were diagnosed at age younger than 50 years, and only 13 patients (72%) met the revised Bethesda guidelines. When these results were added to data on 1,066 previously studied patients, the entire study cohort (N = 1,566) showed an overall prevalence of 44 of 1,566 patients (2.8%; 95% CI, 2.1% to 3.8%) for LS. For each proband, on average, three additional family members carried MMR mutations.
Conclusion
One of every 35 patients with CRC has LS, and each has at least three relatives with LS; all of whom can benefit from increased cancer surveillance. For screening, IHC is almost equally sensitive as MSI, but IHC is more readily available and helps to direct gene testing. Limiting tumor analysis to patients who fulfill Bethesda criteria would fail to identify 28% (or one in four) cases of LS.
INTRODUCTION
Colorectal cancer (CRC) is the third most common type of cancer in the United States, with 145,000 individuals diagnosed each year.1 Lynch syndrome (LS) is the most common hereditary form of colon cancer. Individuals with LS have increased risks for a number of other cancers as well, including endometrial, gastric, ovarian, ureter and renal pelvis, small bowel, bile duct, brain, and certain skin cancers.2 Determining the prevalence of LS among all patients with CRC is an important public health issue. Patients with CRC and LS have an increased risk for second primary cancers, which could be prevented or detected early if the condition is recognized. Maybe more importantly, many of the relatives of those found to have LS will also have inherited LS and high risks for cancer. If these at-risk relatives can be identified early in life, they have the opportunity to prevent CRC through early and more frequent colonoscopy and to detect or prevent the other LS-related cancers through intensive surveillance or risk-reducing surgeries.3-5 At-risk relatives found not to have the proband's mutation do not need intensive surveillance.
Lynch syndrome is due to mutations in at least four DNA mismatch repair (MMR) genes: MLH1, MSH2, MSH6, and PMS2. There are two tumor characteristics that can be screened for in an attempt to identify those patients with CRC who are most likely to have LS. These characteristics are microsatellite instability (MSI)6-8 and loss of one or two of the MMR proteins in the tumor compared with the normal tissue.9,10
Although there is little controversy regarding the value of surveillance for carriers of MMR gene mutations,11,12 there is considerable uncertainty regarding (1) whether the prevalence of MMR mutation carriers is high enough to warrant large-scale screening; (2) the trade-offs between screening all patients with CRC versus targeted screening of some high-risk subgroups as defined by age at diagnosis,13 family history criteria,14-17 or tumor histology;18 and (3) whether to recommend immunohistochemistry (IHC) or MSI as the primary screening tool. To answer these questions, we conducted this study using both the MSI and IHC screening tests and including full sequencing and large deletion testing for all four MMR genes.
We previously reported on the molecular and clinical findings in 1,066 patients.19,20 To compare the suitability of IHC or MSI as a primary screening method, we studied an additional 500 patients with both tests. We also summarize the prevalence of LS and clinical characteristics of the entire cohort of 1,566 patients.
PATIENTS AND METHODS
Patients
Patients with colorectal adenocarcinoma diagnosed at six participating hospitals were eligible for this study, regardless of age at diagnosis or family history of cancer. Patients with a clinical diagnosis of familial adenomatous polyposis were not eligible for this study. These six hospitals perform the vast majority of all operations for CRC in the Columbus metropolitan area (population 1.7 million). The institutional review board at all participating hospitals approved the research protocol and consent form in accordance with assurances filed with and approved by the United States Department of Health and Human Services. The accrual process has been described in detail elsewhere.19,20 Briefly, during the period of January 1999 through August 2004, 1,566 eligible patients with CRC were accrued to the study. Initial results for the first 1,066 patients were reported previously.19 The results of testing in the last 500 patients, which differs because it includes both MSI and IHC, are presented here. These 500 patients had a mean age of 63.6 years, and were largely white (88%) or African American (9%). Approximately one half (48%) were women.
Methods
Sample processing, MSI testing, IHC staining methods, MLH1 promoter methylation testing, and mutation detection methods used in this study have been described in detail previously.19,20
Analytical Strategy
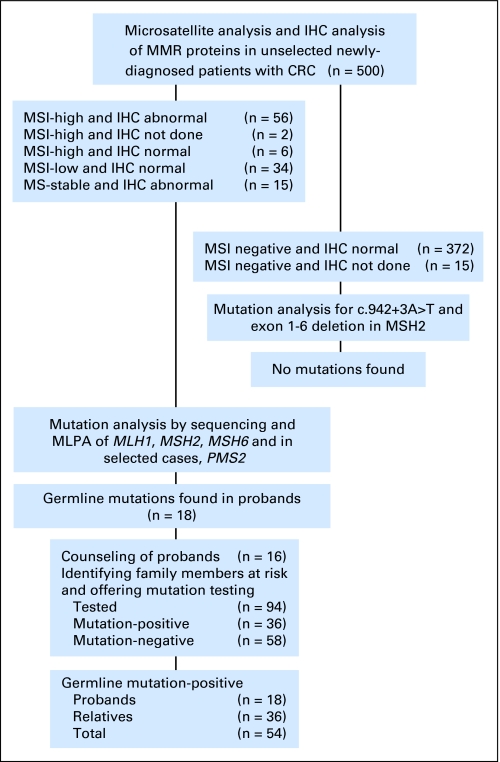
The 500 tumors were analyzed for MSI and 483 of the 500 tumors also had IHC staining for the four MMR proteins (17 cases did not undergo IHC because there was no remaining tumor material after the MSI test). The 387 MSI-negative patients with normal IHC results received a letter explaining that they were not likely to have LS; however, they might still want to be seen for a complete cancer genetic evaluation if they were concerned about their personal or family history. All 113 cases who were MSI positive (MSI-high and MSI-low) or had abnormal IHC underwent full genetic analysis (Fig 1). In the 98 MSI-positive cases, this included resequencing of MLH1, MSH2, MSH6, and, when indicated based on IHC, PMS2; multiplex ligation-dependent probe amplification assay of MLH1, MSH2, MSH6, and PMS2; and methylation analysis of the MLH1 promoter. In the cases who were microsatellite stable (MSS) but had abnormal IHC, the gene(s) corresponding to the protein(s) absent on IHC were tested with both resequencing and multiplex ligation-dependent probe amplification, and in the case of MLH1 absence, MLH1 promoter methylation was also assessed.
Fig 1.
Flow diagram of the analytic strategy and main results of the study. IHC, immunohistochemistry; MMR, mismatch repair; MSI, microsatellite instability; MLPA, multiplex ligation-dependent probe amplification.
In addition, in an effort to determine whether any cases of LS had been missed by both MSI and IHC, the 372 patients with MSS tumors and normal IHC and the 15 cases with MSS tumors in which IHC was not completed received genetic testing for the two most common MMR gene mutations in our series. This includes the c.942 + 3A→T mutation in MSH2 leading to skipping of exon 521 and the American Founder Mutation (AFM), a deletion of exons 1 to 6 in MSH2.22 Together, these mutations account for eight (44.5%) of 18 of all the mutations found in this study. The methods used to detect these mutations are described in the Appendix (online only).
Finally, we combined the 500 patients screened in this study with our prior 1,066 patients to present some new overall results on the entire cohort of 1,566 patients with CRC. This includes the prevalence of LS in the entire cohort, clinical features that were most predictive of finding a germline MMR gene mutation, and the results of genetic testing in the at-risk relatives of everyone diagnosed with LS in this cohort (free genetic counseling and single-mutation analysis was provided as part of this study, as described previously).19
Statistical Methods
Sensitivity,23 specificity23 and positive predictive values24 of clinical parameters were determined. Statistical analysis was conducted using R (http://cran.r-project.org/) software. Two-sided 95% CIs of the proportions were estimated using Wilson score method with continuity correction, as described by Newcombe.25 Differences in proportions were compared using the χ2 test.
RESULTS
MSI
MSI testing was performed for all 500 tumors; 64 tumors were found to be MSI-high (12.8%) and 34 tumors were found to be MSI-low (6.8%). For this study, all 98 patients (19.6%) with MSI-high or MSI-low tumors were considered MSI positive and underwent full genetic analysis. Eighteen (28.1%) of the 64 patients with MSI-high tumors were subsequently found to have LS. No LS gene mutations were found in patients with MSI-low tumors. Furthermore, all 34 MSI-low CRC tumors had normal IHC for all four MMR proteins. There were six tumors that were MSI-high that had normal IHC results. MSI-high tumors accounted for 56 (78.9%) of the 71 tumors with abnormal IHC.
IHC Analysis
IHC was performed on 483 of the 500 tumors. IHC was abnormal in 71 (14.7%) of the 483 CRC tumors. Abnormal results were as follows: 45 patients had MLH1 and PMS2 absent, 12 patients had MSH2 and MSH6 absent, nine patients had MSH6 only absent, two patients had PMS2 only absent, and three patients had other combinations absent (MLH1, MSH6 and PMS2 absent, MLH1 and MSH6 absent, and MLH1 only absent). These 71 patients included 56 (87.5%) of the 62 MSI-high tumors in which IHC was completed. There were 15 MSS tumors in which IHC was abnormal; none were found to have an LS mutation. IHC findings were concordant with the germline mutation findings in 17 of the 18 patients with LS; 17 (23.9%) of the 71 patients whose tumors had abnormal IHC were subsequently found to have LS.
MLH1 Promoter Methylation
Methylation at the proximal region26,27 of the MLH1 promoter was assessed in all 98 MSI-positive tumors and in the six MSS tumors that had absence of MLH1 on IHC. Overall, there were 38 patients with MLH1 promoter methylation; none of whom were found to have LS. Of the 48 patients with MLH1 absent on IHC; 33 patients (68.8%) showed MLH1 promoter methylation, 13 patients did not have MLH1 promoter methylation (four of whom were found to have LS), and two patients had failure of methylation testing.
Probands With LS
Eighteen of the 500 patients with CRC had a deleterious mutation in one of the MMR genes (Table 1). None of these patients had previously been diagnosed with LS. Mutations in MSH2 (n = 10) were more common than mutations in MLH1 (n = 4), MSH6 (n = 3), and PMS2 (n = 1). Five probands had the recurrent c.942 + 3A→T mutation of MSH2, which leads to exon 5 skipping.21 The remaining mutations include six large rearrangements (including three patients with the AFM22), six truncating mutations, and one in-frame deletion that is known to be deleterious (K618del).28,29 The mean age at diagnosis was 50.1 years (range, 28 to 77 years). Only eight (44.4%) of the 18 probands were diagnosed at age younger than 50 years and only 10 (55.6%) of the 18 probands had a first-degree relative with colorectal or endometrial cancer. Regarding published family history criteria, seven of 18 patients fulfilled the Amsterdam II criteria,16,17 13 of 18 patients met the revised Bethesda guidelines,14,15 and five (27.8%) of 18 patients did not meet either.
Table 1.
Patients With Deleterious Mutations
| Patient | Sex | Age at Diagnosis (years) | CRC Site | Amsterdam* | Bethesda† | MSI
|
Gene | Nucleotide Change | Mutation | |
|---|---|---|---|---|---|---|---|---|---|---|
| No. of Unstable Microsatellite Markers | Total No. of Microsatellite Markers Tested | |||||||||
| 57089 | M | 45 | Cecum and rectal | N | Y | 5 | 5 | MLH1 | c.826dupA | p.I276NfsX31 |
| 58443 | F | 48 | Cecum | N | Y | 4 | 5 | MLH1 | c.1852_1854delAAG | p.K618del |
| 68833 | F | 49 | Rectum | Y | Y | 4 | 5 | MLH1 | Del exons 16-19 | Large deletion |
| 67761 | F | 52 | Descending | Y | Y | 4 | 5 | MLH1 | Dup exons 6-12 | Large duplication |
| 61299 | M | 37 | Transverse | N | Y | 5 | 5 | MSH2 | c.830T > G | p. L277X |
| 342 | M | 32 | Cecum | N | Y | 2 | 3 | MSH2 | c.942 + 3A > T | Skip exon 5 |
| 1599 | F | 29 | Colon | N | Y | 5 | 5 | MSH2 | c.942 + 3A > T | Skip exon 5 |
| 1700 | F | 71 | Descending | N | Y | 5 | 5 | MSH2 | c.942 + 3A > T | Skip exon 5 |
| 59233 | F | 28 | Colon | N | Y | 5 | 5 | MSH2 | c.942 + 3A > T | Skip exon 5 |
| 62411 | M | 58 | Transverse | N | Y | 4 | 5 | MSH2 | c.942 + 3A > T | Skip exon 5 |
| 1519 | M | 57 | Sigmoid | N | Y | 3 | 5 | MSH2 | c.2038C > T | p.R680X |
| 1728 | M | 58 | Rectum | N | N | 5 | 5 | MSH2 | Del exons 1-6 | g.26445441_26465484del20045‡ |
| 59400 | M | 60 | Cecum | N | Y | 5 | 5 | MSH2 | Del exons 1-6 | g.26445441_26465484del20045‡ |
| 64763 | F | 45 | Ascending | N | Y | 5 | 5 | MSH2 | Del exons 1-6 | g.26445441_26465484del20045‡ |
| 64754 | M | 54 | Cecum | N | N | 5 | 5 | MSH6 | c.3939_3957dup19” | p.A1320SfsX5 |
| 56838 | F | 58 | Sigmoid | N | N | 2 | 5 | MSH6 | c.3261dupC | p.F1088LfsX5 |
| 57511 | M | 64 | Descending | N | N | 5 | 5 | MSH6 | c.3920_3923dupATCT | p.P1309SfsX11 |
| 62972 | F | 56 | Ascending | N | N | 4 | 5 | PMS2 | Del exon 1 | Large deletion |
Abbreviations: CRC, colorectal cancer; MSI, microsatellite instability; M, male; N, no; Y, yes; F, female.
Yes if patient meets at least Amsterdam II criteria using only first-degree relatives.
Yes if patient meets revised Bethesda guidelines using only first-degree relatives.
Reference sequence: NT 022184.14.
Mutation Analysis in Patients With MSS Tumors
None of the 387 patients with MSS tumors with normal IHC or in which IHC could not be performed were found to have the c.942 + 3A→T mutation in MSH2 or the AFM mutation.
Screening Test Performance
The sensitivity and specificity of screening for patients with LS using four different modalities could be formally determined only if we had performed complete genetic testing in every patient, which would have been a cost-prohibitive and probably unnecessary endeavor. Here we apply the terms sensitivity and specificity to describe these parameters assuming that 100% of all mutations had been detected. This assumption is supported by the fact that all patients with MSS tumors were tested for the two most common LS mutations, and none were found. Both the MSI and IHC screening tests performed well, with sensitivities of 100% and 94.4% and specificities of 90.5% and 88.4%, respectively. On the other hand, the sensitivity of screening for LS based on a diagnosis before age 50 years was 44.4% and the specificity was 85.5%. Positive predictive value of MSI-high, abnormal IHC, diagnosis before age 50 years, and first-degree relative with CRC or endometrial cancer were 28.1%, 23.9%, 10.3%, and 8.8%, respectively. Negative predictive values were 100%, 99.8%, 97.6%, and 97.9% respectively.
Results From Entire Study Cohort of 1,566 Patients With CRC
Overall prevalence of LS.
Notably, after improving our PMS2 mutation detection methods,30 three additional PMS2 mutations have been found among the 1,066 patients described previously. This includes a Lys614X mutation in patient 56850, a c.736_741del6ins11 mutation in patient 1364, and a c.2007-1 G→A in patient 1356. The tumor from patient 1356 was MSI-low, and this is the only case of LS diagnosed in a patient with CRC with an MSI-low tumor on study. This brings the total number of LS cases among the previously reported 1,066 patients with CRC to 26 patients (2.4%). Thus the overall prevalence of LS among all 1,566 CRC cases is 44 of 1,566 (2.8%; 95% CI, 2.1% to 3.8%). The prevalence might be higher if some of the missense mutations (Appendix Table A1, online only) are determined to be deleterious.
Relatives of probands with LS.
Overall, considering the entire series of 1,566 patients, a total of 249 relatives from 33 of the 44 LS families have been counseled and tested; 109 relatives tested positive and 140 relatives tested negative (Table 2). This amounts to more than three relatives per proband being diagnosed with the same mutation as in the proband. Of the 109 mutation-positive relatives, 25 had a prior diagnosis of an LS-related cancer, whereas 84 were unaffected at the time of testing (three relatives have subsequently been diagnosed with cancer). Remarkably, of the 153 individuals identified with LS as part of this study (44 probands and 109 relatives), only one had been previously diagnosed with LS.
Table 2.
Relatives Tested in the Entire Study Cohort
| Relationship | Mutation Positive | Mutation Negative | Total Tested |
|---|---|---|---|
| First degree | 52 | 47 | 99 |
| Second degree | 28 | 36 | 64 |
| Beyond second degree | 29 | 57 | 86 |
| All | 109 | 140 | 249 |
Clinical Findings
The characteristics of the entire cohort of 1,566 patients with CRC have been analyzed with respect to the likelihood of making an LS diagnosis (Appendix Table A2, online only). The features associated with the highest likelihood of finding a germline mutation in an MMR gene include absence of MSH2 with or without absence of MSH6 on IHC (66.7%), absence of MSH6 or PMS2 alone on IHC (23.5% and 55.6%), and absence of MLH1 without MLH1 promoter methylation (33.3%). Patients whose tumors had abnormal IHC had a similar likelihood of having LS as those whose tumors were MSI-high (21.4% v 20.8%; P = .9847, χ2 test). Patients with CRC diagnosed at younger ages are more likely to have LS, with 8.4% of those diagnosed before age 50 years having a mutation as compared with 1.7% of patients diagnosed at age 50 years or older. However, an equal number of patients (n = 22) were found to have LS among those diagnosed at age younger than 50 years and those diagnosed at age ≥ 50 years. Patients with right-sided tumors were twice as likely to have LS (4.0% v 1.9%; P = .0224, χ2 test).
DISCUSSION
Limitations of this study include the fact that we could not perform complete molecular analysis of the MLH1, MSH2, MSH6, and PMS2 genes in all 500 patients to confirm that there were no mutations in the patients who were both MSI-negative and had normal IHC results. In addition, we found many missense mutations in the MMR genes that could not be classified as polymorphisms or deleterious mutations as of yet. Both of these limitations mean that the prevalence we found for LS represents the minimum prevalence.
It is clear that in a typical United States metropolitan area, at least 2.8% of all newly diagnosed patients with CRC have LS. This prevalence translates into a rate of one patient with LS for every 35 individuals diagnosed with CRC. This is a remarkably high prevalence for a highly penetrant, dominantly inherited, potentially lethal condition.
Several sets of criteria for defining high risk have been developed and are widely used in clinical practice. They rely on age at cancer diagnosis and family history of cancer. These criteria could be applied as a prescreen in population-based molecular screening projects. We show here that screening only patients younger than 50 years will leave half of the cases undiagnosed, so we believe this practice should be rejected. The stringent Amsterdam criteria have too low a sensitivity to be suitable (eg, 39% in this study), whereas the sensitivity of the Bethesda guidelines is higher (eg, 72% in this study). Although prescreening using the Bethesda guidelines (or a simpler modification thereof) would dramatically reduce the number of MSI or IHC tests necessary, it would also reduce the number of carriers detected (eg, by 28% in this study). The fact that only one of the 153 individuals identified as having LS in this study had previously been diagnosed or referred to genetics is a sign about how poorly taking and assessing a family history of cancer works in practice.31
We show that IHC and MSI are quite similar in having high sensitivity to detect LS, as has been shown by others.32,33 The overwhelming advantage of IHC over MSI in the setting of large-scale screening is its availability (in principle, wherever there is a pathology laboratory) and the fact that MSI requires microdissection of the tumor and work in a molecular diagnostics laboratory. The use of IHC results in fewer genetic tests than MSI by predicting the responsible gene. Moreover, the costs of IHC screening can be reduced dramatically through the use of tissue microarrays without loss of accuracy.34-36
Four main prerequisites should be fulfilled for any large-scale screening program to be undertaken. First, it should be conceptually and technically feasible. We have endeavored to show that screening patients with CRC for LS is feasible. Second, it should be desirable. We show that LS is relatively prevalent, affecting one in 35 patients with CRC. Empirically, clinical surveillance for CRC in mutation carriers prevents more than 60% of cancers and more than 60% of cancer-related deaths.11 We conclude that early detection of LS is highly desirable. Third, it should be cost-effective. This is currently being explored. Fourth, the screening should not be harmful. We do not know of any circumstances that make large-scale screening more susceptible to harmful effects (psychological or physical) than the less proactive screening that is already practiced in high-risk genetics clinics.
The benefits of any screening program are heavily dependent on the number of at-risk relatives who will receive genetic counseling, undergo genetic testing, and follow appropriate cancer surveillance guidelines. In our study, we achieved a high rate of relatives tested per proband (average, > five per family) and diagnosed 109 at-risk relatives with LS (average, > three per family) and 140 relatives without LS. Compliance to cancer surveillance recommendations on a large scale is yet to be shown. However, single-mutation analysis for the known mutation in the family is extremely simple and inexpensive, which will lead to dramatically more favorable cost effectiveness in family members.
It may be well worth reconsidering current models for providing cancer genetic services, relying on physicians to identify and refer individuals with a positive family history. It is well known that family history is often overlooked.37 In this study where none of 44 probands and only one of 109 mutation-positive relatives had been previously diagnosed with or tested for LS, it is clear that the current approach will only identify a small fraction of all individuals with LS. It will only become more difficult to identify patients with LS on the basis of family history of CRC in the future, given that the average family size is getting smaller and usage of colonoscopy will likely prevent many CRCs through the removal of precancerous polyps. Moving to an active approach of screening for LS among patients with cancer is an attractive and even compelling option.
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Philip Kuebler, Sanofi-Aventis (C), Genentech (C) Stock Ownership: None Honoraria: Philip Kuebler, Genentech Research Funding: None Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Heather Hampel, Albert de la Chapelle
Financial support: Albert de la Chapelle
Administrative support: Heather Hampel, Janet Lockman, Albert de la Chapelle
Provision of study materials or patients: Heather Hampel, Edward Martin, Mark Arnold, Karamjit Khanduja, Philip Kuebler, Ilene Comeras, Albert de la Chapelle
Collection and assembly of data: Heather Hampel, Wendy L. Frankel, Mark Clendenning, Kaisa Sotamaa, Thomas Prior, Judith A. Westman, Jenny Panescu, Dan Fix, Janet Lockman, Jennifer LaJeunesse, Ilene Comeras, Albert de la Chapelle
Data analysis and interpretation: Heather Hampel, Wendy L. Frankel, Mark Clendenning, Kaisa Sotamaa, Jenny Panescu, Dan Fix, Janet Lockman, Jennifer LaJeunesse, Albert de la Chapelle
Manuscript writing: Heather Hampel, Judith A. Westman, Jenny Panescu, Jennifer LaJeunesse, Albert de la Chapelle
Final approval of manuscript: Heather Hampel, Wendy L. Frankel, Edward Martin, Mark Arnold, Karamjit Khanduja, Philip Kuebler, Mark Clendenning, Kaisa Sotamaa, Thomas Prior, Judith A. Westman, Jenny Panescu, Dan Fix, Janet Lockman, Jennifer LaJeunesse, Ilene Comeras, Albert de la Chapelle
Supplementary Material
Acknowledgments
We thank the Ohio State University Comprehensive Cancer Center Pathology Core Facility (Weiqiang Zhao, MD, PhD, Director; Mary Marin, Supervisor, and Susie Jones) and the Ohio State University Comprehensive Cancer Center Tissue Archive Service (Scott Jewell, PhD, Director; Cheryl Reeder, Supervisor) for pathology services.
Appendix
Methods for Detecting Two Widespread Mutations
To detect the c.942 + 3A→T mutation, polymerase chain reactions (PCR) were set up using HotStarTaq (Qiagen GmbH, Hilden, Germany) and primers that flank exon 5 of MSH2 (forward, 5′-ATTGGGAAGGAACACCAAGG-3′; reverse, 5′-GGAGGGGAGAGAAAAATACAGC-3′). Each 15-μL reaction, consisting of 25 ng of genomic DNA and 5 pmoles of each primer, was cycled using the following profile: 15 minutes at 95°C, followed by 30 cycles of 30 seconds at 95°C, 30 seconds at 60°C, and 45 seconds at 72°C; the reaction was finished with a final elongation at 72°C for 9 minutes. PCR products were Exo/SAP-IT (USB Corporation, Cleveland, OH) treated, and 3 μL was subsequently used as the template in a 10-μL primer extension reaction (5′-GTCAGAGCCCTTAACCTTTTTCAGGT-3′; SnaPshot; Applied Biosystems, Foster City, CA). Products were dephosphorylated before being analyzed using an ABI3700 machine (Applied Biosystems).
Test methods for the American Founder Mutation comprising a deletion of exons 1 through 6 of MSH2 included the forward primer (5′-GCCTGGCGTCAAACGTT-3′) and the reverse primer (5′-TGAGTCATTTTGGGGATCAGTT-3′). This produces a breakpoint-specific, 562–base pair amplicon under standard PCR conditions. To ensure that poor DNA quality was not a reason for a negative result, this amplicon was multiplexed with an 811–base pair amplicon (forward, 5′-AAGCATCTCACCTCATCCTAACACA-3′; reverse, 5′-GGATCACACCTGCCTTAAATTGCAT-3′), from the BRAF locus, in a 15-μL PCR reaction. Each 15-μL reaction, containing 7.5 μL of GoTaq master mix (Promega, Madison, WI), 25 ng of genomic DNA, and 5 pmoles of each of the four primers, was cycled under the following conditions: 95°C for 2 minutes, 30 cycles of 95°C for 30 seconds, 60°C for 30 seconds and 72°C for 45 seconds, and a final extension at 72°C for 8 minutes. Samples with a breakpoint-specific product were treated with ExoSAP-IT (USB Corporation) and sequenced with the forward primer to confirm the presence of an American Founder Mutation–specific breakpoint.
Investigators Participating in the Study
In addition to the authors, the following investigators participated in this study: The Ohio State University Medical Center, Columbus, OH (C. Ellison, S. Melvin, J. Winston III); The Human Cancer Genetics Program, The James Cancer Hospital and Solove Research Institute at The Ohio State University, Columbus, OH (W. Burak, K. Jamieson, Y. Zhang); The Ohio State University Hospital East, Columbus, OH (A. Ghany, R. Schlanger); Mount Carmel Health System, Columbus, OH (P. Aguilar, B. Kerner, G. LaValle, M. Lindsey, J. Madhavan, A. Padmanabhan, C. Taylor, T. Vara, L.E. Vassy, J. Zangmeister); St. Ann's Hospital, Westerville, OH (M. Davanzo, B. Grischow, K. Hamelberg, T. Kelly, J. Park, T. Patel); Riverside Methodist Hospital, Columbus, OH (B.C. Behrens, S.C. Blair, M. Brimer, C.S. George, J.K. Hofmeister, P.J. Kourlas, J. Mitchell, T.J. Sweeney, W. Wise, T. Williams); Grant Medical Center, Columbus, OH (S. Miller, T.D. Moore).
Table A1.
Patients With Missense Mutations Not Classified as Disease Causing
| Patient | Sex | Age at Diagnosis (years) | CRC Site | MSI
|
Gene | Nucleotide | Protein | Classification | Family History* | Race | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Unstable Microsatellite Markers | Total No. of Microsatellite Markers Tested | ||||||||||
| 60235 | M | 82 | Cecum | 5 | 5 | MLH1 | c.299G > A | p.Arg100Gln | VUS | N | White |
| 64796 | M | 52 | Transverse | 5 | 5 | MLH1 | c.1852_1853delAAinsGC | p.Lys618Ala | Polymorphism | N | White |
| 67844 | F | 61 | Ascending | 4 | 5 | MLH1 | c.1963A > G | p.Ile655Val | Polymorphism | Y | Black |
| 1373 | F | 85 | Ascending | 1 | 5 | MLH1 | c.2152C > T | p.His718Tyr | VUS | N | Black |
| 62411 | M | 58 | Transverse | 4 | 5 | MLH1 | c.2152C > T | p.His718Tyr | VUS | Y | White |
| 1373 | F | 85 | Ascending | 1 | 5 | MSH2 | c.380A > G | p.Asn127Ser | Polymorphism | N | Black |
| 67844 | F | 61 | Ascending | 4 | 5 | MSH2 | c.380A > G | p.Asn127Ser | Polymorphism | Y | Black |
| 1519 | M | 57 | Sigmoid | 5 | 5 | MSH2 | c.965G > A | p.Gly322Asp | Polymorphism | N | White |
| 68655 | F | 79 | Ascending | 2 | 5 | MSH2 | c.965G > A | p.Gly322Asp | Polymorphism | N | White |
| 57244 | M | 83 | Cecum | 1 | 5 | MSH6 | c.1403G > A | p.Arg468His | VUS | N | White |
| 68947 | M | 59 | Sigmoid | 1 | 5 | MSH6 | c.1729C > T | p.Arg577Cys | VUS | N | White |
| 1766 | M | 55 | Ascending | 0 | 5 | MSH6 | c.2057G > A | p.Gly686Asp | VUS | Y | White |
| 64796 | M | 52 | Transverse | 5 | 5 | MSH6 | c.2633T > C | p.Val878Ala | VUS | N | White |
Abbreviations: MSI, microsatellite instability; CRC, colorectal cancer; M, male; VUS, variant of uncertain significance; N, no; F, female; Y, yes.
Yes if patient has one or more first-degree relative with colorectal or endometrial cancer.
Table A2.
Clinical Findings of Entire Study Cohort
| Clinical Criteria | No. Mutation Positive | Total No. of Patients Meeting Criteria | % | 95% CI |
|---|---|---|---|---|
| Age at diagnosis, years | ||||
| < 30 | 3 | 12 | 25 | 6.7 to 57.2 |
| 30-39 | 7 | 55 | 12.7 | 5.7 to 25.1 |
| 40-49 | 12 | 196 | 6.0 | 3.3 to 10.7 |
| < 50 | 22 | 263 | 8.4 | 5.4 to 12.6 |
| 50-59 | 12 | 330 | 3.6 | 2.0 to 6.4 |
| 60-69 | 4 | 419 | 1.0 | 0.3 to 2.6 |
| 70-79 | 4 | 404 | 1.0 | 0.3 to 2.7 |
| ≥ 80 | 2 | 150 | 1.3 | 0.2 to 5.2 |
| ≥ 50 | 22 | 1303 | 1.7 | 1.1 to 2.6 |
| Family history | ||||
| First-degree relative with CRC or endometrial cancer | 23 | 350 | 6.6 | 4.3 to 9.8 |
| Diagnosis before age 50 years or first-degree relative with CRC or endometrial cancer | 29 | 565 | 5.1 | 3.5 to 7.4 |
| Bethesda criteria | 29 | 419 | 6.9 | 4.8 to 9.9 |
| Amsterdam criteria | 4 | 18 | 22.2 | 7.3 to 48.1 |
| No family history criteria | 11 | 1129 | 1.0 | 0.5 to 1.8 |
| Tumor characteristics | ||||
| MSI-high | 41 | 197 | 20.8 | 15.5 to 27.3 |
| MSI-low | 1 | 107 | 0.9 | 0.05 to 5.8 |
| MSI-stable | 2 | 1262 | 0.2 | 0.03 to 0.6 |
| Abnormal IHC | 43 | 201 | 21.4 | 16.1 to 27.8 |
| Absent MLH1 and/or PMS2 | 9 | 148 | 6.1 | 3.0 to 11.6 |
| Absent MLH1 and no MLH1 promoter methylation | 13 | 39 | 33.3 | 19.6 to 50.3 |
| Absent MSH2± MSH6 | 22 | 33 | 66.7 | 48.1 to 81.4 |
| Absent MSH6 alone | 4 | 17 | 23.5 | 7.8 to 50.2 |
| Absent PMS2 alone | 6 | 9 | 55.6 | 30.9 to 91.0 |
| Normal IHC | 1 | 595 | 0.2 | 0.01 to 1.1 |
| Tumor location | ||||
| Right-sided CRC | 23 | 573 | 4.0 | 2.6 to 6.1 |
| Left-sided CRC | 18 | 942 | 1.9 | 1.2 to 3.1 |
| Colon, NOS | 3 | 40 | 7.5 | 2.0 to 21.5 |
| Appendiceal primary | 0 | 11 | 0 | 0.8 to 32.1 |
Abbreviations: CRC, colorectal cancer; MSI, microsatellite instability; IHC, immunohistochemistry; NOS, not otherwise specified.
published online ahead of print at www.jco.org on September 22, 2008.
Supported by Grants No. CA67941 and CA16058 from the National Institutes of Health. This publication was also prepared under a grant from the State of Ohio Biomedical Research and Technology Transfer Commission.
Authors’ disclosures of potential conflicts of interest and author contributions are found at the end of this article.
REFERENCES
- 1.Society AC: Cancer Facts and Figures 2006. Atlanta, GA, American Cancer Society, 2006
- 2.Lynch H, de la Chapelle A: Genomic medicine: Hereditary colon cancer. N Engl J Med 348:919-932, 2003 [DOI] [PubMed] [Google Scholar]
- 3.Burke W, Petersen G, Lynch P, et al: Recommendations for follow-up care of individuals with an inherited predisposition to cancer: I. Hereditary nonpolyposis colon cancer—Cancer Genetics Studies Consortium. JAMA 277:915-919, 1997 [PubMed] [Google Scholar]
- 4.Lindor NM, Petersen GM, Hadley DW, et al: Recommendations for the care of individuals with an inherited predisposition to Lynch syndrome: A systematic review. JAMA 296:1507-1517, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Vasen HF, de Vos Tot Nederveen Cappel WH: An evidence-based review on surveillance for Lynch syndrome. Dis Colon Rectum 49:1797-1798, 2006; author reply 1799 [DOI] [PubMed] [Google Scholar]
- 6.Aaltonen LA, Peltomaki P, Leach FS, et al: Clues to the pathogenesis of familial colorectal cancer. Science 260:812-816, 1993 [DOI] [PubMed] [Google Scholar]
- 7.Aaltonen LA, Peltomaki P, Mecklin JP, et al: Replication errors in benign and malignant tumors from hereditary nonpolyposis colorectal cancer patients. Cancer Res 54:1645-1648, 1994 [PubMed] [Google Scholar]
- 8.Boland CR, Thibodeau SN, Hamilton SR, et al: A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58:5248-5257, 1998 [PubMed] [Google Scholar]
- 9.Müller W, Burgart LJ, Krause-Paulus R, et al: The reliability of immunohistochemistry as a prescreening method for the diagnosis of hereditary nonpolyposis colorectal cancer (HNPCC): Results of an international collaborative study. Fam Cancer 1:87-92, 2001 [DOI] [PubMed] [Google Scholar]
- 10.Thibodeau SN, French AJ, Roche PC, et al: Altered expression of hMSH2 and hMLH1 in tumors with microsatellite instability and genetic alterations in mismatch repair genes. Cancer Res 56:4836-4840, 1996 [PubMed] [Google Scholar]
- 11.Järvinen HJ, Aarnio M, Mustonen H, et al: Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology 118:829-834, 2000 [DOI] [PubMed] [Google Scholar]
- 12.Schmeler KM, Lynch HT, Chen LM, et al: Prophylactic surgery to reduce the risk of gynecologic cancers in the Lynch syndrome. N Engl J Med 354:261-269, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Barnetson RA, Tenesa A, Farrington SM, et al: Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med 354:2751-2763, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez-Bigas M, Boland C, Hamilton S, et al: National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: Meeting highlights and Bethesda guidelines. J Natl Cancer Inst 89:1758-1762, 1997 [DOI] [PubMed] [Google Scholar]
- 15.Umar A, Boland CR, Terdiman JP, et al: Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst 96:261-268, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasen H, Mecklin J-P, Khan P, et al: The International Collaborative Group on Hereditary Non-polyposis Colorectal Cancer. Dis Colon Rectum 34:424-425, 1991 [DOI] [PubMed] [Google Scholar]
- 17.Vasen H, Watson P, Mecklin J-P, et al: New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative Group on HNPCC. Gastroenterology 116:1453-1456, 1999 [DOI] [PubMed] [Google Scholar]
- 18.Jenkins MA, Hayashi S, O’Shea AM, et al: Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: A population-based study. Gastroenterology 133:48-56, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hampel H, Frankel WL, Martin E, et al: Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 352:1851-1860, 2005 [DOI] [PubMed] [Google Scholar]
- 20.Hampel H, Frankel W, Panescu J, et al: Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res 66:7810-7817, 2006 [DOI] [PubMed] [Google Scholar]
- 21.Desai DC, Lockman JC, Chadwick RB, et al: Recurrent germline mutation in MSH2 arises frequently de novo. J Med Genet 37:646-652, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynch HT, Coronel SM, Okimoto R, et al: A founder mutation of the MSH2 gene and hereditary nonpolyposis colorectal cancer in the United States. JAMA 291:718-724, 2004 [DOI] [PubMed] [Google Scholar]
- 23.Altman DG, Bland JM: Diagnostic tests: 1. Sensitivity and specificity. BMJ 308:1552, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altman DG, Bland JM: Diagnostic tests: 2. Predictive values. BMJ 309:102, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newcombe RG: Two-sided confidence intervals for the single proportion: Comparison of seven methods. Stat Med 17:857-872, 1998 [DOI] [PubMed] [Google Scholar]
- 26.Deng G, Chen A, Hong J, et al: Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res 59:2029-2033, 1999 [PubMed] [Google Scholar]
- 27.Nakagawa H, Nuovo GJ, Zervos EE, et al: Age-related hypermethylation of the 5′ region of MLH1 in normal colonic mucosa is associated with microsatellite-unstable colorectal cancer development. Cancer Res 61:6991-6995, 2001 [PubMed] [Google Scholar]
- 28.Raevaara TE, Vaccaro C, Abdel-Rahman WM, et al: Pathogenicity of the hereditary colorectal cancer mutation hMLH1 del616 linked to shortage of the functional protein. Gastroenterology 125:501-509, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Wijnen J, Khan PM, Vasen H, et al: Majority of hMLH1 mutations responsible for hereditary nonpolyposis colorectal cancer cluster at the exonic region 15-16. Am J Hum Genet 58:300-307, 1996 [PMC free article] [PubMed] [Google Scholar]
- 30.Clendenning M, Hampel H, LaJeunesse J, et al: Long-range PCR facilitates the identification of PMS2-specific mutations. Hum Mutat 27:490-495, 2006 [DOI] [PubMed] [Google Scholar]
- 31.Ramsey SD, Burke W, Pinsky L, et al: Family history assessment to detect increased risk for colorectal cancer: Conceptual considerations and a preliminary economic analysis. Cancer Epidemiol Biomarkers Prev 14:2494-2500, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baudhuin LM, Burgart LJ, Leontovich O, et al: Use of microsatellite instability and immunohistochemistry testing for the identification of individuals at risk for Lynch syndrome. Fam Cancer 4:255-265, 2005 [DOI] [PubMed] [Google Scholar]
- 33.Piñol V, Castells A, Andreu M, et al: Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA 293:1986-1994, 2005 [DOI] [PubMed] [Google Scholar]
- 34.Hardisson D, Moreno-Bueno G, Sanchez L, et al: Tissue microarray immunohistochemical expression analysis of mismatch repair (hMLH1 and hMSH2 genes) in endometrial carcinoma and atypical endometrial hyperplasia: Relationship with microsatellite instability. Mod Pathol 16:1148-1158, 2003 [DOI] [PubMed] [Google Scholar]
- 35.Hendriks Y, Franken P, Dierssen JW, et al: Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumors. Am J Pathol 162:469-477, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svrcek M, Jourdan F, Sebbagh N, et al: Immunohistochemical analysis of adenocarcinoma of the small intestine: A tissue microarray study. J Clin Pathol 56:898-903, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guttmacher AE, Collins FS, Carmona RH: The family history: More important than ever. N Engl J Med 351:2333-2336, 2004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.