Abstract
Lung cancer has become a global public health burden, further substantiating the need for early diagnosis and more effective targeted therapies. The key to accomplishing both these goals is a better understanding of the genes and pathways disrupted during the initiation and progression of this disease. Gene promoter hypermethylation is an epigenetic modification of DNA at promoter CpG islands that together with changes in histone structure culminates in loss of transcription. The fact that gene promoter hypermethylation is a major mechanism for silencing genes in lung cancer has stimulated the development of screening approaches to identify additional genes and pathways that are disrupted within the epigenome. Some of these approaches include restriction landmark scanning, methylation CpG island amplification coupled with representational difference analysis, and transcriptome-wide screening. Genes identified by these approaches, their function, and prevalence in lung cancer are described. Recently, we used global screening approaches to interrogate 43 genes in and around the candidate lung cancer susceptibility locus, 6q23–25. Five genes, TCF21, SYNE1, AKAP12, IL20RA, and ACAT2, were methylated at 14 to 81% prevalence, but methylation was not associated with age at diagnosis or stage of lung cancer. These candidate tumor suppressor genes likely play key roles in contributing to sporadic lung cancer. The realization that methylation is a dominant mechanism in lung cancer etiology and its reversibility by pharmacologic agents has led to the initiation of translational studies to develop biomarkers in sputum for early detection and the testing of demethylating and histone deacetylation inhibitors for treatment of lung cancer.
Keywords: gene promoter hypermethylation, lung cancer, chromosome 6, epigenetics
Lung cancer has become a global public health burden, with 1.5 million deaths expected by 2010. The high mortality from this disease stems from the lack of an effective screening approach for early diagnosis and the refractiveness of advanced cancers to conventional therapies, substantiating the need to develop more effective targeted therapies and chemoprevention. Although smoking cessation does reduce risk for lung cancer, approximately half of lung cancers diagnosed are in former smokers. Adenocarcinoma is the major histologic type of cancer diagnosed in smokers in the United States and now Europe (1, 2). An incidence rate of 40% and up to 80% has been reported for this histologic type of cancer in smokers and never smokers, respectively, diagnosed with lung cancer. Non–small cell lung cancer (NSCLC, comprising mainly adeno, squamous cell, and large cell carcinoma) is diagnosed in approximately 80% of patients, while the remaining 20% of tumors appear to be small cell lung cancer (SCLC).
The detection of numerous cytogenetic changes provided the first link to the molecular pathogenesis of lung cancer. Mapping of chromosomal sites for rearrangement, breakpoints, and losses revealed both common and distinct changes in SCLC and NSCLC. The commonality for specific regions in the genome for allelic loss suggested the presence of tumor suppressor genes (TSGs) within these loci. The retinoblastoma gene was the first TSG linked to lung cancer (3). Loss of function of this gene through either deletion or point mutation occurs in 90% of SCLC, while less than 15% of NSCLCs harbor changes in this TSG (4). The second major TSG inactivated in lung cancer is p53. Although p53 inactivation is common across many malignancies, the mutation spectrum within this gene tracks with specific tumor types. In lung cancer, the most common mutation seen is the G:C to T:A transversion, an alteration potentially stemming from the inability to repair DNA damage caused by polyaromatic hydrocarbons such as benzo[a]pyrene, which is present in tobacco (5, 6). Consistent with this hypothesis, the prevalence for transversion mutations increased in tumors with increasing cumulative exposure to cigarette smoke (7). Mutations in p53 are found in 70% of SCLC, 65% of squamous cell cancer, and 33% of adenocarcinoma. In lung cancer, the search for TSGs inactivated through the two-hit mechanism of loss of one allele and mutation of the remaining allele have not identified any genes whose prevalence for inactivation approaches that seen for the retinoblastoma and p53 genes. The exception to this is LKB gene that is mutated exclusively in approximately one-third of adenocarcinomas (8).
The most commonly mutated oncogene in lung cancer is K-ras with approximately 30 to 40% of adenocarcinomas harboring an activating mutation, while mutations in squamous cell and SCLC are rarely observed (9, 10). Mutations are localized to codons 12, 13, and 61 with the majority (> 85%) occurring within codon 12. Nearly 70% of the mutations seen are G to T transversions within codon 12 that change a glycine codon (GGT) to valine (GTT) or cysteine (TGT) that may reflect DNA adducts formed by metabolism of polyaromatic hydrocarbons in tobacco. Recently, a whole genomic approach was taken to address how many mutations are seen in cancer (11). These studies were focused on breast and colon cancer, but most likely reflect the paradigm seen in lung cancer studies that have evaluated candidate genes discovered through various screening modalities. In this whole genome sequencing study, approximately 80 gene mutations were identified that alter amino acids. What was surprising was that the prevalence of the majority of these mutations in primary tumors was less than 5%. The authors concluded that these minor mutations would each be associated with a “small fitness advantage” that would drive tumor progression, and thus, it is not the most common genetic changes but these rare changes that dominate the cancer genome landscape (11). While this is an interesting hypothesis, the emergence of epigenetic modifications of critical regulatory genes indicates that the epigenome may play an equal, if not greater role in driving cancer initiation and progression than genetic mutations.
The most common epigenetic change in cancer is methylation of DNA at the fifth position of the cytosine ring. Cytosine located 5′ to guanine (CpG) is the prime target of methylation in the mammalian genome and this dinucleotide is concentrated in a much higher frequency than a random genome-wide distribution in regions called CpG islands. About 50% of human promoters contain CpG islands that often extend into exon 1 of many critical regulatory genes (12, 13). When DNA hypermethylation occurs within a CpG island located in the promoter region of a gene, it is also accompanied by histone modifications (such as acetylation, methylation, or phosporylation of histone tails) within the island. Together, these two epigenetic changes create a closed chromatin configuration around the promoter region denying access to RNA polymerase and regulatory proteins needed for transcription (12, 14). The end result of this process is loss of gene transcription and hence “silencing of gene function.” With the development of the methylation-specific PCR assay that can screen for gene methylation in specific promoters, there has been tremendous growth over the past decade in the identification of genes that are silenced in lung cancer through promoter hypermethylation (15). Transcriptional silencing by CpG island hypermethylation now rivals genetic changes that affect coding sequence as a critical trigger for neoplastic development and progression (12, 15). Genes responsible for all types of normal cellular function are targeted for inactivation by methylation at prevalences of 15 to 80% in lung tumors (15). These include genes involved in cell cycle regulation (e.g., p16), apoptosis (e.g., death associated protein kinase), DNA repair (e.g., O6-methylguanine-DNA methyltransferase), cell adhesion (e.g., H-cadherin), signal transduction (e.g., ras effector homolog 1 [RASSF1A]), and cell differentiation (e.g., RAR-β). Importantly, many of these genes appear to be inactivated at the earliest histologic stage of lung cancer and in cytologically normal-appearing bronchial epithelial cells from smokers (15). Understanding which pathways are inactivated in the tumor cell and bronchial epithelium of smokers will be essential for developing targeted therapy for lung cancer and cancer prevention. The realization of gene promoter methylation as a major alteration in the cancer cell has stimulated the development of screening approaches to identify additional genes and pathways that are disrupted within the epigenome. The sections below describe some of the high-throughput genome screening approaches used to identify methylated genes in cancer and a recent study (16) by our group evaluating promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23–25 using a combination of screening approaches.
RESTRICTION LANDMARK GENOME SCANNING
Restriction landmark genome scanning (RLGS) is a two-dimensional gel electrophoresis technique that allows the determination of the methylation status of approximately 2,000 promoter sequences in a single gel (17). High-molecular-weight DNA is digested with methylation-sensitive restriction enzymes such as Not1, end labeled with 32P, and then digested with a second restriction enzyme (EcoRV). The basis for creating different sizes of DNA fragments stems from the specificity of the NotI endonuclease for methylated CpG dinucleotides. DNA fragments are separated in a first dimension followed by a third restriction digestion with Hinf1 and separation on a second dimension polyacrylamide gel followed by exposure to X-ray film. The RLGS profiles between normal and tumor tissue are superimposed to identify differences in intensities and/or presence of the radiolabeled fragments. Global screening studies with RLGS in tumor types that included head and neck estimated that an average of 600 CpG islands of the 45,000 in the genome were aberrantly methylated (17). Subsequent studies have identified specific genes methylated in lung tumors that include bone morphogenesis protein 3B and TCF21 (18, 19). Major limitations of this approach include sensitivity, exclusion of CpG islands that do not contain NotI sites, and the fact that adjacent tumor tissue may contain methylated genes.
METHYLATED CpG ISLAND AMPLIFICATION COUPLED WITH REPRESENTATIONAL DIFFERENCE ANALYSIS
The methylated CpG island amplification (MCA)/representiational difference analysis (RDA) technique was originally developed by Toyota and coworkers (20) and is a PCR/subtraction hybridization-based assay that allows for the rapid amplification and selection of densely methylated CpG-rich regions ranging in size from 200 bp to 2 kb. It has been used most often for the identification of genes methylated in colon and pancreatic cancers (21, 22). Our group used this technique to identify the PAX5 α and PAX5 β genes that are methylated in approximately 50 to 70% of adenocarcinomas and squamous cell carcinomas (23). The PAX5 β gene encodes for the transcription factor B cell–specific activating protein that, in turn, directly regulates CD19, a gene shown to negatively control cell growth. A strong association was observed in this study between PAX5 β methylation and loss of expression of CD19, demonstrating that inactivation of the PAX5 β gene likely contributes to neoplastic development by inhibiting growth regulation through effects on CD19 gene expression. Several other genes identified through this scanning approach are currently being evaluated for methylation in primary lung tumors. Recently, the MCA procedure was coupled to CpG island microarray (MCAM) to increase the throughput for methylation profiling and the sensitivity and specificity to detect hypermethylated loci (24). Those studies identified hundreds of newly methylated genes in colon cancer with a sensitivity and specificity of 88% and 96%. MCAM may represent a high-throughput platform for profiling methylation changes in clinical tumors.
TRANSCRIPTOME-WIDE SCREENING FOR METHYLATION
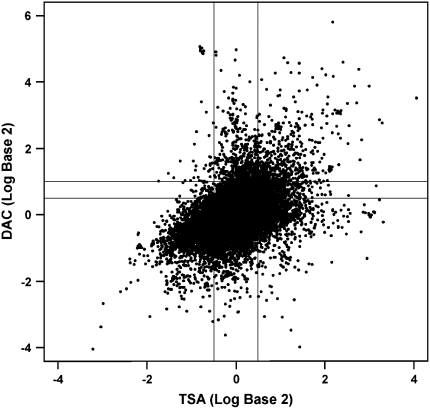
A hallmark of gene silencing by DNA methylation is the presence of heterochromatin over the gene promoter region that denies access to regulatory proteins needed for transcription. The chromatin structure is modified during gene silencing by affecting acetylation, phosphorylation, methylation, and/or ubiquitylation of histone tails (12, 14). Histone H3 is a common target for deacetylation, and this process has been strongly linked to maintaining compacted nucleosomes to block transcription (25). The transcriptome-wide screening approach uses a pharmacologic approach to identify silenced hypermethylated genes in cell lines (26). The DNA demethylating agent 5-aza-2′-deoxycytidine (DAC) can robustly induce gene re-expression and also decrease recruitment of chromatin-modifying proteins to increase transcription of genes without CpG islands. In contrast, the histone deacetylase inhibitor trichostatin A (TSA) alone will not induce re-expression of densely methylated gene promoters, but modulate expression of genes whose transcription is regulated by modification of chromatin structure. Thus, cell lines are treated with vehicle, DAC, or TSA, RNA isolated, and cDNA hybridized to 44K Agilent microarrays. Changes in gene expression between vehicle and TSA, and vehicle and DAC, treatment are characterized and a zone in which TSA treatment does not alter gene expression (−1.4- to 1.4-fold) compared with vehicle is identified. Within this zone, a characteristic spike of DAC-induced gene expression (Figure 1) is seen for genes that fall into two categories: top tier (expression levels of 2-fold or greater) and second tier (expression levels of 1.5- to < 2.0-fold). The initial studies developing this approach were focused on colon cancer and determined that 318 to 532 genes were present in the top-tier response zone in colon cancer cell lines. Importantly, random selection of these genes revealed that 65 to 91% were in fact methylated in the respective cell lines and in primary tumors (26). Furthermore, the second tier contains more than 1,000 genes, and studies to date suggest that up to 50% of these genes could also be methylated. These studies bring to the forefront the magnitude of genes that are silenced through methylation in colon cancer and identify a robust and high-throughput protocol that can be used for all tumor types. Several laboratories (27, 28), including ours, are using this or a similar protocol to identify novel genes methylated in lung cancer, and the genes identified along with prevalence for methylation in primary tumors are shown in Table 1. Our laboratory is asking a critical question: Can transcriptome-wide screening identify genes whose propensity for silencing by promoter hypermethylation differs between adenocarinomas from smokers and never-smokers? Cell lines derived from adenocarcinoma from smokers and never-smokers have been exposed to DAC or TSA, and microarray results indicate 300 to 450 and 150 to 200 genes in the top tier in cell lines from smokers and never-smokers, respectively (M. Tessema, unpublished data). Genes with biological plausibility for contributing to development of lung cancer are currently being validated first in a large series of lung tumor–derived cell lines and then in primary tumors from smokers and never-smokers. These studies will provide a comprehensive characterization of genes and pathways that may be causal for the development of adenocarcinoma.
Figure 1.
Changes in gene expression profile of a lung cancer cell line after treatment with 5-aza-2′-deoxycytidine (DAC) or trichostatin A (TSA). Agilent 44K microarrays were interrogated with cDNA from treated cells, and top-tier genes were identified based on increased expression of twofold or greater after DAC treatment after subtracting the expression change from the control. Next tier genes showed a 1.5- to less than 2.0-fold increase in expression after treatment with DAC.
TABLE 1.
PREVALENCE FOR METHYLATION OF GENES DISCOVERED THROUGH TRANSCRIPTOME PROFILING IN PRIMARY LUNG TUMORS
| Gene | Proposed Function | Prevalence for Methylation (%) | Reference |
|---|---|---|---|
| LOX | Arachidonic acid metabolism | 19/20 (95) | 29 |
| MSX1 | Homeobox transcription factor | 11/20 (55) | 29 |
| BNC1 | Transcription factor for multiple pathways | 18/20 (90) | 29 |
| CTSZ | Cysteine protease | 10/20 (50) | 29 |
| ALDH1A3 | Retinoic acid biosynthesis | 9/20 (45) | 29 |
| CCNA1 | Meiosis regulation | 14/20 (70) | 29 |
| NRCAM | Neuronal cell adhesion | 18/20 (90) | 29 |
| SOX15 | Muscle regeneration | 17/20 (85) | 29 |
| PAK3 | Cytoskeleton organization | 16/34 (47) | 28 |
| NISCH | Imidazine receptor | 12/34 (35) | 28 |
| KIF1A | Organelle transporter | 8/34 (24) | 28 |
| OGDHL | Citric acid cycle | 2/34 (6) | 28 |
IDENTIFICATION AND CHARACTERIZATION OF PROMOTER METHYLATION OF GENES WITHIN CHROMOSOME 6q
A recently conducted genome-wide linkage analysis of 52 extended pedigrees with a minimum of three family members with aerodigestive cancer identified a lung cancer susceptibility locus at chromosome 6q23–25 (29). The high frequency for loss of heterozygosity in this region and the potential existence of a susceptibility locus within 6q23–25 supports the existence of TSGs inactivated through the classical Knudson's two-hit model, in which complete loss of gene function arises through loss of one allele and mutation of the second allele (30). However, only one candidate tumor suppressor gene, p34, localized to 6q25 has been identified, but this gene was not found to be associated with familial lung cancer susceptibility (31). This scenario is reminiscent of the chromosome 3p14–25, in which loss of heterozygosity (LOH) is commonly seen in lung tumors, although no major TSG inactivated by mutation has been identified in this locus. Rather, genes inactivated by promoter hypermethylation at prevalences ranging from 30 to 58% have been identified within this locus. These include RASSF1A, BLU, SEMA38, and retinoic acid receptor β (32).
Chromosome 6 is one of the gene- and CpG island–rich chromosomes that contains about 1,557 genes and 1,070 CpG islands (33, 34). The fact that chromosome 6q accumulates genetic aberrations in the form of LOH could also make this region a hot spot for silencing of genes by promoter hypermethylation. Support for this supposition is growing, as genes silenced by methylation in lung tumors are now being identified within 6q. Estrogen receptor α, which maps to 6q25, has been shown by our laboratory to be silenced by promoter hypermethylation in 20% and 36% of lung tumors from smokers and never-smokers, respectively (35). Recently, Smith and colleagues (19) identified a tumor suppressor gene, TCF21, within the 6q23–24 locus that is normally expressed in lung airway epithelial cells, but silenced in aerodigestive tumors. The goal for our studies on chromosome 6q was to identify novel genes in and around the candidate lung cancer susceptibility locus 6q23–25 that are inactivated by promoter hypermethylation and to compare their prevalence in adenocarcinomas from smokers and never-smokers. A multifaceted strategy was undertaken that used comparative genomic hybridization, a transcriptome microarray, in silico screening, and a candidate approach to select and evaluate the methylation patterns of genes within chromosome 6q (16).
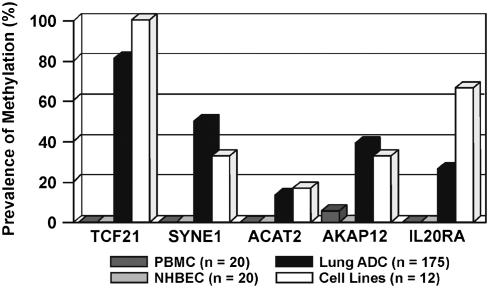
A total of 43 genes with CpG-rich promoters that met the CpG island criteria of Takai and Jones (36) were selected from 6q for methylation analysis. Combined bisulfite restriction analysis (COBRA) was used to screen lung tumor–derived cell lines, normal bronchial epithelial cells (HBECs), and normal peripheral blood lymphocytes (PBMCs) for methylation of gene promoters. The COBRA analysis revealed that eight of 43 genes were methylated in at least one cell line and devoid of methylation in HBECs and PBMCs. Five of these eight genes (TCF21, SYNE1, AKAP12, IL20RA, and ACAT2) were methylated at prevalence greater than 20% in cell lines and thereby selected for further analysis in primary adenocarcinomas from smokers (n = 100) and never-smokers (n = 75). The overall prevalence for methylation of these genes was 81%, 50%, 39%, 26%, and 14%, respectively, and did not differ by smoking status or age at diagnosis (Figure 2). The density of the methylation within the CpG islands of the SYNE1, AKAP12, and IL20RA genes was defined through bisulfite sequencing. Dense methylation was seen for the three gene promoters in cell lines and primary lung adenocarcinomas that correlated with loss of gene expression (16). Similarly, dense methylation of the TCF21 promoter has been described in NSCLC and head and neck tumors (19). Finally, we have also shown that treatment of cell lines with DAC restored expression of the SYNE1, AKAP12, and IL20RA genes, confirming that transcription was being regulated through aberrant promoter hypermethylation.
Figure 2.
Promoter methylation of chromosome 6q genes in primary lung adenocarcinoma. The five genes (TCF21, SYNE1, AKAP12, IL20RA, and ACAT2) that showed greater than 20% prevalence for tumor-specific methylation in lung cancer cell lines were analyzed in 175 primary lung adenocarcinoma samples using MSP. Results are compared with methylation in PBMC, NHBEC, and cell lines.
All five genes identified in this study are candidate tumor-suppressor genes located in and around the candidate 6q23–25 lung cancer susceptibility loci, and were epigenetically silenced in lung cancer. TCF21 is a basic-helix-loop-helix (bHLH) transcription factor that is critical for lung development. The SYNE1 gene is a multifunctional gene involved in cytokinesis, nuclear organization and the structural integrity, and function of the Golgi apparatus (37, 38). AKAP12/Gravin is one of the A kinase–anchoring proteins (AKAPs) that regulates mitogenesis by anchoring key signaling proteins (such as protein kinase A and C) and modulating the expression of genes involved in cell cycle and apoptosis (39). It suppresses tumor cell viability and growth by inducing apoptosis via caspase 3, up-regulation of Bax, and down-regulation of Bcl-2 expression (40). Similarly, IL20RA, which encodes a receptor for interleukin 20 (IL-20) and IL-24, also functions as a tumor suppressor by modulating the bystander tumor-specific cytotoxicity of IL-24 (33). IL-24 (MDA-7) induces G2/M cell cycle arrest and apoptotic cell death through up-regulation of proapoptotic proteins (Bax, Bak) and down-regulation of antiapoptotic proteins (Bcl-2, BCL-xL) (41, 42). A final key question from this study was the contribution of the identified genes silenced in adenocarcinoma to familial susceptibility for lung cancer. It is clear for early-onset cancers (such as breast cancers associated with germ line BRCA1/2 mutations) that the second hit in the affected locus plays a major role in the genesis of the disease. However, the five methylated genes (TCF21, SYNE1, AKAP12, IL20RA, and ACAT2) within the 6q23–25 locus where LOH is frequent in susceptible families did not show an increased prevalence for silencing in patients with early-onset (< 50 yr) lung cancer. Thus, while the genes discovered in this study probably do not predispose one for lung cancer, their high prevalence for silencing likely contributes to the development of sporadic lung cancer in smokers and never-smokers.
FUTURE DIRECTIONS
The realization that methylation is a dominant mechanism in lung cancer etiology, and its reversibility by pharmacologic drugs, has led to the initiation of translational studies in several areas. First, we are assessing whether detection of gene promoter methylation in sputum can be used as a biomarker for early lung cancer detection and/or monitoring for response to therapy or preventive interventions. Methylation of a 6-gene panel in sputum was associated with a 6.5-fold increased risk for lung cancer and a sensitivity and specificity for prediction of incident lung cancer of 65% (43). Studies are ongoing to identify additional genes whose methylation in sputum can distinguish persons with early lung cancer from smokers. Our group is also assessing whether gene promoter methylation in sputum and/or blood can be used to predict tumor recurrence and response to L-selenomethionine in a national Phase III Chemoprevention trial in which stage I resected lung cancer patients are receiving this agent for up to 4 years (44).
The other major area of translation from the bench to the bedside is testing the efficacy of demethylation therapy for treatment of lung cancer. Clinical trials with demethylating agents alone or combined with HDAC inhibitors have shown promising responses in the treatment of myeloid malignancies (45, 46). Approximately 65% of the 30 patients treated with the lowest-dose combination of 5-AZA followed by the HDAC inhibitor MS275 have shown responses that ranged from high lineage recovery to complete response. Treatment was also associated with induction of acetylation of histones H3 and H4. All responders showed cytogenetic effects and demethylation of the p15 or CDH-1 promoters, while nonresponders showed no demethylation (46). The extension of this targeted approach to solid tumors such as those in the lung may also hold promise as a therapy. Our work, in which combined treatment with DAC and sodium phenylbutyrate reduced the number of developing lung tumors in a murine model by over 50%, supports this supposition (47). An NCI-supported Phase I/II trial is now underway in lung cancer at Johns Hopkins and the University of New Mexico. These types of translational studies could ultimately determine if the epigenome is the key target to winning the battle against lung cancer.
Supported by NIH R01 ES008801.
Conflict of Interest Statement: M.T. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.A.B. is a consultant to Oncomethylome Sciences. Under a licensing agreement between Lovelace Respiratory Research Institute and Oncomethylome Sciences, vested methylation-specific PCR was licensed to Oncomethylome Sciences, and the author is entitled to shares of the royalties received by the Institute from sales of the licensed technology. The Institute, in accordance with its conflict of interest policies, is managing the terms of these arrangements.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin 2007;57:43–66. [DOI] [PubMed] [Google Scholar]
- 2.Janssen-Heijnen MLG, Coebergh J-WW. The changing epidemiology of lung cancer in Europe. Lung Cancer 2003;41:245–258. [DOI] [PubMed] [Google Scholar]
- 3.Harbour JW, Lai S-L, Whang-Peng J, Gazdar AF, Minna JD, Kaye FJ. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science 1988;241:353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimizu E, Coxon A, Otterson GA, Steinberg SM, Kratzke RA, Kim YW, Fedorko J, Oie H, Johnson BE, Mulshine JL, et al. RB protein status and clinical correlation from 171 cell lines representing lung cancer, extrapulmonary small cell carcinoma, and mesothelioma. Oncogene 1994;9:2441–2448. [PubMed] [Google Scholar]
- 5.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science 1991;253:49–53. [DOI] [PubMed] [Google Scholar]
- 6.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science 1996;274:430–432. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Christiani DC, Wiencke JK, Fischbein M, Xu X, Cheng TJ, Mark E, Wain JC, Kelsey KT. Mutations in the p53 gene in lung cancer are associated with cigarette smoking and asbestos exposure. Cancer Epidemiol Biomarkers Prev 1995;4:543–548. [PubMed] [Google Scholar]
- 8.Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res 2002;62:3659–3662. [PubMed] [Google Scholar]
- 9.Slebos RJ, Kibbelaar RE, Dalesio O, Kooistra A, Stam J, Meijer CJ, Wagenaar SS, Vanderschueren RG, van Zandwijk N, Mooi WJ. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med 1990;323:561–565. [DOI] [PubMed] [Google Scholar]
- 10.Slebos RJ, Rodenhuis S. The ras gene family in human non-small-cell lung cancer. J Natl Cancer Inst Monogr 1992;13:23–29. [PubMed] [Google Scholar]
- 11.Wood LD, Parsons DW, Jones S, Lin S, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science 2007;318:1108–1113. [DOI] [PubMed] [Google Scholar]
- 12.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415–428. [DOI] [PubMed] [Google Scholar]
- 13.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003;349:2042–2054. [DOI] [PubMed] [Google Scholar]
- 14.Kelly WK, O'Connor OA, Marks PA. Histone deacetylase inhibitors: from target to clinical trials. Expert Opin Investig Drugs 2002;11:1695–1713. [DOI] [PubMed] [Google Scholar]
- 15.Belinsky SA. Gene promoter hypermethylation as a biomarker in lung cancer. Nature Rev Cancer 2004;4:707–717. [DOI] [PubMed] [Google Scholar]
- 16.Tessema M, Willink R, Do K, Ye YY, Baylin SB, Belinsky SA. Promoter hypermethylation of growth regulatory genes in and around the major lung cancer susceptibility locus 6q23–25. Cancer Res 2008;68:1707–1714. [DOI] [PubMed] [Google Scholar]
- 17.Costello JF, Frühwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomäki P, Lang JC. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet 2000;25:132–138. [DOI] [PubMed] [Google Scholar]
- 18.Dai Z, Lakshmanan RR, Zhu W-G, Smiraglia DJ, Rush LJ, Frühwald MC, Brena RM, Li B, Wright FA, Ross P. Global methylation profiling of lung cancer identifies novel methylated genes. Neoplasia 2001;3:314–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith LT, Lin M, Brena RM, Lang JC, Schuller DE, Otterson GA, Morrison CD, Smiraglia DJ, Plass C. Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23-q24 in lung and head and neck cancer. Proc Natl Acad Sci U S A 2006;103:982–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toyota M, Ho C, Ahuja N, Jair KW, Li Q, Ohe-Toyota M, Baylin SA, Issa JP. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res 1999;59:2307–2312. [PubMed] [Google Scholar]
- 21.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal ancer. Proc Natl Acad Sci U S A 1999;96:8681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toyota M, Ho C, Oheo-Toyota M, Baylin SA, Issa JP. Inactivation of CACNAIG, a T-type calcium channel gene, by aberrant methylation of its 5′ CpG island in human tumors. Cancer Res 1999;59:4535–4541. [PubMed] [Google Scholar]
- 23.Palmisano WA, Crume KP, Winters SA, Toyota M, Esteller M, Joste N, Baylin SB, Belinsky SA. Aberrant promoter methylation of the transcription factor genes PAX 5 alpha and beta in human cancers. Cancer Res 2003;63:4620–4625. [PubMed] [Google Scholar]
- 24.Estécio MRH, Yan PS, Ibrahim AEK, Tellez CS, Shen L, Huang TH-M, Issa J-PJ. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res 2007;17:1529–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Natl Rev 2002;1:287–299. [DOI] [PubMed] [Google Scholar]
- 26.Schuebel KE, Chen W, Cope L, Glöckner SC, Suzuki H, Yi JM, Chan TA, Van Neste L, Van Criekinge W, van den Bosch S, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet 2007;3:1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoque MO, Kim MS, Ostrow KL, Liu J, Wisman GBA, Park HL, Poeta ML, Jeronimo C, Henrique R, Lendvai A, et al. Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Res 2008;68:2661–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shames DS, Girard L, Gao B, Sato M, Lewis CM, Shivapurkar N, Jiang A, Perou CM, Kim YH, Pollack JR, et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med 2006;3:2244–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey-Wilson JE, Amos CI, Pinney SM, Petersen GM, de Andrade M, Wiest JS, Fain P, Schwartz AG, You M, Franklin W, et al. A major lung cancer susceptibility locus maps to chromosome 6q23–25. Am J Hum Genet 2004;75:460–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer 2001;1:157–162. [DOI] [PubMed] [Google Scholar]
- 31.Wang M, Vikis HG, Wang Y, Jia D, Wang D, Bierut LJ, Bailey-Wilson JE, Amos CI, Pinney SM, Petersen GM, et al. Identification of a novel tumor suppressor gene p34 on human chromosome 6q25.1. Cancer Res 2007;67:93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito M, Ito G, Kondo M, Uchiyama M, Fukui T, Mori S, Yoshioka H, Ueda Y, Shimokata K, Sekido Y. Frequent inactivation of RASSF1A, BLU, and SEMA3B on 3p21.3 by promoter hypermethylation and allele loss in non-small cell lung cancer. Cancer Lett 2005;225:131–139. [DOI] [PubMed] [Google Scholar]
- 33.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet 2006;38:1378–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mungall AJ, Palmer SA, Sims SK, Edwards CA, Ashurst JL, Wilming L, Jones MC, Horton R, Hunt SE, Scott CE, et al. The DNA sequence and analysis of human chromosome 6. Nature 2003;425:805–811. [DOI] [PubMed] [Google Scholar]
- 35.Issa JP, Baylin SB, Belinsky SA. Methylation of the estrogen receptor CpG island in lung tumors is related to the specific type of carcinogen exposure. Cancer Res 1996;56:3655–3658. [PubMed] [Google Scholar]
- 36.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A 2002;99:3740–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan J, Beck KA. A role for the spectrin superfamily member Syne-1 and kinesin II in cytokinesis. J Cell Sci 2004;117:619–629. [DOI] [PubMed] [Google Scholar]
- 38.Gough LL, Fan J, Chu S, Winnick S, Beck KA. Golgi localization of Syne-1. Mol Biol Cell 2003;14:2410–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon DK, Jeong CH, Jun HO, Chun KH, Cha JH, Seo JH, Lee HY, Choi YK, Ahn BJ, Lee SK, et al. AKAP12 induces apoptotic cell death in human fibrosarcoma cells by regulating CDKI-cyclin D1 and caspase-3 activity. Cancer Lett 2007;254:111–118. [DOI] [PubMed] [Google Scholar]
- 40.Chada S, Mhashilkar AM, Ramesh R, Mumm JB, Sutton RB, Bocangel D, Zheng M, Grimm EA, Ekmekcioglu S. Bystander activity of Ad-mda7: human MDA-7 protein kills melanoma cells via an IL-20 receptor-dependent but STAT3-independent mechanism. Mol Ther 2004;10:1085–1095. [DOI] [PubMed] [Google Scholar]
- 41.Lebedeva IV, Sarkar D, Su ZZ, Kitada S, Dent P, Stein CA, Reed JC, Fisher PB. Bcl-2 and Bcl-xl differentially protect human prostate cancer cells from induction of apoptosis by melanoma differentiation associated gene-7, mda-7/IL-24. Oncogene 2003;22:8758–8773. [DOI] [PubMed] [Google Scholar]
- 42.Lebedeva IV, Su ZZ, Chang Y, Kitada S, Reed JC, Fisher PB. The cancer growth suppressing gene mda-7 induces apoptosis selectively in human melanoma cells. Oncogene 2002;21:708–718. [DOI] [PubMed] [Google Scholar]
- 43.Belinsky SA, Liechty KC, Gentry FD, Wolf HJ, Rogers J, Vu K, Haney J, Kennedy TC, Hirsch FR, Miller Y, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high- risk cohort. Cancer Res 2006;66:3338–3344. [DOI] [PubMed] [Google Scholar]
- 44.Belinsky SA, Klinge DM, Dekker JD, Smith MW, Bocklage TJ, Gilliland FD, Crowell RE, Karp DD, Stidley CA, Picchi MA. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin Cancer Res 2005;11:6505–6511. [DOI] [PubMed] [Google Scholar]
- 45.Yang AS, Doshi KD, Choi SW, Mason JB, Mannari RK, Gharybian V, Luna R, Rashid A, Shen L, Estecio MR, et al. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res 2006;66:5495–5503. [DOI] [PubMed] [Google Scholar]
- 46.Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, Grever M, Galm O, Dauses T, Karp JE, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res 2006;66:6361–6369. [DOI] [PubMed] [Google Scholar]
- 47.Belinsky SA, Klinge DM, Stidley CA, Issa J-P, Herman JG, March TH, Baylin SB. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res 2003;63:7089–7093. [PubMed] [Google Scholar]